Abstract
In Lake Malawi, two ecologically distinct lineages of cichlid fishes (rock- versus sand-dwelling ecotypes, each comprised of over 200 species) evolved within the last million years. The rock-dwelling species (Mbuna) are aggressively territorial year-round and males court and spawn with females over rocky substrate. In contrast, males of sand-dwelling species are not territorial and instead aggregate on seasonal breeding leks in which males construct courtship “bowers” in the sand. However, little is known about how phenotypic variation in aggression is produced by the genome. In this study, we first quantify and compare behavior in seven cichlid species, demonstrating substantial ecotype and species differences in unconditioned mirror-elicited aggression. Second, we compare neural activity in mirror-elicited aggression in two representative species, Mchenga conophoros (sand-dwelling) and Petrotilapia chitimba (rock-dwelling). Finally, we compare gene expression patterns between these two species, specifically within neurons activated during mirror aggression. We identified a large number of genes showing differential expression in mirror-elicited aggression, as well as many genes that differ between ecotypes. These genes, which may underly species differences in behavior, include several neuropeptides, genes involved in the synthesis of steroid hormones, and neurotransmitter activity. This work lays the foundation for future experiments using this emerging genetic model system to investigate the genomic basis of evolved species differences in both brain and behavior.
Keywords: Cichlid fish, aggression, social behavior, RNA-seq, PhosphoTRAP, pS6, neural induction, gene expression, evolution, species difference
1. Introduction:
Animals exhibit aggressive behavior for many reasons, including acquiring and defending limited resources, subduing prey, protecting vulnerable offspring, and competing for mates. However, the specific forms of agonistic behavior vary widely across species and contexts. Aggression, overt behavior with the intention of inflicting harm on another individual, is a polygenic trait with strong environmental components. In both humans and nonhuman animals, aggression is highly heritable, with approximately 50% of variance in aggression being associated with genotype1-9. Unsurprisingly, the number of genes and genetic variants that have been associated with variation in aggressive phenotypes using genome-wide association studies is very large10-12.
The evolution of aggressive behavior and its complex genetic architecture has been the subject of study since the early days of ethology13. Animal studies have implicated several conserved and well-studied neural and physiological pathways involved in aggression, including sex steroid hormones (androgens and estrogens), serotonin (5-HT), nonapeptides (arginine vasopressin, AVP, and oxytocin, OT), γ-aminobutryic-acid (GABA), and dopamine (DA)14,15. Although these neural and molecular systems have been well-characterized, we still have a very poor understanding of how phenotypic variation in aggression is produced by the genome. A better understanding of the genetic basis of aggression would provide insight into the mechanisms of behavioral diversity, as well as point us to genetic and neural networks that may be therapeutic targets in the treatment of behavioral disorders.
Unbiased data-driven approaches in tractable animal systems—such as transcriptomic profiling or quantitative trait loci (QTL) mapping—have the potential to uncover novel genetic mechanisms underlying behavioral variability. Several studies have successfully mapped QTL in inbred lab strains of mice or Drosophila which vary in aggressive behavior16,17. Additionally, transcriptomic analyses have uncovered many differentially expressed genes in rats, canines, and Silver foxes artificially selected for either aggression or tameness18-20. Unbiased methods such as these are critical for discovering genetic variants and relevant molecular pathways, but these studies also have important drawbacks: the limited generalizability of genetic variants found in the context of a lab-adapted, inbred, or artificially selected organisms.
In the current study, we take advantage of a naturally occurring species difference in aggressive behavior in a genetically tractable animal system, Lake Malawi cichlid fish. In Lake Malawi, two ecologically distinct groups of cichlid fish species (rock- versus sand-dwelling ecotypes, each comprising over 200 species) have evolved within the last million years21. The adaptive radiation of Malawi cichlids has led to remarkable phenotypic diversity in both brain and behavior, yet Malawi cichlid species possess remarkably similar genomes and share polymorphism due to both periodic hybridization and retention of ancestral variation22-24. The rock-dwelling species (also known as Mbuna) are aggressively territorial year-round and males court and spawn with females over the rocky substrate. Border contests between territorial Mbuna males involve overt physical aggression, including face-to-face lunges and jaw locking25. In contrast, males of the sand-dwelling species are not territorial and instead aggregate on seasonal breeding leks in which each male constructs a courtship “bower” in the sand where he displays to females26. Sand males exhibit a wide range of agonistic behaviors to defend their bowers from rival males, though it has been hypothesized that bowers and their size, as well as a number of display behaviors, serve to reduce physical aggression among sand species27-29.
Although ecotype differences in aggression have been reported in field studies in Lake Malawi25,30-33, little is known about the neural and genetic basis of this difference. To quantify species differences in aggressive behavior in males under controlled conditions, we employed a classic mirror test assay34. African cichlids, like other fish, do not recognize themselves in the mirror and reliably respond aggressively towards their own reflection35-37. Mirror tests are conceptually similar to resident-intruder tests, very reliably elicit aggressive behaviors, and have been used extensively to measure unconditioned agonistic behavior in fish. A number of researchers have criticized whether both behavior and neural responses in the mirror test are ecologically valid37-41. However, mirror tests have the advantage of eliminating the variance introduced by the opponent and minimizing the risk of injury associated with real-life aggressive encounters. Furthermore, behavior in mirror tests has repeatedly been found to positively correlate with aggression during live agonistic trials37,42,43.
Here, we quantify and compare behavior during the mirror test in seven species of Lake Malawi cichlid (three sand- and four rock-dwelling species) and demonstrate substantial ecotype and species differences in unconditioned mirror-elicited aggression. Second, we compare neural activity in mirror-elicited aggression in two representative species, Mchenga conophoros (MC, sand) and Petrotilapia chitimba (PC, rock). Finally, we compare gene expression patterns between these two species specifically within neurons activated during mirror aggression using PhosphoTRAP44,45. This method uses antibodies to phosphorylated ribosomal protein S6 (pS6) to enrich for transcripts bound to phosphorylated ribosomes. In neurons, this phosphorylation occurs downstream of the binding of neurotransmitters. Thus, pS6 antibodies are increasingly being used to label neurons activated by a stimulus, similar to immediate early genes (IEGs) like c-fos or egr-1, especially in non-traditional or non-mammalian systems46-50. Using whole brain samples, we use this unbiased RNA-sequencing approach and identify a large number of genes associated with species differences in aggressive behaviors. Several of these candidate genes are in peptide and hormonal signaling pathways. In addition, we consider the analytical challenges of PhosphoTRAP data sets and suggest next steps for future analyses.
2. Materials & Methods:
2.1. Animals
Adult Malawi cichlids were housed in a recirculating aquarium system at 28°C. Water quality monitoring, feedings of spirulina flakes (Pentair ZSF25, 2x daily), and husbandry were provided by the Georgia Tech Physiological Research Laboratory staff. All animal experiments were approved by the Georgia Tech Institutional Animal Care and Use Committee (Protocol # A100028).
2.2. Mirror Test Assay & Behavior Scoring
The following protocol was used for all mirror assays. Mature, mirror-naïve males observed were removed from mixed-sex social tanks and were housed alone in a 95-liter tank (0.44m x 0.44m x 0.41m) for 6 days. Each experimental housing tank was equipped with a single PVC pipe on a randomly selected side to serve as a shelter and facilitate territory establishment. Each tank was surrounded on 3 sides with white plastic, to prevent visual interaction between adjacent tanks.
On the seventh day, a 0.3m x 0.3m square mirror was introduced to the tank against the wall of same side of the tank as the PVC pipe shelter (Fig. 1a). Behavior was filmed using a GoPro 4.0 HD video camera for 15 min, after which the mirror was removed from the tank. In the control condition, the same mirror was introduced backwards, with the opaque gray backing of the mirror facing the tank. This resulted in a similar disturbance, but there was no reflective surface with which the fish could interact. All mirror tests were conducted between 11:00 and 16:00.
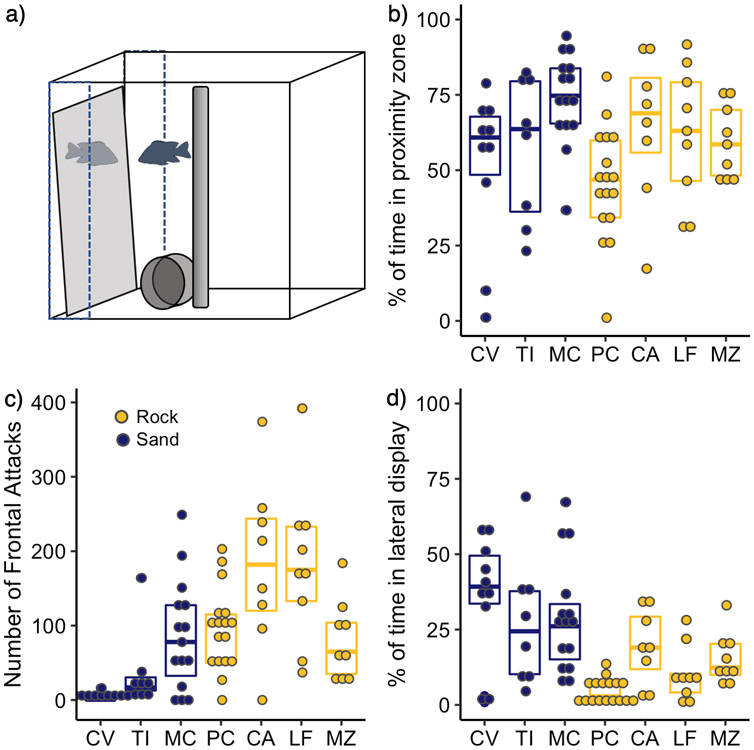
Figure 1:
a) Diagram of mirror test aggression assay. Dotted lines delineate the zone of proximity within 5cm of the mirror surface. b) Boxplot of % of 15min assay spent (within 5cm of the mirror surface (Rock vs. sand, Z = −1.98, p = 0.047) c) Number of frontal attacks (Z = 4.20, p < 0.0001) d) Time (s) performing lateral displays during (Z = 4.67, p < 0.0001). Sand species are shown in blue; rock species are in yellow. Species: C. virginalis, CV (n = 10); T. intermedius, TI (n = 8); M. conophoros, MC (n = 15); P. chtimba, PC (n = 17), C. afra, CA (n = 8); L. fuelleborni, LF (n = 9); M. zebra, MZ (n = 9).
Behavior was analyzed by a trained observer using Noldus Observer XT. We recorded the amount of time subjects spent in proximity to the mirror (within 5cm of the mirror surface), performing lateral displays (body oriented parallel to the mirror with fins extended), and with their nuptial colors ‘on’. In addition, we scored the occurrence of frontal attacks (i.e. bites and/or open mouth contact with the mirror). Although other aggressive behaviors such as body quivering and operculum flaring displays were occasionally observed, these occurred at very low frequencies in a small number of individuals and were thus excluded from the analyses.
