Abstract
Abstract
Desirable features of exosomes have made them a suitable manipulative platform for biomedical applications, including targeted drug delivery, gene therapy, cancer diagnosis and therapy, development of vaccines, and tissue regeneration. Although natural exosomes have various potentials, their clinical application is associated with some inherent limitations. Recently, these limitations inspired various attempts to engineer exosomes and develop designer exosomes. Mostly, designer exosomes are being developed to overcome the natural limitations of exosomes for targeted delivery of drugs and functional molecules to wounds, neurons, and the cardiovascular system for healing of damage. In this review, we summarize the possible improvements of natural exosomes by means of two main approaches: parental cell-based or pre-isolation exosome engineering and direct or post-isolation exosome engineering. Parental cell-based engineering methods use genetic engineering for loading of therapeutic molecules into the lumen or displaying them on the surface of exosomes. On the other hand, the post-isolation exosome engineering approach uses several chemical and mechanical methods including click chemistry, cloaking, bio-conjugation, sonication, extrusion, and electroporation. This review focuses on the latest research, mostly aimed at the development of designer exosomes using parental cell-based engineering and their application in cancer treatment and regenerative medicine.
Graphic Abstract
Key Points
| Designer exosomes could erase the limitations of natural exosomes, including low targeting capability, low half-life in circulation, and low concentration of functional molecules. |
| A designer exosome benefits simultaneously from its own functional molecules and the newly loaded molecules. |
| Anti-cancer designer exosomes could be engineered for targeted delivery of chemotherapeutic drugs and short interfering RNAs to tumor tissue and requires further development of highly specific targeting peptides. |
| Regenerative designer exosomes from engineered mesenchymal stem cells (MSCs) could be used for healing of wounds, and neural and cardiac tissue healing and regeneration. |
Introduction
Exosomes comprise a small fraction of extracellular vesicles (EVs), which are produced by all types of cells and secreted into the extracellular environment [1]. Since EVs consist of different heterogeneous populations of vesicles, isolation of a pure population of exosomes requires several purification and characterization steps [2]. The three main populations of EVs have been categorized as exosomes, microvesicles, and apoptotic bodies. These populations of vesicles are different in their origin and size. Exosomes originate from multivesicular bodies (MVBs), which are derived from the budding of the plasma membrane of cells, and apoptotic bodies are formed from cell fragmentation through apoptosis. For the most detailed information on EVs and exosome populations, readers are referred to a recently published review by Willms et al. [3].
Among EVs, exosomes have been widely studied due to their potential application in medicine. Thus far, exosomes have been used in many studies for tissue regeneration, delivery of drugs and genes, and diagnosis of diseases. Naturally, in addition to the well-known cell-to-cell communication, new studies have shown other roles for exosomes, including spreading of various infectious agents, such as the human immunodeficiency virus (HIV) [4, 5], Epstein–Barr virus (EBV) [6], and prions [7]. Exosomes derived from different cells have specific features, composition, and effects on their target cells [8, 9]. In addition, exosomes naturally exhibit the characteristics of their parental cells [10, 11]. These two main features of exosomes could have many biomedical applications. For instance, researchers use mesenchymal stem cell (MSC)-derived exosomes in regenerative medicine for tissue regeneration and wound healing [12, 13]. On the other hand, exosomes are a new source and platform for the discovery of biomarkers in biofluids for the diagnosis and screening of cancer and other disorders [14, 15].
Structurally, the surfaces of exosomes are rich in trans-membrane proteins, receptors, and other functional molecules. In the lumen, there is a vast range of different proteins, lipids, RNAs, DNAs, and metabolites, which vary widely between exosomes derived from different sources [16–18]. Readers are referred to our most recent published review article, in which the molecular composition of exosomes is discussed in depth [19].
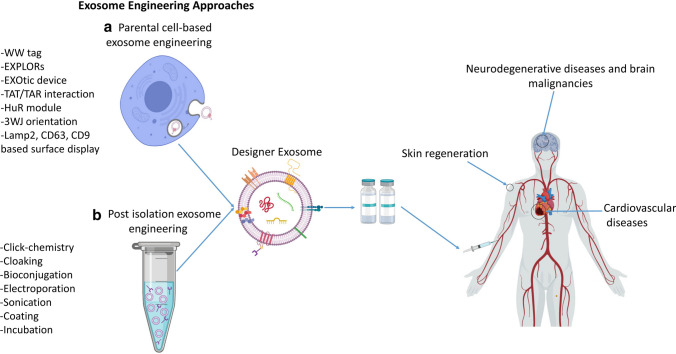
Although, extensive research in the recent 2 decades has enriched our knowledge of the biology, chemistry, and physiology of exosomes, a Food and Drug Administration (FDA)-approved exosomal-based therapeutic or diagnostic platform is yet to be established. This is mainly due to the fact that exosomes, as natural vesicles, lack some features for therapeutic development. For instance, for cancer treatment using exosomes, targeting of therapeutic exosomes specifically to cancer cells is essential. In addition, in many applications, researchers need to load a completely new drug, a protein, or RNA into exosomes to impart a therapeutic effect. Besides the challenges of large-scale manufacturing of exosomes, these limitations of natural exosomes have challenged the translational development of exosomes. Fortunately, various biotechnological approaches have led to the emergence of the new field of exosome engineering. Exosome engineering aims to achieve the following goals: (1) targeting the exosomes to a particular type of tissue or cell; (2) loading the exogenous molecules, drugs, proteins, or nucleic acids into exosomes or onto their surface; and (3) enrichment of an endogenous molecule into the lumen of exosomes or on their surface (Fig. 1). Most recently, advanced studies have combined several exosome engineering approaches in order to design highly specialized exosomes called designer exosomes.
Fig. 1.
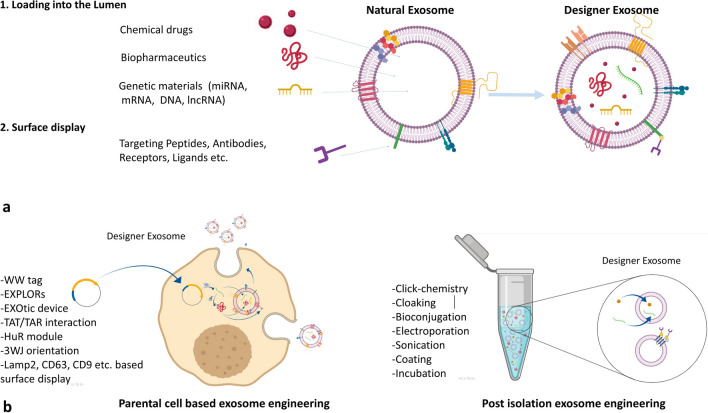
Exosome engineering approaches. a Different therapeutic molecules can be either loaded into lumen or displayed on the exosome surface for clinical purposes. b Two main approaches of exosome engineering, parental cell-based and post-isolation engineering, and their different methods. EXPLORs Exosomes for Protein Loading via Optically Reversible protein-protein interactions, EXOtic Exosomal Transfer Into Cells, 3WJ 3-way junction, Lamp2 lysosome-associated membrane protein 2, lncRNA long non-coding RNA, TAR trans-activating response element, TAT trans-activator of transcription
These engineered exosomes offer several novel features for special therapeutic applications. Herein, we aimed to classify exosome engineering approaches: parental cell-based and direct exosome engineering. However, different methods have been used in each class, and this could help researchers in various fields of biomedical sciences to recognize possible approaches for creating or improving therapeutic features of exosomes. In this review, we begin with natural exosomes and their potential and limitations in medicine. In the next sections, we focus on the necessity of exosome engineering, different engineering approaches, and applications of designer exosomes.
Natural Exosomes Versus Designer Exosomes
In the last 2 decades, synthetic nanoparticles such as liposomes (synthetic lipid bilayer used as a carrier for drugs and genes), aptamers (short single-stranded RNAs or DNAs that strongly bind to specific ligands), dendrimers (highly branched polymers used as carriers of drugs and genes), and many other biomolecules have provided useful tools for selective delivery of drugs and genes to target cells and tissues [1]. Despite this progress, multiple challenges still remain in using these nanoparticles including, bioavailability (systemic circulation access of administered carrier-loaded therapeutics), cost, biocompatibility, undesirable effects such as cytotoxic effects on some cells [1, 20], and short half-life in vivo, resulting in quick elimination from the systemic circulation [21, 22].
Since their introduction in the early 1980s by Pan and Johnstone, exosomes have been considered carriers that transport cell waste to the extracellular environment [23]. However, through vast studies on EVs, their various crucial functions have been discovered and experimentally approved for many physiological and pathophysiological conditions. These critical roles of exosomes consist of extensive cell-to-cell communications [24], cancer progression and metastasis [25, 26], embryogenesis [27], organ development [28], wound healing and tissue regeneration [29], cell differentiation [30], immunomodulation [31], antigen presentation [32], and pathogenesis [33–35]. These functions have led to intensive research on their biomedical applications, including tissue regeneration [36], cancer treatment and diagnosis [37], drug delivery [38], gene therapy [39], and vaccine development [40, 41].
Exosomes are natural vesicles with 50–150 nm size and a bilayer membrane, and their luminal contents consist of surface ligands and receptors [20, 22]. Exosomes can be detected and isolated from cell culture supernatants and all body fluids [1, 21]. Compared to synthetic nanovesicles such as liposomes, exosomes have higher stability and bioavailability [42], have a natural origin [43, 44], longer in vivo circulation half-life [45], a role as endogenous functional biomolecules, lack of toxicity, low to no immunogenicity, biocompatibility (because of their natural origin) [46], and ability to pass biological barriers such as the blood–brain barrier (BBB) [47, 48], penetrating massive tissues with their surface ligands and receptors [49]. As a result, exosomes have the potential to be a suitable platform for delivering drugs, genes, and functional biomolecules to target tissues or cells for different applications [21].
However, the application of exosomes in their natural form and composition as therapeutic agents is challenging. Generally, a natural exosome lacks the specific cell- or tissue-targeting feature. The therapeutic application of exosomes as a delivery system necessitates the design of controlled and targeted exosomes. Furthermore, many clinical applications involve the loading of certain additional therapeutic molecules alongside exosomes’ own content, which demands different loading techniques to be developed. In this regard, targeted designer exosomes or designer exosomes loaded with tested molecules could potentially address these limitations. In addition to the functional contents, ligands, and receptors of natural exosomes, designer exosomes also benefit from other engineered features [50, 51]. In the following sections, different approaches implemented for the development of designer exosomes are discussed.
Exosome Engineering Approaches
The functional molecule can be either loaded into the lumen or displayed on the surface of exosomes for a therapeutic purpose (Fig. 1a). The two major strategies, including parental cell-based and direct exosome engineering, are routinely employed for loading and displaying functional molecules, and mainly differ in the level of engineering applied for their development. In the parental cell-based approach, the starting material is the cells that are the source of the exosomes, and engineering occurs before exosome isolation from cells. In contrast, in the second approach, exosomes are the direct starting material for engineering, and the engineering procedure occurs after exosome isolation. The next two subsections separately discuss different methods employed in the two main exosome engineering approaches (Fig. 1b).
