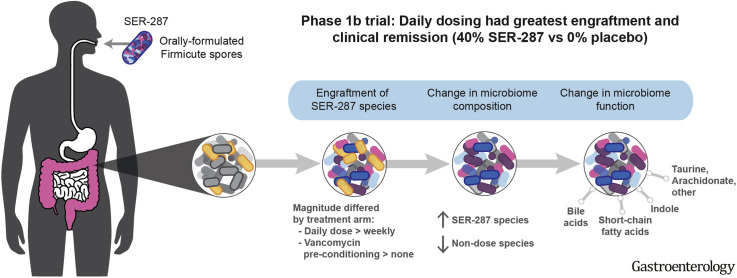
Abstract
Background & Aims
Firmicutes bacteria produce metabolites that maintain the intestinal barrier and mucosal immunity. Firmicutes are reduced in the intestinal microbiota of patients with ulcerative colitis (UC). In a phase 1b trial of patients with UC, we evaluated the safety and efficacy of SER-287, an oral formulation of Firmicutes spores, and the effects of vancomycin preconditioning on expansion (engraftment) of SER-287 species in the colon.
Methods
We conducted a double-blind trial of SER-287 in 58 adults with active mild-to-moderate UC (modified Mayo scores 4–10, endoscopic subscores ≥1). Participants received 6 days of preconditioning with oral vancomycin (125 mg, 4 times daily) or placebo followed by 8 weeks of oral SER-287 or placebo. Patients were randomly assigned (2:3:3:3) to groups that received placebo followed by either placebo or SER-287 once weekly, or vancomycin followed by SER-287 once weekly, or SER-287 once daily. Clinical end points included safety and clinical remission (modified Mayo score ≤2; endoscopic subscores 0 or 1). Microbiome end points included SER-287 engraftment (dose species detected in stool after but not before SER-287 administration). Engraftment of SER-287 and changes in microbiome composition and associated metabolites were measured by analyses of stool specimens collected at baseline, after preconditioning, and during and 4 weeks after administration of SER-287 or placebo.
Results
Proportions of patients with adverse events did not differ significantly among groups. A higher proportion of patients in the vancomycin/SER-287 daily group (40%) achieved clinical remission at week 8 than patients in the placebo/placebo group (0%), placebo/SER-287 weekly group (13.3%), or vancomycin/SER-287 weekly group (17.7%) (P = .024 for vancomycin/SER-287 daily vs placebo/placebo). By day 7, higher numbers of SER-287 dose species were detected in stool samples from all SER-287 groups compared with the placebo group (P < .05), but this difference was not maintained beyond day 7 in the placebo/SER-287 weekly group. In the vancomycin groups, a greater number of dose species were detected in stool collected on day 10 and all subsequent time points through 4 weeks post dosing compared with the placebo group (P < .05). A higher number of SER-287 dose species were detected in stool samples on days 7 and 10 from subjects who received daily vs weekly SER-287 doses (P < .05). Changes in fecal microbiome composition and metabolites were associated with both vancomycin/SER-287 groups.
Conclusions
In this small phase 1b trial of limited duration, the safety and tolerability of SER-287 were similar to placebo. SER-287 after vancomycin was significantly more effective than placebo for induction of remission in patients with active mild to moderate UC. Engraftment of dose species was facilitated by vancomycin preconditioning and daily dosing of SER-287. ClinicalTrials.gov ID NCT02618187.
Keywords: Inflammatory Bowel Disease, Gastrointestinal Microbiome, Microbe-Associated Metabolites, Microbiome Therapeutics
Abbreviations used in this paper: BA, bile acid; DCA, deoxycholic acid; FMT, fecal microbiota transplantation; HMP2, Integrative Human Microbiome Project; IBD, inflammatory bowel disease; LCA, lithocholic acid; SCFA, short-chain fatty acid; TMMS, total modified Mayo score; UC, ulcerative colitis; UDCA, ursodeoxycholic acid
Graphical abstract
What You Need to Know.
Background and Context
Firmicutes bacteria produce metabolites that help maintain the intestinal barrier and mucosal immunity. Firmicutes and their associated metabolites are reduced in intestinal microbiota of patients with ulcerative colitis (UC).
New Findings
In a phase 1b trial, administration of SER-287 (Firmicutes spores) following vancomycin preconditioning induced clinical remission in a significantly higher proportion of patients with mild to moderately active UC than placebo. The safety and tolerability of SER-287 were similar to those of placebo.
Limitations
This was a small study of limited duration (12 weeks)—larger studies are needed.
Impact
SER-287 may induce remission by changing the composition and function of the gut microbiome.
The gastrointestinal microbiome is a diverse ecosystem that provides essential functions for the host and represents a novel therapeutic target for UC. In “healthy” adults in the Human Microbiome Project, the microbiome is dominated by bacteria in 2 phyla, the Firmicutes and Bacteroidetes.1, 2, 3 Spore-forming Firmicutes, particularly the families Clostridiaceae, Lachnospiraceae, and Ruminococcaceae, have an important role in gut homeostasis through production of metabolites, which enhance gastrointestinal barrier and mucosal immune functions.4 Although the pathogenesis of disease remains unclear, changes in the gastrointestinal microbiome and associated metabolites may be important.5, 6, 7 UC patients with active disease exhibit lower microbial diversity with depletion of beneficial Firmicutes and relative expansion of pro-inflammatory Enterobacteriaceae.8 Inflammatory responses are elicited by Enterobacteriaceae via lipopolysaccharide signaling though Toll-like receptors.9 Microbiome compositional changes are associated with alterations in microbe-associated products, such as short-chain fatty acids (SCFA), secondary bile acids (BAs), and tryptophan metabolites, implicated in barrier function, mucin production, and colonic inflammation.5 Temporal shifts in microbial composition and their metabolites have been postulated to precipitate disease flares.6 , 10 , 11 Modulating the microbiome and the microbial triggers of inflammation and immune activation may provide an improved therapeutic option with limited off-target effects.
Although microbiome modulation with antibiotics or probiotics has not been an effective therapeutic strategy in UC,12 data from placebo-controlled trials support a benefit for multidose fecal microbiota transplantation (FMT),13 although the mechanism of action is not fully understood. Patients randomized to FMT had increased microbial diversity and a shift toward similarity to the donor microbiome13, 14, 15 compared with placebo recipients. Importantly, the microbial taxa associated with clinical remission included spore-forming Firmicutes, such as Lachnospiraceae, Ruminococcaceae, and Eubacteriaceae.8 , 13 , 15
The potential role of the microbiome in UC pathogenesis, coupled with FMT efficacy data, motivated our development of SER-287 as a novel therapeutic. Although FMT contains the full spectrum of stool components to modify the microbiome, our approach fractionates spore-forming bacteria to specifically target Firmicutes relevant to gastrointestinal homeostasis.4 Gram-positive Firmicute spores are isolated and enriched from stool of rigorously screened healthy donors by ethanol treatment and purification. This process also removes potential pathogens, such as vegetative bacteria, viruses, and 99% of other nonspore components, producing a more refined and safer therapeutic. Spores, which are intrinsically resistant to gastric acid, are formulated into oral capsules. Bacteria that germinate from these spores develop into metabolically active vegetative bacteria and colonize the colon, a process termed engraftment.
Our hypothesis was that SER-287 may reduce colonic inflammation by inducing compositional changes in the microbiome and related metabolites favorable to gastrointestinal homeostasis. We conducted a phase 1b randomized double-blind, placebo-controlled trial to evaluate the efficacy and safety of SER-287 in patients with active mild to moderate UC. To further evaluate the mechanism of action, the engraftment of SER-287 bacteria and their relationship to broader changes in the gastrointestinal microbiome and related metabolites was evaluated. We also investigated whether vancomycin preconditioning of the gastrointestinal microbiome could enhance engraftment of SER-287 bacteria. We hypothesized that clearance of Gram-positive bacteria with vancomycin can enhance engraftment of SER-287 bacteria by decreasing competition for nutrients and space.
Methods
Study Design
We conducted a randomized, double-blind, placebo-controlled, multiple-dose trial at 20 US sites. The protocol was approved by Investigational Review Boards and patients gave written informed consent. The trial was funded by Seres Therapeutics and designed in conjunction with the academic advisors.
Patient Population
Eligible patients were ≥18 years of age with active mild to moderate disease as defined by a total modified Mayo score (TMMS) of 4–10 and an endoscopic subscore ≥1 (mild disease TMMS 4–6; moderate disease TMMS 7–10). During the screening period, medical history and symptom severity were collected. Disease activity and severity was assessed by the TMMS, which includes endoscopy, patient-reported outcomes of stool frequency and rectal bleeding, and a physician global assessment.
Treatment-naïve patients and those who had failed mesalamine formulations, thiopurines, or biologics were eligible to enroll. After randomization, patients were required to remain on a stable dose of mesalamine, oral corticosteroids (≤15 mg daily of prednisone or equivalent), or thiopurines. Biologic therapies, probiotics, or treatments delivered via enema or suppository were not permitted. The washout period for biologics was 3 months and for probiotics and rectal therapies was approximately 14 days. Inclusion and exclusion criteria and allowable medications are provided in the Supplementary Material.
Randomization and Treatments
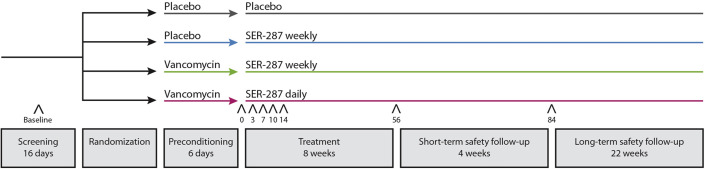
The trial design is shown in Supplementary Figure 1. Patients were randomly assigned, in a 2:3:3:3 ratio, to 4 arms that incorporated 2 treatment phases. The first phase consisted of preconditioning with vancomycin (125 mg 4 times daily) or matching placebo for 6 days. The second phase consisted of administration of SER-287 (4 capsules containing 1 × 107 colony-forming units) once daily or once weekly or matching placebo once daily for 8 weeks. Subjects who received once weekly SER-287 took placebo capsules on days 2–7 weekly to maintain the blind. Randomization was performed centrally with an interactive web response system and was balanced across drug product lots (Supplementary Material).
Manufacture and Characterization of SER-287
Our donor screening and manufacturing program has been reviewed by the US Food and Drug Administration and is consistent with their recent publication.16 Three donors underwent extensive health examination, including personal and family medical history and laboratory screening for chemistries; hematology; urinalysis; and blood and stool viral, bacterial, and parasite testing before donating stool. Donors completed physical examinations, questionnaires, and laboratory testing during and after the donation period before material was released for manufacturing.
Stool donations underwent Good Manufacturing Process–compliant manufacturing steps, including clearance of nonspore forms of bacteria, fungi, parasites, and viruses via solvent treatment and by sequential purification steps. Three lots of SER-287 were prepared from 3 different donors’ materials. All stool donations for a lot came from a single donor and all subjects received SER-287 made from a single lot. Nonspore matter was reduced by >200-fold during purification to <5 mg/dose, with final product purity approximately 10%–20% spores by weight. No contaminating bioburden was detected in the final product (assay limit of detection 20 colony-forming units/g). The SER-287 lots used in the trial were characterized by direct whole metagenomic shotgun sequencing and taxonomic profiling (Supplementary Material). Firmicutes genera identified in lots are in Supplementary Table 1.
Clinical End Points
Clinical evaluations were performed during the screening phase and at the end of the 8-week treatment phase (Supplementary Figure 1). Endoscopy was scored by central readers who were unaware of the treatment assignment, study visit number, or clinical information (https://www.robartsinc.com/). We evaluated clinical remission (TMMS ≤2 and an endoscopic subscore ≤1), endoscopic improvement (decrease in the endoscopic subscore ≥1), and clinical response (decrease of ≥3 points in TMMS from baseline plus either a decrease of ≥1 point in rectal bleeding subscore or an absolute rectal bleeding subscore of 0 or 1 at week 8.
Safety was assessed from the preconditioning phase through the end of the long-term safety phase. Assessments performed from randomization and for 4 weeks after completion of study treatment included laboratory values and adverse events classified by system organ class and preferred term using the Medical Dictionary for Regulatory Activities.
Statistical Methods
Primary end points were safety and changes in microbiome composition. Clinical remission, endoscopic improvement, and clinical response were secondary end points. The safety population comprised all patients who received at least 1 dose of study drug (vancomycin, SER-287, or placebo). The intent-to-treat population comprised all patients who were randomized. Descriptive statistics were used to summarize baseline characteristics. For the analysis of clinical remission, the proportions of patients achieving clinical remission at week 8 was compared between each SER-287 arm and placebo using a 2-sided Fisher exact test; a P value <.05 was considered significant. Asymptotic 95% confidence intervals for the rate differences were also estimated. In these analyses, an outcome of “failure” was assigned to patients who discontinued treatment before study day 48, received a rescue medication for UC flare, or who lacked the necessary data for determination of the relevant end point. No adjustment for multiple comparisons was made, as the secondary efficacy end points were intended to provide only preliminary data on efficacy. A similar approach was taken for analysis of the proportions of patients with clinical and endoscopic response. The rates of treatment-emergent adverse events were described by treatment group. The randomization plan, which included 15 patients into each of the SER-287 arms and 10 patients into the placebo/placebo arm, was determined to be sufficient to provide initial assessment of a potential treatment effect on efficacy, safety, and the microbiome.
Microbiome and Metabolomics Evaluations
We evaluated baseline microbiome composition, engraftment of SER-287 bacteria, and broader changes in the gastrointestinal microbiome and metabolites using metagenomic shotgun sequencing and mass spectrometry (metabolomics) of stool samples collected before treatment and post treatment. Details are provided in the Supplementary Material; the number of patients with evaluable metagenomic shotgun sequencing data from both baseline and post-treatment samples is reported in Supplementary Table 2.
Baseline microbiome composition and post-treatment changes were characterized by calculating α-diversity and β-diversity. α-Diversity is reported as the number of unique species detected in a single sample.17 β-diversity, which measures compositional similarity between samples, was calculated using the binary Jaccard index.17 , 18 The binary Jaccard metric measures similarity based on presence or absence of shared species, and ranges in value from 0 to 1, with 1 indicating 2 samples are identical.
No correction for multiple hypothesis testing was performed as this phase 1b trial was exploratory; the microbiome data collected were intended to provide preliminary insights on engraftment and microbiome change.
Baseline microbiome composition
For baseline composition, we compared 2 summary measures, α-diversity and Enterobacteriaceae abundance, in our study population to an external cohort of UC patients and controls from the Integrative Human Microbiome Project (HMP2).6 , 19 Controls included people who presented for age-related colorectal cancer screening or patients with gastrointestinal symptoms without inflammatory bowel disease (IBD).
Engraftment of drug product species
Two approaches were taken to assess engraftment of drug product species (ie, “dose-species”).
In the first prespecified analysis, we evaluated the prevalence of SER-287 dose-species in patients at baseline (screening endoscopy visit before treatment) and on day 0 (post-vancomycin preconditioning, before SER-287 dosing) and days 3, 7, 10, 14, 56, and 84 (post treatment with SER-287). We defined engrafting species as SER-287 dose-species detected post treatment that were not detected at baseline. A Mann-Whitney 2-sided test was performed comparing the number of engrafting species in patients of 1 arm with patients of another.
In the second post-hoc analysis, the change in compositional similarity between all spore-forming species in the patient microbiomes and the SER-287 dose from baseline to post treatment was quantified using the Jaccard β-diversity metric. This second complementary approach was performed to avoid underestimation of engraftment by not including species overlapping with drug product, which were present at baseline.
Changes in overall microbiome composition
The impact of SER-287 on overall changes in microbiome composition was assessed in a prespecified analysis by evaluating the distance between patients’ baseline and post-treatment samples using a Jaccard β-diversity metric. A Mann-Whitney 2-sided test was performed comparing patients across arms.
Changes in microbiome composition were additionally assessed post-hoc by evaluating the proportion of patients in whom species were gained or lost at 8 weeks post treatment relative to baseline after SER-287 or placebo. These compositional changes represent engrafting species from the dose, in addition to species not detected in the dose.
The impact of SER-287 treatment on Enterobacteriaceae species was evaluated post-hoc. Individual species abundances were summed to determine the relative abundance of Enterobacteriaceae as a fraction of all species detected. Patients with paired baseline and 8-week stool samples were assessed, and fold-changes in Enterobacteriaceae relative abundance post treatment were tested using a Wilcoxon signed-rank test.