2.3. Behavioral Experiments
To test the hypothesis that males of rock species and sand species differ in aggressive behavior, we tested seven species of Lake Malawi cichlid (3 sand and 4 rock species) in the mirror test assay: Copadichromis virginalis (CV, sand, n = 8), Tramitichromis intermedius (TI, sand, n = 8), Mchenga conophoros (MC, sand, n = 15), Petrotilapia chitimba (PC, rock, n = 17), Cynotilapia afra cobue (CA, rock, n = 8), Labeotropheus fuelleborni (LF, rock, n = 9), and Metriaclima zebra (MZ, rock, n = 9). In this behavioral comparison, all subjects were in the mirror condition.
In order to identify the brain regions which exhibited differential activation during mirror-elicited aggression, we compared pS6 immunostaining between two representative species, MC and PC, in either the mirror test (MC, n = 7; PC, n = 6) or in the control condition (MC, n = 6; PC, n = 6). These species were chosen because they are representative of the ecotype difference (particularly in terms of lateral display behavior), well-studied, and breed readily in the lab. In the pS6 RNA-Immunoprecipitation experiment, we compared behaviorally relevant gene expression in the same two species (MC and PC) in either the mirror test or in the control condition, with 2 males per group (n = 8 total).
2.4. Tissue Collection
Ninety-minutes after the introduction of the mirror or control stimulus (75 min after the end of the stimulus), the subjects were sacrificed by rapid decapitation. For tissue processed for immunohistochemistry, the top of the skull was removed and the whole head was immersed overnight in 4% paraformaldehyde (PFA) in 1x PBS, pH 7.4 at 4°C. After fixation, the tissue was washed in 1x PBS and carefully removed from the skull. The brain was then cryoprotected in 30% sucrose in 1x PBS at 4°C overnight and frozen in embedding medium and stored at −80°C until sectioning. The brain was sectioned in the coronal plane at 20μm in four series, thaw-mounted onto positively-charged slides, and stored at −80°C until immunohistochemistry. For tissue used for pS6-immunoprecipitation, the brain was fully extracted from the skull in under 3min and flash frozen in cold 2-methylbutane and stored at −80°C until immunoprecipitation.
2.5. Immunohistochemistry
Sectioned brains were labeled with an antibody to neural activation marker phospho-S6-ribsomal protein (pS6). Slides were air-dried at room temperature for 40min. The tissue was fixed a second time on the slides for 5min in chilled 4% PFA and then rinsed in 1x PBS. To inactivate endogenous peroxidases, the slides were incubated in 1% hydrogen peroxide in 1x PBS for 20min and then rinsed in 1x PBS 3 times for 5min each. To block nonspecific binding, the slides were incubated for 1 hr in a 5% blocking solution (5% normal goat serum and 0.3% Triton X-100 in 1x PBS) followed by 1x PBS for 2min. The sections were then incubated in the same pS6 primary antibody used in the pS6-immunoprecipitation (1:300, Cell Signal #2215 pS6 ribosomal protein S240/244 antibody, 2% NGS, 0.3% Triton X-100 in 1x PBS) overnight at room temperature. Slides were then rinsed in 1x PBS 3 times for 5min each. Next, sections were stained using the Vectastain ABC Anti-Rabbit IgG Kit (VectorLabs PK-6101) per manufacturer’s instructions and visualized by reaction with DAB for ~15min. Slides were rinsed in distilled water and mounted with an aqueous mounting medium (Vector Labs H-5501). Tissue was processed in two batches.
Slides were analyzed using a Zeiss AxioObserver Z1 brightfield microscope. We quantified the number of cells in five brain regions previously implicated in aggressive behavior in fish: the granular zone of lateral zone of the dorsal telencephalon, Dl-g; ventral part of the lateral zone of the dorsal telencephalon, subdivision 2, Dl-v2; medial part of the dorsal telencephalon, subdivision 1, Dm-1; medial part of the dorsal telencephalon, subdivision 3, Dm-3; and the anterior parvocellular preoptic nucleus, which is homologous in part to the preoptic area (POA) in other vertebrates51. Images were captured from 1 to 6 sections per side, depending on structure size and tissue quality. Cells were quantified by hand, using the Image J Cell Counter plugin to keep track of cell counts within fixed counting frames which varied depending on the shape of the structure: 180 x 360 μm counting frame for Dm-1 (oval) and POA (rectangle), a 360 μm circle for Dl-g, and a 360 x 360 μm square for both Dm-3 and Dl-v2.
2.6. pS6 RNA-Immunoprecipitation (i.e. PhosphoTRAP)
The following methods are adapted from 44 and 45. Prior to the immunoprecipitation (IP), magnetic Protein A Dynabeads (Life Technologies 10001D) were loaded with pS6 antibody (Cell Signal #2215). Note that the same pS6 antibody was used for both the IHC and the pS6-immunoprecipitation. For each IP, we rinsed 100μL of Dynabeads in 0.15 KCl wash buffer (10mM HEPES,150mM KCl, 5mM MgCl2, and 1% NP40 in RNAse-free water prepared in 500mL sterile disposable filter units). Beads were then resuspended in 250μL of 0.15 KCl buffer for each IP. Then 20μL of antibody and 30μL of BSA was added per IP. The antibody-bound beads were mixed end-over-end overnight at 4°C.
The following homogenization steps were performed on ice in the fume hood. All procedures in the homogenization and immunoprecipitation steps were performed with low-bind microcentrifuge tubes. Samples were processed in two batches, each representing a balanced experimental block. Each batch was processed on a separate day. Whole brains were homogenized in 2mL of homogenization buffer [10 mM HEPES, 150mM KCl, 5mM MgCl2 in RNAse-free water prepared in advance in 500mL sterile disposable filter units with the following supplements added per 2 mL aliquot within 1 hour: 5μL of 1M DTT, 1/2 EDTA-free protease inhibitor tablet (Roche 11836170001), 5μL RNAsin (Promega N2611), 5μL cycloheximide (100mg/mL stock in DMSO), 10μL phosphatase inhibitor cocktail (ThermoScientific 78440), and 2μL of Calyculin A (1mg/mL in ethanol stock].
Immediately after removal from the −80°C, the frozen brain was homogenized in 1.4mL homogenization buffer on ice using a RNAse Away-treated glass pestle and tube. The supernatant was then transferred to two microcentrifuge tubes (~700μL each) and each were topped with 300μL of additional homogenization buffer to ~1mL. The homogenized lysate was then centrifuged at 2000xg for 10 min at 4°C. The supernatant from each tube was transferred to new tubes and 70μL of 10% NP40 and 70μL DHPC (300 mM stock in chloroform) was added to each ~1mL tube. The tubes were mixed briefly and then incubated for 2 min on ice. The tubes were then spun at Rmax for 10 min at 4°C. The supernatant was transferred again to two new tubes. For the input RNA sample, 25μL from each tube (50μL total) of this supernatant was added to 350μL of buffer RLT (Qiagen RNeasy Micro Kit) and stored on ice until purification
Next, we rinsed the pS6-loaded magnetic beads 2x in 0.15M KCl buffer and distributed the beads into two microcentrifuge tubes for each IP. After the second wash, the 0.15M KCl buffer was removed and then ~1mL of clarified supernatant was added to each tube and mixed to resuspend the beads. The tubes were incubated with end-over-end mixing for 10 min at 4°C, which allows for antibody binding. After a brief spin, the tubes were placed on the magnet, and the supernatant was removed. The beads were then rinsed 4x in 0.9mL of cold 0.35M KCl buffer (10mM HEPES, 350mM KCl, 5mM MgCl2, 1% NP40, plus the following supplements added per 4mL: 5μL 1M DTT, 10μL RNAsin, 5μL cycloheximide, and 2μL Calyculin A). After the 3rd wash, the samples incubated at room temperature for 5 min, which allows for the dissociation of some of the non-specific RNA. After the 4th wash, the supernatant was removed, and the beads were resuspended in 350μL of buffer RLT and incubated on ice for 5 min.
RNA from both the input and IP samples was purified using the Qiagen RNeasy Micro Kit (Qiagen 74004), with two minor modifications to the protocol. First, we added 350μL of 70% ethanol to the IP RNA in RLT and 375μL of 70% ethanol to the input RNA in RLT. Second, in order to maximize yield, we used 14μL of RNase-free water which had been heated to 65°C during the final elution step. RNA samples were analyzed using the Agilent RNA 6000 Pico kit. Input RNA had RNA integrity number (RIN) scores ranged 7.1-8.6 and concentrations of 658-4,571pg/μL. IP RNA had RIN scores of 6.8-8.8 and concentrations of 22-100pg/μL.
cDNA synthesis was performed using the SMART-Seq v4 Ultra Low Input RNA kit (Takara Bio 634888). We used 2μL of input RNA and the 10.5μL of the remaining IP RNA for cDNA synthesis and the maximum number of PCR cycles (18) during the cDNA amplification step. We analyzed cDNA samples using the Agilent DNA High Sensitivity kit. cDNA samples had concentrations of 10 - 75ng/μL for input samples and 0.340-1.5ng/μL for IP samples as measured by DeNovix Microvolume Spectrophotometer.
2.7. Library Preparation & Sequencing
Library preparation was performed by the Molecular Evolution core facility at GT using NEBNext Ultra II FS DNA library Prep Kit for Illumina (NEB E7805S). Either 1ng (IP samples), or 5ng (input samples) of cDNA were used as a starting material for the library preparation. The enzymatic fragmentation was done by adding 7μL of Ultra II FS Buffer and 2μL Ultra II FS Enzyme mix to each sample. The samples were then incubated at 37°C for 15min and 65°C for 30min in a thermocycler. Adaptor ligation was performed in accordance to the manual for NEB Ultra II FS DNA library Prep Kit. The adaptors were diluted 25-fold for IP cDNA samples and 10-fold for all other samples. A mix of 30μL of NEBNext Ultra II Ligation Master Mix, 1μL NEBNext Ligation Enhancer and 2.5μL NEBNext diluted Adaptor for Illumina was added to the FS reaction mixture and incubated at 20°C for 15min a thermocycler. 3μl of USER® Enzyme were added to the ligation mixture and incubated at 37°C for 15min with the heated lid set to ≥ 47°C.
Adaptor clean-up was performed without size selection because of the low initial DNA quantity. NEBNext Sample purification beads were used in this process by adding 57μL (0.8X) of the beads to the adaptor ligation reaction. The samples were incubated at room temperature for 5min, then placed on magnetic stand for 5min to separate the beads from the supernatant. Supernatant was discarded. The beads were washed by adding 200μL freshly prepared 80% ethanol to the tubes while still on the magnetic plate; incubated for 30sec, and then discarded. This step was repeated once. The beads then were air dried for 5min while still standing on the magnetic plate. The DNA was than eluted by adding 17μL of 0.1X TE buffer directly to the beads. After mixing the beads with TE buffer, the samples were incubated at room temperature for 2min, then placed back on the magnetic stand until the supernatant was cleared.