Parental Cell-Based Exosome Engineering
The parental cell-based approach of exosome engineering, which was first used for discovering the biology of exosomes and not for the purpose of manipulation, consists of genetically engineering cells to produce specifically fabricated exosomes (Fig. 1a). Here, the basics of this approach are discussed via previously published examples, to demonstrate how a functional molecule can be directed to either exosome lumen (loaded) or surface (displayed) using genetic engineering of the parental cells.
Exosomal Surface Display Using Parental Cell-Based Approach
Using the parental cell-based approach of exosome engineering, the most common method for directing a protein to the surface of exosomes uses a exosomal signal peptide. For example, lysosome-associated membrane protein 2b (Lamp2b) is an exosomal surface protein with an exosomal signal peptide. Fusion of a protein of interest to Lamp2b is very common for displaying the protein on the surface of exosomes as a targeting moiety, ligand, or receptor. A Lamp2b-based fusion protein can be used to display rabies viral glycoprotein (RVG), a neuron-specific targeting peptide, on the surface. Expression of Lamp2b-RVG fusion proteins in dendritic cells (DCs) displays RVG on the surface of DC-derived exosomes (Fig. 2a). When these exosomes are systemically administered, surface displayed RVG leads to the accumulation of these exosomes in the neurons and brain of mice [43]. Removal of the signal peptide from Lamp2b-RVG significantly reduces surface display of RVG on the engineered exosomes. Altogether, the signal peptide of Lamp2b can be used to display any fusion protein on the surface of exosomes. In addition, introduction of a glycosylation motif into Lamp2b-RVG fusion proteins could further enhance exosome delivery to the neurons and glial cells by protecting the surface-displayed fusion protein from enzymatic degradation. Therefore, this method allows simultaneous glyco-engineering and display of a protein on the surface of exosomes [52, 53].
Fig. 2.
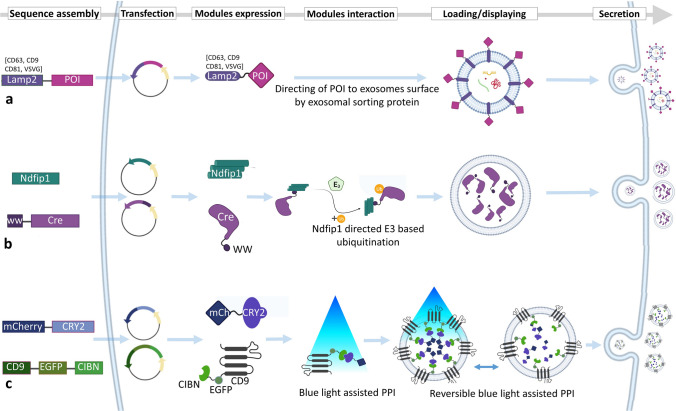
Parental cell-based exosome engineering for loading proteins into the lumen of exosomes or displaying them on their surface. a Surface display of a POI using different sorting modules. The fusion protein composed of the POI and the sorting module is directed to the surface of exosomes by an exosomal signal peptide. b Use of WW tag and Ndfip1 for loading Cre enzyme into the exosomes. Ndfip1 recognizes the WW tag and activates the E3 ubiquitin ligases, leading to the ubiquitination of Cre and its subsequent loading into the exosomes. c The EXPLOR method for loading mCherry reporter protein into the exosomes. Blue light-assisted PPI between CRY2 and CIBN results in the formation of a complex between two modules. Sorting of the whole complex into the exosomes is assisted by the CD9 sorting module. After elimination of the blue light, the two parts of the complex separate and mCherry-CRY2 is released to the lumen of exosomes. CRY2 photoreceptor cryptochrome 2, EGFP enhanced green fluorescent protein, EXPLOR exosomes for protein loading via optically reversible protein-protein interactions, Lamp2 lysosome-associated membrane protein 2, mCh mCherry, POI protein of interest, PPI protein–protein interaction, VSVG vesicular stomatitis virus glycoprotein
Other commonly used molecules for exosomal surface display of fusion proteins include tetraspanins (CD63, CD9, CD81) [54], glycosylphosphatidylinositol (GPI) [55], platelet-derived growth factor receptors (PDGFRs) [56], and lactadherin (C1C2 domain) [57]. In a very similar method to Lamp2b fusion proteins, the NH2-terminal of CD63 can be fused to a protein of interest for the same purpose [58].
Other non-cellular and exosomal proteins such as vesicular stomatitis virus glycoprotein (VSVG) have also been used for surface display of proteins on exosomes. VSVG is a key protein involved in virus envelope formation during microdomain budding from the surface of the plasma membrane. Since a similar domain to VSVG participates in the formation of endosomes and exosomes in the cell, researchers used VSVG to express proteins on the exosomal surface. VSVG contains ectoplasmic, transmembrane, and cytoplasmic domains [59]. Replacement of the ecto- and cytoplasmic domains of VSVG with another protein (e.g., green and red fluorescent proteins [GFPs and RFPs] or luciferase), without any alteration in the signal peptide and the transmembrane domain, results in proper exosomal surface display of the protein. Improved protein surface display and improved exosome uptake to the target cells are two advantages of using VSVG fusion proteins for exosomal surface display [60]. The stated methods allow proper display of a protein on the surface of exosomes, while some parental cell-based exosome engineering techniques have been developed recently for loading proteins and RNAs into the lumen of exosomes. These methods are discussed in the next subsection.
Loading Proteins into the Lumen of Exosomes Using Parental Cell-Based Engineering
The loading of therapeutic molecules is a common requirement for the production of therapeutic exosomes. The methods of parental cell-based engineering for loading molecules into the lumen of exosomes were developed very recently. These methods recruit the molecule sorting modules (MSMs) for sorting of proteins and RNAs into the lumen. These modules are able to bind to the protein or RNA of interest and direct them to exosomes. However, different MSMs or other modules have been used in different methods.
Two new methods for loading of proteins into the exosomes mimic the natural protein sorting system of cells based on ubiquitination. In the first method, an engineered ubiquitin tag (the last two glycine residues in the C-terminal are removed) has been developed. Removal of the two glycine residues results in enhanced ubiquitinated protein half-life. Fusion of this ub-tag to the proteins of interest, such as Ag85B and ESAT6 (Mycobacterium tuberculosis proteins) or enhanced green fluorescent protein (EGFP) and nHer2 (tumor antigens), leads to the loading of the proteins into the lumen of exosomes in human embryonic kidney (HEK293) cells. A recent study showed that the ubiquitin tag acts as a sorting sequence, resulting in efficient loading of proteins into exosomes [61]. In the second method, a short tag (WW tag) with the ability to bind specifically to the L-domain motif of Ndfip1 (which activates HECT domain-containing E3 ubiquitin-protein ligases) is used. The WW tag method is another system that uses ubiquitination for the loading of proteins into exosomes. Fusion of Cre recombinase (as a protein of interest) to the WW tag and simultaneous expression with Ndfip1 leads to recognition of the WW tag by Ndfip1 and activation of E3 ubiquitin ligases and ubiquitination of Cre. Subsequently, the ubiquitinated Cre-WW are loaded into the exosomes. Ndfip1 protein induces the molecular switch for ubiquitination and helps with the exosomal packaging of the Cre-WW fusion protein (Fig. 2b). The validation results of the mentioned method showed that when engineered exosomes containing the Cre-WW fusion protein are taken up by the target cells, Cre-WW is capable of DNA recombination [62].
Another method was recently used for the loading of specific proteins into the exosomes by a non-functional mutant of the HIV-I Nef protein. This engineered mutant of Nef (Nefmut) lacks the enzymatic activity and most functions of the intact Nef protein. The Nef protein is associated with lipid-raft microdomains of the plasma membrane [63] and exosomes. Similar to Lamp2b, CD63, and VSVG fusion proteins, Nefmut is developed for sorting of proteins to the exosomes [64]. Nefmut-GFP fusion protein was successfully loaded into exosomes for monitoring transfection and loading efficiency [65]. Moreover, fusing Nefmut to antigens of several pathogens, including Ebola virus VP24, VP40, and NP, influenza virus NP, Crimean-Congo hemorrhagic fever NP, West Nile virus NS3, and hepatitis C virus NS3, resulted in the expression of stable fusion proteins and their efficient loading into exosomes [66].
A recent attempt to load proteins into the lumen of exosomes was reported by Yim et al. [67]. They employed optically reversible protein–protein interaction (PPI) for the development of EXPLORs (exosomes for protein loading via optically reversible PPIs) using photoreceptor cryptochrome 2 (CRY2) and CIBN PPI modules. This method depends on intracellular delivery of proteins by reversible PPIs and their co-localization into exosomes by irradiation with a blue light. In the first step, a reporter protein was fused to CRY2 and CIBN protein was fused to the exosomal membrane CD9 protein (a representative marker of exosomes) to obtain two fusion proteins (CIBN-EGFP-CD9 and mCherry-CRY2). Blue light induced reversible PPI between CIBN and CRY2 indicates the reporter fusion protein (mCherry-CRY2) direction to the inner surface of exosomes through interaction with CIBN in the CIBN-EGFP-CD9 complex (Fig. 2c). The results of the study showed that protein-loaded EXPLORs increased intracellular levels of reporter proteins in the recipient cells [67].
Loading of RNAs into Exosomes Using Parental Cell-Based Engineering
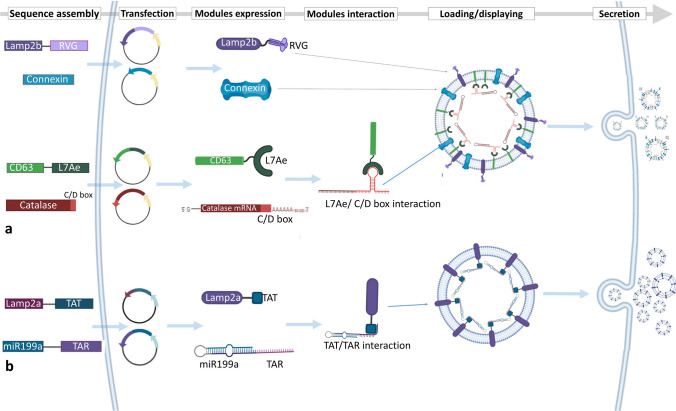
In addition to the parental cell-based exosome engineering methods and devices developed for loading of proteins into exosomes, similar methods have been developed for loading of RNAs. Recently, a novel parental cell-based strategy was established for loading mRNAs, called the EXOtic (Exosomal Transfer Into Cells) device. In the EXOtic device, in addition to the RNA packaging system (CD63-L7Ae, an archaeal ribosomal protein, which can bind to the C/Dbox in the 3′ untranslated region [UTR] of any RNA structure), two other devices known as cytosolic delivery helper (Cx43 S368A, a mutated connexin 43) and targeting module (RVG-Lamp2b) also contribute, which together provide a new method for loading therapeutic mRNAs into exosomes. Using this device, the mRNA of a catalase enzyme carrying a C/Dbox sequence on its 3′ UTR was loaded into exosomes, which are simultaneously targeted to brain tissue by the Lamp2b-RVG targeting module. Briefly, as we discussed earlier, since CD63 is an exosomal surface protein, the binding of CD63-L7Ae to the C/Dbox of catalase mRNA (through the L7Ae protein) directs the whole complex (CD63-L7Ae-mRNA) to exosomes. This system loads catalase mRNA into exosomes. The validation of this method showed that Lamp2b-RVG on the surface of these exosomes targets them to brain tissue and subsequently reduces neuroinflammation and neurotoxicity in the mouse model of Parkinson’s disease [68]. This is one of the most complex reports on parental cell-based exosome engineering that displays a neuron-targeting protein (RVG) on exosomes and loads a therapeutic mRNA into their lumen simultaneously (Fig. 3a).