Metabolic changes in stool
Using both targeted metabolomics of SCFAs and BAs and global metabolomics, we evaluated post-hoc how microbe-associated metabolites correlated with SER-287 treatment and clinical remission. Analyses were performed on 5 SCFAs and 9 BAs from the targeted panels and 1328 metabolites identified by global metabolomics. Changes in metabolite abundance post treatment were assessed based on fold-changes between patients’ baseline and post-treatment samples at 8 weeks post dosing using a Wilcoxon signed-rank test. Direct statistical comparison with placebo was not performed due to the low number of patients with paired samples. Association of metabolite concentrations with clinical remission was assessed at the clinical efficacy end point 8 weeks post dosing. A 2-sided Mann-Whitney test was performed to assess whether metabolites differed in remitters vs nonremitters across arms.
Results
Patient Demographics and Baseline Characteristics
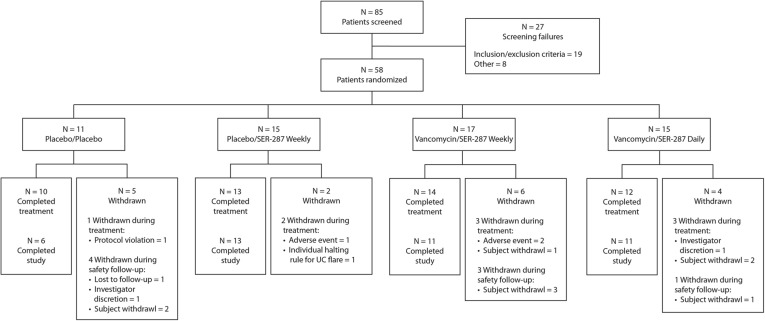
Table 1 and Supplementary Figure 2 show the patient disposition and demographics. Eighty-five patients were screened and 58 enrolled at 20 US sites within 17 months. Thirty-nine patients (67.2%) had left-sided disease and 19 patients (32.8%) had extensive disease. Mean time since diagnosis for the overall population was 12.2 years.
Table 1.
Demographic and Baseline Characteristics
| Characteristic | Placebo/placebo (n = 11) | Placebo/SER-287 weekly (n = 15) | Vancomycin/ SER-287 weekly (n = 17) | Vancomycin/ SER-287 daily (n = 15) | All subjects (n = 58) |
|---|---|---|---|---|---|
| Age, y, mean | 45.8 | 46.5 | 47.9 | 47.8 | 47.1 |
| Female sex, n (%) | 7 (63.6) | 9 (60.0) | 7 (41.2) | 8 (53.3) | 31 (53.4) |
| Extent of disease, n (%) | |||||
| Left-sided UC | 8 (72.7) | 10 (66.7) | 12 (70.6) | 9 (60.0) | 39 (67.2) |
| Extensive UC | 3 (27.3) | 5 (33.3) | 5 (29.4) | 6 (40.0) | 19 (32.8) |
| Smoking history, n (%) | 3 (27.3) | 5 (33.3) | 5 (29.4) | 6 (40.0) | 19 (32.8) |
| Severity of UC, n (%) | |||||
| Mild | 3 (27.3) | 6 (40.0) | 9 (52.9)a | 6 (40.0) | 24 (41.4) |
| Moderate | 8 (72.7) | 9 (60.0) | 7 (41.2)a | 9 (60.0) | 33 (56.9) |
| Mayo score at study entry, mean | 7.3 | 6.8 | 6.4 | 6.9 | 6.8 |
| UC medication(s) at study entry, n (%) | 9 (81.8) | 13 (86.7) | 11 (64.7) | 12 (80.0) | 45 (77.6) |
| Mesalamine | 7 (63.6) | 11 (73.3) | 9 (52.9) | 11 (73.3) | 38 (65.5) |
| Immunomodulator | 2 (18.2) | 4 (26.7) | 2 (11.8) | 1 (6.7) | 9 (15.5) |
| Steroid | 3 (27.3) | 2 (13.3) | 0 | 3 (20.0) | 8 (13.8) |
| Combination therapy | 3 (27.3) | 3 (20.0) | 0 | 3 (20.0) | 9 (15.5) |
| Endoscopy score at baseline, n (%) | |||||
| 1 | 1 (9.1) | 3 (20.0) | 5 (29.4) | 3 (20.0) | 12 (20.7) |
| 2 | 5 (45.4) | 7 (46.7) | 7 (41.2) | 9 (60.0) | 28 (48.3) |
| 3 | 5 (45.4) | 5 (33.3) | 5 (29.4) | 3 (20.0) | 18 (31.3) |
| Time since UC diagnosis, y, mean | 11.52 | 12.43 | 11.84 | 12.74 | 12.17 |
One subject in vancomycin/SER-287 weekly group with TMMS = 3 at entry is not included.
Most patients (91.4%) had failed at least 1 UC therapy. Forty-five (77.6%) were receiving stable UC medications at study entry, which included mainly mesalamine formulations (65.5%); 9 patients (15.5%) were receiving more than 1 medication (eg, mesalamine, an immunonomodulator, and/or a corticosteroid). Six patients (10.3%) had prior exposure to biologic therapy (ie, tumor necrosis factor antagonists, anti-integrins, or both). Thirteen patients (22.4%) were not receiving UC medications.
Safety
SER-287 was generally safe and well-tolerated across treatment arms (Table 2 ). Adverse events were mild (67.0%) or moderate (33.0%). One subject from the vancomycin/SER-287 daily dosing arm with a history of depression had a serious adverse event of worsening depression, deemed unrelated to treatment, which did not lead to treatment discontinuation. During the preconditioning phase, 7 patients had adverse events, most of which were gastrointestinal: 3 received placebo and 4 received vancomycin (Supplementary Table 3).
Table 2.
Adverse Events
| Variable | Placebo/placebo (n = 11) | Placebo/SER-287 weekly (n = 15) | Vancomycin/SER-287 weekly (n = 17) | Vancomycin/SER-287 daily (n = 15) |
|---|---|---|---|---|
| All AEs | 7 (63.6) | 9 (60.0) | 14 (82.4) | 8 (53.3) |
| Serious AEs | 0 | 0 | 0 | 1 (6.7) |
| AEs of special interest | 0 | 1 (6.7) | 0 | 0 |
| Severity of AEs | ||||
| Mild | 3 (27.3) | 7 (46.7) | 6 (35.3) | 3 (20.0) |
| Moderate | 4 (36.4) | 2 (13.3) | 8 (47.1) | 5 (33.3) |
| Treatment-related AEs | 1 (9.1) | 4 (26.7) | 7 (41.2) | 2 (13.3) |
| AEs leading to discontinuation of study drug | 0 | 1 (6.7) | 2 (11.8) | 0 |
NOTE. Values are n (%).
AE, adverse event.
During the treatment phase and the subsequent 4 weeks, the most commonly reported adverse events were gastrointestinal disorders, including abdominal pain, constipation, diarrhea, flatulence, and nausea. Rates of gastrointestinal adverse events were lowest among patients who received daily dosing of SER-287 (6.7%) compared with placebo/SER-287 weekly (46.7%), vancomycin/SER-287 weekly (41.2%), and placebo/placebo (45.5%). The rate of infections among those who received daily dosing of SER-287 was similar to placebo (Supplementary Table 4).
Treatment discontinuation due to gastrointestinal adverse events occurred in 1 patient (6.7%) in the placebo/SER-287 weekly arm; 2 patients (11.8%) in the vancomycin/SER-287 weekly arm, and none of the patients on the SER-287 daily dosing or placebo arms (Table 2). One patient in the placebo/SER-287 weekly arm (6.7%) discontinued treatment due to a UC flare (deemed unrelated to study drug by the investigator), and 1 patient (5.9%) in the vancomycin/SER-287 weekly arm and 1 patient in the vancomycin/SER-287 daily arm (6.7%) discontinued the study prematurely due to perceived lack of efficacy (Supplementary Figure 2).
Disposition of the Patients and Treatment Adherence
Overall, 84.5% of patients completed induction therapy, including 10 of 11 (90.9%) who received placebo/placebo; 13 of 15 (86.7%) who received placebo/SER-287 weekly; 14 of 17 (82.3%) who received vancomycin/SER-287 weekly; and 12 of 15 (80.0%) who received vancomycin/SER-287 daily. Mean adherence with study drug ranged from 84.5% to 86.9% of all dispensed capsules taken across treatment arms.
Efficacy
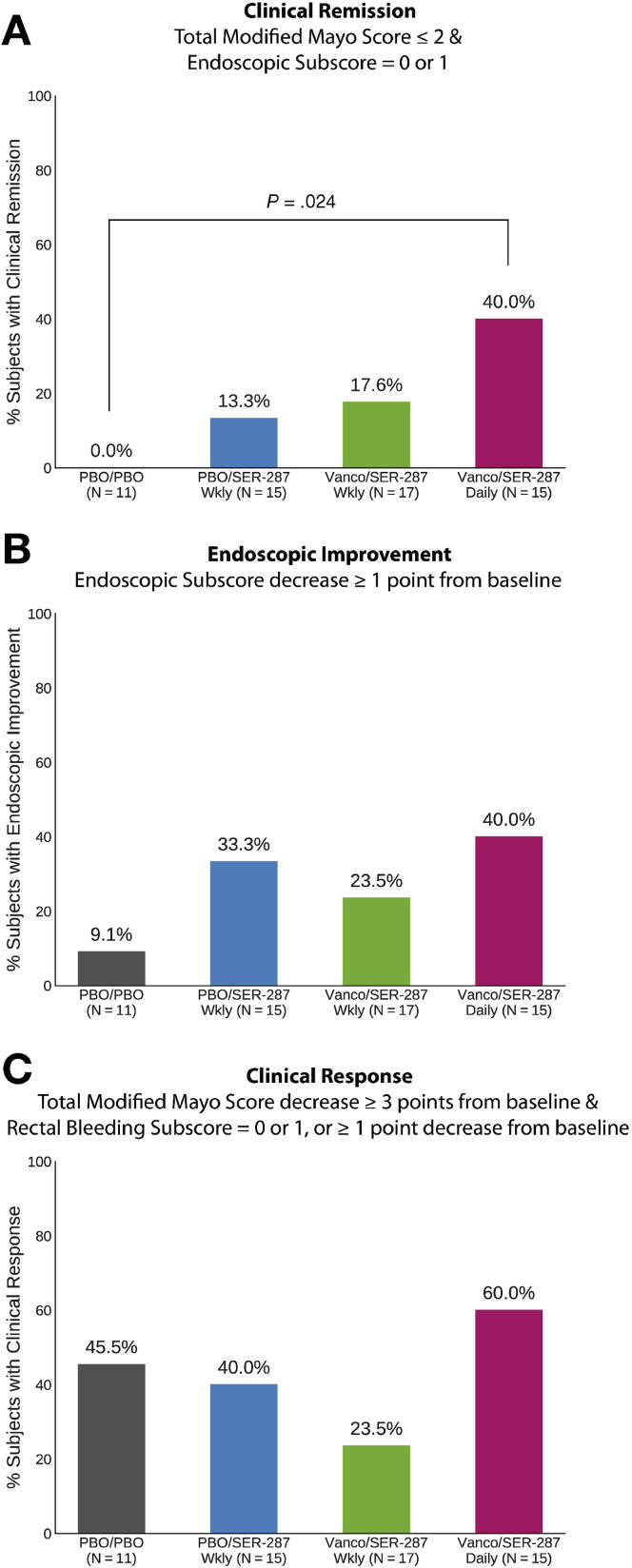
Patients assigned to the vancomycin/SER-287 daily arm achieved a higher clinical remission rate at week 8 (40%) than those assigned to placebo/placebo (0%), placebo/SER-287 weekly (13.3%), and vancomycin/SER-287 weekly (17.7%) (Figure 1 A). Remission in the vancomycin/SER-287 daily arm was significantly higher than the placebo/placebo arm (Fisher exact test, P = .024). Greater remission was observed in vancomycin/SER-287 daily vs vancomycin/SER-287 weekly, suggesting dose-dependence. Endoscopic improvement rates were numerically greater in the SER-287 arms but not significantly different than placebo (Figure 1 B). The outcomes for the 2 weekly dosing regimens of SER-287 were similar. No significant differences in clinical response (Figure 1 C) were observed in any of the 3 SER-287 treatment arms compared with placebo. No significant differences in fecal calprotectin and C-reactive protein data were observed across arms at 8 weeks.
Figure 1.
Proportion of patients achieving end points of clinical remission (A), endoscopic improvement (B), and clinical response (C) at week 8 in intent-to-treat population. Two-sided Fisher exact P value indicated when <.05. PBO, placebo; Vanco, vancomycin; Wkly, weekly.
Characteristics of the Baseline Microbiome
In comparing the baseline values of our study population with controls from HMP2,7 no significant differences in α-diversity were seen (Kruskal-Wallis, P > .05). Median abundance of Enterobacteriaceae in our study patients trended higher than the UC cohort in HMP2 and significantly higher than the non-IBD healthy cohort (median 0.39%, 0.17%, and 0.02%, respectively; Mann-Whitney P = .003).
In comparing patients with mild or moderate disease, α-diversity did not significantly differ. In moderate patients, Enterobacteriaceae abundance was higher than in mild patients (Mann-Whitney P = .038); moderate patients also had more genera considered proinflammatory (eg, Klebsiella, Proteus). Twelve of 51 patients (24%) had no detectable Enterobacteriaceae; most (75%) had mild UC. In contrast, of the 39 patients (76%) with detectable Enterobacteriaceae, most (63%) had moderate disease.
Engraftment of SER-287 Drug Product Species
Engraftment occurred rapidly by day 7, with a significant increase in engrafting species in all SER-287 arms compared with placebo (Figure 2 , Supplementary Table 5). Beyond day 7, the distinction between the placebo/placebo and placebo/SER-287 weekly arms was not significant. In contrast, the favorable effect of antibiotic preconditioning on engraftment was clearly apparent by day 10 for both weekly and daily SER-287 administration and sustained through 4 weeks post-dosing (Figure 2, P < .05 for all comparisons, Supplementary Table 5). In the vancomycin preconditioning arms, a significantly greater number of engrafting species were observed at days 7 and 10 in the SER-287 daily dosing arm compared with the SER-287 weekly dosing arm (P < .05, Supplementary Table 5). This observation suggests that engraftment is both facilitated by vancomycin preconditioning and dose-dependent (Figure 2, Supplementary Table 5). Engraftment at days 7 and 10 was also greater in remitters than nonremitters in all arms combined (Mann-Whitney, P = .001 and P = .025); however, this association was not significant within any individual arm or at later time points.
Figure 2.
Engraftment of SER-287 species is dose-dependent and facilitated by vancomycin preconditioning. Boxplots display median (horizontal line), 25th and 75th percentiles (box edges), range of nonoutlier observations (whiskers), and outlier observations (dots; >1.5 times interquartile range). Significance values are in Supplementary Table 5.
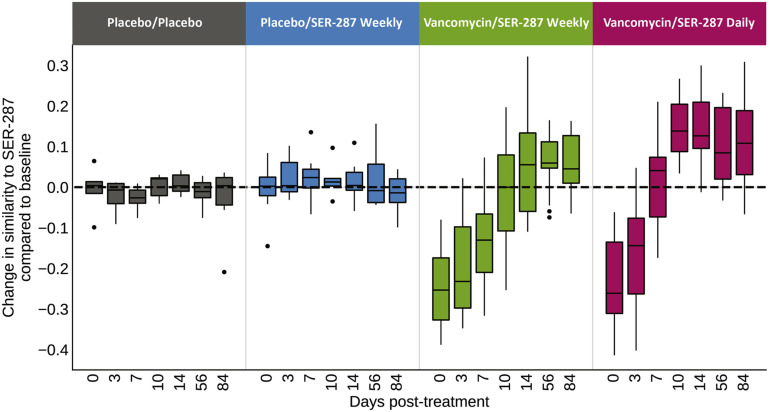
In the second assessment of engraftment, based on β-diversity, we observed a similar favorable impact of vancomycin-preconditioning and SER-287 dosing frequency as patient spore composition became most similar to SER-287 drug product species in the vancomycin/SER-287 daily arm (Supplementary Figure 3). We saw no difference in engraftment as assessed by β-diversity between the placebo/placebo and placebo/SER-287 weekly arms. We also did not observe significant differences in engraftment across drug lots by either measure of engraftment.