PCR enrichment of Adaptor-ligated DNA was performed with the use of 15μL of eluted DNA for each sample. A mix of the following components was added to each 15μL: 25μL NEBNext Ultra II Q5 Master Mix, 5μL Index Primer/i7 Primer and 5μL Universal PCR Primer/i5 Primer (Total reaction volume 50μL). The samples were pooled by equimolar normalization and sequenced on together in a single run on an Illumina NextSeq 500 instrument with 76 cycles (single-end).
2.8. RNA-sequencing Analyses
Raw FASTQ files were processed using cutadapt to 1) trim sequences from the SmartSeq kit, Illumina TruSeq Indexed adapter, and both poly-A and poly-T contamination and 2) exclude sequences shorter than 25bp. The remaining sequence reads were then aligned to the Metriaclima zebra (Malawi cichlid) reference genome UMD2A23,52 using HiSat253. Input samples had a slightly higher alignment rates on average than IP samples (85.5% versus 70.5%). Although PC samples had a higher alignment rate than MC samples (85.2% versus 70.7%), this was not driven by sequence divergence between the species and the MZ reference genome, but rather by sample quality. There is no statistically significant difference between the alignment rate between the two species to the M. zebra (reference) genome versus ‘consensus’ genome sequences generated for those species (χ2(2) = 2.09, p = 0.351) (Streelman lab, unpublished data). Transcripts were quantified using StringTie54 and differential expressional analyses were performed using the Bioconductor package DESeq255.
2.9. Statistical Analysis
All statistical analyses were performed with R software (R Development Core Team 2007). To analyze behavioral data, which were not normally distributed, we used nonparametric tests to separately evaluate the effects of ecotype (Wilcoxon signed rank test) and of species (Kruskal-Wallis test followed by posthoc tests using Dunn’s test for multiple comparisons).
To analyze cell count data in the pS6-immunohistochemistry, we used generalized linear mixed models (GLMM) with a Poisson link function using raw cell counts from each brain region using the R package lme4. Condition (mirror test versus control), species (MC versus PC), and their interactions were included as main effects predicting the number of pS6+ cells. Individual ID nested within IHC-processing batch was included as a random effect, allowing us to control for batch effects and variability in staining across individual slides. In addition, we tested whether any of the measured behaviors (time in proximity to the mirror, duration of lateral displays, and the number of frontal attacks) were predictive of the average number of pS6+ cells within any brain region using simple linear models. In these models, the average number of pS6+ cells within a brain region was the dependent variable and the behavior was the independent variable, using a Bonferroni correction to adjust the p-value to take into account multiple testing. To perform model comparisons for both the linear models and the GLMM models, we used likelihood ratio tests to compare the full model to a reduced null model with the factor of interest removed using the anova function to perform a chi-square test.
For the RNA-seq analyses in the pS6-RNA immunoprecipitation experiment, we used the DESeq2 package to compare transcript abundance in whole-brain input RNA to pS6-IP RNA from two species (PC, rock and MC, sand) exposed to either a mirror or control stimulus (Table 1). DESeq2 uses a negative binomial generalized linear modeling approach, followed by likelihood ratio tests to compare a full model to a reduced model with the variable of interest removed.55 We examined three separate models, each testing separate hypotheses. We first aimed to identify genes that were differentially expressed between input and IP samples, controlling for batch, species, and condition (Model 1). For models 2 & 3, we consider the interaction terms using likelihood ratio tests to identify genes for which the effects of condition (Model 2) and of the effects of both species and condition (Model 3) differ between input and IP samples. The R code and input files, have been made publicly available at: https://github.com/nmbaran/CichlidAggression.
Table 1:
Numbers of statistically significant genes (adjusted p-value < 0.05) found using each model, assessed using the likelihood ratio test to compare the full model to a reduced model without the bolded variable. Lists the number of differentially expressed genes with positive fold-change (FC), negative fold change, and total, with the number of each set successfully matched to a human homolog (with HGNC).
| Model | PosFC | NegFC | Total | PosFC (with HCNC) |
NegFC (with HGNC) |
Total (with HGNC) |
|---|---|---|---|---|---|---|
| Input vs IP Only, LRT | ||||||
| ~ Batch + Species + Condition + SampleType IP x MT Interaction, LRT | 1,443 | 14,786 | 16,229 | 476 | 9,661 | 10,137 |
| IP x MT Interaction, LRT | ||||||
| ~ Batch + SampleType + Condition + Species + SampleType x Condition | 307 | 475 | 782 | 222 | 359 | 581 |
| IP x MT x Species Interaction, LRT | ||||||
| ~ Batch + SampleType + Condition + Species + SampleType x Species + Condition x Species + SampleType x Condition + SampleType x Condition x Species | 392 | 601 | 993 | 307 | 479 | 786 |
Data using the PhosphoTRAP approach differ from traditional RNA-seq data in the both 1) the lower detection rate of many genes (i.e. a larger number of zeroes, especially in IP samples) and 2) some genes with very high read counts (i.e. high variance). These issues, which have also emerged in the analysis of single-cell RNA-sequencing data and in 16S microbial abundance data, present special challenges for statistical analysis56-59. It is important to note, however, that the lower detection rate (i.e. the average proportion of non-zero counts per gene for each sample, 54.1% for input and 12.6% for IP samples) and high read count variance are in this case the result of both technical and biological factors. The higher number of ‘dropouts’ may be due to either low concentration RNA samples or because those genes were not expressed in the neurons activated during aggression. Similarly, high read counts could be a function of PCR amplification bias introduced during the cDNA synthesis step or because those genes are in fact highly expressed in activated neurons. Of interest here is the differential enrichment of genes in pS6 immunoprecipitate across species and conditions, where the ratio of fold-enrichment value for each gene is the IP sample read count divided by the input sample read count for each subject (IP/input).
We explored a number of options for using an unbiased differential expression analysis for this ‘zero-inflated’ data. DESeq2’s default normalization procedure is based on the geometric means of counts, where any gene for which any samples have a zero read count are excluded from normalization. This is problematic when there are a large number of zeroes and when the frequency of zeroes vary systematically by sample, as is the case with PhosphoTRAP data. It is worth noting that the DESeq2 vignette recommendation for single-cell analysis using the zinbwave package does not apply well to our data set either56. PhosphoTRAP data in fact have a lower number of ‘samples’ (i.e. sixteen samples in our case versus the several thousand cells now typical of single-cell sequencing data sets). In addition, we have a larger number of genes expressed per sample than are typically uncovered per cell in single cell sequencing experiments.
After evaluating multiple options, we chose to use the DESeq2 default settings with a pseudocount of 1 added to all reads to statistically test for differential expression. This is in fact a standard approach for analyzing 16S microbial abundance data, though some have criticized that such approaches may lead to a high false discovery rate60-64. Adding a pseudocount of 1 to our raw data also facilitated the calculation of fold-enrichment for each gene in each sample. Although reporting fold-enrichment values and calculating differential fold enrichment has been standard in previous PhosphoTRAP studies44-46,65,66, the fold-enrichment value is not itself amenable to unbiased statistical testing. Thus, all analyses were performed with the read count (with an added pseudocount of 1) as the dependent variable and SampleType (input versus IP) as an independent variable. For the purposes of visualization and as a sanity-check, the results in the figures are presented in terms of the calculated fold-enrichment values, as well as the log10(Fold Change) and p-values standard for differential expression analyses.
To identify gene ontology categories and KEGG pathway enrichment based on lists of differentially expressed genes, we used ToppFun using the human homolog HGNC gene names67-69. We present Bonferroni corrected p-values, which accounts for multiple testing.
3. Results:
3.1. Species comparison of aggressive behavior in the mirror test assay
Sand species spent a slightly higher proportion of time within 5cm of the mirror surface than rock species [0.66 (IQR 0.57 to 0.79) vs 0.59 (IQR 0.43 to 0.70), Z = −1.98, p = 0.047, Fig. 1b]. This effect was largely driven by the difference between two species, MC & PC (Kruskal Wallis test of effect of species, χ2(6) = 17.63, p = 0.007 followed by Dunn Test: Z = 4.01, padj = 0.001 for PC-MC comparison). No other species difference in time in proximity to the mirror was significant.
Rock species performed more frontal attacks than sand species [107 (IQR 53 to 170) vs 17 (IQR 1 to 78), Z = −4.20, p < 0.0001, Fig. 1c]. Rock species also had a shorter attack latency, as well [51s (IQR 26s to 86s) vs 287s (IQR 136s to 696s), Z = 4.82, p < 0.0001]. Once approaching the mirror, rock species escalate to a frontal attack in a median of 14s (IQR 3s to 35s) whereas sand species took substantially longer [115s (IQR 51s to 179s), Z = 5.696, p <0.0001]. In contrast, sand species spent a greater proportion of time performing lateral displays than rock species [0.40 (IQR 0.30 to 0.69) vs 0.15 (IQR 0.08 to 0.35), Z = 4.68, p < 0.0001, Fig. 1d].
In each of these two aggressive behaviors, there is substantial variability across species, as well. CA and LF performed the most frontal attacks whereas CV and TI perform the fewest (Kruskal-Wallis χ2(6) = 33.12, p < 0.0001). PC perform the fewest lateral displays closely followed by LF, whereas the sand species all perform a similarly high number of lateral displays (Kruskal-Wallis χ2(6) = 33.15, p <0.0001). MC and PC exhibit a similar number of frontal attacks (Kruskal-Wallis χ2 (6) = −0.57, p = 0.66), but MC perform a substantially higher number of lateral displays (Kruskal-Wallis χ2 (6) = 4.53, p < 0.0001).