Fig. 3.
Parental cell-based exosome engineering for loading RNAs into the lumen of exosomes. a The different modules of the EXOtic device for loading catalase mRNA into the lumen of exosomes using the CD63-L7Ae sorting module. Connexin is a packaging helper module, and Lamp2b-RVG is a targeting module. b Loading of miR-199a into exosomes using TAT–TAR interaction. Interaction of Lamp2a-TAT as the sorting module with the TAR sequence fused to miR-199a results in the loading of miR-199a into the exosomes. EXOtic EXOsomal Transfer Into Cells, Lamp2 lysosome-associated membrane protein 2, RVG rabies viral glycoprotein, TAR trans-activating response element, TAT trans-activator of transcription
Sutaria et al. [69] designed a system for loading miR-199a into exosomes. They fused a modified miR-199a composed of pre-miR-199a, to a trans-activating response element (TAR) sequence (trans-activation response RNA loop) and separately fused a trans-activator of transcription (TAT) peptide (transcription activator peptide of HIV-1) to Lamp2a (Lamp2a-TAT). When expressed in HEK293 cells, miR-199a-TAR binds to Lamp2a-TAT, and the interaction on the luminal C-terminal of Lamp2a leads to the effective loading of miR-199a into the lumen of exosomes (Fig. 3b). This TAT-TAR protein–RNA interaction strategy can be an efficient tool for loading any therapeutic microRNAs (miRNAs) into exosomes and microvesicles [69].
In this context, using of RNA binding modules seems to be an efficient method for loading RNA into exosomes. Similar to TAT and L7Ae, HuR is an RNA binding protein that binds strongly to miR-155. In this regard, HuR protein was used as an RNA sorting module for exosome engineering. In one study, HuR was fused to the C-terminal of CD9 in order to be localized in the exosomal lumen. Binding to miR-155 and simultaneous localization of the HuR module inside the exosomes results in the loading of miR-155 into the exosomes. HuR can also bind to adenylate uridylate-rich elements of engineered RNAs [70].
Besides the specific loading of molecules into exosomes by means of MSMs, one approach for nonspecific enrichment of exosomes with a molecule is based on the transfection of parental cells with a gene of interest without any other modules for packaging and sorting. Expression of this gene in the cells results in a general enrichment of the molecules in the exosomes. For instance, when cells are transfected with miRNA mimics, after expression of these miRNAs, they are significantly enriched in both MSCs and their derived EVs [9].
Direct or Post-Isolation Exosome Engineering Approach
Small nucleic acid molecules (e.g., miRNAs and short interfering RNAs [siRNAs]) and therapeutic molecules such as anti-cancer drugs can be encapsulated in exosomes by direct exosome engineering. This common type of exosome engineering is technically less complex compared with the parental cell-based methods, and has been widely employed for the application of exosomes as drug delivery or gene delivery vehicles in the last 2 decades. In this approach, common manipulation methods of electroporation [20, 71], extrusion, sonication, incubation, freeze-thaw, bio-conjugation, click chemistry, and cloaking [55, 61] can be used for direct engineering of exosomes after isolation from cells. Given that these methods do not provide a continuous source of engineered exosomes, in contrast to the parental cell-based engineering methods, concise protocols for frequent engineering procedures are warranted for clinical applications. Several informative review articles have been published that describe direct exosome engineering; thus, we refer the readers to these reviews for more information [72–74]. However, a brief review of these methods is presented here in the next sections and in Table 1.
Table 1.
Studies that used direct or post-isolation exosome engineering approach
| EVs source | Loaded molecule | Loading method | Displayed molecule | Displaying method | Mechanism of action | Result | Target cell/organ/tissue | References |
|---|---|---|---|---|---|---|---|---|
| A549 | Pd catalyst | Incubation | – | – | Pd induce dealkylation and prodrug activation | Tropism for A549 cells and activation of HDAC prodrug | A549 | [75] |
| HFL-1 | Erastin | Sonication | FA | Incubation | FA bound to FA-R | Suppress proliferation and migration | MDA-MB-231 | [76] |
| Raw264.7 | – | – | CHP | Incubation | Exosome was coated with CHP nanogel | CHP systems induce transporting and effective delivery by exosomes | HeLa | [77] |
| MCF-7 | DOX | Electroporation | FA-EV [TA-Fe3+] | Clocking | EV-[TA-Fe3+] as protective layer contain less negative charge | Tumor targeting and pH-controlled release of drugs | MCF-7 | [78] |
| LIM1215 | DOX | Incubation | A33 antibody | By iron oxide nanoparticles | Attachment of A33Ab to A33+ exosomes | Targeting of Ab-exosomes-DOX to A33+ cells | LIM1215 | [79] |
| Macrophage | PTX | Sonication | PEG-AA | Incubation | AA binds to sigma receptor | Targeted drug delivery and accumulated in cancer cells | NSCLC | [80] |
| HEK293T | Survivin-siRNA | Incubation | PSMAapt, EGFRapt, FA | Decoration by 3WJ | Aptamers and FA bound to receptors on the cells | Knockdown of survivin gene, inhibits tumor growth | Prostate, breast and colorectal cancer cells | [81] |
| HUVECs | PTX | Incubation | Biotin-avidin | Incubation | Facilitates PTX loaded exosomes isolation by microfluidics device | As a drug delivery vehicle, accumulates in cancer tissue | HepG2 | [82] |
| DCs | let-7 and VEGF siRNA | Electroporation | AS1411 aptamer | Incubation | AS1411 binding to nucleolin | AS1411-EVs act as a potential drug delivery vehicle for treat cancers | MDA-MB-231 | [15] |
| Neuro2A | – | – | EGa1 nanobody and PEG | Bioconjugation | EGa1 bound to the EGFR and PEG enhance EVs characteristics | Prolonged circulation times, EV accumulation in targeted tissues and improving cargo delivery. | Neuro2A and A431 | [83] |
| H1299 and YRC9 cells | GNPs and DOX | Bioconjugation | – | – | DOX-GNP complex by pH-sensitive linker induce pH-sensitive drug release | Low toxicity and efficient drug delivery to cancer cells and pH-sensitive drug release | H1299 | [84] |
3WJ 3-way junction, A431 epidermoid carcinoma cell line, A549 human lung cancer cell line, CHP cardiac homing peptide, DC dendritic cell, DOX doxorubicin, EGFR epidermal growth factor receptor, EV extracellular vesicle, FA folic acid, FA-R folic acid receptor, GNP gold nanoparticle, H1299 human non-small cell lung carcinoma cell, HDAC histone deacetylase inhibitor, HeLa immortalized human adenocarcinoma of the cervix cells, HepG2 human hepatoma cell line, HFL-1 human fetal lung fibroblast cells, HUVEC human umbilical vein endothelial cell, LIM1215 human colorectal carcinoma cell line, MCF-7 breast cancer cell line, MDA-MB-231 human breast cancer cell line, MDA-MB-241 triple negative breast cancer cell line, Neuro2A murine neuroblastoma cell line, NSCLC non-small cell lung cancer, Pd palladium, PEG polyethylene glycol, PTX paclitaxel, Raw264.7 murine macrophage tumor cell lines, siRNA short interfering RNA, VEGF vascular endothelial growth factor, YRC9 lung cancer cell line
Clinical Applications of Designer Exosomes
In the previous section, we described how researchers can engineer exosomes, and the basics of the methods and underlying mechanisms were discussed. In this section, we discuss the latest research on designer exosomes for specific clinical purposes. Indeed, cancer treatment and regenerative medicine seem to be the most important fields in which designer exosomes are being used.
Targeted Designer Exosomes
Tetraspanins on the surface of exosomes indicate both the origin of exosomes and their desired cells. These proteins cause a general slight attraction between exosomes and specific cells [85]. However, this general attraction of exosomes is not enough for targeted delivery of molecules to specific cells, and it requires the existence of targeting peptides or proteins on the surface of exosomes. In recent years, a substantial number of studies have focused on using exosomes for gene and drug delivery, while only a few of them exploited targeted designer exosomes. In the previous sections, we briefly discussed the basics of exosome targeting approaches. Here, recent studies aimed at developing targeted designer exosomes will be discussed.
In an example of an immunotherapeutic application of designer exosomes, transfection of murine melanoma cells by the CIITA gene led to the overexpression of MHC-II on their surfaces, hence, producing exosomes enriched in MHC-II. Next, the engineered exosomes were targeted toward T cells and increased the type 1 T helper cell (TH1) response against cancer cells. Therefore, the enriched MHC-II molecule in the engineered exosomes acted as a targeting peptide and a therapeutic agent [86]. Alternatively, an antibody light chain can be attached to the exosomal surface to direct exosomes toward T cells [87].
Due to the distinct ability of exosomes to cross the BBB and deliver drugs to the brain, treatment of neurodegenerative disorders and related malignancies remains one of the main goals for targeted designer exosomes. Using targeting modules on the surface of exosomes specific for neurons and brain tissue can efficiently target the engineered exosomes toward brain to deliver therapeutic molecules. In one study, overexpression of folate receptor α (FRα) on the surface of exosomes permitted them to specifically bind to the brain parenchyma [88]. Another study used RVG surface-displayed designer exosomes for the delivery of siRNAs to knockdown β-secretase 1 (BACE1), an essential factor of Alzheimer’s disease, which suppressed BACE1 by ~ 40% [43]. Other designer exosomes developed against neurodegenerative diseases will be discussed in the regenerative designer exosomes section.