Effects of SER-287 Treatment on Overall Microbiome Composition
Across all species, α-diversity (the number of unique species) did not significantly increase in any arm (P > .05 for all arms). However, α-diversity of spore-forming species significantly increased, and was durable in the vancomycin/SER-287 daily dosing arm, consistent with greater engraftment (Supplementary Table 6). Through 8 weeks, β-diversity was significantly different from baseline only in the arms with SER-287/vancomycin preconditioning indicating changes in overall microbiome composition (Supplementary Table 7).
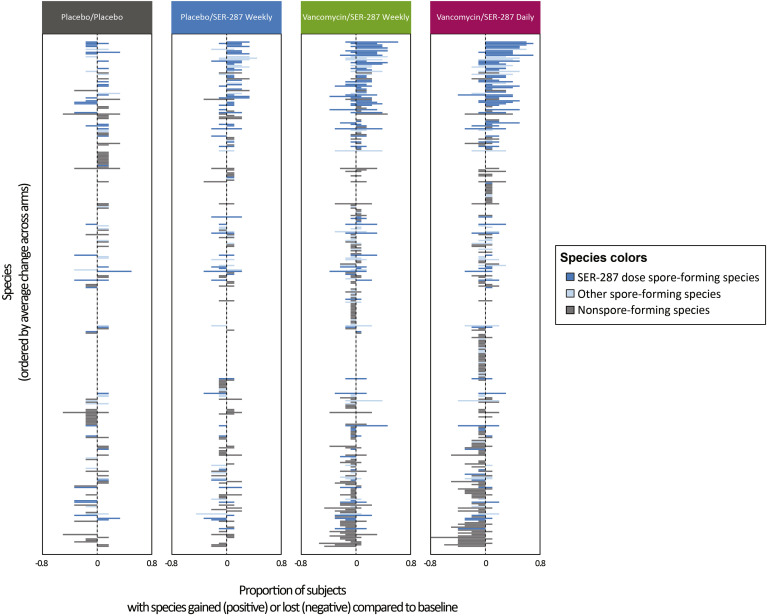
We next sought to characterize compositional changes in the microbiome at finer resolution based on gain and loss of individual species post treatment (Figure 3 ). In the placebo/placebo arm, microbiome changes are due to natural fluctuations.19 Compared with placebo/placebo, minimal compositional shifts were observed in the placebo/SER-287 weekly arm consistent with low magnitude engraftment. In contrast, significant changes were observed in both vancomycin preconditioning arms with a trend toward greater change in the SER-287 daily-dosing arm. To further characterize these changes, we separately evaluated the proportion of species increasing or decreasing in prevalence. More species were gained (Mann-Whitney, P = .022 and P = .014 for vancomycin/SER-287 weekly and vancomycin/SER-27 daily arms, respectively) and lost (Mann-Whitney, P = .004 and P = .006 for vancomycin/SER-287 weekly and vancomycin/SER-287 daily arms, respectively) in the vancomycin-preconditioning arms than in the placebo/placebo arm.
Figure 3.
Vancomycin preconditioning followed by SER-287 results in a broad shift in overall composition of spore and nonspore microbial species 8 weeks post treatment. The proportion of patients in each arm that gained (positive values) or lost (negative values) a given species compared with baseline is shown (x-axis). Species (y-axis; n = 344) are ordered by average change across arms; species without change in any subject are not depicted (n = 15). Bar colors indicate spore-forming species detected in SER-287 (dark blue), other spore-forming species not detected in SER-287 (light blue), and other non–spore-forming species (gray). SER-287 dose species gained compared with baseline are the same species represented in Figure 2.
Genera of newly detected species were predominantly spore-formers found in SER-287, such as Clostridium, Gemminger, Dorea, Roseburia, Blautia, and Faecalibacterium (Figure 3, blue bars). Genera decreasing post treatment were predominantly nonspore formers, such as Veillonella, Streptococcus, and Bacteroides (Figure 3, gray bars). Taken together, global changes in the microbiome were associated with engraftment of drug product species (Supplementary Table 1) and additional changes related to nondrug product species.
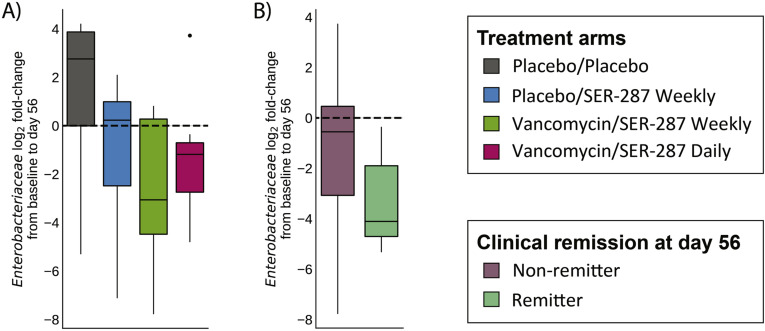
We evaluated the effect of SER-287 on Enterobacteriaceae abundance at 8 weeks post treatment among patients with Enterobacteriaceae detected at baseline (n = 29). We observed the greatest decrease of Enterobacteriaceae relative abundance compared with baseline in vancomycin preconditioning arms (median decreases –0.09% and –0.58% for SER-287 weekly and daily, respectively) and a slight increase in placebo preconditioning arms (median increases +0.36% and +0.03% for placebo and SER-287 weekly, respectively) (Supplementary Figure 4 and Supplementary Table 8). Furthermore, an association was observed between a decline in Enterobacteriaceae and clinical remission (P =.02; vancomycin/SER-287 weekly + daily arms, Supplementary Table 9).
Effect of SER-287 Treatment on Microbe-Associated Metabolites
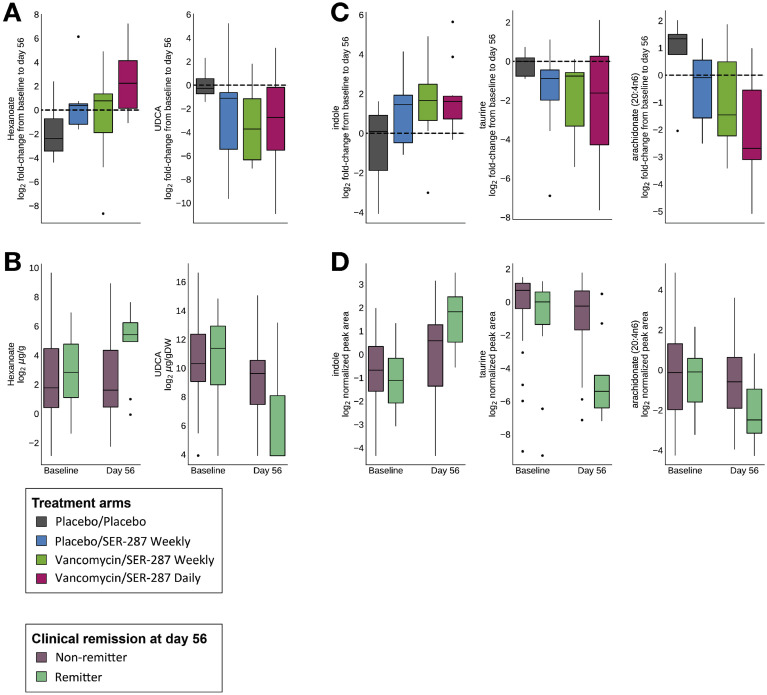
For SCFA and BA concentrations we assessed change post treatment within arms and association with remission across arms (see Methods; Supplementary Tables 10 and 11). The SCFA hexanoate was positively associated with both SER-287 (P = .02 in vancomycin/daily dosing arm; Figure 4 A) and clinical remission (P = .05; Figure 4 B). Butyrate did not significantly change post treatment, but there was a trend for an increase in the ratio of butyrate to propionate concentrations in the vancomycin/daily dosing arm (P = .07). We also examined secondary BA lithocholic acid (LCA), deoxycholic acid (DCA), and ursodeoxycholic acid (UDCA). We did not observe a significant change in LCA and DCA post treatment, but UDCA was negatively associated with both SER-287 treatment (P = .047 in the vancomycin/daily dosing arm; Figure 4 A) and clinical remission (P = .043; Figure 4 B). LCA and DCA increases tended to positively associate with clinical remission (P = .053 and P = .09, respectively), with an observed increase in the ratio of LCA/UDCA.
Figure 4.
Microbe-associated metabolites associated with SER-287 treatment and clinical outcome. Metabolite levels in fecal samples were assessed via targeted (A, B) and global (C, D) metabolomics. In (A) and (C), metabolite fold-changes are shown by treatment arm; black horizontal dashed line indicates whether the metabolite is higher or lower compared with baseline. For (B) and (D), metabolite abundance is shown by remission status. Significance values are provided in Supplementary Table 10, Supplementary Table 11, Supplementary Table 12, Supplementary Table 13.
Global liquid chromatography-tandem mass spectrometry metabolomics were used to identify metabolites associated with SER-287 treatment and remission. Of the 1328 metabolites in our data set, we conducted 3 analyses. First, we examined changes in metabolite concentrations relative to pretreatment baseline (Supplementary Table 12). We focused our analysis on the vancomycin-preconditioning arms where SER-287 engraftment was observed at the 8-week time point; in total, 166 metabolites changed (13 increased; 153 decreased; P < .05). Second, we examined the association of metabolites with outcome (Supplementary Table 13); 186 metabolites differentiated remitters from nonremitters (57 increased; 129 decreased; P < .05). Lastly, 38 metabolites were associated with both vancomycin/SER-287 treatment and remission; of these, 3 metabolites increased and 35 decreased. All 3 metabolites that increased are amino-acid derivatives: N-acetylarginine and 3 derived from tryptophan (indole and 3-methylindole). The 35 metabolites that decreased include lipids (eg, sphingolipids, acyl-carnitines, and arachidonate), dinucleotides, taurine-derived metabolites (taurine and N-acetyltaurine), and 5 unidentified compounds. Three of the 38 metabolites are microbe-associated and affect inflammatory pathways, specifically indole, taurine, and arachidonate (Figure 4 C and D), and several others are host-derived lipids and acyl-carnitines known to be perturbed in IBD.6 , 20, 21, 22
Discussion
This first clinical trial of an oral microbiome drug, SER-287, provides evidence of a favorable safety profile and preliminary insights into drug pharmacology and multiple mechanisms of action. In keeping with our hypothesis, SER-287 bacteria engrafted and modulated the microbiome and microbe-associated metabolites of functional relevance to UC. In this small study, we also have preliminary data of an efficacy signal of SER-287 supporting a role for microbiome therapeutics in the treatment of active mild to moderate UC.
The safety profile of SER-287, which was the primary safety end point, was similar to placebo. The most commonly reported adverse events were gastrointestinal disorders, which were less frequent in the SER-287 daily dosing arm relative to other arms, including placebo. Favorable tolerability might be expected given that SER-287 is composed of Firmicutes, considered important to gastrointestinal health.1 , 3 Safety confirmation in larger trials would establish this therapeutic approach as particularly suitable for monotherapy for milder UC patients or in combination with other agents. Oral vancomycin preconditioning was well-tolerated relative to placebo, consistent with its well-known safety profile.
In this small phase 1b study, the daily SER-287 regimen was the most effective regimen as assessed by all prespecified efficacy end points. The use of centrally read endoscopy, a modality that reduces bias in assessment of clinical remission, and was likely critical to determining a benefit of SER-287.23 Although clinical response rates did not significantly differ between placebo and SER-287 arms, the placebo response rate was high, typical of UC trials conducted in mild to moderately active disease24; it is noteworthy that this highly subjective outcome is no longer recommended as a primary end point.24 , 25 Clinical remission and endoscopic improvement were highly similar between the 2 weekly SER-287 regimens, with and without vancomycin preconditioning, suggesting that the direct therapeutic effect of vancomycin was limited. Likewise, we suspect that the efficacy of the daily dosing arm was driven by greater SER-287 drug exposure, although we cannot definitively rule out any impact of vancomycin without a placebo comparator.
We propose engraftment as a key mechanism of action of a live microbiome biotherapeutic. This proposal is based on the hypothesis that a microbiome drug is more likely to have a therapeutic effect if there is measurable engraftment of drug product, which evokes measurable changes in the overall composition of the gut microbiome, associated metabolites, and other functional changes in the host. We propose that engraftment kinetics of a microbiome therapeutic are comparable to small molecule drug pharmacokinetics.
Most ecosytems are stable and generally resistant to colonization. Thus, the ability to engraft is dependent on disruption of the microbial environment.26 , 27 The decision to use an antibiotic as preconditioning therapy was informed by human studies showing the disruptive impact of these drugs on the gut microbial community28 and preclinical studies supporting increased engraftment after antibiotic treatment. For example, in the context of recurrent Clostridium difficile infection, antibiotic-induced reductions in gut microbial diversity29 can be restored through engraftment of orally dosed bacteria.30 Oral vancomycin was specifically evaluated as a preconditioning regimen based on its well-known safety profile; lack of systemic absorption; its spectrum of antibacterial activity; and the hypothesis that reduction of pre-existing Gram-positive bacteria would create an ecological niche for engraftment of Gram-positive SER-287 species. The 2 SER-287 weekly dosing arms affirm that vancomycin preconditioning facilitated engraftment, although we were not able to definitively link engraftment to efficacy. The rules of engraftment may also be influenced by drug dosing frequency.27 Daily dosing of SER-287 led to greater engraftment at earlier time points compared with weekly dosing, consistent with a dose-dependent effect. Although, we did not observe differences in engraftment or clinical outcome across drug product lots, further evaluation in future trials will confirm whether or not SER-287 is associated with a donor effect.
SER-287 engraftment was associated with gains and losses of both spore-forming and non–spore-forming species. Post-treatment genera gained included Firmicutes related to dose-species, such as Clostridium, Gemminger, Dorea, Roseburia, Blautia, and Faecalibacterium, implicated in colonocyte health, epithelial gut integrity, and regulation of immune tolerance.31 We only observed a significant increase in the α-diversity of spore-forming Firmicutes in the SER-287 daily dosing arm. Additionally, declines in Enterobacteriaceae abundance, including pro-inflammatory species, such as Klebsiella pneumoniae and Proteus mirabilis,9 , 32 were associated with remission in the 2 SER-287 treatment arms after vancomycin preconditioning. The clinical significance of this finding is unclear, given the limited sample size, the heterogeneity of Enterobacteriaceae detection at baseline and the magnitude of the changes. Collectively, these observations support that modulation of the microbiome was due to replacement of pre-existing spore-formers with drug product species in addition to changes in species not in the dose. These data suggest that total diversity may be less informative than the specific species that are interacting and functioning within the microbial network.33 , 34 In support of that concept, baseline α-diversity was not associated with disease severity, whereas baseline Enterobacteriaceae abundance was higher in moderate subjects. We propose that these compositional changes in non–dose-related species represent 1 significant aspect of the mechanism of action and pharmacodynamics of SER-287.
Changes in microbe-associated metabolites are another dimension of SER-287 pharmacodynamics. In post-hoc analyses, we focused on metabolites that impact epithelial barrier integrity and signaling of anti-inflammatory pathways. Consistent with the HMP2 IBD cohort,6 we did not observe significant increases in abundances of butyrate or secondary BAs DCA and LCA in SER-287 recipients. Notably, we did see changes in ratios of metabolites, such as butyrate to propionate and LCA/UDCA, suggesting that SER-287 can lead to shifts in the functional pathways utilized with clinical benefit. The metabolites with the strongest associations with both SER-287 treatment and clinical remission are microbe-associated metabolites with relevance to gut homeostasis, such as hexanoate, indole, taurine, and arachidonate. Microbial products of tryptophan metabolism, such as indole, can signal aryl hydrocarbon receptor in the gut and impact mucin production, epithelial barrier function, and immune signaling.5 Arachidonic acid is a precursor of prostaglandins that can impact neutrophil migration across the epithelium.35 , 36 Notably, the constellation of metabolite associations with SER-287 or clinical remission aligns with emerging data from several UC cohort studies6 , 20, 21, 22 (Supplementary Table 10, Supplementary Table 11, Supplementary Table 12, Supplementary Table 13). We hypothesize that these associations reflect modulation of multiple metabolic pathways by SER-287 and contribute to its mechanism of action. Although we did not observe a direct overall correlation of SER-287 engraftment with clinical outcomes in this small safety study, we suspect that metabolic correlations are likely a more sensitive indicator of the pharmacodynamics of a microbiome drug; this may become clearer in our phase 2 trial.