3.2. pS6 Immunohistochemistry
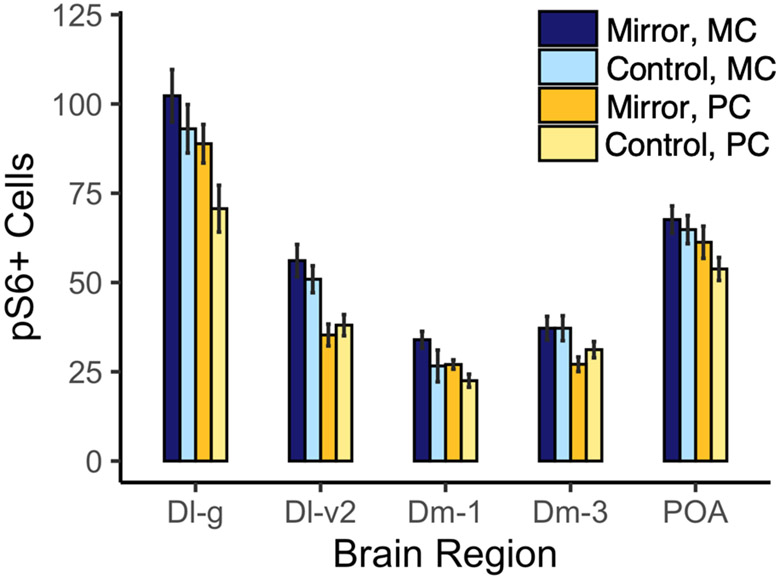
To determine whether any brain regions exhibited differential activation during mirror-elicited aggression, we examined pS6 staining in several regions previously implicated in aggressive behavior (Fig. 2). In the Dl-g, we found a higher number of pS6+ cells per counting frame in sand species (χ2(1) = 8.00, p = 0.0047), but there was no effect of condition on the number of pS6+ cells (χ2(1) = 1.065, p = 0.30) and no interaction between condition and species (χ2(1) = 0.60, p = 0.44). In the Dl-v2, we found a higher number of pS6+ cells per counting frame in sand species (χ2(1) = 6.96, p = 0.0083), but similarly no effect of condition alone (χ2(1) = 0.091, p = 0.76) and no interaction between condition and species (χ2(2) = 0.78, p = 0.68). In the Dm-1, there is a nearly significant effect of both species (χ2(1) = 3.7889, p = 0.052) and condition (χ2(1) = 3.48, p = 0.062) on the number of pS6+ cells. However, there was no statistically significant interaction between species and condition on the number of pS6+ cells in the Dm-1 (χ2(1) = 0.018, p = 0.89). In the Dm-3, there was no significant effect of either species (χ2(1) 1.14, p = 0.29) or condition (χ2(1) = 0.03, p = 0.86) on the number of pS6+ cells. There was also not a significant interaction between species and condition (χ2(1) = 0.22, p = 0.64). In the parvocellular POA, there was no effect of either species (χ2(1) = 1.14, p = 0.29) or condition (χ2(1) = 0.031, p = 0.86) on the number of pS6+ cells, and no effect of interaction between the two (χ2(1) = 0.22, p = 0.64). The number of pS6+ cells in the Dl-v2 and in Dm-3 was positively predicted by lateral display behavior independent of species (Dl-v2: χ2(1) = 2343, padj = 0.027 and Dm-3: χ2(1) = 1074, padj = 0.046). All associations between behavior and pS6-labeling are shown in Supplementary Fig. 1.
Figure 2:
Mean ± within-subject SE of cells immunopositive for pS6 in M. conophoros (sand) versus P. chtimba (rock). Granular zone of lateral zone of the dorsal telencephalon, Dl-g; ventral part of the lateral zone of the dorsal telencephalon, subdivision 2, Dl-v2; medial part of the dorsal telencephalon, subdivision 1, Dm-1; medial part of the dorsal telencephalon, subdivision 3, Dm-3; and preoptic area, POA.
3.3. RNA-immunoprecipitation quality control and analysis
The average RNA concentration for input samples was 1812.5pg/μL and only 56.4pg/μL for IP samples, resulting in an average yield of 0.15% from the immunoprecipitation. The average RNA quality between input and IP samples was comparable: 7.9 RIN score for input samples versus 7.6 RIN score for IP samples. RNA-sequencing yielded an average of 16 million reads per sample (21 million for input and 11 million for IP samples). The average alignment rate was 85.5% for input samples and 70.5% for IP samples. Detailed quality control information can be found in the Supplementary Materials.
3.4. Differential Expression in Input versus IP samples
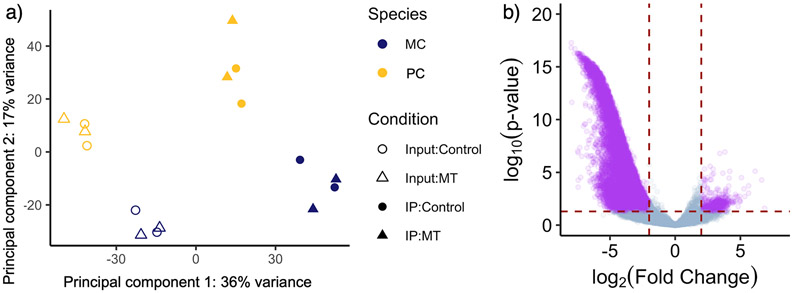
PhosphoTRAP RNA-seq analyses from 8 individuals revealed a large number of genes which showed differential expression between input and IP samples, controlling for differences in batch, species, and condition (Table 1, Model 1). We used DESeq2 the evaluate the following model: SampleType + Batch + Condition + Species. We then used a likelihood ratio test to evaluate the effect of SampleType, an independent variable for whether the sample was input or IP. We found 16,229 genes [10,137 with human homologs, i.e. HUGO Gene Nomenclature Committee (HGNC) names] which were differentially expressed between input and IP samples (Fig. 3), accounting for 45.1% of annotated genes in the genome. Only 1,443 (8.9%) were more highly expressed in IP samples. Genes involved in the MAPK (p = 1.312 x 10−8), PI3K/Akt (p = 9.418 x 10−3), cAMP (p = 9.368 x 10−5), and mTOR (p = 5.855 x 10−5) pathways were overrepresented in the list of differentially expressed genes (S6 is phosphorylated downstream of these four pathways).
Figure 3:
a) Scatterplot of principal component 1 vs principal component 2 reveals clustering of overall expression profile by sample type and species, but not by condition. Principal component 1 accounts for 36% of all gene expression variation and, by inspection, separates the Input (open circles & triangles) and IP samples (closed circles and triangles). Principal component 2 accounts for 17% of the variation in gene expression and varies significantly with species. b) Volcano plot showing gene expression fold-difference and significance between input and IP samples (Model 1). We found that 1,443 genes are up-regulated in the IP samples while 14,786 genes are up-regulated in the input samples. Genes for which the adjusted p-value < 0.05 and where the Log2 Fold Change > 2 are shown in purple.
3.4. Differential expression in IP samples in the Mirror Test
Next, we sought to identify the genes that were differentially expressed in IP samples during mirror aggression independent of species. To do this, we used the following model: Batch + SampleType + Condition + Species + SampleType x Condition, using a likelihood ratio test to evaluate the effect of the interaction term (Table 1, Model 2). This approach identifies either 1) the genes that are expressed in the population of cells that are activated during mirror aggression in both species or, alternatively, 2) the genes whose expression is altered within the population of cells that are activated during mirror aggression.
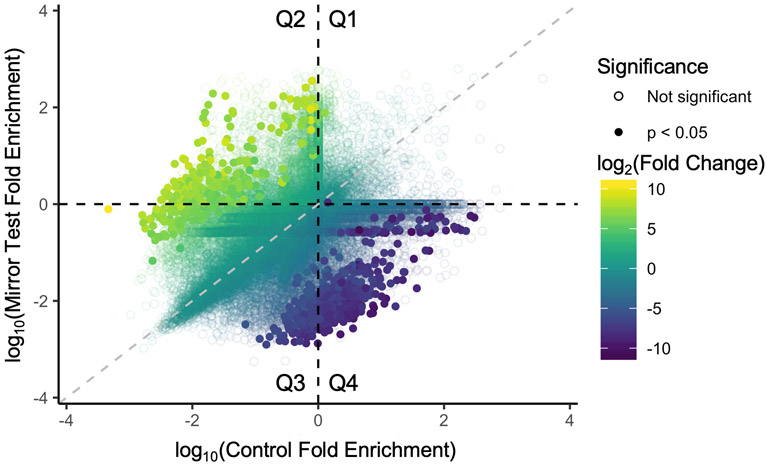
We found 782 genes (581 with HGNC names) that exhibited this pattern of differential expression: 307 with positive fold-change (FC) (i.e. up in mirror test condition and in IP samples) and 475 with negative FC. For all genes that were found to be statistically significant, the interaction term effect is at least 16 FC (i.e. log2FC > 4). Results from this analysis are shown on the scatterplot as the average fold-enrichment value (IP/input) for Control versus Mirror Test samples for each gene (Fig. 4). Genes that emerged as significant are clustered away from the identity line. Genes with a positive FC are also clustered primarily in Q2 and, to a lesser extent in Q3. Thus, genes with a significant positive FC have a higher fold-enrichment value in mirror test samples relative to the control samples. The top three most statistically significant gene ontology molecular function categories for positive FC genes are “DNA-binding transcription factor binding” (p = 2.51 x 10−3), as well as the related sub-categories “RNA polymerase II-specific DNA-binding transcription factor binding” (p = 1.10 x 10−2) and “RNA polymerase II repressing transcription factor binding” (p = 3.34 x 10−2). Among the genes exhibiting this pattern of expression are several less investigated genes involved in the hypothalamic-pituitary-gonadal axis, including HSD17B (hydroxysteroid 17-β dehydrogenase 8) and GNRH2 (gonadotropin-releasing hormone 2). A complete list of differentially expressed genes can be found in the Supplementary Materials.
Figure 4:
Log10 average fold-enrichment values of each gene for Control versus Mirror Test samples. Color is the Log2 Fold Change for IP & Mirror Test samples (Model 2). Solid circles have an adjusted p-value < 0.05. Gray dashed line is the identity (1:1) line.
3.4. Differential expression in IP samples in the Mirror Test by Species
Finally, we aimed to identify the genes that showed a pattern of differential expression in activated neurons during mirror aggression and which varied by species. To do this, we used the following three-way interaction model: Batch + SampleType + Condition + Species + SampleType x Condition + Sampletype x Condition x Species (Table 1, Model 3). These are genes for which their expression in activated cells during mirror-elicited aggression varies by species and, thus, candidate genes that underlie species differences in mirror-elicited aggression.
We found 993 genes (786 genes with HGNC names) that exhibited this pattern of expression, 392 (307) with positive FC and 601 (479) with negative FC. The three significant gene ontology biological function categories for all differentially expressed genes were ‘response to peptide hormone’ (p = 8.347 x 10−4), ‘response to peptide’ (p = 1.146 x 10−2), and ‘response to insulin’ (p = 3.161 x 10−2). Two of these categories, ‘response to peptide hormone’ and ‘response to insulin’ were significant when considering just negative fold change genes.