Exosome targeting can also be accomplished by surface chemical modification in a direct engineering approach. In a study by Nag and Awasthi [89], a method of direct engineering called “cloaking” was applied, which is a membrane anchoring platform and consists of three components: a DMPE phospholipid membrane anchor, a polyethylene glycol (PEG) spacer, and a streptavidin molecule. Briefly, DMPE-PEG-streptavidin inserts into the exosomal membrane via hydrophobic interaction and provides an anchor for the connection of biotinylated ligand proteins [89]. A recent study used both the cloaking platform and parental cell-based surface display methods to develop targeted designer exosomes. These exosomes were engineered to target the injured myocardium via chemical engineering or surface display of an ischemic homing peptide. Comparative analysis revealed the same results of in vivo biodistribution to injured myocardium for both methods [90], verifying the potential of the two different approaches for targeted designer exosome development. Although chemical-based engineering methods such as cloaking are more straightforward than the parental cell-based methods, they need to be carried out for every isolated exosome. Overall, the cloaking technology has been recognized as a versatile exosome surface engineering method for development of targeted designer exosomes [78]. Targeting moieties are one of the most important parts of targeted therapy, and Table 2 provides the targeting moieties used for targeted designer exosome development.
Table 2.
Developed targeted designer exosomes using different methods and targeting molecules
| Targeting molecule | Target to | Target cell | Display method | Cargo | Loading method | EVs source | Aim | References |
|---|---|---|---|---|---|---|---|---|
| RVG | Acetylcholine receptor | Neurons | Fusion to lamp-2 | GAPDH siRNA | Electroporation | DCs | Knockdown of BACE1 in Alzheimer’s disease | [43] |
| Pullulan | Asialoglycoprotein-R | Hepatocytes | Incubation | – | – | MSCs | Targeting regenerative exosomes to injured liver | [91] |
| 4F-LDL peptide | LDL-R | Glioma cell line U87 | Decoration | Methotrexate | Incubation | L929 cells | Crossing BBB and distribution in glioma site | [92] |
| FA | Folate-R | TNBC | Coating with DSPE‐PEG2000‐FA | Erastin | Sonication | HFL‐1 cells | Suppression of GPX4/upregulation of CDO1 | [76] |
| GEa1 | EGFR | Neuro2A | Bioconjugation | – | - | Neuro2A | Targeting tumor cells | [83] |
| cRGD | αvβ3 integrin | Neurons | Bio-orthogonal copper-free azide alkyne cyclo-addition | Curcumin | Incubation | MSCs | Suppression of the inflammatory response and apoptosis | [93] |
| c-Met binding peptide | c-Met | TNBC/MDA-MB-231 | Decoration | DOX | Incubation | Macrophage | Release of the encapsulated DOX, which will accumulate in the nuclei of cancer cells | [94] |
| IL-3 | IL-3-R | CML | Fusion to lamp-2 | Imatinib and BCR-ABL siRNA | Transfection | HEK293T | Targeting and inhibition of tumor growth | [95] |
| iRGD | av integrin | MDA-MB-231 and MCF-7 | Fusion to lamp-2 | DOX | Electroporation | imDCs | Targeting and inhibition of tumor growth | [96] |
| Tf | Tf-R | Neurons | Natural | Dopamine | Incubation | HeLa and DCs | Binding to Tf-R, receptor on cells | [97] |
| cRGD | αvβ3 integrin | Neurons | DBCO-NHS | miR-210 | Incubation | MSCs | Treat ischemic stroke | [98] |
| AS1411 aptamer | Nucleolin | MDA-MB-231 | Conjugating polypeptides with cholesterol | let-7 and VEGF siRNA | Electroporation | DCs | Targeting tumor tissues | [99] |
| RVG | Acetylcholine receptor | Neurons | Fusion to Lamp2b | AuNPs | Extrusion | HEK293T | Targeting to the brain | [100] |
| Darpins | Her-2 | SK-BR-3 and BT-474 | Fusion to Lamp2b | DOX | Electroporation | MSCs | Targeted delivery of DOX to Her-2+ cells | [101] |
| Anti-HIV scFvs | Env | Env+ CHO cells | 10E8-pDisplay fusion protein | Curcumin and miR-143 | Saponin | HEK293T | Targeting Env+ cells | [102] |
| Anti-Her2 scFvs | Her-2 | HeLa, SK-BR-3 | Fusion with C1C2 domain of lactadherin | CFSE | Incubation | HEK293T | Targeting to cells | [103] |
| RGD | αvβ3 integrin | MCF-7 and HUVECs | DSPE-PEG-RGD assemble | Ac4ManNAz | Reversible permeabilization with streptolysin O | K562 cells | Angiogenesis therapy and imaging | [104] |
| Darpins | Her-2 | SK-BR-3 and BT-474 | Fusion to Lamp2b | TPD52 siRNA | Electroporation | HEK293T | Delivering siRNA and downregulating of TPD52 | [105] |
| Apo-A1´ | Scavenger receptor class B | HepG2 cells | Fusion to CD63 | miR-26a | Electroporation | HEK293T | Upregulation of miR-26a and decrease the cell migration and proliferation | [106] |
| AA-PEG | Sigma receptor | Lung cancer cells |
DSPE-PEG-AA assemble |
PTX | Sonication | RAW264.7 and BMM | Targeting cancer cells upon systemic administration | [80] |
| FA | FA-R | MDA-MB-231 | DSPE-PEG-FA assemble | Bcl-2 siRNA and PTX | Electroporation | Cal-27 | Targeting tumor cells with the aid of FA-R binding | [107] |
| FA | FA-R | Tumors MCF-7 and HeLa | DSPE-PEG assemble | DOX | Electroporation | Macrophage | Targeting FA-R expressing cells | [21] |
| Anti-HPV16-E7 scFv | HPV16-E7 | HPV16-E7+ cells | Fusion to Nefmut | – | – | HEK293T | Binding to HPV16-E7+ cells, and inhibition of the proliferation | [108] |
| CHP | – | Cardiomyocytes | Incubation with DOPE-peptide | – | – | CDCs | Targeted delivery to myocardial infarction | [109] |
| RVG | Acetylcholine receptor | Neurons | Fusion to Lamp2b | Catalase Mrna | Packaging device | HEK293T | Attenuation of neurotoxicity Parkinson’s disease | [68] |
| RVG | MOR | Neurons | Fusion to Lamp2b | MOR siRNA | Transfection | HEK293T | Downregulating the expression of MOR | [110] |
| GE11 | EGFR | Hcc70 | Fusion to PDGFR | Let7 miRNA | Electroporation | Hek293 | Binding to EGFR in Rag2-/- mice model of breast cancer | [56] |
| RGE | NRP-1 | U251 | Phosphatidylethanolamine | SPIONs and curcumin | Electroporation | Raw264.7 | Crossing the BBB, targeted imaging | [111]. |
Apo-A1 apolipoprotein A1, BACE1 β-secretase 1, BBB blood–brain barrier, BMM bone marrow-derived macrophages, BT-474 human breast tumor cell line, Cal-27 oral adenosquamous cell carcinoma, CDC cardiosphere-derived cell, CFSE carboxyfluorescein succinimidyl ester, CHO Chinese hamster ovary, CHP cardiac homing peptide, DC dendritic cell, DOX doxorubicin, EGFR epidermal growth factor receptor, Env envelope protein, EV extracellular vesicle, FA folic acid, FA-R folic acid receptor, HEK293T human embryonic kidney cell line, HeLa immortalized human adenocarcinoma of the cervix cells, HepG2 human hepatoma cell line, Her-2 human epidermal growth factor receptor 2, HFL-1 human fetal lung fibroblast cells, HPV human papilloma virus, HUVEC human umbilical vein endothelial cell, K562 human leukemia cell line, L929 mouse fibroblast cells, Lamp2 lysosome-associated membrane protein 2, LDL-R low-density lipoprotein receptor, MCF-7 breast cancer cell line, MOR mu-opioid receptor, MSC mesenchymal stem cell, Nefmut mutant of Nef, Neuro2A murine neuroblastoma cell line, NRP-1 neuropilin-1, PDGFR platelet-derived growth factor receptor, PEG polyethylene glycol, PTX paclitaxel, RAW264.7 murine macrophage tumor cell lines, RVG rabies virus glycoprotein, scFv single-strain variable fragment, siRNA short interfering RNA, SK-BR-3 human breast cancer cell line, SPION superparamagnetic iron oxide nanoparticle, Tf transferrin, TPD52 tumor protein D52, U251 human glioma cell line, VEGF vascular endothelial growth factor
Anti-cancer Designer Exosomes
As described in the beginning of the previous section, in some cases, exosomes have intrinsic homing ability toward certain cells. For instance, exosomes derived from Raw264.7 (macrophage; Abelson murine leukemia virus-transformed) and U937 (of the myeloid lineage) cells show targeting ability toward cancerous endothelial cells [112]. Jang et al. hypothesized that (at least for the exosome-mimetic nanovesicles) the presence of lymphocyte function-associated antigen 1 (LFA-1) inherited from the donor cell line facilitated binding of the exosomes to endothelial cell adhesion molecules (CAMs), and in turn directed them to the growing tumors with widespread neovascularization [112]. However, significant adverse side effects for healthy proliferative cells, especially in the case of anti-cancer chemotherapeutic agents, have encouraged scientists to design targeted vehicles [101, 105]. In this section, we focus on targeted designer exosomes specifically designed for cancer therapy.
The GE11 peptide has a strong affinity for epidermal growth factor receptor (EGFR). A recent study developed a GE11-based targeted designer exosome for breast cancer treatment. They used a GE11 targeting peptide, hemagglutinin, and myc-tag, fused to the PDGFR, in order to be expressed on the exosome surface [56]. EGFR upregulation in several types of solid cancer represents great potential for cancer treatment through targeting this receptor [113]. GE11 binds to EGFR without any receptor activation; thus, it makes a potential candidate for drug delivery to tumor tissue [56]. In this study, EGFR targeting by systematically injected targeted designer exosomes in a Rag2−/− mice model of breast cancer significantly decreased the tumor size via accumulation in tumor tissue. In this regard, the main step prior to the development of targeted designer exosomes is finding suitable and specific targeting peptides. Table 1 summarizes the targeting peptides previously used for designing targeted designer exosomes or for detecting exosome-producing cancer cells [114].
RGE is a specific targeting peptide for NRP-1 on glioma cells. Following the click chemistry-assisted surface display of RGE peptide, targeted designer exosomes were loaded with curcumin and superparamagnetic iron oxide nanoparticles (SPIONs) (RGE-Exo-SPION/Cur). Binding of RGE peptide to the exosome’s surface was carried out by a cycloaddition reaction of sulfonyl azide. Briefly, the targeted designer exosomes were generated in two steps: conjugation of the alkyne group with the protein (the phosphatidylethanolamine) on the exosomal membrane with an EDC-NHS condensation reaction and conjugation of RGE with the azido group to the alkyne group by triazole linkages. Parental cell-based assessment showed that systemic administration of these targeted designer exosomes can potentially cross the BBB in a mouse glioma model and accumulate in the tumor tissue. These targeted exosomes can also be used for targeted imaging and diagnosis [111].