SER-287 offers advantages over other microbiome-modulating approaches, such as single-strain therapeutics and FMT. Single-strain therapeutics are unlikely to provide the spectrum of functional activity necessary for efficacy. In contrast, FMT represents a broad spectrum of microbes, but due to minimal processing can also include harmful pathogens. Bacterial infections transmitted through contaminated FMT have, uncommonly, led to deaths and hospitalizations.37 , 38 However, because most FMT use has been outside the clinical trial setting, the safety profile of FMT remains unknown.39 The US Food and Drug Administration has also alerted providers of the potential risk of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) transmission via FMT due to prolonged fecal shedding.40 The COVID-19 (coronavirus disease 2019) pandemic is a reminder that FMT will always be vulnerable to emerging infections when pathogens are unrecognized until after transmission.41 , 42 In addition, FMT carries risk of transmission of proinflammatory strains and potentially carcinogenic bacteria.9 , 43 In contrast, SER-287 is a multifunctional consortia of bacterial spores purified under stringent manufacturing processes. These processes isolate the microbial components essential to gut homeostasis, inactivating bacteria, parasites, fungi, and viruses, providing a reliable safety net to mitigate risk.30
Strengths of this trial included its placebo-controlled design, affording an opportunity to examine the impact of a therapeutic intervention on the UC microbiome. Blinded central reads of endoscopic assessments enhance confidence in the outcomes.23 The collective and consistent observations relative to clinical outcomes, engraftment, microbiome composition, and microbe-associated metabolites are consistent with the biologic activity of SER-287. One limitation was its relatively small sample size, which precludes definitive conclusions about efficacy and safety. A further limitation of our trial was the absence of vancomycin preconditioning followed by placebo. This arm was not included due to concerns about a potential bloom of proinflammatory Enterobactericeae after antibiotics alone,44 , 45 and the potential risk of Clostridioides difficile infection.46 One study of 27 UC subjects who were randomly assigned to antibiotic preconditioning followed by FMT compared with antibiotics alone is particularly informative.46 C difficile infection occurred in 30% of subjects in the antibiotic alone arm, which the authors attributed to the low diversity state that ensued. In addition, reduction of beneficial Firmicutes and expansion of proinflammatory Proteobacteria were observed, which might explain the absence of any clinical remitters in this arm compared with 24% randomized to FMT. Because patients with UC are at increased risk of C difficile infection with worse clinical outcomes than those without underlying UC, we chose not to include a vancomycin preconditioning/placebo arm in our phase 2 trial based on these concerning data. In addition, vancomycin monotherapy is not recommended by Society guidelines.12
These prospective trial data support the favorable impact of SER-287 on the gut microbiota and their associated metabolites through multiple physiologic pathways, with important implications for UC management. SER-287 may represent a new therapeutic paradigm for UC, with potential advantages with respect to safety and convenience of oral dosing. Based on these results, daily SER-287 after vancomycin preconditioning is undergoing additional evaluation in a phase 2b trial (ClinicalTrials.gov ID NCT03759041).
Acknowledgments
The authors thank all of the patients and clinical collaborators that participated in this study, including Raj Bandari, Alan Moss, Hans Herfarth John Curran, Kieran Jagarlamudi, Katharina Oneto, Alan Sadfi, Caterina Oneto, Gregory Wiener, and David Kerman; Kevin Litcofsky, Chris McChalicher, Jonathan Winkler, Jose Otero, and Nedim Altaras for the manufacture of SER-287; Carol Lewis-Cullinan, Gabrielle Glick, and Kostia Franklin for clinical operations; Kevin Horgan for review of earlier drafts of the manuscript.
Patricia Bernardo’s and Amelia Tomlinson’s current affiliation is ResTORbio Boston MA; Peng Zhao current affiliation is Axcella Health, Cambridge, MA; Sheri Simmons’s current affiliation is Janssen Human Microbiome Institute, Cambridge, MA; David Cook’s current affiliation is DN Cook Consulting, Brookline, MA; Roger Pomerantz’s current affiliation is Contrafect, Yonkers, NY; and Michele Trucksis’s current affiliation is LLC, Wayland, MA.
CRediT Authorship Contributions
Matthew R. Henn, PhD (Conceptualization: Equal; Data curation: Supporting; Formal analysis: Supporting; Methodology: Equal; Software: Supporting; Supervision: Equal; Visualization: Supporting; Writing – original draft: Lead; Writing – review & editing: Lead). Edward J O'Brien, PhD (Data curation: Equal; Formal analysis: Equal; Methodology: Equal; Software: Equal; Visualization: Equal; Writing – original draft: Supporting; Writing – review & editing: Supporting). Liyang Diao, PhD (Data curation: Equal; Formal analysis: Equal; Methodology: Equal; Software: Equal; Visualization: Equal; Writing – original draft: Supporting; Writing –review & editing: Supporting). Brian G. Feagan, MD (Methodology: Supporting; Writing – original draft: Supporting; Writing – review & editing: Equal). Curtis Huttenhower, PhD (Methodology: Supporting; Software: Supporting; Writing – review & editing: Supporting) William J. Sanborn (Methodology: Supporting; Writing – review & editing: Supporting). Jennifer R. Wortman, MS (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Software: Supporting; Supervision: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting). Barbara McGovern, MD (Formal analysis: Supporting; Writing – original draft: Equal; Writing – review & editing: Supporting). Sherry Wang-Weigand, MD, PhD (Methodology: Supporting; Writing – review & editing: Supporting). David Lichter, MS (Writing – review & editing: Supporting). Meghan Chafee, PhD (Formal analysis: Supporting; Methodology: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting). Chris B. Ford, PhD (Methodology: Equal; Writing – review & editing: Supporting). Patricia Bernardo, PhD (Conceptualization: Equal; Formal analysis: Equal; Methodology: Equal). Peng Zhao, PhD (Conceptualization: Supporting; Formal analysis: Equal; Methodology: Equal). Sheri Simmons, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Software: Supporting). Amelia Tomlinson, PhD (Data curation: Supporting; Formal analysis: Supporting). David N. Cook (Conceptualization: Supporting; Methodology: Supporting). Roger J. Pomerantz, MD (Conceptualization: Supporting). Bharat K Misra, MD (Investigation: Equal). John G. Auninš, PhD (Conceptualization: Supporting; Investigation: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting). Michele Trucksis, MD (Conceptualization: Lead; Data curation: Supporting; Formal analysis: Supporting; Investigation: Lead; Methodology: Supporting; Supervision: Equal; Writing – original draft: Supporting; Writing – review & editing: Supporting).
ClinicalTrials.gov ID NCT02618187.
Footnotes
Conflicts of interest These authors disclose the following: M. Henn, E. O’Brien, L. Diao, C. Huttenhower, J. Wortman, B. McGovern, S. Wang-Weigand, D. Lichter, M. Chaffee, C. Ford, P. Bernardo, P. Zhao, S. Simmons, A. Tomlinson, D. Cook, R. Pomerantz, J. Aunins, and M. Trucksis own stock in Seres Therapeutics. C. Huttenhower is a member of the Scientific Advisory Board of Seres Therapeutics. A Tomlinson is a past employee of Seres Therapeutics. W.J. Sandborn reports: research grants from Atlantic Healthcare Limited, Amgen, Genentech, Gilead Sciences, Abbvie, Janssen, Takeda, Lilly, Celgene/Receptos,Pfizer, Prometheus Laboratories (now Prometheus Biosciences); consulting fees from Abbvie, Allergan, Amgen, Arena Pharmaceuticals, Avexegen Therapeutics, BeiGene, Boehringer Ingelheim, Celgene, Celltrion, Conatus, Cosmo, Escalier Biosciences, Ferring, Forbion, Genentech, Gilead Sciences, Gossamer Bio, Incyte, Janssen, Kyowa Kirin Pharmaceutical Research, Landos Biopharma, Lilly, Oppilan Pharma, Otsuka, Pfizer, Progenity, Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories), Reistone, Ritter Pharmaceuticals, Robarts Clinical Trials (owned by Health Academic Research Trust, HART), Series Therapeutics, Shire, Sienna Biopharmaceuticals, Sigmoid Biotechnologies, Sterna Biologicals, Sublimity Therapeutics, Takeda, Theravance Biopharma, Tigenix, Tillotts Pharma, UCB Pharma, Ventyx Biosciences, Vimalan Biosciences, Vivelix Pharmaceuticals; and stock or stock options from BeiGene, Escalier Biosciences, Gossamer Bio, Oppilan Pharma, Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories), Progenity, Ritter Pharmaceuticals, Ventyx Biosciences, Vimalan Biosciences. Spouse: Opthotech–consultant, stock options; Progenity–consultant, stock; Oppilan Pharma–employee, stock options; Escalier Biosciences–employee, stock options; Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories) –employee, stock options; Ventyx Biosciences–employee, stock options; Vimalan Biosciences–employee, stock options. The remaining authors disclose no conflicts.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://doi.org/10.1053/j.gastro.2020.07.048.
Supplementary Methods
Complete Inclusion and Exclusion Criteria
To be eligible for enrollment, a subject was required meet all of the following criteria before undergoing any study-related procedures:
-
1.
Signed informed consent;
-
2.
Male or female, 18 years or older;
-
3.
UC diagnosed by routine clinical, radiographic, endoscopic and pathologic criteria;
-
4.Active mild to moderate UC as determined by lower endoscopy (flexible sigmoidoscopy or colonoscopy) within approximately 3 days of randomization to study, defined as:
-
a.TMMS of 4–10, inclusive
-
b.Modified Mayo endoscopic subscore of ≥1, with evidence of mucosal lesions
-
c.At least 15 cm of disease from anal verge
-
a.
-
5.If female, subject is nonlactating, and either:
-
a.Not of childbearing potential, defined as postmenopausal for at least 1 year or surgically sterile due to bilateral tubal ligation, bilateral oophorectomy, or hysterectomy.
-
b.Of childbearing potential and is practicing at least 1 highly effective method of birth control, including the barrier method; oral or parenteral contraceptives; a vasectomized partner; or abstinence from sexual intercourse. The investigator will discuss with the subject the option of practicing more than 1 of these methods for the duration of the study.
-
a.
-
6.
If male and partner is of childbearing potential, subject agrees to practice at least 1 highly effective method of birth control for the duration of the study.
Any of the following was regarded as a criterion for exclusion from the trial:
-
1.
Fever >38.3°C;
-
2.
Known or suspected toxic megacolon and/or known small bowel ileus;
-
3.
Known history of Crohn’s disease;
-
4.
Subjects with serum albumin ≤2.5 g/dL at baseline;
-
5.
Cytomegalovirus polymerase chain reaction positive from blood plasma at screening;
-
6.
Known stool tests positive for ova and/or parasites, or positive stool culture, within the 30 days before enrollment;
-
7.
Subjects on cyclosporine or triple immunosuppression. Triple immunosuppression will include any 3 of the following classes of drugs taken in combination: steroids (ie, prednisone, budesonide, budesonide MMX), immunosuppressant (ie, methotrexate, azathioprine, 6-mercaptopurine), and/or other immunosuppressant (ie, tacrolimus, mycophenolate mofetil [CellCept]);
-
8.
Biologic medication (infliximab, adalimumab, golimumab, certolizumab, vedolizumab, ustekinumab, or natalizumab) use within 3 months before screening;
-
9.
Known active malignancy, except for basal cell skin cancer or squamous cell skin cancer;
-
10.
Subjects with previous colectomy, ostomy, J-pouch, or previous intestinal surgery (excluding cholecystectomy, appendectomy);
-
11.
Subjects with known history of celiac disease or gluten enteropathy;
-
12.
Subjects with C difficile–positive stool toxin test performed by the central laboratory at screening visit;
-
13.
Antibiotic use within 1 month before randomization;
-
14.
Expected to receive antibiotics within 8 weeks of signing the informed consent form (ie, for planned/anticipated procedure).
-
15.
Received an investigational drug within 1 month before study entry;
-
16.
Received an investigational antibody or vaccine within 3 months before study entry;
-
17.
Previously enrolled in a SER-109 or SER-287 study;
-
18.
Received an FMT within 6 months before study entry;
-
19.Poor concurrent medical risks with clinically significant comorbid disease such that, in the opinion of the investigator, the subject should not be enrolled including:
-
a.Subjects with decompensated liver cirrhosis (Child-Pugh class B or C) or uncontrolled liver disease
-
b.History of bone marrow transplantation
-
c.Known hypogammaglobulinemia
-
d.Known severe immunodeficiency
-
e.Underlying liver function test (screening alanine aminotransferase or aspartate aminotransferase) abnormalities >2× upper limit of normal
-
f.Absolute neutrophil count <500 cells/mm3.
-
a.
-
20.
Subjects with anatomic or medical contraindications to lower endoscopy (flexible sigmoidoscopy or colonoscopy), including but not necessarily limited to toxic megacolon; gastrointestinal fistulas; immediate postoperative status from abdominal surgery; severe coagulopathy; large or symptomatic abdominal aortic aneurysm; or any subject where study physician deems subject at significant risk of complications of lower endoscopy (flexible sigmoidoscopy or colonoscopy);
-
21.
Unable to stop steroid or mesalamine enemas or suppositories before screening visit;
-
22.
Unable to stop opiate treatment, unless on a stable dose with no increase in dose planned for the duration of the study;
-
23.
Unable to stop probiotics before screening visit;
-
24.
Concurrent intensive induction chemotherapy, radiotherapy, or biologic treatment for active malignancy (subjects on maintenance chemotherapy may only be enrolled after consultation with medical monitor);
-
25.
Unable to comply with the protocol requirements;
-
26.
Any condition that, in the opinion of the investigator, might interfere with study objectives;
-
27.
Known allergy or intolerance to oral vancomycin; and
-
28.
Known active intravenous drug or alcohol abuse or use of other drugs of abuse.
Permitted Concomitant Medications
The complete list of concomitant medications permitted on study included:
-
•
Oral aminosalicylates (aminosalicylates taken for at least 6 weeks, with a stable dose for ≥2 weeks before screening required)
-
•
Immunomodulators: 6-mercaptopurine, azathioprine, and methotrexate (stable dose for ≥12 weeks before screening required)
-
•
Prednisone ≤15 mg (stable dose for ≥2 weeks before screening required)
-
•
Budesonide ≤6 mg (stable dose for ≥2 weeks before screening required)
-
•
Budesonide MMX ≤9 mg (stable dose for ≥2 weeks before screening required)
-
•
Opiate treatment (stable dose or short-term use is permitted)
-
•
Short term use of nonsteroidals/nonsteroidal anti-inflammatory drugs
-
•
Long-term use of low-dose aspirin (81 mg)
Subjects receiving any of the above permitted concomitant medications at the time of study entry were required to maintain a stable dosage throughout the study (except for drugs allowed for short term use as noted above), unless investigator judgment required increase, reduction, or discontinuation for safety concerns or medical necessity. Dosing changes due to safety concerns or medical necessity were a cause to discontinue subjects from the study.
Clinical End Points and Statistics: Ulcerative Colitis Flares
UC flares were considered adverse events of special interest defined as a 2-point increase in the partial Mayo score, documented at 2 contiguous visits after initiation of study drug plus worsening clinical status warranting a change in UC treatment, as determined by the principal investigator. During the subsequent 22-week follow-up period, occurrence of a UC flare was defined by a change in UC medication, as reported by the patient.
Stool Sample Collection
Subject stool samples were collected at baseline (before vancomycin preconditioning); on day 0 (post-vancomycin preconditioning, before SER-287 dosing); and on days 3, 7, 10, 14, 56, and 84 (post treatment with SER-287). For analyses involving comparisons of baseline with post-treatment microbiome, all subjects with available paired samples are included (Supplementary Table 2).
Whole Metagenomic Shotgun Sequencing
Bacterial DNA was extracted from 200 μL or 0.2 g of subject stool and dose samples using the MagAttract PowerSoil DNA Kit (Qiagen, Hilden, Germany). Libraries were sequenced on the Illumina HiSeq to a target depth of 5 gigabases for subject samples and 10 gigabases for SER-287 lots.
Samples were processed per standard HMP-2 data processing guidelines (https://hmpdacc.org/hmp/resources/). Read pair deduplication was implemented using a custom Python function to remove polymerase chain reaction and sequencing artifacts. Adapter sequences and low-quality sequence data was trimmed using trimmomatic.1 Finally, reads were aligned to human and artificial reference sequences using bowtie22 to remove unwanted sequence data (eg, human DNA or artificial DNA). Raw sequencing data have not been deposited in public databases to protect the intellectual property of Seres Therapeutics.