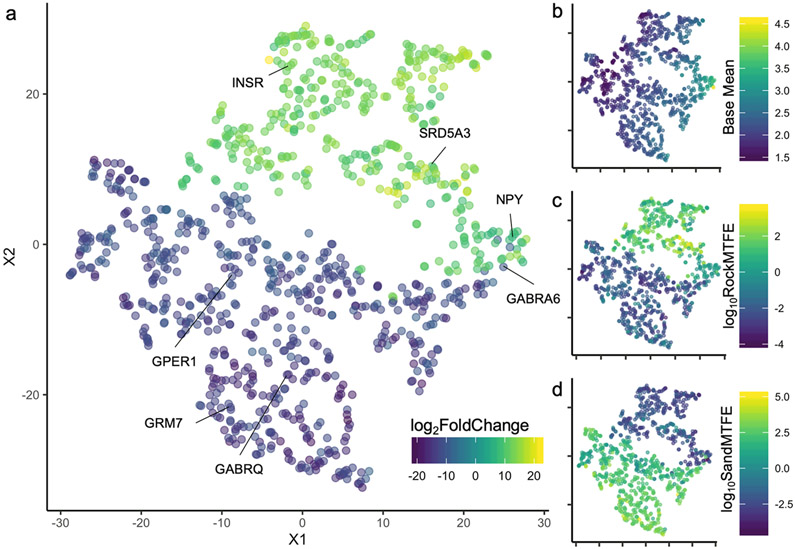
Three-way interactions can be difficult to interpret in gene expression analyses, thus we performed t-Distributed Stochastic Neighbor Embedding (t-SNE) using the R package Rtsne to identify genes that exhibited similar patterns of gene expression to each other. t-SNE is non-linear technique for dimensionality reduction that is used for the visualization of high-dimensional datasets. In this case, the input for the t-SNE algorithm was the raw count reads for the 993 significant genes identified by DESeq2. This analysis results in a plot in low-dimensional space which we found retains the features of the two major outputs of the DESeq2 analysis: log2FoldChange and base mean (the mean of normalized read count of all samples) (Fig. 5a, 5b). In this case, genes with a positive FC in the three-way interaction, in general, have higher fold-enrichment values in individuals of the rock species in the mirror test condition compared to rock individuals in the control condition (Fig. 5a, 5c). In contrast, genes with a negative FC are genes that have higher fold-enrichment values in sand individuals in the mirror test condition compared to controls (Fig. 5a, 5d).
Figure 5:
t-SNE plot showing the overall relationship between the 993 genes significant in the three-way interaction with 8 immunoprecipitated and 8 input samples per gene. Genes that are clustered closer together exhibit a similar pattern of expression across samples. a) Color scale is the log2FoldChange for each gene in the three-way interaction. b) Color scale depicts the base mean, i.e. the mean of normalized counts of all samples, normalizing for sequencing depth. c) Color scale depicts the log10 average fold-enrichment values for the rock species (PC) in the mirror test relative to rock species in the control condition. This pattern is highly similar to the log2FoldChange. d) Color scale depicts the log10 average fold-enrichment values for the sand species (MC) in the mirror test relative to sand species in the control condition.
Among the genes that exhibit a positive log2FoldChange are neuropeptide Y (NPY), insulin receptor A (INSR), and steroid 5 alpha-reductase 3 (SRD5A3). Among the genes that have a negative log2FoldChange are G-protein coupled estrogen receptor 1 (GPER1), gamma-aminobutyric acid (GABA) A receptor, theta (GABRA6), gamma-aminobutyric acid (GABA) A receptor, alpha 6 (GABRQ), and glutamate receptor, metabotropic 7 (GRM7).
Finally, we compared the list of genes that are differentially expressed across MC and PC in the mirror test (Model 3) to the genes that either contain or are near genomic variants that differ between rock & sand species (Streelman lab, unpublished data). We found that 49 of the 778 differentially expressed genes with a human homolog (~6.3%) contain some kind of alternately-fixed variant between rock and sand genomes within 10kB of the gene, either a single-nucleotide polymorphism (SNP) (38/778), an alternately-fixed insertion-deletion (InDel, 11/778), or both (9/778) (see Supplementary Materials for gene list).
4. Discussion:
Our results confirmed that ecotype differences in aggression reported in field studies can be reliably assayed in the lab. We found that males of rock-dwelling species were faster to attack the mirror and attacked a greater number of times, whereas males of sand-dwelling species performed more lateral displays. This observation is consistent with ecological pressures experienced by these species in the wild. Mbuna males must aggressively defend their rocky territories against intruders, as it is both where they feed and their spawning site30,70. Border contests between territorial males are described as including face-to-face lunges and jaw locking25. In contrast, the sand species are not territorial, though they will defend their bower against rival males. However, there is support for the hypothesis that sand species evolved bower building to reduce the likelihood of agonistic encounters, because the complex extended phenotype allows males to more accurately evaluate their rival’s size, quality, and species27,29. The higher time spent performing lateral displays and increased latency to frontal attack in sand species is consistent with this hypothesis.
By comparing neural activity in two closely related species which differ in aggressive phenotype, we hoped to identify brain regions that were differentially engaged in agonistic interactions across species. We found that sand species had more pS6+ cells than did the rock species in two brain regions in the dorsolateral telencephalon (Dl-g and Dl-v2), which together comprise the putative teleost homolog of the mammalian hippocampus71. Contrary to our expectations, we did not observe any differences in pS6+ staining in across conditions or any Condition x Species interactions. It is possible that species differences in neural induction during mirror-elicited aggression are subtle and that our sample size was not large enough to detect small changes. Alternatively, it’s possible that the differential activity is occurring within different sub-populations of cells within the same region or different brain regions altogether.
Previous studies in other closely-related cichlid species have reported a “dissociation between the behavioral and the physiological responses towards mirror images”35,36. This has led a number of researchers to be skeptical about the validity and reliability of mirror-elicited aggression and whether the mirror tests should be used to investigate the physiological and neural correlates of aggression in the brain37,38,40. The mirror image is a highly unusual stimulus, and mirror fights lack the sensory richness of real-life encounters, which include auditory, olfactory, and tactile cues. Additionally, cichlids encounter reversed image of their rival in the mirror. Normally, cichlids will perform lateral displays to each other nose-to-tail and aggressive display behaviors are highly lateralized, as they are in many species39,41,43,72-74. Mangrove rivulus fish males (K. marmoratus) were found to be more aggressive in their behavior when the experimenters used a non-reversing mirror and the fish’s behavior in this apparatus was more predictive of real-life contests than a normal mirror75. Thus, we may have failed to find a difference across species in our pS6+ staining because of the use of a mirror induced a lesser or distorted behavioral response compared to real-life aggressive behaviors.
However, we nevertheless find substantial differences in gene expression in response to mirror-elicited aggression, suggesting that there is indeed a robust transcriptional response to the stimulus. The pattern of differentially expressed genes we observed in both MC and PC, suggests that mirror aggression results in widespread changes to the regulation of gene expression at the level of transcription factor binding. In addition, we noted several interesting genes in the hormone pathways that may play under-explored roles in fish aggression. The gene HSD17B (hydroxysteroid (17-beta) dehydrogenase 8, 17β-HSD) encodes a steroidogenic enzyme responsible for the last step in the synthesis of 11-ketotestosterone, the primary fish androgen. Surprisingly little is known about the role of 17β-HSD in the brains of fish, but 17β-HSD type 8 is ubiquitously expressed in the related fish species Nile tilapia, Oreochromis niloticus76,77. Another gene of interest is gonadotropin-releasing hormone 2 (GnRH-2). GnRH-2 is less well-understood than GNRH-1. Across vertebrates, GnRH-2 is expressed exclusively in the midbrain tegmentum and is thought to play roles in the control of reproductive motor behavior and feeding78-84. However, it has not previously been linked to aggression and two previous studies failed to find an association between aggressive behavior and GnRH-2 in fish85,86.
Finally, we identified a large number of genes that show differential patterns of activation across species during mirror-elicited aggression, many of which are specifically involved in peptide hormone signaling pathways. Several of the genes that emerged from the analysis also play important roles in feeding and metabolism, including a number of genes in the insulin pathway and neuropeptide Y. There is, in fact, a large literature linking metabolic syndromes and insulin resistance to aggression87-89. For example, diabetic rats show a loss of aggression and an increase in submissive displays87. Several researchers have theorized that the overlap between the same signaling molecules that are associated with insulin resistance are also associated with reduced aggression, likely because of the evolutionary origins of aggression in the service of acquiring food or resources, as well as the metabolic changes needed to fight or recover from agonistic encounters88,89. Neuropeptide Y, an abundantly expressed peptide with well-known roles in regulating food intake, growth, and reproduction, is also similarly linked to aggression90. For example, the knockout of Y1 receptors for NPY was found to substantially increase in aggression in mice91. NPY has also been associated with aggression in zebrafish (Danio rerio)85 and NPY mRNA levels were found to be substantially higher in the POA of socially subordinate versus dominant rainbow trout (Oncorhynchus mykiss)92. In the Lake Malawi cichlids, the gene for NPY is near a highly divergent region between rock and sand species, which may also account for the observed pattern of differential expression.
Intriguingly, we found that two different 5α-reductase genes were differentially expressed in the two species: SRD5A3 was more highly expressed in PC during mirror-elicited aggression, whereas SRD5A1 was more highly expressed in MC. The inferred evolutionary history for the SRD5α family of genes suggests that SRD5A1 and SRD5A3 diverged early in the evolution of eukaryotes93. The SRD5A1 gene encodes an enzyme that catalyzes the reduction of testosterone, progesterone, corticosterone, and estrone into the more active 5α-reduced derivatives, including the conversion of testosterone (T) to 5α-dihydrotestosterone (5α-DHT), a non-aromatizable, potent androgen93. In mice, 5α-reductase type 1 was found to mediate testosterone enhancement of aggression during a resident-intruder test94. 5α-DHT has been found to induce territorial aggression in both mice and songbirds95-97, as well as male-typical social signals in weakly electric fish98. It was only recently discovered that SRD5A3 is likely not a steroidogenic enzyme, but it is instead involved in glycosylation99,100. Humans with mutations of the SRD5A3 gene typically present with severe ocular issues, cerebellar malformations, and cognitive impairment101. SRD5A3 is known to be widely expressed in fish brains in a region-specific manner102, but it remains unclear whether it plays a role in aggression103,104.
Another interesting candidate gene for further exploration is GPER1, which encodes the gene for the G-protein coupled estrogen receptor. A number of studies have shown rapid, non-genomic effects of estradiol on aggression across taxa, but the specific role of GPER1 remains poorly understood105. In fish, rapid effects of estrogens have been reported in Plainfin midshipman fish106 and the GPER1 gene was implicated in the pacemaker region of weakly electric fish107. This suggests that evolution of the G-protein coupled estrogen receptor may underly differences in behavioral aggression in fish, especially in the production of male-typical social signals. Furthermore, G-protein coupled receptors are hypothesized to be particularly evolvable neuromodulatory systems, as a result of their compartmentalization and subfunctionalization within the brain108.
We also observed differences in several neurotransmitter receptors. In rodents, aggressive behavior is thought to arise as a result of an imbalance between glutamatergic excitation and GABA-ergic inhibition109. GABAA receptors have previously been linked to aggressive behaviors (see 110,111 for review). In addition, we also identified differential expression of metabotropic glutamate receptor subtype 7 (mGluR7). mGluR7 in the medial bed nucleus of the hypothalamus has been shown to be essential for male-male aggression in mice112 and agonists to mGluR7 significantly reduce both threat and attack behaviors113. Both pharmacological and genetic studies have shown that almost all subtypes of glutamate (NMDA, AMPA, kainate receptors, and mGluRs) and GABA (GABAA and GABAB) receptors are involved in aggression in some capacity. However, the role of each of the receptors may vary depending on the composition of receptor subunits, their localization, and the type of aggressive behavior studied, all of which can vary across species.