Regenerative Designer Exosomes
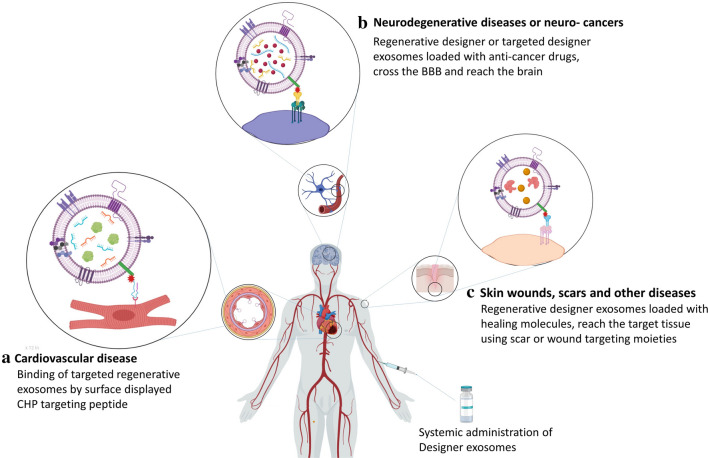
Various previously published studies corroborate the regenerative effects of exosomes isolated from progenitor and stem cells. MSC-derived exosomes are well studied, and their regenerative potentials have been experimentally approved in regenerative medicine for the treatment of various diseases. However, the contents of exosomes as the key factor of their regenerative effects are not yet fully identified. RNAs and proteins are the main effectors of the regenerative potential of the exosomes, while miRNAs and mRNAs are the most critical bioactive elements in these exosomes. The content of each exosome depends on the cell type and its physiological state. Recent studies have demonstrated that the exosomes secreted by differentiated cells such as induced pluripotent stem cell-derived cardiomyocytes (iCMs) contain a distinct set of miRNAs, which can modify the post-transcriptional and translational profile and phenotype of the recipient cells more than exosomes secreted by undifferentiated cells. Eventually, exosomes derived from these cells have a substantial effect on the promotion of infarcted heart recovery by providing cardiac protective factors such as clusterin, miR-21, miR-126 [115], and miR-210, resulting in reduced infarct size, hypertrophy, apoptosis, and stimulation of angiogenesis [116]. The exosomes isolated from MSCs of different tissues have been extensively used for regeneration of tissues and wound healing. This approach is a safe and non-immunogenic cell-free alternative to cell-based therapies [117]. However, the lack of targeted therapy and the complexity of exosomal contents are still the main challenges for tissue repairing by exosomes (Fig. 4). Wang et al. [119] developed an efficient method eliminating the risk of the accumulation of exosomes in non-specific organs. Similar to the previously described methods involving targeted designer exosomes, they reconstructed exosomal Lamp2b protein to be fused to ligands or homing peptides to target a specific organ. Fusing ischemic myocardium-targeting peptide (IMTP) to Lamp2b (IMPT-Lamp2b) in the treatment of acute myocardial infarction (AMI) resulted in accumulation of engineered exosomes in the myocardium. In this study, by using IMTP, and in another study, by attaching glycosylation sequences to a cardiac-targeting peptide (CTP-Lamp2b [118]), an increase was recorded in the retention of engineered exosomes in the myocardium of mice in a mouse myocardial infarction (MI) model ex vivo and in vitro. Furthermore, high specificity and efficiency of targeting myocardium using the engineered exosomes resulted in the reduction of inflammation and apoptosis, induced angiogenesis, decreased infarct size, and refined cardiac function [109, 119].
Fig. 4.
Application of regenerative designer exosomes for cardiovascular and neurodegenerative disorders and malignancies. a Targeted designer exosomes loaded with healing molecules reach the injury site of MI by injury-targeting specific peptides after intravenous injection. b Use of neuron- and brain-targeting moieties on exosomes allows the exosomes to cross the BBB and reach the injury site or tumor tissue in the brain and deliver healing molecules or anti-cancer drugs and c regenerative designer exosomes containing wound healing factors and anti-scarring and anti-inflammatory factors targeting the scar tissue for treatment of wounds and dermal diseases. BBB blood–brain barrier, CHP cardiac homing peptide, MI myocardial infarction
A real challenge for targeting different tissue injuries is the development of tissue-specific targeting moieties. For instance, cardiomyocyte-specific cTnI, is involved in the regulation of cardiac muscle contraction and exists in higher concentrations in the microenvironment of the infarct area. In this regard, cTnI is a potential ligand for targeted delivery of functional regenerative molecules to the MI site. When a targeting peptide that specifically targets cTnI is fused with a regenerative protein, this protein is delivered to the MI site. For targeting exosomes to the MI site, a cTnI-targeting small peptide “STSMLKA” was fused to the N-terminal of Lamp2b and expressed in MSCs. P1–8 BM-MSC was used as the source of designer exosomes. In addition, hsa-miR-590-3p, which downregulates the expression of cell proliferation inhibitor genes, was loaded into the lumen of exosomes after isolation. Treatment of MI models of rats showed that after systemic administration, designer exosomes are localized to the MI site through the concentration gradient of cTnI. As a result, these designer exosomes enhanced cardiomyocyte proliferation in the peri-infarct area and improved cardiac function [4].
For cerebral ischemia treatment, the main obstacle is crossing the BBB. Mesenchymal stromal cell-derived exosomes fused to a cyclo (Arg-Gly-Asp-D-Tyr-Lys) peptide [c(RGDyK)] showed high affinity for integrin αvβ3 in reactive cerebral vascular endothelial cells. The c(RGDyK)-conjugated exosomes (cRGD-Exo) loaded with various drugs, such as curcumin, suppressed the inflammation and cellular apoptosis in the lesion region in the mouse model of transient middle cerebral artery occlusion (MCAO) [93].
In another study, using cRGD peptide, the RGD-displaying exosomes loaded with miR-210 were systemically administered to localize in the injury site of the ischemic brain. Following intravenous administration, the miR-210 level significantly increased at the site of lesion in a mouse model of transient MCAO. Intravenous administration of these designer exosomes once per day for 14 days resulted in upregulation of integrin β3, vascular endothelial growth factor (VEGF), and CD34 and enhanced the survival rate of animals (5). RGD surface functionalized regenerative exosomes also can be used for angiogenesis therapy (3).
In a study using designer exosomes for renal regeneration, MSCs were transfected with multiple miRNA mimics (miR-10a-5p, miR-127-3p, miR-29a-3p, let-7a, miR-30a-5p, and miR-486-5p) and their significant enrichment in both MSCs and their EVs confirmed. These miRNAs were selected based on their involvement in tissue regeneration and cell proliferation pathways, and were enriched in MSC-derived EVs to promote renal regeneration. Results demonstrated that EVs loaded with these miRNAs maintain their pro-regenerative effects, and verified the potential application of a low dose of the engineered EVs (82.5 × 106) for the promotion of renal regeneration [9].
Polyethylenimine (PEI) is a polymer capable of forming non-covalent complexes with DNA, making it a suitable carrier for DNA and RNA delivery. In order to promote bone regeneration, EVs derived from human periodontal ligament stem cells were engineered with PEI polymer. The PEI-EVs may act through enhancement and promotion of EVs’ content delivery to the target cells. The in vivo analysis of PEI-EVs in rat models of calvarial defect showed significant expression of osteogenic markers, increased VEGF and VEGF receptor 2 (VEGFR2), increased vascularization, and consequently enhanced bone regeneration [120].
Finally, using previously described parental cell-based and direct methods, therapeutic molecules including drugs, mRNAs, miRNAs, siRNAs, peptides, growth factors, anti-scarring factors (transforming growth factor [TGF]β1 inhibitors such as decorin and TGFβ3 [121, 122]), and wound healing factors (SDF-1, which has a chemotactic function for bone marrow MSCs [BMSCs] and endothelial progenitor cells [EPCs] in vascularization and healing [123]) can be efficiently loaded (as fully described in Sect. 3.1) into the regenerative engineered exosomes. This kind of regenerative exosome loaded with effective wound healing, regenerative, and anti-scarring factors can be targeted to the wound and scar tissues for regenerative purposes, using targeting peptides such as cardiac homing peptide (CHP) [109] and SDF-1 (with CXCR4 receptor on BMSCs). The recently reported targeting peptide, CAR (CARSKNKDC), showed a potential capacity for homing and penetration into injured and diseased tissues, and can be expressed on the surface of regenerative exosomes for targeted regeneration [124].
Companies Developing Engineered Exosome-Based Therapeutics
Like other areas of basic science, exosomes have been favored by many biotechnology companies because of their predicted lucrative future, which has encouraged companies to invest in exosomes research and development. Consequently, various companies have been funded to promote the commercialization of a particular exosomal product. One of these companies is Codiak Bioscience, founded in 2015, which is arguably the largest developer of exosome-based therapeutics. This company seeks to produce engineered exosomes for the development of advanced targeted drug delivery platforms for neuronal cells. In addition, developing the large-scale production systems of exosomes is one of the main goals of this company (http://www.codiakbio.com/). Evox Therapeutics is pursuing a completely biotechnological approach to produce new platforms for engineered exosomes with novel features in the loading of drugs and targeting them to specific cancers and tissues such as the brain and the central nervous system (https://www.evoxtherapeutics.com/). Exopharm works on the commercialization of exosome-based products for regenerative medicine, currently including a platform for downstream analysis and two products. The downstream technology developed by Exopharm is called LEAP technology, which is based on chromatography for the separation of ultrapure exosomes for pharmaceutical use (https://exopharm.com/). With a long history in stem cell production, Aruna Bio Company has commercialized the first neuronal stem cell or neuronal precursor from human pluripotent stem cells. Recently, the company decided to commercialize pharmaceutical exosomes for drug delivery to the brain and neurons. This company is also seeking to provide a solution for the large-scale production of exosomes (https://arunabio.com/). In 2018, at the Gordon Scientific Conference, Capricor Therapeutics reported successful development of an exosome-based product called Cap-2003 from cardiosphere-derived EVs that can be used for immunomodulation in immune-related diseases (http://capricor.com/). Many other companies are also active in the development of exosome therapeutics, including Exogenus (www.Exogenus-t.com), Versatope Therapeutics (www.versatope.com), and VivaZome Therapeutics (www.vivazome.com).
Future Perspectives and Conclusions
Exosomes are fascinating biological materials. Promising recently published research and developments indicate that most of the current cell therapies could be replaced by their exosomes. The exosomes produced by MSCs, which were widely noticed after the publication of an article by Lai et al. [125], encouraged thousands of other studies on exosomes in the field of regenerative medicine. Interestingly, these studies showed that exosomes could exhibit MSCs’ potentials. Therefore, exosomes have been introduced as an advantageous alternative for whole MSCs in cell therapies and regeneration medicine.