Taxonomic Profiling of Metagenomic Shotgun Sequencing Samples
Species taxonomic profiling of metagenomic shotgun sequencing reads for both SER-287 lots and subject stools was performed using the MetaPhlAn2 software.3 , 4 The curated public reference database used for MetaPhlAn2 includes species-level genomic markers. In addition to these publicly available bacterial species genomes, we added spore-forming genomes from our proprietary Seres strain collection to increase detection of spore-forming species underrepresented in public databases. For all analyses, species were classified as being spore-forming or not spore-forming according to a proprietary, curated annotation of species experimentally demonstrated to form spores or identified in the literature as spore-forming species.
Before generation of final taxonomic profiles, all samples were subsampled to the same sequencing depth to account for any correlation between sequencing depth and observed species richness. A subsampling depth of 150,000 reads mapped to the MetaPhlAn2 reference database was used. Any samples with <150,000 mapped reads were omitted from downstream analysis. The ensuing output from the MetaPhlAn2 taxonomic profiling pipeline is referred to as the “microbiome profile” of a sample.
Sample Preparation for Metabolomics
Short-chain fatty acids
Sample preparation and data generation was performed by Metabolon Inc (Durham, NC). Samples were maintained at –80°C until processed. Samples were analyzed for SCFAs by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Metabolon Method TAM135). Human stool samples are spiked with stable labeled internal standards and are subjected to protein precipitation with methanol. After centrifugation, an aliquot of the supernatant is derivatized to form the corresponding SCFA hydrazides. The reaction mixture is diluted, and an aliquot is injected onto an Agilent 1290/AB Sciex QTrap 5500 LC-MS/MS system (Agilent, Santa Clara, CA) equipped with a C18 reverse-phase ultra high-performance liquid chromatography (UPLC) column. The mass spectrometer is operated in negative mode using electrospray ionization (ESI). The peak area of the individual analyte product ions is measured against the peak area of the product ions of the corresponding internal standards. Quantitation is performed using a weighted linear least squares regression analysis generated from fortified calibration standards prepared immediately before each run. Metabolite concentrations are reported in units of microgram of metabolite per gram of stool.
Bile acids
Sample preparation and data generation was performed at Seres Therapeutics. Samples were maintained at –80°C until processed. Three sample aliquots were prepared from thawed samples. Aliquot 1 was weighed and spiked with labeled standards at a final concentration of 5 μM/mg before any further extraction. Aliquots 1 and 2 were homogenized in 10× v/w 50% methanol, incubated on ice for 1 hour and further extracted with an equal volume of cold acetonitrile. Extracted samples were spun down, filtered, and analyzed by targeted liquid chromatography mass spectrometry. Aliquot 2 was spiked with labeled BA standards post extraction, before analysis by LC-MS. Aliquot 3 was weighed, dried overnight in an oven at 105°C, and weighed again to determine the water content of the sample. LC-MS analysis was carried out using an Agilent 1260 series Liquid Chromatograph coupled to a Bruker Compact qTOF mass spectrometer (Bruker, Billerica, MA). BAs were separated using a Microsolv C18 bidentate column (Microsolv, Eatontown, NJ) with a gradient of water and acetonitrile. Data were processed using Bruker Compass DataAnalysis software and quantified using calibration curves generated with commercially available pure standards. All data were expressed relative to sample dry weight (ng/mg). Spike in controls were used to determine extraction efficiency of bile acids of interest. Additional controls to ensure quality and consistency across samples and runs were included with every batch analysis on the LC-MS.
Global metabolomics
Sample preparation and data generation was performed by Metabolon Inc. Samples were maintained at –80°C until processed. Proteins were precipitated with methanol (Glen Mills GenoGrinder 2000; Glen Mills Inc, Clifton, NJ) followed by centrifugation to remove protein, dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites. The resulting extract was divided into 5 fractions: 2 for analysis by 2 separate reverse-phase UPLC-MS/MS methods with positive ion mode ESI; 1 for analysis by reverse-phase/UPLC-MS/MS with negative ion mode ESI; 1 for analysis by hydrophilic interaction UPLC-MS/MS with negative ion mode ESI, and 1 sample was reserved for backup. All methods utilize a Waters ACQUITY UPLC and a Thermo Scientific (Waltham, MA) Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization source and Orbitrap mass analyzer operated at 35,000 mass resolution. The scan range varies slightly between methods, but covers approximately 70–1000 m/z. Compounds were identified based on 3 distinct parameters: Retention index, accurate mass, and MS/MS spectrum in comparison with Metabolon’s proprietary library of purified standards and recurrent unknown entities. Peaks were quantified using area under the curve, adjusted for variations across runs and scaled as necessary for further analysis. Missing values were assumed to be below the level of detection for that biochemical with the instrumentation used and were imputed to the observed minimum for that particular biochemical across all samples.
Sample Selection and Taxonomic Profiling of Integrative Human Microbiome Project Data
Metadata for the Integrative Human Microbiome Project were downloaded from the link https://ibdmdb.org/tunnel/products/HMP2/Metadata/hmp2_metadata.csv. Data were accessed March 14, 2018. Only metagenomics samples with available sequencing files were considered (N = 1338). These represented multiple visits from 106 unique subjects, including non-IBD (n = 50), UC (n = 30), and Crohn’s disease (n = 50) cohorts. For each unique subject, the sample corresponding to the earliest visit number was selected. In cases where multiple samples were attributed the same visit number, the sample with the lowest human read count was selected.
Each sample was then processed using the same steps as the SERES-101 study samples, including all preprocessing steps (read deduplication, trimming, and human read filtering; see whole metagenomic shotgun sequencing). Samples were also subsampled to the same depth as the SERES-101 study samples for comparability (150,000 mapped reads) before taxonomic profiling with MetaPhlAn2. Samples with <150,000 mapped reads were omitted from further analyses.
After all processing steps, a total of 97 samples were available for HMP2 comparative analyses, including samples from 25 non-IBD, 26 UC, and 46 Crohn’s disease unique subjects.
Long-Term Ulcerative Colitis Flares
In a post-hoc assessment, the number of flares was low in all arms. The following number of reported UC flares were observed during a 26-week period by treatment arm: 5 (33.3%) in the placebo/weekly SER-287 arm; 1 (5.9%) in the vancomycin/weekly SER-287 arm; 2 (13.3%) in the vancomycin/daily SER-287 arm; and 3 (27.3%) in the placebo/placebo arm. Among the 11 patients who achieved clinical remission on the SER-287 dosing arms, there were no reported UC flares, as assessed by a change of UC medications during the 26-week long-term safety follow-up phase.
Supplementary Table 10. Bile Acid and Short-Chain Fatty Acid Change From Baseline to 8 Weeks Post Treatment.
NOTE. Change in metabolite abundance post treatment was assessed based on log2 fold-change between subjects’ baseline and post-treatment samples; a Wilcoxon signed-rank test was performed, to assess whether the fold-changes are centered around 0. The number of subjects with baseline and post-treatment SCFA data is: n = 4 for placebo/placebo, n = 8 for placebo/SER-287 weekly, n = 11 for vancomycin/SER-287 weekly, n = 11 for vancomycin/SER-287 daily. The number of subjects with baseline and post-treatment BA data is: n = 3 for placebo/placebo, n = 8 for placebo/SER-287 weekly, n = 9 for vancomycin/SER-287 weekly, n = 11 for vancomycin/SER-287 daily.
Supplementary Table 11. Bile Acid and Short-Chain Fatty Acid Association With Clinical Remission
NOTE. Association of metabolite abundance with clinical remission was assessed at 8 weeks post treatment. A 2-sided Mann-Whitney U test was performed to assess whether the abundance of metabolites significantly differed in remitters compared with nonremitters; subjects in all treatment arms are considered. There are n = 29 nonremitters and n = 9 remitters with SCFA data post treatment. There are n = 27 nonremitters and n = 9 remitters with BA data post treatment.
Supplementary Table 12. Global Metabolomic Change From Baseline to 8 Weeks Post Treatment
Change in metabolite abundance post-treatment was assessed based on log2 fold-change between subjects’ baseline and post-treatment samples; a Wilcoxon signed-rank test was performed to assess whether the fold-changes were centered around zero. All metabolites are reported with P < .05 for vancomycin/SER-287 daily dosing and vancomycin/SER-287 weekly dosing arms combined; metabolites with names starting with “X – “ indicate compounds of unknown identity; asterisks indicate putative metabolite identities. The number of subjects with baseline and post-treatment data is: n = 4 for placebo/placebo, n = 8 for placebo/SER-287 weekly, n = 11 for vancomycin/SER-287 weekly, n = 11 for vancomycin/SER-287 daily.
Supplementary Table 13. Global Metabolomics Association With Clinical Remission
NOTE. Association of metabolite abundance with clinical remission was assessed at 8 weeks post treatment. A 2-sided Mann-Whitney U test was performed to assess whether the abundance of metabolites significantly differed in remitters (n = 9) compared with nonremitters (n = 29); subjects in all treatment arms are considered. No correction for multiple hypothesis testing was performed. All metabolites are reported with P < .05; metabolites with names starting with “X –“ indicate compounds of unknown identity; asterisks indicate putative metabolite identities.
Supplementary Figure 1.
Study Schematic. Carats (ˆ) indicate approximate times of stool sampling at baseline (screening endoscopy visit before any treatment) and on days 0 (post-vancomycin preconditioning, before SER-287 dosing), 3, 7, 10, 14, 56, and 84 post-treatment with SER-287. Final samples were collected at the end of treatment, at the end of the short-term safety follow-up period and/or at any early termination visit.
Supplementary Figure 2.
Consolidated Standards of Reporting Trials diagram.
Supplementary Figure 3.
Engraftment of SER-287 species is dose-dependent and facilitated by vancomycin preconditioning; preconditioned patient microbiome samples become more similar to SER-287 dose composition after rebound from vancomycin. Similarity between a subject’s microbiome and SER-287 dose composition is quantified with a binary Jaccard similarity coefficient of spore-forming species in subject samples and SER-287 doses; the change in similarity for each subject and timepoint is relative to that subjects’ baseline sample (y-axis) across time (x-axis). Black horizontal dashed line indicates the threshold where a subjects’ spore fraction is more (>0) or less (<0) similar to SER-287 post-treatment. Initial preconditioning with vancomycin moves patient samples further from SER-287 dose composition; by day 10 in the vancomycin/SER-287 daily dosing arm, the patient spore-forming microbiome becomes more similar to SER-287 and remains so through the 8-week dosing period (day 56) and the long-term follow-up at day 84 (P < .05, Wilcoxon signed-rank test, all time points); in the vancomycin/SER-287 weekly dosing arm, only the day 56 time point was statistically significant P < .05, consistent with a dose-dependent effect.
Supplementary Figure 4.
Enterobacteriaceae species abundance tends to decrease with vancomycin pre-conditioning and SER-287 with remitters having a larger decrease in Enterobacteriaceae compared with non-remitters. (A) Fold-change in Enterobacteriaceae species abundance relative to baseline is shown 8 weeks post treatment (day 56) for subjects in each arm; boxplots are colored by treatment arm. (B) Enterobacteriaceae fold-change is shown across remitters and nonremitters for subjects in vancomycin preconditioning arms only; boxplots are colored by clinical remission status.
Supplementary Table 1.
Spore-Forming Bacterial Taxa Identified in SER-287 Lots
| Phylum | Class | Order | Family | Genus | Drug lots, n |
|---|---|---|---|---|---|
| Firmicutes | Clostridia | Clostridiales | Christensenellaceae | Unclassified Christensenellaceae | 2 |
| Clostridiaceae | Clostridium | 3 | |||
| Eubacteriaceae | Anaerofustis, Eubacterium | 1 | |||
| [Eubacterium] | 2 | ||||
| Lachnospiraceae | Acetatifactor, Coprococcus | 1 | |||
| Desulfotomaculum | 2 | ||||
| [Clostridium], [Coprococcus], [Eubacterium], [Faecalicatena], [Ruminococcus], Anaerostipes, Blautia, Dorea, Eisenbergiella, Fusicatenibacter, Lachnospira, unclassified Lachnospiraceae, Lactonifactor, Marvinbryantia, Roseburia | 3 | ||||
| Peptostreptococcaceae | Intestinibacter | 3 | |||
| Ruminococcaceae | Harryflintia | 1 | |||
| [Clostridium], [Ruminococcus], Agathobaculum, Faecalibacterium, Fusicatenibacter, Gemmiger, Massilimaliae, unclassified Ruminococcaceae, Ruminococcus, Subdoligranulum | 3 | ||||
| Erysipelotrichia | Erysipelotrichales | Erysipelotrichaceae | Catenibacterium, Holdemanella, Solobacterium | 1 | |
| Erysipelatoclostridium | 2 | ||||
| [Clostridium], Holdemania, Turicibacter | 3 |
NOTE. The 3 lots of SER-287 that were used in the clinical trial were characterized by direct whole metagenomic shotgun sequencing. The list of genera with spore-forming species identified in any of the 3 lots is indicated; bracketed genera are pending reassignment in National Center for Biotechnology Information taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy); unclassified family names within the genera column indicate species without genus assignments. Rows in the table are organized according to the hierarchical taxonomy. The last column indicates the number of drug lots in which the genera were identified.
Supplementary Table 2.
Number of Subjects With Evaluable Metagenomic Shotgun Sequencing Data From Both Baseline and Post-Treatment Samples
| Days post treatment | Placebo/ placebo | Placebo/SER-287 Weekly | Vancomycin/SER-287 Weekly | Vancomycin/SER-287 Daily |
|---|---|---|---|---|
| 0 | 5 | 8 | 12 | 10 |
| 3 | 7 | 7 | 12 | 8 |
| 7 | 7 | 11 | 14 | 10 |
| 10 | 7 | 5 | 12 | 10 |
| 14 | 7 | 10 | 13 | 10 |
| 56 | 6 | 9 | 13 | 10 |
| 84 | 6 | 9 | 8 | 8 |
NOTE. Indicated is the number of subjects within each arm (columns) at the indicated time point (rows) with both baseline and post-treatment samples with metagenomic shotgun sequencing data.
Supplementary Table 3.
Treatment-Emergent Adverse Events by System Organ Class and Preferred Term: Preconditioning Phase, Safety Population
| Variable | Placebo pretreatment (n = 26) | Vancomycin pretreatment (n = 32) |
|---|---|---|
| Subjects with at least 1 TEAE | 3 (11.5) | 4 (12.5) |
| Gastrointestinal disorders | 2 (7.7) | 3 (9.4) |
| Constipation | 1 (3.8) | 1 (3.1) |
| Diarrhea | 1 (3.8) | 1 (3.1) |
| Fecal incontinence | 0 | 1 (3.1) |
| Flatulence | 0 | 1 (3.1) |
| Infections and infestations | 0 | 1 (3.1) |
| Candida infection | 0 | 1 (3.1) |
| Musculoskeletal and connective tissue disorders | 1 (3.8) | 0 |
| Back pain | 1 (3.8) | 0 |
| Muscle spasms | 1 (3.8) | 0 |
| Psychiatric disorders | 1 (3.8) | 0 |
| Insomnia | 1 (3.8) | 0 |
| Skin and subcutaneous tissue disorders | 0 | 1 (3.1) |
| Rash | 0 | 1 (3.1) |
TEAE, treatment-emergent adverse event.
Supplementary Table 4.