We were surprised that we did not observe any differences in expression in either the gene for arginine vasotocin (homolog of arginine vasopressin, AVP) or in the vasopressin-1a receptor (V1aR), both of which have previously been linked to aggression in other species, including dominance behaviors in the African cichlid Astatotilapia burtoni114-116. V1aR contains an alternately fixed 11-base pair deletion in sand species, which is predicted to alter the C-terminal domain of the G-protein coupled receptor via a frameshift, removing multiple palmitoylation sites117. However, this mutation is predicted to alter protein functions like the trafficking of the receptor to the membrane, which may not be observable at the level of gene expression and further exploration of the consequences of this variant is warranted. We were also surprised to not observe any genes related to serotonin (5-HT) or in dopamine (DA) emerge from these analyses, as both have been frequently linked to aggressive behaviors.
In our analyses, we identified a large number of genes that exhibited patterns of expression consistent with differential involvement in aggressive behaviors across two closely related cichlid species. In general, there are two ways to interpret our PhosphoTRAP data. First, the data may reveal specific neuronal cell types activated during mirror-elicited aggression, in the cases in which the differentially expressed genes have been identified as cell-type specific markers, such as NPY. Alternatively, PhosphoTRAP can reveal stimulus- or behavior-driven changes in gene expression within the population of activated cells. However, it is impossible to distinguish between these possibilities with PhosphoTRAP data alone. Additionally, one drawback of using whole-brain samples versus punches of tissue from a single brain region pooled across multiple samples for PhosphoTRAP (as is more common), is that different populations of cells of the same type may show opposite patterns of activation. Although our goal was examine differences in gene expression throughout the brain, this approach may lead us to fail to identify region-specific patterns of differential expression, which are undoubtedly important in the context of complex social phenotypes. Future studies using single-cell RNA sequencing technologies, coupled with activation-based cell sorting, would enable us to parse these two different interpretations of our data and provide more detailed information about cell types118,119.
Here we used the immunoprecipitation of translational ribosomes via phosphorylated ribosomal protein S6 to produce a rich set of candidate genes involved in aggression in two species of Lake Malawi cichlid fish which differ in aggressive behaviors. To our knowledge, this study is the first time that PhosphoTRAP has been performed on whole-brain tissue samples, rather than brain-region specific punches pooled from multiple individuals (see44,46,65,66). We used an unbiased approach, with the aim of discovering novel genes and pathways involved in a complex social behavior to provide a foundation for further exploration. Future studies can interrogate the causal relationship between specific genomic variants and aggressive behavior using genetic approaches such as genetic mapping or CRISPR-Cas9 genome editing120-124. This work thus lays the foundation for future experiments using this emerging genetic model system to investigate the genomic basis of evolved species differences in both brain and behavior.
Supplementary Material
6. Acknowledgements
We would like to thank C. Patil, Z. Johnson, P.T. McGrath, and members of the Streelman lab for hands-on assistance and helpful feedback throughout. We thank animal care staff Physiological Research Laboratory, as well as the Molecular Evolution Core, Parker H. Petit Institute Histology Core, and the Parker H. Petit Institute Optical Microscopy Core at Georgia Tech. This work was supported by NIH Grants R01GM101095 (to J.T.S.) and F32GM125496 (to N.M.B.)
Footnotes
Data Availability Statement
The data that support the findings of this study are openly available in the Gene Expression Omnibus (GEO) database (accession number GSE140690) at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140690. In addition, the R code and input files, have been made publicly available at: https://github.com/nmbaran/CichlidAggression.
5. References:
- 1.van Oortmerssen GA, Bakker ThCM. Artificial selection for short and long attack latencies in wild Mus musculus domesticus. Behav Genet. 1981;11 (2):115–126. doi: 10.1007/BF01065622 [DOI] [PubMed] [Google Scholar]
- 2.Drent PJ, van Oers K, van Noordwijk AJ. Realized heritability of personalities in the great tit (Parus major). Proc R Soc B. 2003;270(1510):45–51. doi: 10.1098/rspb.2002.2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudziak JJ, van Beijsterveldt CEM, Bartels M, et al. Individual differences in aggression: Genetic analyses by age, gender, and informant in 3-, 7-, and 10-year-old Dutch Twins. Behav Genet. 2003;33(5):575–589. doi: 10.1023/A:1025782918793 [DOI] [PubMed] [Google Scholar]
- 4.Fairbanks LA, Newman TK, Bailey JN, et al. Genetic contributions to social impulsivity and aggressiveness in vervet monkeys. Biol Psychiatry. 2004;55(6):642–647. doi: 10.1016/j.biopsych.2003.12.005 [DOI] [PubMed] [Google Scholar]
- 5.Saetre P, Strandberg E, Sundgren P-E, Pettersson U, Jazin E, Bergström TF. The genetic contribution to canine personality. Genes Brain Behav. 2006;5(3):240–248. doi: 10.1111/j.1601-183X.2005.00155.x [DOI] [PubMed] [Google Scholar]
- 6.Yeh MT, Coccaro EF, Jacobson KC. Multivariate behavior genetic analyses of aggressive behavior subtypes. Behav Genet. 2010;40(5):603–617. doi: 10.1007/s10519-010-9363-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuvblad C, Baker LA. Human aggression across the lifespan: Genetic propensities and environmental moderators. Adv Genet. 2011;75:171–214. doi: 10.1016/B978-0-12-380858-5.00007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariyomo TO, Carter M, Watt PJ. Heritability of boldness and aggressiveness in the zebrafish. Behav Genet. 2013;43(2):161–167. doi: 10.1007/s10519-013-9585-y [DOI] [PubMed] [Google Scholar]
- 9.Araya-Ajoy YG, Dingemanse NJ. Repeatability, heritability, and age-dependence of seasonal plasticity in aggressiveness in a wild passerine bird. J Anim Ecol. 2017;86(2):227–238. doi: 10.1111/1365-2656.12621 [DOI] [PubMed] [Google Scholar]
- 10.Craig IW, Halton KE. Genetics of human aggressive behaviour. Hum Genet. 2009;126(1):101–113. doi: 10.1007/s00439-009-0695-9 [DOI] [PubMed] [Google Scholar]
- 11.Fernàndez-Castillo N, Cormand B. Aggressive behavior in humans: Genes and pathways identified through association studies. Am J Med Genet B Neuropsychiatry Genet. 2016;171(5):676–696. doi: 10.1002/ajmg.b.32419 [DOI] [PubMed] [Google Scholar]
- 12.Zhang-James Y, Fernàndez-Castillo N, Hess JL, et al. An integrated analysis of genes and functional pathways for aggression in human and rodent models. Mol Psychiatry. June 2018:1–13. doi: 10.1038/s41380-018-0068-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenz K On Aggression. New York: Harcourt, Brace & World; 1966. [Google Scholar]
- 14.Nelson RJ, ed. Biology of Aggression. 1 edition. Oxford ; New York: Oxford University Press; 2005. [Google Scholar]
- 15.Nelson RJ, Chiavegatto S. Molecular basis of aggression. Trends Neurosci. 2001;24(12):713–719. doi: 10.1016/S0166-2236(00)01996-2 [DOI] [PubMed] [Google Scholar]
- 16.Brodkin ES, Goforth SA, Keene AH, Fossella JA, Silver LM. Identification of quantitative trait loci that affect aggressive behavior in mice. J Neurosci. 2002;22(3):1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards AC, Mackay TFC. Quantitative trait loci for aggressive behavior in Drosophila melanogaster. Genetics. 2009;182(3):889–897. doi: 10.1534/genetics.109.101691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyne HO, Lautenschläger S, Nelson R, et al. Genetic influences on brain gene expression in rats selected for tameness and aggression. Genetics. 2014;198(3):1277–1290. doi: 10.1534/genetics.114.168948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kukekova AV, Johnson JL, Teiling C, et al. Sequence comparison of prefrontal cortical brain transcriptome from a tame and an aggressive silver fox (Vulpes vulpes). BMC Genomics. 2011;12:482. doi: 10.1186/1471-2164-12-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, vonHoldt BM, Reynolds A, et al. Artificial selection on brain-expressed genes during the domestication of dog. Mol Biol Evol. 2013;30(8):1867–1876. doi: 10.1093/molbev/mst088 [DOI] [PubMed] [Google Scholar]
- 21.Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 2004;5(4):288–298. doi: 10.1038/nrg1316 [DOI] [PubMed] [Google Scholar]
- 22.Loh Y-HE, Katz LS, Mims MC, Kocher TD, Yi SV, Streelman JT. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;9(7):R113. doi: 10.1186/gb-2008-9-7-r113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brawand D, Wagner CE, Li YI, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513(7518):375–381. doi: 10.1038/nature13726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh Y-HE, Bezault E, Muenzel FM, et al. Origins of shared genetic variation in African cichlids. Mol Biol Evol. 2013;30(4):906–917. doi: 10.1093/molbev/mss326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holzberg S A field and laboratory study of the behaviour and ecology of Pseudotropheus zebra (Boulenger), an endemic cichlid of Lake Malawi (Pisces; Cichlidae). J Zool Syst Evol Res. 1978;16(3):171–187. doi: 10.1111/j.1439-0469.1978.tb00929.x [DOI] [Google Scholar]
- 26.York RA, Patil C, Hulsey CD, Anoruo O, Streelman JT, Fernald RD. Evolution of bower building in Lake Malawi cichlid fish: phylogeny, morphology, and behavior. Front Ecol Evol. 2015;3:18. doi: 10.3389/fevo.2015.00018 [DOI] [Google Scholar]
- 27.Martin CH, Genner MJ. A role for male bower size as an intrasexual signal in a Lake Malawi cichlid fish. Behaviour. 2009;146(7):963–978. doi: 10.1163/156853908X396836 [DOI] [Google Scholar]
- 28.Young KA, Genner MJ, Joyce DA, Haesler MP. Hotshots, hot spots, and female preference: exploring lek formation models with a bower-building cichlid fish. Behav Ecol. 2009;20(3):609–615. doi: 10.1093/beheco/arp038 [DOI] [Google Scholar]