Exosome engineering and the production of designer exosomes are entering a new phase. The previous phase of exosome engineering was based on the curiosity of the researchers about the engineering of exosomes. In recent years, development of biological therapeutics has been steadily pursued, and some companies have been founded to manufacture the engineered exosome therapeutics.
As discussed in various sections of this review article, some of the undesirable features of natural exosomes seem to prevent their extension to clinical applications. In this regard, the problems associated with the production and manufacture of exosomes on a large scale and their purification and heterogeneity need to be addressed. It is expected that with the development of large-scale production methods for exosomes as well as engineered cell lines for the production of improved engineered exosomes, the establishment of therapeutic and diagnostic platforms associated with EVs may be possible in the near future. Designer exosomes have been shown to have a high potential, while methods of exosome engineering for improving the effectiveness of natural exosomes expand rapidly. For example, chemotherapeutic drugs [126, 127] commonly used for cancer treatment are associated with significant adverse effects due to undifferentiated cytotoxicity for tumor and healthy cells. In this regard, specifically targeted designer exosomes displaying specific targeting moieties on their surface and loaded with anti-cancer drugs in their lumen are a versatile platform that meet several challenges together [80].
Finally, another highly promising application of designer exosomes is in vaccine development. Studies on vaccine development showed that host cells infected with parasites, viruses, or bacterial pathogens secrete exosomes filled with several antigens of those pathogens [128]. Experiments in vaccine development showed that cocktail and multi-epitopic vaccines (MEVs) could overcome antigenic escape of pathogens from the immune system [129, 130]. Hopefully, using exosome engineering methods, exosomes could be recruited to present multiple antigens as designer vaccine exosomes.
Acknowledgements
The authors acknowledge support from Iran University of Medical Sciences.
Declarations
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing Interests
The authors declare that they have no competing interests.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Materials
Further information is available from the first author and corresponding author on request.
Code Availability
Not applicable.
Author contributions
DJ: conceptualization, data acquisition, writing, reviewing, and editing, artwork preparation, and validation; SS: writing, original draft preparation, and artwork preparation; RJ: writing, original draft preparation, and reviewing; NM: writing, original draft preparation; HG: writing, original draft preparation; FG: writing, original draft preparation; MFM: reviewing and validation; AS: supervision, conceptualization, validation, reviewing, and editing.
Contributor Information
Mehdi Forouzandeh Moghadam, Email: foroz@modares.ac.ir.
Ali Samadikuchaksaraei, Email: Samadikuchaksaraei@yahoo.com.
References
- 1.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasser C, Jang SC, Lotvall J. Subpopulations of extracellular vesicles and their therapeutic potential. Mol Aspects Med. 2018;60:1–14. doi: 10.1016/j.mam.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Willms E, Cabanas C, Mager I, Wood MJA, Vader P. Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol. 2018;9:738. doi: 10.3389/fimmu.2018.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welch JL, Stapleton JT, Okeoma CM. Vehicles of intercellular communication: exosomes and HIV-1. J Gen Virol. 2019;100(3):350–366. doi: 10.1099/jgv.0.001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheruiyot C, Pataki Z, Ramratnam B, Li M. Proteomic analysis of exosomes and its application in HIV-1 infection. Proteom Clin Appl. 2018;12(5):e1700142. doi: 10.1002/prca.201700142. [DOI] [PubMed] [Google Scholar]
- 6.Teow S-Y, Liew K, Khoo AS-B, Peh S-C. Pathogenic Role of exosomes in Epstein-Barr Virus (EBV)-associated cancers. Int J Biol Sci. 2017;13(10):1276–1286. doi: 10.7150/ijbs.19531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, et al. Cells release prions in association with exosomes. Proc Natl Acad Sci USA. 2004;101(26):9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancho-Albero M, Navascués N, Mendoza G, Sebastián V, Arruebo M, Martín-Duque P, et al. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J Nanobiotechnol. 2019;17(1):16. doi: 10.1186/s12951-018-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collino F, Pomatto M, Bruno S, Lindoso RS, Tapparo M, Sicheng W, et al. Exosome and microvesicle-enriched fractions isolated from mesenchymal stem cells by gradient separation showed different molecular signatures and functions on renal tubular epithelial cells. Stem Cell Rev. 2017;13(2):226–243. doi: 10.1007/s12015-016-9713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oksvold MP, Kullmann A, Forfang L, Kierulf B, Li M, Brech A, et al. Expression of B-cell surface antigens in subpopulations of exosomes released from B-cell lymphoma cells. Clin Ther. 2014;36(6):847-62.e1. doi: 10.1016/j.clinthera.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Savvateeva EN, Tikhonov AA, Butvilovskaya VI, Tsybulskaya MV, Rubina AY. Exosomal surface protein markers in diagnosis of colorectal cancer. Mol Biol. 2017;51(5):752–760. doi: 10.7868/s0026898417050019. [DOI] [PubMed] [Google Scholar]
- 12.Zhu F, Shin OLSCL, Pei G, Hu Z, Yang J, Zhu H, et al. Adipose-derived mesenchymal stem cells employed exosomes to attenuate AKI-CKD transition through tubular epithelial cell dependent Sox9 activation. Oncotarget. 2017;8(41):70707–70726. doi: 10.18632/oncotarget.19979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan T, Jia P, Chen N, Fang Y, Liang Y, Guo M, et al. Delayed remote ischemic preconditioning confers renoprotection against septic acute kidney injury via exosomal miR-21. Theranostics. 2019;9(2):405–423. doi: 10.7150/thno.29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen F, Li Q. Exosomes as diagnostic biomarkers in cardiovascular diseases. Adv Exp Med Biol. 2017;998:61–70. doi: 10.1007/978-981-10-4397-0_4. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez S, Suazo C, Boltansky A, Ursu M, Carvajal D, Innocenti G, et al. Urinary exosomes as a source of kidney dysfunction biomarker in renal transplantation. Transpl Proc. 2013;45(10):3719–3723. doi: 10.1016/j.transproceed.2013.08.079. [DOI] [PubMed] [Google Scholar]
- 16.Bhome R, Goh R, Pickard K, Mellone M, Sayan AE, Mirnezami A. Profiling the microRNA payload of exosomes derived from ex vivo primary colorectal fibroblasts. Methods Mol Biol (Clifton, NJ). 2017;1509:115–122. doi: 10.1007/978-1-4939-6524-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eirin A, Riester SM, Zhu XY, Tang H, Evans JM, O’Brien D, et al. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene. 2014;551(1):55–64. doi: 10.1016/j.gene.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eirin A, Zhu X-Y, Puranik AS, Woollard JR, Tang H, Dasari S, et al. Integrated transcriptomic and proteomic analysis of the molecular cargo of extracellular vesicles derived from porcine adipose tissue-derived mesenchymal stem cells. PloS One. 2017;12(3):e0174303. doi: 10.1371/journal.pone.0174303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jafari D, Malih S, Eslami SS, Jafari R, Darzi L, Tarighi P, et al. The relationship between molecular content of mesenchymal stem cells derived exosomes and their potentials: opening the way for exosomes based therapeutics. Biochimie. 2019;165:76–89. doi: 10.1016/j.biochi.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Hood JL. Post isolation modification of exosomes for nanomedicine applications. Nanomedicine. 2016;11(13):1745–1756. doi: 10.2217/nnm-2016-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Dong Y, Li Y, Li W, Cheng K, Qian Y, et al. Designer exosomes for active targeted chemo-photothermal synergistic tumor therapy. Adv Funct Mater. 2018;28(18):1707360. doi: 10.1002/adfm.201707360. [DOI] [Google Scholar]
- 22.Hood JL, Wickline SA. A systematic approach to exosome-based translational nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4(4):458–467. doi: 10.1002/wnan.1174. [DOI] [PubMed] [Google Scholar]
- 23.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 24.Paolicelli RC, Bergamini G, Rajendran L. Cell-to-cell communication by extracellular vesicles: focus on microglia. Neuroscience. 2018 doi: 10.1016/j.neuroscience.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Tai Y-L, Chen K-C, Hsieh J-T, Shen T-L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018;109(8):2364–2374. doi: 10.1111/cas.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weston WW, Ganey T, Temple HT. The relationship between exosomes and cancer: implications for diagnostics and therapeutics. BioDrugs. 2019;33(2):137–158. doi: 10.1007/s40259-019-00338-5. [DOI] [PubMed] [Google Scholar]
- 27.Almiñana C, Tsikis G, Labas V, Uzbekov R, da Silveira JC, Bauersachs S, et al. Deciphering the oviductal extracellular vesicles content across the estrous cycle: implications for the gametes-oviduct interactions and the environment of the potential embryo. BMC Genom. 2018;19(1):622. doi: 10.1186/s12864-018-4982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang N, Xiang L, He L, Yang G, Zheng J, Wang C, et al. Exosomes mediate epithelium-mesenchyme crosstalk in organ development. ACS Nano. 2017;11(8):7736–7746. doi: 10.1021/acsnano.7b01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun B, Peng J, Wang S, Liu X, Zhang K, Zhang Z, et al. Applications of stem cell-derived exosomes in tissue engineering and neurological diseases. Rev Neurosci. 2018;29(5):531–546. doi: 10.1515/revneuro-2017-0059. [DOI] [PubMed] [Google Scholar]
- 30.Huang C-C, Narayanan R, Alapati S, Ravindran S. Exosomes as biomimetic tools for stem cell differentiation: applications in dental pulp tissue regeneration. Biomaterials. 2016;111:103–115. doi: 10.1016/j.biomaterials.2016.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toh WS, Lai RC, Zhang B, Lim SK. MSC exosome works through a protein-based mechanism of action. Biochem Soc Trans. 2018;46:843–853. doi: 10.1042/BST20180079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Utsugi-Kobukai S, Fujimaki H, Hotta C, Nakazawa M, Minami M. MHC class I-mediated exogenous antigen presentation by exosomes secreted from immature and mature bone marrow derived dendritic cells. Immunol Lett. 2003;89(2):125–131. doi: 10.1016/S0165-2478(03)00128-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Jiang X, Bao J, Wang Y, Liu H, Tang L. Exosomes in pathogen infections: a bridge to deliver molecules and link functions. Front Immunol. 2018;9:90. doi: 10.3389/fimmu.2018.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sampaio NG, Cheng L, Eriksson EM. The role of extracellular vesicles in malaria biology and pathogenesis. Malar J. 2017;16(1):245. doi: 10.1186/s12936-017-1891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Liu Y, Xiu F, Wang J, Cong H, He S, et al. Characterization of exosomes derived from Toxoplasma gondii and their functions in modulating immune responses. Int J Nanomed. 2018;13:467–477. doi: 10.2147/ijn.s151110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jing H, He X, Zheng J. Exosomes and regenerative medicine: state of the art and perspectives. Transl Res. 2018;196:1–16. doi: 10.1016/j.trsl.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Lan FM, Qing Q, Pan Q, Hu M, Yu HM, Yue X. Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol. 2018;41(1):25–33. doi: 10.1007/s13402-017-0355-3. [DOI] [PubMed] [Google Scholar]