Treatment-Emergent Adverse Events by System Organ Class and Preferred Term: Treatment Phase and Short-Term Safety Phase, Safety Population
| Variable | Placebo/placebo (n = 11) | Placebo/SER-287 weekly (n = 15) | Vancomycin/SER-287 weekly (n = 17) | Vancomycin/SER-287 daily (n = 15) | All subjects receiving any dose of SER-287 (n = 47) |
|---|---|---|---|---|---|
| Subjects with at least 1 TEAE | 7 (63.6) | 9 (60.0) | 13 (76.5) | 7 (46.7) | 29 (61.7) |
| Gastrointestinal disorders | 5 (45.5) | 7 (46.7) | 7 (41.2) | 1 (6.7) | 15 (31.9) |
| Abdominal distension | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Abdominal pain | 1 (9.1) | 3 (20.0) | 4 (23.5) | 0 | 7 (14.9) |
| Abdominal pain upper | 0 | 1 (6.7) | 0 | 0 | 1 (2.1) |
| Abnormal feces | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Bowel movement irregularity | 0 | 0 | 2 (11.8) | 0 | 2 (4.3) |
| Colitis ulcerative | 1 (9.1) | 1 (6.7) | 1 (5.9) | 0 | 2 (4.3) |
| Constipation | 0 | 1 (6.7) | 0 | 1 (6.7) | 2 (4.3) |
| Diarrhea | 2 (18.2) | 1 (6.7) | 1 (5.9) | 0 | 2 (4.3) |
| Diarrhea hemorrhagic | 1 (9.1) | 0 | 0 | 0 | 0 |
| Dyspepsia | 1 (9.1) | 0 | 0 | 0 | 0 |
| Flatulence | 0 | 0 | 2 (11.8) | 0 | 2 (4.3) |
| Frequent bowel movements | 0 | 1 (6.7) | 0 | 0 | 1 (2.1) |
| Gastroesophageal reflux disease | 0 | 1 (6.7) | 0 | 0 | 1 (2.1) |
| Mucous stools | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Nausea | 0 | 3 (20.0) | 1 (5.9) | 1 (6.7) | 5 (10.6) |
| Esophagitis | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Rectal discharge | 0 | 1 (6.7) | 0 | 0 | 1 (2.1) |
| Vomiting | 0 | 1 (6.7) | 1 (5.9) | 0 | 2 (4.3) |
| General disorders and administration site conditions | 1 (9.1) | 0 | 3 (17.6) | 1 (6.7) | 4 (8.5) |
| Asthenia | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Chest pain | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Influenza-like illness | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Edema peripheral | 0 | 0 | 0 | 1 (6.7) | 1 (2.1) |
| Pyrexia | 1 (9.1) | 0 | 0 | 0 | 0 |
| Immune system disorders | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Hypersensitivity | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Infections and infestations | 3 (27.3) | 1 (6.7) | 5 (29.4) | 4 (26.7) | 10 (21.3) |
| Cellulitis | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Nasopharyngitis | 0 | 0 | 1 (5.9) | 1 (6.7) | 2 (4.3) |
| Oral candidiasis | 1 (9.1) | 0 | 0 | 0 | 0 |
| Otitis externa | 1 (9.1) | 0 | 0 | 0 | 0 |
| Pneumonia | 0 | 0 | 0 | 1 (6.7) | 1 (2.1) |
| Pyelonephritis | 0 | 1 (6.7) | 0 | 0 | 1 (2.1) |
| Rhinitis | 0 | 1 (6.7) | 0 | 1 (6.7) | 2 (4.3) |
| Sinusitis | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Tooth infection | 1 (9.1) | 0 | 0 | 0 | 0 |
| Upper respiratory tract infection | 0 | 0 | 1 (5.9) | 2 (13.3) | 3 (6.4) |
| Urinary tract infection | 0 | 1 (6.7) | 0 | 0 | 1 (2.1) |
| Viral upper respiratory tract infection | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Injury, poisoning and procedural complications | 2 (18.2) | 0 | 0 | 0 | 0 |
| Contusion | 1 (9.1) | 0 | 0 | 0 | 0 |
| Limb injury | 1 (9.1) | 0 | 0 | 0 | 0 |
| Muscle injury | 1 (9.1) | 0 | 0 | 0 | 0 |
| Investigations | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Nitrite urine present | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| White blood cells urine positive | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Metabolism and nutrition disorders | 0 | 0 | 1 (5.9) | 1 (6.7) | 2 (4.3) |
| Decreased appetite | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Hypokalemia | 0 | 0 | 0 | 1 (6.7) | 1 (2.1) |
| Musculoskeletal and connective tissue disorders | 0 | 2 (13.3) | 1 (5.9) | 2 (13.3) | 5 (10.6) |
| Back pain | 0 | 2 (13.3) | 1 (5.9) | 1 (6.7) | 4 (8.5) |
| Muscle spasms | 0 | 0 | 0 | 1 (6.7) | 1 (2.1) |
| Nervous system disorders | 0 | 0 | 1 (5.9) | 3 (20.0) | 4 (8.5) |
| Headache | 0 | 0 | 1 (5.9) | 3 (20.0) | 4 (8.5) |
| Psychiatric disorders | 0 | 0 | 0 | 1 (6.7) | 1 (2.1) |
| Depression | 0 | 0 | 0 | 1 (6.7) | 1 (2.1) |
| Reproductive system and breast disorders | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Pelvic pain | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Respiratory, thoracic and mediastinal disorders | 0 | 1 (6.7) | 2 (11.8) | 1 (6.7) | 4 (8.5) |
| Cough | 0 | 1 (6.7) | 0 | 1 (6.7) | 2 (4.3) |
| Nasal congestion | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Rhinitis allergic | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Skin and subcutaneous tissue disorders | 0 | 0 | 1 (5.9) | 2 (13.3) | 3 (6.4) |
| Hidradenitis | 0 | 0 | 0 | 1 (6.7) | 1 (2.1) |
| Skin reaction | 0 | 0 | 1 (5.9) | 0 | 1 (2.1) |
| Swelling face | 0 | 0 | 0 | 1 (6.7) | 1 (2.1) |
NOTE. Values are n (%).
TEAE, treatment-emergent adverse event.
Supplementary Table 5.
Comparison of Number of Engrafting SER-287 Dose Species Across Arms
| Days post treatment | P value (unadjusted) | Placebo/placebo mean | Placebo/SER-287 weekly mean |
|---|---|---|---|
| 0 | .768 | 4.600 | 3.875 |
| 3 | .104 | 2.000 | 5.571 |
| 7 | .009 | 1.857 | 6.091 |
| 10 | .805 | 4.000 | 6.200 |
| 14 | .730 | 4.143 | 4.800 |
| 56 | .515 | 4.333 | 7.222 |
| 84 | .372 | 4.333 | 5.889 |
| Days post treatment | P value (unadjusted) | Placebo/placebo mean | Vancomycin/SER-287 weekly mean |
|---|---|---|---|
| 0 | .008 | 4.600 | 0.417 |
| 3 | .863 | 2.000 | 2.583 |
| 7 | .018 | 1.857 | 6.143 |
| 10 | .001 | 4.000 | 12.583 |
| 14 | .001 | 4.143 | 15.385 |
| 56 | .002 | 4.333 | 13.615 |
| 84 | .011 | 4.333 | 13.375 |
| Days post treatment | P value (unadjusted) | Placebo/placebo mean | Vancomycin/SER-28 daily mean |
|---|---|---|---|
| 0 | 0.006 | 4.600 | 0.100 |
| 3 | 0.411 | 2.000 | 3.375 |
| 7 | 0.001 | 1.857 | 13.500 |
| 10 | 0.001 | 4.000 | 20.600 |
| 14 | 0.001 | 4.143 | 21.300 |
| 56 | 0.002 | 4.333 | 16.700 |
| 84 | 0.007 | 4.333 | 18.000 |
| Days post treatment | P value (unadjusted) | Placebo/SER-287 weekly mean | Vancomycin/SER-287 daily mean |
|---|---|---|---|
| 0 | <.001 | 3.875 | 0.100 |
| 3 | .172 | 5.571 | 3.375 |
| 7 | .002 | 6.091 | 13.500 |
| 10 | .017 | 6.200 | 20.600 |
| 14 | <.001 | 4.800 | 21.300 |
| 56 | .012 | 7.222 | 16.700 |
| 84 | .007 | 5.889 | 18.000 |
| Days post treatment | P value (unadjusted) | Placebo/SER-287 weekly mean | Vancomycin/SER-287 weekly mean |
|---|---|---|---|
| 0 | .001 | 3.875 | 0.417 |
| 3 | .049 | 5.571 | 2.583 |
| 7 | .804 | 6.091 | 6.143 |
| 10 | .044 | 6.200 | 12.583 |
| 14 | .001 | 4.800 | 15.385 |
| 56 | .020 | 7.222 | 13.615 |
| 84 | .018 | 5.889 | 13.375 |
| Days post treatment | P value (unadjusted) | Vancomycin/SER-287 weekly mean | Vancomycin/SER-287 daily mean |
|---|---|---|---|
| 0 | .658 | 0.417 | 0.100 |
| 3 | .285 | 2.583 | 3.375 |
| 7 | .003 | 6.143 | 13.500 |
| 10 | .012 | 12.583 | 20.600 |
| 14 | .067 | 15.385 | 21.300 |
| 56 | .335 | 13.615 | 16.700 |
| 84 | .317 | 13.375 | 18.000 |
NOTE. Pairwise arm comparisons are indicated. P value indicates 2-sided Mann-Whitney U test for the indicated treatment arm compared with placebo. See Supplementary Table 2 for the number of subjects.
Supplementary Table 6.
Change in Spore-Forming Species Richness (α-Diversity) Post Treatment Compared With Placebo
| Treatment group compared with placebo/placebo | Days post treatment | P value (unadjusted) | Placebo/placebo mean | Treatment group mean |
|---|---|---|---|---|
| Placebo/SER-287 weekly | 0 | .769 | 1.800 | –1.500 |
| 3 | .158 | –4.000 | 1.571 | |
| 7 | .093 | –3.143 | 1.545 | |
| 10 | .622 | 1.286 | 0.000 | |
| 14 | .219 | 2.714 | 0.000 | |
| 56 | .766 | 0.167 | 2.333 | |
| 84 | .935 | –2.600 | –3.286 | |
| Vancomycin/SER-287 daily | 0 | .008 | 1.800 | –27.667 |
| 3 | .027 | –4.000 | –20.875 | |
| 7 | .305 | –3.143 | 4.600 | |
| 10 | .001 | 1.286 | 21.556 | |
| 14 | .002 | 2.714 | 17.900 | |
| 56 | .037 | 0.167 | 12.100 | |
| 84 | .048 | –2.600 | 13.625 | |
| Vancomycin/SER-287 weekly | 0 | .012 | 1.800 | –27.400 |
| 3 | .005 | –4.000 | –25.545 | |
| 7 | .074 | –3.143 | –11.846 | |
| 10 | .821 | 1.286 | –1.636 | |
| 14 | .361 | 2.714 | 8.769 | |
| 56 | .313 | 0.167 | 5.615 | |
| 84 | .065 | –2.600 | 8.000 |
NOTE. P value indicates 2-sided Mann-Whitney U test for the indicated treatment arm compared with placebo. See Supplementary Table 2 for the number of subjects.
Supplementary Table 7.
Change in Microbiome Composition (β-Diversity) Post-Treatment Compared With Placebo
| Treatment group compared with placebo/placebo | Days post treatment | P value (unadjusted) | Placebo/placebo mean | Treatment group mean |
|---|---|---|---|---|
| Placebo/SER-287 weekly | 0 | .769 | 0.330 | 0.329 |
| 3 | .701 | 0.317 | 0.296 | |
| 7 | .469 | 0.288 | 0.339 | |
| 10 | .871 | 0.265 | 0.354 | |
| 14 | .733 | 0.326 | 0.342 | |
| 56 | .517 | 0.340 | 0.400 | |
| 84 | .626 | 0.421 | 0.443 | |
| Vancomycin/SER-287 daily | 0 | .003 | 0.330 | 0.865 |
| 3 | .001 | 0.317 | 0.775 | |
| 7 | .001 | 0.288 | 0.747 | |
| 10 | .001 | 0.265 | 0.663 | |
| 14 | .001 | 0.326 | 0.672 | |
| 56 | .002 | 0.340 | 0.650 | |
| 84 | .067 | 0.421 | 0.680 | |
| Vancomycin/SER-287 weekly | 0 | .002 | 0.330 | 0.906 |
| 3 | <.001 | 0.317 | 0.841 | |
| 7 | <.001 | 0.288 | 0.775 | |
| 10 | .001 | 0.265 | 0.718 | |
| 14 | <.001 | 0.326 | 0.654 | |
| 56 | .001 | 0.340 | 0.601 | |
| 84 | .092 | 0.421 | 0.606 |
NOTE. β-diversity distance (binary Jaccard) from baseline for subjects in each treatment group at the given time point, compared with placebo. P value indicates 2-sided Mann Whitney U test for the indicated treatment arm compared with placebo. See Supplementary Table 2 for the number of subjects.
Supplementary Table 8.
Change in Enterobacteriaceae Relative Abundance From Baseline to 8 Weeks Post Treatment Across Arms
| Arms | No. of subjects | Median Enterobacteriaceae log2FC | P value |
|---|---|---|---|
| Placebo/placebo | 4 | 2.758 | .875 |
| Placebo/SER-287 weekly | 8 | 0.229 | .945 |
| Vancomycin/SER-287 weekly | 10 | –3.061 | .064 |
| Vancomycin/SER-287 daily | 7 | –1.183 | .156 |
| Vancomycin/SER-287 weekly + daily | 17 | –1.377 | .009 |
NOTE. The change in Enterobacteriaceae abundance was assessed at 8 weeks post treatment. Only subjects with baseline and 8-week stool samples and subjects with Enterobacteriaceae at baseline (n = 29) were included in analyses. Change in Enterobacteriaceae abundance post treatment was assessed using log2 fold-change between subjects' baseline and 8-week post-treatment samples. A 2-sided Wilcoxon signed-rank test was performed to assess whether the fold-changes were centered around 0 for each arm individually and for vancomycin/SER-287 weekly and daily treatment arms combined.
Supplementary Table 9.
Decrease In Enterobacteriaceae Abundance Is Associated With Clinical Remission
| Arms | Clinical remission | No. of subjects | Median Enterobacteriaceae log2 FC | P value |
|---|---|---|---|---|
| Vancomycin/SER-287 weekly + daily | Y | 7 | –4.108 | .016 |
| Vancomycin/SER-287 weekly + daily | N | 10 | –0.552 | .322 |
NOTE. The association Enterobacteriaceae abundance with clinical remission was assessed at 8 weeks post treatment. Only subjects with baseline and 8-week stool samples and subjects with Enterobacteriaceae at baseline (n = 29) were included in analyses. Change in Enterobacteriaceae abundance post treatment was assessed using log2 fold-change between subjects' baseline and 8-week post-treatment samples. A 2-sided Wilcoxon signed-rank test was performed to assess whether the fold-changes are centered around 0 for vancomycin/SER-287 weekly and daily treatment arms combined in remitters vs nonremitters.
Supplementary Table 10.
| Mean log2 fold-change |
p-value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Placebo/Placebo | Placebo/SER-287 weekly | Vancomycin/SER-287 weekly | Vancomycin/SER-287 daily | Vancomycin/SER-287 weekly + daily | Placebo/Placebo | Placebo/SER-287 weekly | Vancomycin/SER-287 weekly | Vancomycin/SER-287 daily | Vancomycin/SER-287 weekly + daily | |
| Glycocholic acid | 0.530 | 2.101 | -2.254 | -0.310 | -1.184 | 1.000 | 0.208 | 0.139 | 0.721 | 0.398 |
| Taurocholic acid | -0.772 | -0.669 | -1.792 | -2.117 | -1.971 | 0.593 | 0.463 | 0.515 | 0.214 | 0.157 |
| Glycochenodeoxycholic acid | 0.079 | 1.721 | -1.999 | -0.848 | -1.366 | 1.000 | 0.263 | 0.214 | 0.959 | 0.355 |
| Taurochenodeoxychoic acid | 4.798 | -0.041 | -3.557 | -1.066 | -2.187 | 0.109 | 1.000 | 0.214 | 1.000 | 0.287 |
| Cholic acid | 0.583 | -1.843 | -1.693 | -1.374 | -1.518 | 1.000 | 0.484 | 0.314 | 0.477 | 0.232 |
| Chenodeoxycholic acid | 0.223 | -1.724 | -2.714 | -1.015 | -1.779 | 1.000 | 0.161 | 0.086 | 0.594 | 0.058 |
| Deoxycholic acid | -1.172 | -1.286 | -0.593 | 0.693 | 0.114 | 0.109 | 0.161 | 0.767 | 0.477 | 0.823 |
| Lithocholic acid | -1.256 | -0.419 | -0.733 | 0.859 | 0.143 | 1.000 | 0.889 | 0.441 | 0.285 | 0.687 |
| Ursodeoxycholic acid | 0.283 | -2.280 | -3.436 | -3.106 | -3.255 | 1.000 | 0.093 | 0.021 | 0.047 | 0.003 |
| Acetate | -0.185 | 0.124 | -0.180 | 0.173 | -0.003 | 0.465 | 0.779 | 0.445 | 1.000 | 0.543 |
| Butyrate | -1.074 | 0.067 | -0.597 | 1.065 | 0.234 | 0.144 | 0.779 | 0.075 | 0.328 | 0.067 |
| Propionate | -0.480 | 0.288 | -0.456 | -0.520 | -0.488 | 0.273 | 0.263 | 0.155 | 0.131 | 0.042 |
| Valerate | -1.406 | 0.460 | -0.599 | -0.016 | -0.307 | 0.715 | 0.575 | 0.534 | 0.131 | 0.168 |
| Hexanoate | -1.274 | 0.505 | -0.316 | 2.447 | 1.065 | 0.465 | 1.000 | 0.790 | 0.026 | 0.095 |
Supplementary Table 11.