- 29.Magalhaes IS, Croft GE, Joyce DA. Altering an extended phenotype reduces intraspecific male aggression and can maintain diversity in cichlid fish. PeerJ. 2013;1:e209. doi: 10.7717/peerj.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genner MJ, Turner GF, Hawkins SJ. Resource control by territorial male cichlid fish in Lake Malawi. J Anim Ecol. 1999;68(3):522–529. doi: 10.1046/j.1365-2656.1999.00301.x [DOI] [Google Scholar]
- 31.Pauers MJ, Kapfer JM, Fendos CE, Berg CS. Aggressive biases towards similarly coloured males in Lake Malawi cichlid fishes. Biol Lett. 2008;4(2):156–159. doi: 10.1098/rsbl.2007.0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danley PD. Aggression in closely related Malawi cichlids varies inversely with habitat complexity. Environ Biol Fish. 2011;92(3):275. doi: 10.1007/s10641-011-9838-7 [DOI] [Google Scholar]
- 33.Pauers MJ, Kapfer JM, Doehler K, Lee JT, Berg CS. Gross colour pattern is used to distinguish between opponents during aggressive encounters in a Lake Malawi cichlid. Ecol Freshw Fish. 2012;21(1):34–41. doi: 10.1111/j.1600-0633.2011.00520.x [DOI] [Google Scholar]
- 34.Tinbergen N The Study of Instinct. Vol xii New York, NY, US: Clarendon Press/Oxford University Press; 1951. [Google Scholar]
- 35.Oliveira RF, Carneiro LA, Canário AVM. Behavioural endocrinology: No hormonal response in tied fights. Nature. 2005;437(7056):207–208. doi: 10.1038/437207a [DOI] [PubMed] [Google Scholar]
- 36.Desjardins JK, Fernald RD. What do fish make of mirror images? Biol Lett. 2010;6(6):744–747. doi: 10.1098/rsbl.2010.0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balzarini V, Taborsky M, Wanner S, Koch F, Frommen JG. Mirror, mirror on the wall: the predictive value of mirror tests for measuring aggression in fish. Behav Ecol Sociobiol. 2014;68(5):871–878. doi: 10.1007/s00265-014-1698-7 [DOI] [Google Scholar]
- 38.Earley RL, Hsu Y, Wolf LL. The use of standard aggression testing methods to predict combat behaviour and contest outcome in Rivulus marmoratus dyads (Teleostei: Cyprinodontidae). Ethology. 2000;106(8):743–761. doi: 10.1046/j.1439-0310.2000.00586.x [DOI] [Google Scholar]
- 39.Arnott G, Ashton C, Elwood RW. Lateralization of lateral displays in convict cichlids. Biol Lett April 2011:rsbl20110328. doi: 10.1098/rsbl.2011.0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliveira RF, Canário AVM. Nemo through the looking-glass: a commentary on Desjardins & Fernald. Biol Lett. 2011;7(4):487–488. doi: 10.1098/rsbl.2010.0760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elwood RW, Stoilova V, McDonnell A, Earley RL, Arnott G. Do mirrors reflect reality in agonistic encounters? A test of mutual cooperation in displays. Anim Behav. 2014;97(Supplement C):63–67. doi: 10.1016/j.anbehav.2014.07.028 [DOI] [Google Scholar]
- 42.Ariyomo TO, Watt PJ. Aggression and sex differences in lateralization in the zebrafish. Animal Behaviour. 2013;86(3):617–622. doi: 10.1016/j.anbehav.2013.06.019 [DOI] [Google Scholar]
- 43.Scherer U, Buck M, Schuett W. Lateralisation in agonistic encounters: do mirror tests reflect aggressive behaviour? A study on a West African cichlid. Journal of Fish Biology. 2016;89(3):1866–1872. doi: 10.1111/jfb.13069 [DOI] [PubMed] [Google Scholar]
- 44.Knight ZA, Tan K, Birsoy K, et al. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell. 2012;151(5):1126–1137. doi: 10.1016/j.cell.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y, Gong NN, Hu XS, Ni MJ, Pasi R, Matsunami H. Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat Neurosci. 2015; 18(10):1446–1454. doi: 10.1038/nn.4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer EK, Roland AB, Moskowitz NA, et al. Mechanisms of convergent egg-provisioning in poison frogs. bioRxiv. May 2019:653501. doi: 10.1101/653501 [DOI] [PubMed] [Google Scholar]
- 47.Fischer EK, Westrick SE, Hartsough L, Hoke KL. Differences in neural activity, but not behavior, across social contexts in guppies, Poecilia reticulata. Behav Ecol Sociobiol. 2018;72(8):131. doi: 10.1007/s00265-018-2548-9 [DOI] [Google Scholar]
- 48.Butler JM, Whitlow SM, Roberts DA, Maruska KP. Neural and behavioural correlates of repeated social defeat. Sci Rep. 2018;8(1):1–13. doi: 10.1038/s41598-018-25160-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmadiantehrani S, London SE. Bidirectional manipulation of mTOR signaling disrupts socially mediated vocal learning in juvenile songbirds. PNAS. 2017;114(35):9463–9468. doi: 10.1073/pnas.1701829114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renier N, Adams EL, Kirst C, et al. Mapping of brain activity by automated volume analysis of immediate early genes. Cell. 2016;165(7):1789–1802. doi: 10.1016/j.cell.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forlano PM, Bass AH. Neural and hormonal mechanisms of reproductive-related arousal in fishes. Horm Behav. 2011;59(5):616–629. doi: 10.1016/j.yhbeh.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conte MA, Kocher TD. An improved genome reference for the African cichlid, Metriaclima zebra. BMC Genomics. 2015;16(1). doi: 10.1186/s12864-015-1930-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016; 11(9):1650–1667. doi: 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Risso D, Perraudeau F, Gribkova S, Dudoit S, Vert J-P. A general and flexible method for signal extraction from single-cell RNA-seq data. Nat Commun. 2018;9(1):284. doi: 10.1038/s41467-017-02554-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss S, Xu ZZ, Peddada S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5(1):27. doi: 10.1186/s40168-017-0237-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50(8):1–14. doi: 10.1038/s12276-018-0071-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L, Reeve J, Zhang L, Huang S, Wang X, Chen J. GMPR: A robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ. 2018;6:e4600. doi: 10.7717/peerj.4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters BA, McCullough ML, Purdue MP, et al. Association of coffee and tea intake with the oral microbiome: Results from a large cross-sectional study. Cancer Epidemiol Biomarkers Prev. 2018;27(7):814–821. doi: 10.1158/1055-9965.EPI-18-0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar MS, Slud EV, Okrah K, Hicks SC, Hannenhalli S, Bravo HC. Analysis and correction of compositional bias in sparse sequencing count data. BMC Genomics. 2018; 19(1):1–23. doi: 10.1186/s12864-018-5160-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsilimigras MCB, Fodor AA. Compositional data analysis of the microbiome: fundamentals, tools, and challenges. Ann Epidemiol. 2016;26(5):330–335. doi: 10.1016/j.annepidem.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 63.Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11(5):e1004226. doi: 10.1371/journal.pcbi.1004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9):e1002687. doi: 10.1371/journal.pcbi.1002687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Z-D, Yang WZ, Gao C, et al. A hypothalamic circuit that controls body temperature. PNAS. 2017;114(8):2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan CL, Cooke EK, Leib DE, et al. Warm-sensitive neurons that control body temperature. Cell. 2016;167(1):47–59.e15. doi: 10.1016/j.cell.2016.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8(1):392. doi: 10.1186/1471-2105-8-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J, Aronow BJ, Jegga AG. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics. 2009;10(1):73. doi: 10.1186/1471-2105-10-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(suppl_2):W305–W311. doi: 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hert E Factors in habitat partitioning in Pseudotropheus aurora (Pisces: Cichlidae), an introduced species to a species-rich community of Lake Malawi. J Fish Biol. 1990;36(6):853–865. doi: 10.1111/j.1095-8649.1990.tb05633.x [DOI] [Google Scholar]
- 71.Rodríguez F, López JC, Vargas JP, Gómez Y, Broglio C, Salas C. Conservation of spatial memory function in the pallial forebrain of reptiles and ray-finned fishes. J Neurosci. 2002;22(7):2894–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeuchi Y, Hori M, Myint O, Kohda M. Lateral bias of agonistic responses to mirror images and morphological asymmetry in the Siamese fighting fish (Betta splendens). Behav Brain Res. 2010;208(1):106–111. doi: 10.1016/j.bbr.2009.11.021 [DOI] [PubMed] [Google Scholar]
- 73.Arnott G, Beattie E, Elwood RW. To breathe or fight? Siamese fighting fish differ when facing a real opponent or mirror image. Behav Process. 2016;129(Supplement C):11–17. doi: 10.1016/j.beproc.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 74.Forsatkar MN, Dadda M, Nematollahi MA. Lateralization of aggression during reproduction in male Siamese fighting Fish. Ethology. 2015;121(11):1039–1047. doi: 10.1111/eth.12418 [DOI] [Google Scholar]
- 75.Li C-Y, Curtis C, Earley RL. Nonreversing mirrors elicit behaviour that more accurately predicts performance against live opponents. Anim Behav. 2018;137:95–105. doi: 10.1016/j.anbehav.2018.01.010 [DOI] [Google Scholar]
- 76.Diotel N, Do-Rego J-L, Anglade I, et al. The brain of teleost fish, a source, and a target of sexual steroids. Front Neurosci. 2011. ;5. doi: 10.3389/fnins.2011.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou LY, Wang DS, Senthilkumaran B, et al. Cloning, expression and characterization of three types of 17β-hydroxysteroid dehydrogenases from the Nile tilapia, Oreochromis niloticus. J Mol Endocrinol. 2019;35(1):103–116. doi: 10.1677/jme.1.01801 [DOI] [PubMed] [Google Scholar]
- 78.White SA, Kasten TL, Bond CT, Adelman JP, Fernald RD. Three gonadotropin-releasing hormone genes in one organism suggest novel roles for an ancient peptide. PNAS. 1995;92(18):8363–8367. doi: 10.1073/pnas.92.18.8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fernald RD, White RB. Gonadotropin-releasing hormone genes: phylogeny, structure, and functions. Front Neuroendocrinol. 1999;20(3):224–240. doi: 10.1006/frne.1999.0181 [DOI] [PubMed] [Google Scholar]
- 80.Uchida H, Ogawa S, Harada M, et al. The olfactory organ modulates gonadotropin-releasing hormone types and nest-building behavior in the tilapia Oreochromis niloticus. J Neurobiol. 2005;65(1):1–11. doi: 10.1002/neu.20156 [DOI] [PubMed] [Google Scholar]