- 38.Gomari H, Moghadam MF, Soleimani M. Targeted cancer therapy using engineered exosome as a natural drug delivery vehicle. OncoTargets Ther. 2018;11:5753. doi: 10.2147/OTT.S173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathiyalagan P, Sahoo S. Exosomes-based gene therapy for microRNA delivery. Methods Mol Biol. 2017;1521:139–152. doi: 10.1007/978-1-4939-6588-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aline F, Bout D, Amigorena S, Roingeard P, Dimier-Poisson I. Toxoplasma gondii antigen-pulsed-dendritic cell-derived exosomes induce a protective immune response against T. gondii infection. Infect Immun. 2004;72(7):4127–4137. doi: 10.1128/iai.72.7.4127-4137.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan A, De La Peña H, Seifalian AM. The application of exosomes as a nanoscale cancer vaccine. Int J Nanomed. 2010;5:889–900. doi: 10.2147/IJN.S13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bell BM, Kirk ID, Hiltbrunner S, Gabrielsson S, Bultema JJ. Designer exosomes as next-generation cancer immunotherapy. Nanomedicine. 2016;12(1):163–169. doi: 10.1016/j.nano.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 44.Viaud S, Ullrich E, Zitvogel L, Chaput N. Exosomes for the treatment of human malignancies. Horm Metab Res. 2008;40(02):82–88. doi: 10.1055/s-2007-1022548. [DOI] [PubMed] [Google Scholar]
- 45.Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles—endogenous nanocarriers for targeted cancer therapy. Biochimica et Biophysica Acta Rev Cancer. 2014;1846(1):75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Shahabipour F, Banach M, Sahebkar A. Exosomes as nanocarriers for siRNA delivery: paradigms and challenges. Arch Med Sci. 2016;12(6):1324. doi: 10.5114/aoms.2016.62911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J Control Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andaloussi SE, Lakhal S, Mäger I, Wood MJ. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65(3):391–397. doi: 10.1016/j.addr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Headland SE, Jones HR, Norling LV, Kim A, Souza PR, Corsiero E, et al. Neutrophil-derived microvesicles enter cartilage and protect the joint in inflammatory arthritis. Sci Transl Med. 2015;7(315):315ra190. doi: 10.1126/scitranslmed.aac5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharmaceutica Sinica B. 2016;6(4):287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aryani A, Denecke B. Exosomes as a nanodelivery system: a key to the future of neuromedicine? Mol Neurobiol. 2016;53(2):818–834. doi: 10.1007/s12035-014-9054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 53.Hung ME, Leonard JN. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290(13):8166–8172. doi: 10.1074/jbc.M114.621383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stickney Z, Losacco J, McDevitt S, Zhang Z, Lu B. Development of exosome surface display technology in living human cells. Biochem Biophys Res Commun. 2016;472(1):53–59. doi: 10.1016/j.bbrc.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 55.Kooijmans SA, Aleza CG, Roffler SR, van Solinge WW, Vader P, Schiffelers RM. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J Extracell Vesicles. 2016;5(1):31053. doi: 10.3402/jev.v5.31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohno S-I, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rountree RB, Mandl SJ, Nachtwey JM, Dalpozzo K, Do L, Lombardo JR, et al. Exosome targeting of tumor antigens expressed by cancer vaccines can improve antigen immunogenicity and therapeutic efficacy. Cancer Res. 2011;71(15):5235–5244. doi: 10.1158/0008-5472.CAN-10-4076. [DOI] [PubMed] [Google Scholar]
- 58.Mentkowski KI, Snitzer JD, Rusnak S, Lang JK. Therapeutic potential of engineered extracellular vesicles. AAPS J. 2018;20(3):50. doi: 10.1208/s12248-018-0211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Dongen HM, Masoumi N, Witwer KW, Pegtel DM. Extracellular vesicles exploit viral entry routes for cargo delivery. Microbiol Mol Biol Rev. 2016;80(2):369–386. doi: 10.1128/MMBR.00063-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meyer C, Losacco J, Stickney Z, Li L, Marriott G, Lu BJI. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int J Nanomed. 2017;12:3153. doi: 10.2147/IJN.S133430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng Y, Schorey JS. Targeting soluble proteins to exosomes using a ubiquitin tag. Biotechnol Bioeng. 2016;113(6):1315–1324. doi: 10.1002/bit.25884. [DOI] [PubMed] [Google Scholar]
- 62.Sterzenbach U, Putz U, Low LH, Silke J, Tan SS, Howitt J. Engineered exosomes as vehicles for biologically active proteins. Mol Ther. 2017;25(6):1269–1278. doi: 10.1016/j.ymthe.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Gassart A, Géminard C, Février B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102(13):4336–4344. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- 64.Manfredi F, Di Bonito P, Arenaccio C, Anticoli S, Federico M. Incorporation of heterologous proteins in engineered exosomes. Methods Mol Biol. 2016;1448:249–260. doi: 10.1007/978-1-4939-3753-0_18. [DOI] [PubMed] [Google Scholar]
- 65.McNamara RP, Costantini LM, Myers TA, Schouest B, Maness NJ, Griffith JD, et al. Nef secretion into extracellular vesicles or exosomes is conserved across human and simian immunodeficiency viruses. MBio. 2018;9(1):e02344-17. doi: 10.1128/mbio.02344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anticoli S, Manfredi F, Chiozzini C, Arenaccio C, Olivetta E, Ferrantelli F, et al. An exosome-based vaccine platform imparts cytotoxic T lymphocyte immunity against viral antigens. Biotechnol J. 2018;13(4):e1700443. doi: 10.1002/biot.201700443. [DOI] [PubMed] [Google Scholar]
- 67.Yim N, Ryu SW, Choi K, Lee KR, Lee S, Choi H, et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat Commun. 2016;7:12277. doi: 10.1038/ncomms12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kojima R, Bojar D, Rizzi G, Hamri GC, El-Baba MD, Saxena P, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat Commun. 2018;9(1):1305. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sutaria DS, Jiang J, Elgamal OA, Pomeroy SM, Badawi M, Zhu X, et al. Low active loading of cargo into engineered extracellular vesicles results in inefficient miRNA mimic delivery. J Extracell Vesicles. 2017;6(1):1333882. doi: 10.1080/20013078.2017.1333882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, Zhou X, Wei M, Gao X, Zhao L, Shi R, et al. In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019;19(1):19–28. doi: 10.1021/acs.nanolett.8b02689. [DOI] [PubMed] [Google Scholar]
- 71.Naseri Z, Oskuee RK, Jaafari MR, Forouzandeh Moghadam M. Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int J Nanomed. 2018;13:7727–7747. doi: 10.2147/ijn.S182384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38(6):754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richardson JJ, Ejima H. Surface engineering of extracellular vesicles through chemical and biological strategies. Chem Mater. 2019;31(7):2191–2201. doi: 10.1021/acs.chemmater.9b00050. [DOI] [Google Scholar]
- 74.Murphy DE, de Jong OG, Brouwer M, Wood MJ, Lavieu G, Schiffelers RM, et al. Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Exp Mol Med. 2019;51(3):1–12. doi: 10.1038/s12276-019-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sancho-Albero M, Rubio-Ruiz B, Pérez-López AM, Sebastián V, Martín-Duque P, Arruebo M, et al. Cancer-derived exosomes loaded with ultrathin palladium nanosheets for targeted bioorthogonal catalysis. Nat Catal. 2019;2(10):864–872. doi: 10.1038/s41929-019-0333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu M, Gai C, Li Z, Ding D, Zheng J, Zhang W, et al. Targeted exosome-encapsulated erastin induced ferroptosis in triple negative breast cancer cells. Cancer Sci. 2019;110(10):3173–3182. doi: 10.1111/cas.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sawada SI, Sato YT, Kawasaki R, Yasuoka JI, Mizuta R, Sasaki Y, et al. Nanogel hybrid assembly for exosome intracellular delivery: effects on endocytosis and fusion by exosome surface polymer engineering. Biomater Sci. 2020;8(2):619–630. doi: 10.1039/c9bm01232j. [DOI] [PubMed] [Google Scholar]
- 78.Kumar S, Michael IJ, Park J, Granick S, Cho YK. Cloaked exosomes: biocompatible, durable, and degradable encapsulation. Small. 2018;14(34):e1802052. doi: 10.1002/smll.201802052. [DOI] [PubMed] [Google Scholar]
- 79.Li Y, Gao Y, Gong C, Wang Z, Xia Q, Gu F, et al. A33 antibody-functionalized exosomes for targeted delivery of doxorubicin against colorectal cancer. Nanomedicine. 2018;14(7):1973–1985. doi: 10.1016/j.nano.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 80.Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen I, Klyachko NL, et al. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine. 2018;14(1):195–204. doi: 10.1016/j.nano.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 81.Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol. 2018;13(1):82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J, Li W, Zhang L, Ban L, Chen P, Du W, et al. Chemically edited exosomes with dual ligand purified by microfluidic device for active targeted drug delivery to tumor cells. ACS Appl Mater Interfaces. 2017;9(33):27441–27452. doi: 10.1021/acsami.7b06464. [DOI] [PubMed] [Google Scholar]
- 83.Kooijmans SAA, Fliervoet LAL, van der Meel R, Fens M, Heijnen HFG, van Henegouwen PMPBE, et al. PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. J Control Release. 2016;224:77–85. doi: 10.1016/j.jconrel.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 84.Srivastava A, Amreddy N, Babu A, Panneerselvam J, Mehta M, Muralidharan R, et al. Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells. Sci Rep. 2016;6(1):38541. doi: 10.1038/srep38541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rana S, Yue S, Stadel D, Zöller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol. 2012;44(9):1574–1584. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 86.Lee YS, Kim SH, Cho JA, Kim CW. Introduction of the CIITA gene into tumor cells produces exosomes with enhanced anti-tumor effects. Exp Mol Med. 2011;43(5):281. doi: 10.3858/emm.2011.43.5.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bryniarski K, Ptak W, Jayakumar A, Püllmann K, Caplan MJ, Chairoungdua A, et al. Antigen-specific, antibody-coated, exosome-like nanovesicles deliver suppressor T-cell microRNA-150 to effector T cells to inhibit contact sensitivity. J Allergy Clin Immunol. 2013;132(1):170.e9–181.e9. doi: 10.1016/j.jaci.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grapp M, Wrede A, Schweizer M, Hüwel S, Galla H-J, Snaidero N, et al. Choroid plexus transcytosis and exosome shuttling deliver folate into brain parenchyma. Nat Commun. 2013;4:2123. doi: 10.1038/ncomms3123. [DOI] [PubMed] [Google Scholar]