| Mean log2 fold-change |
p-value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Placebo/Placebo | Placebo/SER-287 weekly | Vancomycin/SER-287 weekly | Vancomycin/SER-287 daily | Vancomycin/SER-287 weekly + daily | Placebo/Placebo | Placebo/SER-287 weekly | Vancomycin/SER-287 weekly | Vancomycin/SER-287 daily | Vancomycin/SER-287 weekly + daily | |
| (15:0)-anacardic acid | 0.00 | 0.31 | -1.57 | -0.62 | -1.10 | 1.00000 | 0.31731 | 0.10881 | 0.17971 | 0.04311 |
| (15:2)-anacardic acid | 0.12 | 1.37 | -3.83 | -1.40 | -2.61 | 0.65472 | 0.17971 | 0.04311 | 0.28505 | 0.01729 |
| (16 or 17)-methylstearate (a19:0 or i19:0) | 0.39 | 0.36 | -0.86 | -0.92 | -0.89 | 0.46521 | 0.57540 | 0.09116 | 0.05046 | 0.00985 |
| (3'-5')-adenylyladenosine* | -0.34 | -0.05 | -1.08 | -1.04 | -1.06 | 1.00000 | 1.00000 | 0.03815 | 0.13864 | 0.00740 |
| (3'-5')-adenylylcytidine | -0.20 | 0.25 | -0.65 | -1.44 | -1.04 | 0.31731 | 0.86577 | 0.26262 | 0.06632 | 0.02772 |
| (3'-5')-adenylylguanosine* | -1.23 | 0.23 | -1.30 | -0.81 | -1.05 | 0.10881 | 0.73532 | 0.02088 | 0.21352 | 0.01387 |
| (3'-5')-adenylyluridine | -0.91 | 0.37 | -0.83 | -0.92 | -0.87 | 0.28505 | 0.88864 | 0.13864 | 0.13864 | 0.03858 |
| (3'-5')-guanylylcytidine | 1.25 | 0.00 | -1.28 | -1.33 | -1.30 | 0.17971 | 1.00000 | 0.13864 | 0.12349 | 0.03516 |
| (3'-5')-guanylyluridine | 0.00 | 0.54 | -1.39 | -0.89 | -1.14 | 1.00000 | 0.75315 | 0.10974 | 0.17295 | 0.04683 |
| (3'-5')-uridylylcytidine* | -0.15 | -0.70 | -0.59 | -1.27 | -0.93 | 0.31731 | 0.12349 | 0.26039 | 0.03815 | 0.02223 |
| (3'-5')-uridylyluridine | -1.11 | -0.32 | -0.53 | -1.14 | -0.84 | 0.17971 | 0.48384 | 0.37426 | 0.03815 | 0.04753 |
| 1,2-dipalmitoyl-GPC (16:0/16:0) | 0.97 | -0.82 | -0.78 | -1.57 | -1.17 | 0.71500 | 0.20758 | 0.28600 | 0.09116 | 0.04245 |
| 1-margaroylglycerol (17:0) | 0.82 | -0.38 | -0.65 | -1.17 | -0.91 | 0.14413 | 0.16143 | 0.07537 | 0.00444 | 0.00138 |
| 1-oleoyl-GPG (18:1)* | 1.21 | 0.09 | 0.58 | 1.17 | 0.88 | 0.27332 | 0.71500 | 0.44127 | 0.02840 | 0.04753 |
| 1-palmitoyl-2-linoleoyl-galactosylglycerol (16:0/18:2)* | -1.41 | -0.32 | 1.93 | 0.40 | 1.17 | 0.28505 | 0.49896 | 0.02842 | 0.72128 | 0.04380 |
| 1-palmitoyl-2-stearoyl-GPC (16:0/18:0) | 0.53 | -0.34 | -0.59 | -1.79 | -1.19 | 0.46521 | 0.23672 | 0.20262 | 0.04685 | 0.02277 |
| 1-ribosyl-imidazoleacetate* | 0.30 | -0.36 | -0.25 | -0.87 | -0.56 | 0.65472 | 0.65472 | 0.28505 | 0.07962 | 0.03569 |
| 1-stearoyl-GPC (18:0) | 0.98 | -0.43 | -1.01 | -1.78 | -1.39 | 0.46521 | 0.32699 | 0.21322 | 0.05046 | 0.02405 |
| 1-stearoyl-GPG (18:0) | 0.19 | -0.79 | -1.13 | -0.72 | -0.92 | 0.71500 | 0.26262 | 0.10951 | 0.16881 | 0.03255 |
| 1-stearoyl-GPI (18:0) | 0.02 | -0.47 | -1.19 | -1.68 | -1.44 | 1.00000 | 0.26262 | 0.15486 | 0.07537 | 0.01702 |
| 2'-deoxyadenosine | 0.49 | -0.49 | -0.87 | -1.57 | -1.22 | 0.46521 | 0.75315 | 0.17307 | 0.01660 | 0.00791 |
| 2'-deoxyadenosine 5'-monophosphate | 0.92 | 0.95 | -1.10 | -1.63 | -1.37 | 0.10881 | 0.27332 | 0.13864 | 0.04086 | 0.01237 |
| 2'-deoxyinosine | 1.37 | -0.13 | -0.45 | -1.22 | -0.84 | 0.14413 | 0.88864 | 0.32806 | 0.05046 | 0.03346 |
| 2-hydroxyoleate | -0.81 | -0.56 | -2.12 | -0.67 | -1.39 | 0.10881 | 0.67442 | 0.06188 | 0.33288 | 0.04202 |
| 2-stearoyl-GPE (18:0)* | 0.33 | -0.20 | -0.83 | -2.00 | -1.41 | 1.00000 | 0.71500 | 0.23672 | 0.04685 | 0.02168 |
| 3-(4-hydroxyphenyl)propionate | -0.34 | -1.05 | -1.18 | -1.89 | -1.54 | 0.46521 | 0.48384 | 0.15486 | 0.02080 | 0.00815 |
| 3-aminoisobutyrate | -0.16 | -2.10 | -0.72 | -1.40 | -1.06 | 0.71500 | 0.01796 | 0.28600 | 0.05934 | 0.02983 |
| 3-carboxy-4-methyl-5-propyl-2-furanpropanoate (CMPF) | 0.91 | 0.16 | -0.76 | -1.24 | -1.00 | 0.59298 | 0.89274 | 0.16143 | 0.04995 | 0.01306 |
| 3-hydroxy-2-ethylpropionate | -0.41 | 0.72 | -1.20 | -1.02 | -1.11 | 1.00000 | 0.49896 | 0.17307 | 0.05061 | 0.03467 |
| 3-hydroxyphenylacetate | 0.68 | -0.77 | -2.03 | -1.10 | -1.56 | 0.31731 | 0.07474 | 0.02840 | 0.12349 | 0.00748 |
| 3-sulfo-L-alanine | 0.38 | -1.54 | -1.46 | -0.77 | -1.11 | 0.71500 | 0.01729 | 0.02842 | 0.01516 | 0.00128 |
| 4-hydroxychlorothalonil | 0.53 | -0.81 | -0.83 | -0.39 | -0.61 | 0.27332 | 0.02771 | 0.06298 | 0.49896 | 0.04799 |
| 4-hydroxycinnamate | 1.10 | 0.37 | -0.74 | -1.43 | -1.08 | 0.14413 | 0.48384 | 0.21322 | 0.03285 | 0.01558 |
| 5-dodecenoate (12:1n7) | 2.89 | 1.15 | -0.66 | -1.14 | -0.90 | 0.06789 | 0.23672 | 0.26039 | 0.04086 | 0.03334 |
| 5alpha-androstan-3alpha,17alpha-diol monosulfate | -0.37 | -0.45 | -0.40 | -0.58 | -0.49 | 0.59298 | 0.50018 | 0.05934 | 0.16881 | 0.01688 |
| 5alpha-androstan-3alpha,17beta-diol monosulfate (1) | -0.15 | -0.77 | -1.14 | -0.82 | -0.98 | 0.59298 | 0.34545 | 0.06870 | 0.09289 | 0.01306 |
| 5alpha-androstan-3beta,17alpha-diol disulfate | -0.31 | -0.29 | -0.27 | -0.22 | -0.24 | 0.28505 | 0.50018 | 0.10974 | 0.16143 | 0.04947 |
| 5alpha-pregnan-3beta,20alpha-diol disulfate | -0.02 | -0.92 | -0.93 | -2.06 | -1.50 | 1.00000 | 0.32699 | 0.38627 | 0.01660 | 0.02063 |
| N,N-dimethyl-5-aminovalerate | 1.20 | -1.57 | -0.10 | -2.29 | -1.19 | 0.71500 | 0.09289 | 0.32806 | 0.05046 | 0.04245 |
| N-acetyl-beta-alanine | -0.55 | -0.91 | -1.46 | -0.38 | -0.92 | 0.46521 | 0.49896 | 0.00765 | 0.50762 | 0.01427 |
| N-acetyl-beta-glucosaminylamine | 0.51 | 0.26 | -0.28 | -0.62 | -0.45 | 0.46521 | 1.00000 | 0.32806 | 0.09260 | 0.04955 |
| N-acetylarginine | -0.47 | -0.60 | 0.49 | 1.29 | 0.89 | 0.14413 | 0.26262 | 0.18231 | 0.07537 | 0.01558 |
| N-acetylaspartate (NAA) | -0.58 | 0.22 | -1.07 | -0.60 | -0.83 | 0.46521 | 0.48384 | 0.09116 | 0.18231 | 0.04586 |
| N-acetylhistidine | 0.77 | 0.10 | 0.35 | 0.83 | 0.59 | 0.46521 | 0.88864 | 0.42360 | 0.03285 | 0.03925 |
| N-acetylserine | 0.80 | 0.09 | -0.29 | -1.01 | -0.65 | 0.27332 | 0.77943 | 0.32806 | 0.07537 | 0.03925 |
| N-acetyltaurine | 0.31 | -2.20 | -2.35 | -2.00 | -2.17 | 1.00000 | 0.03569 | 0.02080 | 0.00691 | 0.00054 |
| N-oleoyltaurine | -1.01 | -0.93 | -2.00 | -0.65 | -1.32 | 0.28505 | 0.34523 | 0.05061 | 0.50018 | 0.03033 |
| N-propionylalanine | 0.39 | -0.58 | -0.89 | -0.80 | -0.84 | 0.46521 | 0.40081 | 0.16881 | 0.07537 | 0.02731 |
| N-stearoyltaurine | -1.24 | -1.39 | -1.67 | -1.39 | -1.53 | 0.10881 | 0.20758 | 0.01252 | 0.02799 | 0.00118 |
| N1-Methyl-2-pyridone-5-carboxamide | 0.28 | -0.24 | -0.98 | -1.40 | -1.19 | 0.59298 | 0.32699 | 0.24775 | 0.00765 | 0.01082 |
| N6-carboxymethyllysine | -0.52 | -1.89 | -1.14 | -0.97 | -1.05 | 0.14413 | 0.09289 | 0.13067 | 0.13067 | 0.03626 |
| STVLT | 0.80 | -0.46 | -0.86 | -1.18 | -1.02 | 0.31731 | 0.31731 | 0.06789 | 0.17630 | 0.02623 |
| TMP | 1.74 | 0.92 | -0.72 | -1.89 | -1.31 | 0.10881 | 0.68583 | 0.39802 | 0.05061 | 0.04373 |
| X - 11299 | 0.11 | 0.00 | -0.45 | -0.28 | -0.36 | 0.65472 | 1.00000 | 0.31731 | 0.06789 | 0.04311 |
| X - 12849 | 1.16 | 0.16 | -0.51 | -1.09 | -0.80 | 0.17971 | 0.31731 | 0.17971 | 0.10881 | 0.04311 |
| X - 13255 | -0.77 | 0.00 | -0.54 | -1.67 | -1.11 | 0.31731 | 1.00000 | 0.10881 | 0.06789 | 0.01796 |
| X - 13703 | 0.13 | 0.17 | -0.34 | -1.04 | -0.69 | 0.31731 | 0.31731 | 0.17971 | 0.13801 | 0.04252 |
| X - 13835 | -1.50 | -0.86 | -1.28 | -0.66 | -0.97 | 0.14413 | 0.12349 | 0.02080 | 0.59371 | 0.04586 |
| X - 14254 | 1.28 | 0.82 | -1.54 | -1.43 | -1.49 | 0.46521 | 0.57540 | 0.04086 | 0.05046 | 0.00366 |
| X - 14263 | 0.46 | -0.56 | -0.31 | -0.65 | -0.48 | 1.00000 | 0.32699 | 0.47691 | 0.01279 | 0.02616 |
| X - 14416 | -0.03 | 0.54 | -1.46 | -1.92 | -1.69 | 0.65472 | 0.46307 | 0.24112 | 0.05046 | 0.04202 |
| X - 14454 | -1.30 | 0.24 | -1.32 | -1.24 | -1.28 | 0.27332 | 0.61209 | 0.13067 | 0.02623 | 0.01558 |
| X - 14662 | 2.07 | 0.00 | -0.44 | -0.77 | -0.60 | 0.17971 | 1.00000 | 0.10881 | 0.17971 | 0.04311 |
| X - 15806 | 0.90 | 0.12 | -0.85 | -1.16 | -1.01 | 0.31731 | 0.89274 | 0.09097 | 0.13864 | 0.02618 |
| X - 17162 | -0.19 | 0.08 | 3.22 | 1.66 | 2.44 | 0.65472 | 0.86577 | 0.16881 | 0.21352 | 0.04862 |
| X - 17869 | -0.13 | 0.23 | -0.36 | -0.97 | -0.67 | 0.65472 | 0.68583 | 0.31049 | 0.04640 | 0.03924 |
| X - 17877 | 0.54 | 0.22 | -1.16 | -1.22 | -1.19 | 0.65472 | 1.00000 | 0.02088 | 0.03569 | 0.00193 |
| X - 17960 | -1.21 | 0.33 | -1.09 | -0.33 | -0.71 | 0.46521 | 0.77943 | 0.06188 | 0.42360 | 0.04951 |
| X - 17983 | 0.88 | 0.83 | -1.53 | -2.38 | -1.95 | 0.59298 | 0.34523 | 0.17630 | 0.01252 | 0.00748 |
| X - 18938 | 1.07 | -0.56 | -1.22 | -0.72 | -0.97 | 0.06789 | 0.48384 | 0.09116 | 0.24775 | 0.03626 |
| X - 19746 | 0.91 | 0.00 | -0.33 | -0.95 | -0.64 | 0.31731 | 1.00000 | 0.17971 | 0.06789 | 0.02771 |
| X - 19751 | -0.63 | -0.37 | -0.28 | -2.42 | -1.35 | 0.46521 | 0.77943 | 0.78968 | 0.03285 | 0.03626 |
| X - 20172 | 0.74 | 0.92 | -1.29 | -2.46 | -1.87 | 0.71500 | 0.57540 | 0.15486 | 0.05046 | 0.01702 |
| X - 20197 | 0.95 | 0.48 | -1.20 | -2.37 | -1.78 | 0.46521 | 0.57540 | 0.13067 | 0.02623 | 0.00896 |
| X - 20624 | 0.33 | -1.58 | -2.81 | -1.13 | -1.97 | 1.00000 | 0.12349 | 0.00691 | 0.06789 | 0.00123 |
| X - 21467 | 1.17 | 0.60 | -0.95 | -1.75 | -1.35 | 0.28505 | 0.31731 | 0.06789 | 0.06789 | 0.01172 |
| X - 21788 | 0.93 | -2.38 | -3.16 | -1.98 | -2.57 | 0.46521 | 0.06870 | 0.01637 | 0.01172 | 0.00084 |
| X - 22030 | 0.20 | 0.00 | -1.51 | -0.50 | -1.00 | 0.65472 | 0.91651 | 0.11585 | 0.34545 | 0.04986 |
| X - 23115 | -3.07 | 0.33 | -1.63 | -1.08 | -1.35 | 0.06789 | 1.00000 | 0.09116 | 0.10951 | 0.02027 |
| X - 23581 | 0.88 | -0.32 | 0.84 | 0.79 | 0.82 | 0.46521 | 0.57540 | 0.18231 | 0.13067 | 0.04245 |
| X - 23585 | -0.52 | 1.13 | 1.22 | 2.56 | 1.89 | 0.10881 | 0.09289 | 0.13067 | 0.05934 | 0.01897 |
| X - 23639 | 0.90 | -0.04 | 0.96 | 0.50 | 0.73 | 0.06789 | 0.88864 | 0.03285 | 0.38627 | 0.04202 |