- 81.Miyamoto K, Hasegawa Y, Nomura M, Igarashi M, Kangawa K, Matsuo H. Identification of the second gonadotropin-releasing hormone in chicken hypothalamus: evidence that gonadotropin secretion is probably controlled by two distinct gonadotropin-releasing hormones in avian species. PNAS. 1984;81(12):3874–3878. doi: 10.1073/pnas.81.12.3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoskins LJ, Xu M, Volkoff H. Interactions between gonadotropin-releasing hormone (GnRH) and orexin in the regulation of feeding and reproduction in goldfish (Carassius auratus). Horm Behav. 2008;54(3):379–385. doi: 10.1016/j.yhbeh.2008.04.011 [DOI] [PubMed] [Google Scholar]
- 83.Kang KS, Shimizu K, Azuma M, et al. Gonadotropin-releasing hormone II (GnRH II) mediates the anorexigenic actions of α-melanocyte-stimulating hormone (α-MSH) and corticotropin-releasing hormone (CRH) in goldfish. Peptides. 2011;32(1):31–35. doi: 10.1016/j.peptides.2010.10.013 [DOI] [PubMed] [Google Scholar]
- 84.Marvel M, Spicer OS, Wong T-T, Zmora N, Zohar Y. Knockout of the GnRH genes in zebrafish: effects on reproduction and potential compensation by reproductive and feeding-related neuropeptides. Biol Reprod. 2018;99(3):565–577. doi: 10.1093/biolre/ioy078 [DOI] [PubMed] [Google Scholar]
- 85.Filby AL, Paull GC, Hickmore TF, Tyler CR. Unravelling the neurophysiological basis of aggression in a fish model. BMC Genomics. 2010;11(1):498. doi: 10.1186/1471-2164-11-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kagawa N, Hirose S, Fujimoto K, et al. Social rank-dependent expression of gonadotropin-releasing hormones and kisspeptin in the medaka brain. Gen Comp Endocrinol. 2017;249:48–54. doi: 10.1016/j.ygcen.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 87.Leedom LJ, Meehan WP. The psychoneuroendocrinology of diabetes mellitus in rodents. Psychoneuroendocrinology. 1989;14(4):275–294. doi: 10.1016/0306-4530(89)90030-9 [DOI] [PubMed] [Google Scholar]
- 88.Watve MG, Yajnik CS. Evolutionary origins of insulin resistance: a behavioral switch hypothesis. BMC Evolutionary Biology. 2007;7(1):61. doi: 10.1186/1471-2148-7-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Belsare PV, Watve MG, Ghaskadbi SS, Bhat DS, Yajnik CS, Jog M. Metabolic syndrome: Aggression control mechanisms gone out of control. Med Hypotheses. 2010;74(3):578–589. doi: 10.1016/j.mehy.2009.09.014 [DOI] [PubMed] [Google Scholar]
- 90.Emeson RB, Morabito MV. Food fight: the NPY-serotonin link between aggression and feeding fehavior. Sci Signal. 2005;2005(277):pe12–pe12. doi: 10.1126/stke.2772005pe12 [DOI] [PubMed] [Google Scholar]
- 91.Karl T, Lin S, Schwarzer C, et al. Y1 receptors regulate aggressive behavior by modulating serotonin pathways. PNAS. 2004;101(34):12742–12747. doi: 10.1073/pnas.0404085101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Doyon C, Gilmour KM, Trudeau VL, Moon TW. Corticotropin-releasing factor and neuropeptide Y mRNA levels are elevated in the preoptic area of socially subordinate rainbow trout. Gen Comp Endocrinol. 2003;133(2):260–271. doi: 10.1016/S0016-6480(03)00195-3 [DOI] [PubMed] [Google Scholar]
- 93.Langlois VS, Zhang D, Cooke GM, Trudeau VL. Evolution of steroid-5α-reductases and comparison of their function with 5β-reductase. Gen Comp Endocrinol. 2010;166(3):489–497. doi: 10.1016/j.ygcen.2009.08.004 [DOI] [PubMed] [Google Scholar]
- 94.Frye CA, Rhodes ME, Walf A, Harney JP. Testosterone enhances aggression of wild-type mice but not those deficient in type I 5α-reductase. Brain Res. 2002;948(1):165–170. doi: 10.1016/S0006-8993(02)03076-7 [DOI] [PubMed] [Google Scholar]
- 95.Sperry TS, Wacker DW, Wingfield JC. The role of androgen receptors in regulating territorial aggression in male song sparrows. Horm Behav. 2010;57(1):86–95. doi: 10.1016/j.yhbeh.2009.09.015 [DOI] [PubMed] [Google Scholar]
- 96.Finney HC, Erpino MJ. Synergistic effect of estradiol benzoate and dihydrotestosterone on aggression in mice. Horm Behav. 1976;7(4):391–400. doi: 10.1016/0018-506X(76)90010-6 [DOI] [PubMed] [Google Scholar]
- 97.Archawaranon M, Wiley RH. Control of aggression and dominance in white-throated sparrows by testosterone and its metabolites. Horm Behav. 1988;22(4):497–517. doi: 10.1016/0018-506X(88)90054-2 [DOI] [PubMed] [Google Scholar]
- 98.Bass AH. A hormone-sensitive communication system in an electric fish. J Neurobiol. 1986;17(3):131–155. doi: 10.1002/neu.480170303 [DOI] [PubMed] [Google Scholar]
- 99.Cantagrel V, Lefeber DJ, Ng BG, et al. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell. 2010;142(2):203–217. doi: 10.1016/j.cell.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chávez B, Ramos L, García-Becerra R, Vilchis F. Hamster SRD5A3 lacks steroid 5α-reductase activity in vitro. Steroids. 2015;94:41–50. doi: 10.1016/j.steroids.2014.11.005 [DOI] [PubMed] [Google Scholar]
- 101.Kara B, Ayhan Ö, Gökçay G, Başboğaoğlu N, Tolun A. Adult phenotype and further phenotypic variability in SRD5A3-CDG. BMC Med Genet. 2014;15(1):10. doi: 10.1186/1471-2350-15-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martyniuk CJ, Bissegger S, Langlois VS. Current perspectives on the androgen 5 alpha-dihydrotestosterone (DHT) and 5 alpha-reductases in teleost fishes and amphibians. Gen Comp Endocrinol. 2013;194:264–274. doi: 10.1016/j.ygcen.2013.09.019 [DOI] [PubMed] [Google Scholar]
- 103.Schlinger BA, Callard GV. Aromatization mediates aggressive behavior in quail. Gen Comp Endocrinol. 1990;79(1):39–53. doi: 10.1016/0016-6480(90)90086-2 [DOI] [PubMed] [Google Scholar]
- 104.Soma KK, Schlinger BA, Wingfield JC, Saldanha CJ. Brain aromatase, 5α-reductase, and 5β-reductase change seasonally in wild male song sparrows: Relationship to aggressive and sexual behavior. J Neurobiol. 2003;56(3):209–221. doi: 10.1002/neu.10225 [DOI] [PubMed] [Google Scholar]
- 105.Heimovics SA, Trainor BC, Soma KK. Rapid effects of estradiol on aggression in birds and mice: The fast and the furious. Integr Comp Biol. 2015;55(2):281–293. doi: 10.1093/icb/icv048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Remage-Healey L, Bass AH. Rapid, hierarchical modulation of vocal patterning by steroid hormones. J Neurosci. 2004;24(26):5892–5900. doi: 10.1523/JNEUROSCI.1220-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smith GT, Proffitt MR, Smith AR, Rusch DB. Genes linked to species diversity in a sexually dimorphic communication signal in electric fish. J Comp Physiol A. 2018;204(1):93–112. doi: 10.1007/s00359-017-1223-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Katz PS, Lillvis JL. Reconciling the deep homology of neuromodulation with the evolution of behavior. Curr Opin Neurobiol. 2014;29:39–47. doi: 10.1016/j.conb.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 109.Takahashi A, Miczek KA. Neurogenetics of aggressive behavior: Studies in rodents In: Miczek KA, Meyer-Lindenberg A, eds. Neuroscience of Aggression. Berlin: Springer; 2014:3–44. doi: 10.1007/7854_2013_263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miczek KA, Fish EW, De Bold JF. Neurosteroids, GABAA receptors, and escalated aggressive behavior. Horm Behav. 2003;44(3):242–257. doi: 10.1016/j.yhbeh.2003.04.002 [DOI] [PubMed] [Google Scholar]
- 111.Miczek KA, Fish EW. Monoamines, GABA, glutamate, and aggression In: Nelson RJ, ed. Biology of Aggression. New York, NY: Oxford University Press; 2006:114–149. [Google Scholar]
- 112.Masugi-Tokita M, Flor PJ, Kawata M. Metabotropic glutamate receptor subtype 7 in the bed nucleus of the stria terminalis is essential for intermale aggression. Neuropsychopharmacol. 2016;41(3):726–735. doi: 10.1038/npp.2015.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Navarro JF, de Castro V, Martín-López M. Behavioural profile of selective ligands for mGlu7 and mGlu8 glutamate receptors in agonistic encounters between mice. Psicothema. 2009;21(3):475–479. [PubMed] [Google Scholar]
- 114.Greenwood AK, Wark AR, Fernald RD, Hofmann HA. Expression of arginine vasotocin in distinct preoptic regions is associated with dominant and subordinate behaviour in an African cichlid fish. Proc R Soc B. 2008;275(1649):2393–2402. doi: 10.1098/rspb.2008.0622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Almeida O, Oliveira RF. Social status and arginine vasotocin neuronal phenotypes in a cichlid fish. Brain Behav Evol. 2015;85(3):203–213. doi: 10.1159/000381251 [DOI] [PubMed] [Google Scholar]
- 116.Huffman LS, Hinz FI, Wojcik S, Aubin-Horth N, Hofmann HA. Arginine vasotocin regulates social ascent in the African cichlid fish Astatotilapia burtoni. Gen Comp Endocrinol. 2015;212:106–113. doi: 10.1016/j.ygcen.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 117.Schappaugh NA. Aggressive phenotypes in Malawi cichlids associated with V1aR variant. July 2017. https://smartech.gatech.edu/handle/1853/58482. Accessed October 28, 2019.
- 118.Lacar B, Linker SB, Jaeger BN, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7(1):1–13. doi: 10.1038/ncomms11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu YE, Pan L, Zuo Y, Li X, Hong W. Detecting activated cell populations using single-cell RNA-seq. Neuron. 2017;96(2):313–329.e6. doi: 10.1016/j.neuron.2017.09.026 [DOI] [PubMed] [Google Scholar]
- 120.Streelman JT, Albertson RC, Kocher TD. Genome mapping of the orange blotch colour pattern in cichlid fishes. Molecular Ecology. 2003;12(9):2465–2471. doi: 10.1046/j.1365-294X.2003.01920.x [DOI] [PubMed] [Google Scholar]
- 121.Parnell NF, Streelman JT. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity. 2013;110(3):239–246. doi: 10.1038/hdy.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Juntti SA, Hilliard AT, Kent KR, et al. A neural basis for control of cichlid female reproductive behavior by prostaglandin F2α. Current Biology. 2016;26(7):943–949. doi: 10.1016/j.cub.2016.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kratochwil CF, Liang Y, Gerwin J, et al. Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science. 2018;362(6413):457–460. doi: 10.1126/science.aao6809 [DOI] [PubMed] [Google Scholar]
- 124.York RA, Patil C, Abdilleh K, et al. Behavior-dependent cis regulation reveals genes and pathways associated with bower building in cichlid fishes. PNAS. October 2018:201810140. doi: 10.1073/pnas.1810140115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.