- 89.Nag O, Awasthi V. Surface engineering of liposomes for stealth behavior. Pharmaceutics. 2013;5(4):542–569. doi: 10.3390/pharmaceutics5040542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Antes TJ, Middleton RC, Luther KM, Ijichi T, Peck KA, Liu WJ, et al. Targeting extracellular vesicles to injured tissue using membrane cloaking and surface display. J Nanobiotechnol. 2018;16(1):61. doi: 10.1186/s12951-018-0388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamura R, Uemoto S, Tabata Y. Augmented liver targeting of exosomes by surface modification with cationized pullulan. Acta Biomater. 2017;57:274–284. doi: 10.1016/j.actbio.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 92.Ye Z, Zhang T, He W, Jin H, Liu C, Yang Z, et al. Methotrexate-loaded extracellular vesicles functionalized with therapeutic and targeted peptides for the treatment of glioblastoma multiforme. ACS Appl Mater Interfaces. 2018;10(15):12341–12350. doi: 10.1021/acsami.7b18135. [DOI] [PubMed] [Google Scholar]
- 93.Tian T, Zhang H-X, He C-P, Fan S, Zhu Y-L, Qi C, et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. 2018;150:137–149. doi: 10.1016/j.biomaterials.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 94.Li S, Wu Y, Ding F, Yang J, Li J, Gao X, et al. Engineering macrophage-derived exosomes for targeted chemotherapy of triple-negative breast cancer. Nanoscale. 2020;12(19):10854–10862. doi: 10.1039/d0nr00523a. [DOI] [PubMed] [Google Scholar]
- 95.Bellavia D, Raimondo S, Calabrese G, Forte S, Cristaldi M, Patinella A, et al. Interleukin 3-receptor targeted exosomes inhibit in vitro and in vivo chronic myelogenous leukemia cell growth. Theranostics. 2017;7(5):1333–1345. doi: 10.7150/thno.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 97.Qu M, Lin Q, Huang L, Fu Y, Wang L, He S, et al. Dopamine-loaded blood exosomes targeted to brain for better treatment of Parkinson’s disease. J Control Release. 2018;287:156–166. doi: 10.1016/j.jconrel.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 98.Zhang H, Wu J, Wu J, Fan Q, Zhou J, Wu J, et al. Exosome-mediated targeted delivery of miR-210 for angiogenic therapy after cerebral ischemia in mice. J Nanobiotechnol. 2019;17(1):29. doi: 10.1186/s12951-019-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Y, Chen X, Tian B, Liu J, Yang L, Zeng L, et al. Nucleolin-targeted extracellular vesicles as a versatile platform for biologics delivery to breast cancer. Theranostics. 2017;7(5):1360–1372. doi: 10.7150/thno.16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khongkow M, Yata T, Boonrungsiman S, Ruktanonchai UR, Graham D, Namdee K. Surface modification of gold nanoparticles with neuron-targeted exosome for enhanced blood–brain barrier penetration. Sci Rep. 2019;9(1):8278. doi: 10.1038/s41598-019-44569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gomari H, Moghadam MF, Soleimani M, Ghavami M, Khodashenas S. Targeted delivery of doxorubicin to HER2 positive tumor models. Int J Nanomed. 2019;14:5679–5690. doi: 10.2147/IJN.S210731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zou X, Yuan M, Zhang T, Wei H, Xu S, Jiang N, et al. Extracellular vesicles expressing a single-chain variable fragment of an HIV-1 specific antibody selectively target Env(+) tissues. Theranostics. 2019;9(19):5657–5671. doi: 10.7150/thno.33925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Longatti A, Schindler C, Collinson A, Jenkinson L, Matthews C, Fitzpatrick L, et al. High affinity single-chain variable fragments are specific and versatile targeting motifs for extracellular vesicles. Nanoscale. 2018;10(29):14230–14244. doi: 10.1039/c8nr03970d. [DOI] [PubMed] [Google Scholar]
- 104.Wang J, Li W, Lu Z, Zhang L, Hu Y, Li Q, et al. The use of RGD-engineered exosomes for enhanced targeting ability and synergistic therapy toward angiogenesis. Nanoscale. 2017;9(40):15598–15605. doi: 10.1039/c7nr04425a. [DOI] [PubMed] [Google Scholar]
- 105.Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F. Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl Biochem Biotechnol. 2019;187(1):352–364. doi: 10.1007/s12010-018-2813-4. [DOI] [PubMed] [Google Scholar]
- 106.Liang G, Kan S, Zhu Y, Feng S, Feng W, Gao S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int J Nanomed. 2018;13:585–599. doi: 10.2147/ijn.S154458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhu L, Dong D, Yu ZL, Zhao YF, Pang DW, Zhang ZL. Folate-engineered microvesicles for enhanced target and synergistic therapy toward breast cancer. ACS Appl Mater Interfaces. 2017;9(6):5100–5108. doi: 10.1021/acsami.6b14633. [DOI] [PubMed] [Google Scholar]
- 108.Ferrantelli F, Arenaccio C, Manfredi F, Olivetta E, Chiozzini C, Leone P, et al. The intracellular delivery of anti-HPV16 E7 scFvs through engineered extracellular vesicles inhibits the proliferation of HPV-infected cells. Int J Nanomed. 2019;14:8755–8768. doi: 10.2147/ijn.S209366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vandergriff A, Huang K, Shen D, Hu S, Hensley MT, Caranasos TG, et al. Targeting regenerative exosomes to myocardial infarction using cardiac homing peptide. Theranostics. 2018;8(7):1869–1878. doi: 10.7150/thno.20524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu Y, Li D, Liu Z, Zhou Y, Chu D, Li X, et al. Targeted exosome-mediated delivery of opioid receptor Mu siRNA for the treatment of morphine relapse. Sci Rep. 2015;5(1):17543. doi: 10.1038/srep17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jia G, Han Y, An Y, Ding Y, He C, Wang X, et al. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials. 2018;178:302–316. doi: 10.1016/j.biomaterials.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 112.Jang SC, Kim OY, Yoon CM, Choi D-S, Roh T-Y, Park J, et al. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano. 2013;7(9):7698–7710. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- 113.Nedergaard MK, Hedegaard CJ, Poulsen HS. Targeting the epidermal growth factor receptor in solid tumor malignancies. BioDrugs. 2012;26(2):83–99. doi: 10.2165/11599760-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 114.Carney RP, Hazari S, Rojalin T, Knudson A, Gao T, Tang Y, et al. Targeting tumor-associated exosomes with integrin-binding peptides. Adv Biosyst. 2017 doi: 10.1002/adbi.201600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luo Q, Guo D, Liu G, Chen G, Hang M, Jin MJCP, et al. Exosomes from mir-126-overexpressing adscs are therapeutic in relieving acute myocardial ischaemic injury. Cell Physiol Biochem. 2017;44(6):2105–2116. doi: 10.1159/000485949. [DOI] [PubMed] [Google Scholar]
- 116.Xitong D, Xiaorong ZJG. Targeted therapeutic delivery using engineered exosomes and its applications in cardiovascular diseases. Gene. 2016;575(2):377–384. doi: 10.1016/j.gene.2015.08.067. [DOI] [PubMed] [Google Scholar]
- 117.Liu B, Lee BW, Nakanishi K, Villasante A, Williamson R, Metz J, et al. Cardiac recovery via extended cell-free delivery of extracellular vesicles secreted by cardiomyocytes derived from induced pluripotent stem cells. Nat Biomed Eng. 2018;2(5):293. doi: 10.1038/s41551-018-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim H, Yun N, Mun D, Kang JY, Lee SH, Park H, et al. Cardiac-specific delivery by cardiac tissue-targeting peptide-expressing exosomes. Biochem Biophys Res Commun. 2018;499(4):803–808. doi: 10.1016/j.bbrc.2018.03.227. [DOI] [PubMed] [Google Scholar]
- 119.Wang X, Chen Y, Zhao Z, Meng Q, Yu Y, Sun J, et al. Engineered exosomes with ischemic myocardium-targeting peptide for targeted therapy in myocardial infarction. J Am Heart Assoc. 2018;7:15. doi: 10.1161/jaha.118.008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pizzicannella J, Gugliandolo A, Orsini T, Fontana A, Ventrella A, Mazzon E, et al. Engineered extracellular vesicles from human periodontal-ligament stem cells increase VEGF/VEGFR2 expression during bone regeneration. Front Physiol. 2019;10:512. doi: 10.3389/fphys.2019.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jarvinen TA, Ruoslahti E. Targeted antiscarring therapy for tissue injuries. Adv Wound Care. 2013;2(2):50–54. doi: 10.1089/wound.2011.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Samadikuchaksaraei A, Mehdipour A, Habibi Roudkenar M, Verdi J, Joghataei MT, Aśadi K, et al. A dermal equivalent engineered with TGF-β3 expressing bone marrow stromal cells and amniotic membrane: cosmetic healing of full-thickness skin wounds in rats. Artif Org. 2016;40(12):E266–E279. doi: 10.1111/aor.12807. [DOI] [PubMed] [Google Scholar]
- 123.Tang T, Jiang H, Yu Y, He F, Ji SZ, Liu YY, et al. A new method of wound treatment: targeted therapy of skin wounds with reactive oxygen species-responsive nanoparticles containing SDF-1alpha. Int J Nanomed. 2015;10:6571–6585. doi: 10.2147/ijn.S88384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Toba M, Alzoubi A, O’Neill K, Abe K, Urakami T, Komatsu M, et al. A novel vascular homing peptide strategy to selectively enhance pulmonary drug efficacy in pulmonary arterial hypertension. Am J Pathol. 2014;184(2):369–375. doi: 10.1016/j.ajpath.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem cell Res. 2010;4(3):214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 126.Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. 2015;32(6):2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ramezani A, Haddad R, Sedaghati B, Jafari DJSA. Effects of fungal extracts on vinblastine and vincristine production and their biosynthesis pathway genes in Catharanthus roseus. S Afr J Bot. 2018;119:163–171. doi: 10.1016/j.sajb.2018.08.015. [DOI] [Google Scholar]
- 128.Schorey JS, Cheng Y, Singh PP, Smith VL. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015;16(1):24–43. doi: 10.15252/embr.201439363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Davod J, Fatemeh DN, Honari H, Hosseini R. Constructing and transient expression of a gene cassette containing edible vaccine elements and shigellosis, anthrax and cholera recombinant antigens in tomato. Mol Biol Rep. 2018;45(6):2237–2246. doi: 10.1007/s11033-018-4385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jafari D, Dehghan NF, Honari H, Hoseini R, Jafari R. Bioinformatic analysis of different fusions of ipaD, PA20 and CTxB antigens: a preliminary analysis for vaccine design. Genetics. 2016;14:4234–4241. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Further information is available from the first author and corresponding author on request.