| X - 24243 | -0.58 | -0.89 | -1.47 | -0.70 | -1.08 | 0.27332 | 0.40081 | 0.00335 | 0.28450 | 0.00784 |
| X - 24608 | 0.77 | 0.15 | -0.37 | -0.46 | -0.41 | 0.14413 | 0.57540 | 0.21322 | 0.01637 | 0.04245 |
| X - 24699 | 1.32 | -1.13 | -1.02 | -2.24 | -1.63 | 0.71500 | 0.27332 | 0.67840 | 0.02840 | 0.03858 |
| X - 24728 | 0.82 | 0.67 | -0.28 | -1.87 | -1.07 | 0.10881 | 0.65472 | 0.17971 | 0.06789 | 0.02771 |
| X - 24952 | -0.17 | 0.20 | -0.23 | -0.26 | -0.25 | 0.46521 | 0.32699 | 0.28600 | 0.06188 | 0.03085 |
| X - 24983 | 0.80 | -0.53 | -0.69 | -0.95 | -0.82 | 0.14413 | 0.23672 | 0.06188 | 0.24775 | 0.03346 |
| X - 24996 | -1.17 | 0.51 | -0.77 | -0.44 | -0.60 | 0.17971 | 0.31731 | 0.10881 | 0.10881 | 0.02771 |
| X - 25053 | -0.78 | -0.27 | 0.05 | -2.44 | -1.19 | 0.46521 | 0.73532 | 0.95935 | 0.01279 | 0.04955 |
| X - 25264 | 0.43 | 0.30 | -0.72 | -1.33 | -1.03 | 0.65472 | 0.89274 | 0.17295 | 0.13941 | 0.03861 |
| acetylagmatine | -0.06 | 0.90 | 1.71 | 1.58 | 1.65 | 0.59298 | 0.34545 | 0.10974 | 0.10974 | 0.03110 |
| adenine | -0.50 | -0.47 | -0.79 | -0.83 | -0.81 | 0.27332 | 0.32699 | 0.06188 | 0.15486 | 0.02842 |
| adrenate (22:4n6) | -0.98 | -0.51 | -1.27 | -1.70 | -1.49 | 0.27332 | 0.17295 | 0.05046 | 0.00691 | 0.00187 |
| alpha-ketoglutarate | 1.51 | -0.21 | -0.56 | -0.62 | -0.59 | 0.06789 | 0.77943 | 0.32806 | 0.04086 | 0.04245 |
| arachidate (20:0) | -0.72 | 0.19 | -1.38 | -0.82 | -1.10 | 0.06789 | 0.32699 | 0.01637 | 0.07537 | 0.00297 |
| arachidonate (20:4n6) | 0.51 | -0.38 | -1.01 | -2.08 | -1.54 | 0.71500 | 0.40081 | 0.09116 | 0.01279 | 0.00267 |
| arachidoylcarnitine (C20)* | 0.56 | -0.01 | -1.14 | -1.06 | -1.10 | 0.27332 | 0.67442 | 0.00444 | 0.02623 | 0.00038 |
| arginine | 0.92 | 0.31 | 1.11 | 1.83 | 1.47 | 0.46521 | 0.67442 | 0.28600 | 0.02623 | 0.02027 |
| behenate (22:0)* | -1.44 | 0.30 | -1.81 | -1.20 | -1.50 | 0.06789 | 0.88864 | 0.01279 | 0.00765 | 0.00029 |
| behenoyl sphingomyelin (d18:1/22:0)* | 1.06 | 0.35 | -0.64 | -1.18 | -0.91 | 0.28505 | 0.89274 | 0.10974 | 0.06632 | 0.01387 |
| behenoylcarnitine (C22)* | 0.61 | 0.07 | -0.72 | -0.61 | -0.66 | 0.06789 | 0.48384 | 0.00585 | 0.03285 | 0.00069 |
| betaine | 2.07 | -0.07 | -1.42 | -1.62 | -1.52 | 0.14413 | 0.26262 | 0.09116 | 0.05046 | 0.01082 |
| cadaverine | -0.46 | 1.94 | -1.99 | -1.70 | -1.85 | 0.14413 | 0.12349 | 0.07537 | 0.13067 | 0.02616 |
| citrulline | 0.35 | -0.27 | -0.40 | -0.85 | -0.63 | 1.00000 | 0.16143 | 0.32806 | 0.00993 | 0.00450 |
| cytidine | -1.51 | 0.20 | 1.89 | 0.95 | 1.42 | 0.46521 | 0.88864 | 0.02623 | 0.28600 | 0.01558 |
| dCMP | 1.39 | 1.09 | -1.35 | -2.14 | -1.75 | 0.10881 | 0.50018 | 0.07446 | 0.04086 | 0.00707 |
| dihomolinoleate (20:2n6) | -0.98 | 0.02 | -0.54 | -1.25 | -0.89 | 0.27332 | 1.00000 | 0.42360 | 0.01637 | 0.03085 |
| dihomolinolenate (20:3n3 or 3n6) | -1.06 | -0.54 | -1.48 | -2.23 | -1.86 | 0.27332 | 0.32699 | 0.09116 | 0.00335 | 0.00098 |
| dihydroorotate | 1.81 | -1.40 | -1.15 | -1.39 | -1.27 | 0.06789 | 0.02506 | 0.28450 | 0.10951 | 0.04566 |
| docosahexaenoate (DHA; 22:6n3) | 0.68 | 0.05 | -0.20 | -2.09 | -1.15 | 0.46521 | 1.00000 | 0.92915 | 0.00993 | 0.04586 |
| docosapentaenoate (DPA; 22:5n3) | -0.33 | -0.46 | -0.88 | -2.32 | -1.60 | 0.71500 | 0.48384 | 0.28600 | 0.00444 | 0.00740 |
| eicosanoylsphingosine (d20:1)* | -0.65 | -1.28 | -1.08 | -0.69 | -0.88 | 0.14413 | 0.04995 | 0.04086 | 0.15486 | 0.01187 |
| eicosenoate (20:1n9 or 1n11) | -0.82 | 0.35 | -0.94 | -0.94 | -0.94 | 0.06789 | 0.88864 | 0.03285 | 0.05046 | 0.00366 |
| eicosenoylcarnitine (C20:1)* | -0.64 | -0.57 | -1.01 | -0.43 | -0.72 | 0.27332 | 0.26262 | 0.02080 | 0.37394 | 0.01858 |
| enterodiol | -0.14 | -1.68 | -4.71 | -2.00 | -3.35 | 0.59298 | 0.17630 | 0.00506 | 0.13864 | 0.00170 |
| erucate (22:1n9) | -1.10 | 0.29 | -0.36 | -1.06 | -0.71 | 0.06789 | 0.57540 | 0.32806 | 0.06188 | 0.04245 |
| erucoylcarnitine (C22:1)* | 0.19 | -0.35 | -1.33 | -0.98 | -1.15 | 0.46521 | 0.48384 | 0.00506 | 0.04086 | 0.00102 |
| gamma-glutamyl-epsilon-lysine | 0.52 | 0.60 | -0.46 | -1.34 | -0.90 | 0.46521 | 0.67442 | 0.21322 | 0.04086 | 0.01424 |
| gamma-glutamylglycine | 0.97 | -0.82 | -1.17 | -0.73 | -0.95 | 0.17971 | 0.17971 | 0.06298 | 0.13801 | 0.01860 |
| gamma-glutamylmethionine | 0.80 | -1.25 | -0.28 | -1.59 | -0.93 | 0.59298 | 0.09289 | 0.65664 | 0.00993 | 0.02842 |
| gamma-glutamylvaline | 0.52 | -0.29 | -0.67 | -0.85 | -0.76 | 0.10881 | 0.71500 | 0.20262 | 0.12349 | 0.03110 |
| hydantoin-5-propionate | 0.80 | -1.83 | -0.97 | -1.69 | -1.33 | 0.46521 | 0.02771 | 0.15486 | 0.02842 | 0.00869 |
| hypoxanthine | -1.01 | -0.03 | -0.62 | -1.24 | -0.93 | 0.27332 | 1.00000 | 0.28600 | 0.06188 | 0.03346 |
| indole | 0.18 | 1.08 | 1.45 | 1.78 | 1.62 | 0.71500 | 0.09289 | 0.04086 | 0.00585 | 0.00069 |
| lactosyl-N-behenoyl-sphingosine (d18:1/22:0)* | 0.80 | -1.10 | -0.71 | -1.43 | -1.07 | 0.71500 | 0.20758 | 0.13067 | 0.05061 | 0.01524 |
| lactosyl-N-nervonoyl-sphingosine (d18:1/24:1)* | 1.37 | -0.72 | -1.36 | -2.05 | -1.70 | 0.46521 | 0.20758 | 0.09116 | 0.03285 | 0.00551 |
| lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) | 1.18 | -0.65 | -1.28 | -1.76 | -1.52 | 0.46521 | 0.26262 | 0.10951 | 0.04086 | 0.00896 |
| leucylglycine | 0.11 | -0.58 | -1.36 | -1.32 | -1.34 | 0.71500 | 0.46307 | 0.02799 | 0.07537 | 0.00740 |
| lignoceroyl sphingomyelin (d18:1/24:0) | 1.21 | 0.41 | -0.39 | -1.72 | -1.05 | 0.59298 | 0.50018 | 0.51467 | 0.02182 | 0.02977 |
| linolenoyl ethanolamide | -0.66 | -1.28 | -0.25 | -2.34 | -1.29 | 0.10881 | 0.26262 | 0.72128 | 0.00506 | 0.02063 |
| linoleoyl ethanolamide | -0.91 | -1.40 | -0.97 | -2.71 | -1.84 | 0.14413 | 0.09289 | 0.28600 | 0.00506 | 0.00784 |
| linoleoyl-arachidonoyl-glycerol (18:2/20:4) [2]* | 2.44 | -0.71 | -1.23 | -1.47 | -1.35 | 0.14413 | 0.67442 | 0.11413 | 0.21322 | 0.04955 |
| lysylleucine | -1.18 | 0.32 | -1.04 | -1.54 | -1.29 | 0.10881 | 1.00000 | 0.21322 | 0.10951 | 0.04245 |
| margarate (17:0) | 0.05 | 0.27 | -1.35 | -1.35 | -1.35 | 1.00000 | 1.00000 | 0.02623 | 0.03285 | 0.00215 |
| margaroylcarnitine (C17)* | 1.13 | -0.12 | -0.67 | -0.90 | -0.79 | 0.06789 | 0.88864 | 0.04086 | 0.09116 | 0.01424 |
| nervonate (24:1n9)* | -1.21 | 0.12 | -1.30 | -1.11 | -1.20 | 0.14413 | 0.88864 | 0.07537 | 0.04086 | 0.00671 |
| nervonoylcarnitine (C24:1)* | 0.26 | 0.00 | -0.55 | -0.37 | -0.46 | 0.14413 | 0.88864 | 0.10951 | 0.16881 | 0.04202 |
| nonadecanoate (19:0) | -0.14 | 0.64 | -0.95 | -1.28 | -1.12 | 0.71500 | 0.16143 | 0.05046 | 0.01279 | 0.00155 |
| oleate/vaccenate (18:1) | -0.51 | -0.07 | -1.12 | -0.83 | -0.98 | 0.46521 | 0.67442 | 0.03285 | 0.05046 | 0.00450 |
| oleoyl ethanolamide | -1.08 | -0.47 | -0.99 | -1.82 | -1.40 | 0.14413 | 0.40081 | 0.18231 | 0.02080 | 0.00985 |
| oleoylcarnitine (C18) | 0.96 | -0.61 | -0.43 | -1.60 | -1.02 | 0.28505 | 0.57540 | 0.32806 | 0.01637 | 0.01702 |
| orotidine | 1.16 | 0.79 | -1.09 | -1.32 | -1.20 | 0.10881 | 0.22492 | 0.04311 | 0.03569 | 0.00373 |
| palmitate (16:0) | -0.10 | 0.18 | -1.10 | -0.75 | -0.92 | 0.71500 | 0.77943 | 0.02080 | 0.06188 | 0.00173 |
| palmitoleoyl ethanolamide* | -1.48 | -0.92 | -0.65 | -1.78 | -1.21 | 0.27332 | 0.17295 | 0.28450 | 0.01516 | 0.01260 |
| palmitoyl sphingomyelin (d18:1/16:0) | 0.96 | -0.55 | -0.65 | -1.52 | -1.09 | 0.46521 | 0.16143 | 0.15486 | 0.01637 | 0.00740 |
| palmitoyl-arachidonoyl-glycerol (16:0/20:4) [2]* | 1.13 | -0.80 | -0.95 | -0.78 | -0.86 | 0.46521 | 0.31049 | 0.05934 | 0.28600 | 0.04566 |
| palmitoylcarnitine (C16) | 1.70 | -0.67 | -0.44 | -1.62 | -1.03 | 0.06789 | 0.40081 | 0.32806 | 0.06188 | 0.03346 |
| phenethylamine | 2.86 | -0.81 | -0.90 | -2.06 | -1.48 | 0.14413 | 0.31049 | 0.50762 | 0.02842 | 0.03334 |
| phenylpyruvate | 2.67 | -0.51 | -0.96 | -1.48 | -1.22 | 0.06789 | 0.20758 | 0.24775 | 0.03285 | 0.02616 |
| phytosphingosine | -0.71 | -1.95 | -0.84 | -1.31 | -1.07 | 0.27332 | 0.01172 | 0.06188 | 0.02080 | 0.00240 |
| prolylglycine | 1.44 | -0.91 | -0.52 | -2.05 | -1.29 | 0.06789 | 0.40081 | 0.47691 | 0.02623 | 0.03085 |
| skatol | -0.11 | 1.37 | 0.61 | 0.74 | 0.68 | 0.31731 | 0.27332 | 0.10881 | 0.22492 | 0.03569 |
| sphingomyelin (d18:1/24:1, d18:2/24:0)* | 1.35 | -0.70 | -0.78 | -1.77 | -1.27 | 0.28505 | 0.23672 | 0.28600 | 0.03666 | 0.03255 |
| sphingomyelin (d18:2/16:0, d18:1/16:1)* | 1.49 | -0.36 | -0.80 | -1.59 | -1.20 | 0.17971 | 0.34545 | 0.18231 | 0.08583 | 0.02762 |
| sphingomyelin (d18:2/24:1, d18:1/24:2)* | 0.47 | -1.21 | -0.84 | -2.20 | -1.52 | 0.59298 | 0.17630 | 0.20262 | 0.02842 | 0.01524 |
| stearate (18:0) | -0.29 | 0.32 | -1.00 | -0.75 | -0.88 | 0.46521 | 0.57540 | 0.02080 | 0.05046 | 0.00267 |
| stearoyl-arachidonoyl-glycerol (18:0/20:4) [2]* | 1.53 | -0.73 | -1.24 | -1.83 | -1.53 | 0.27332 | 0.67442 | 0.09260 | 0.16881 | 0.04786 |
| stearoylcarnitine (C18) | 0.70 | -0.25 | -1.03 | -1.30 | -1.16 | 0.06789 | 0.67442 | 0.01637 | 0.01637 | 0.00069 |
| taurine | 0.03 | -1.61 | -1.82 | -2.10 | -1.96 | 1.00000 | 0.09289 | 0.00765 | 0.07537 | 0.00267 |
| trans-nonadecenoate (tr 19:1)* | -2.32 | -1.10 | -0.24 | -1.34 | -0.79 | 0.27332 | 0.09097 | 0.48384 | 0.00444 | 0.01959 |
| tryptamine | 1.08 | -0.64 | -1.45 | -2.40 | -1.93 | 0.46521 | 0.57540 | 0.42360 | 0.03285 | 0.04245 |
| tryptophan | 0.87 | -0.16 | -0.59 | -0.57 | -0.58 | 0.14413 | 0.32699 | 0.10951 | 0.13067 | 0.02405 |
| tryptophylglycine | 0.54 | 0.46 | -0.43 | -1.45 | -0.94 | 0.46521 | 0.61209 | 0.51467 | 0.02842 | 0.04014 |
| tyrosol | -0.35 | 0.92 | -1.30 | -0.75 | -1.02 | 1.00000 | 0.16143 | 0.06188 | 0.13067 | 0.01858 |
| uridine-2',3'-cyclic monophosphate | -0.09 | -0.53 | -0.94 | -1.72 | -1.33 | 0.71500 | 0.26262 | 0.07537 | 0.02840 | 0.00359 |
Supplementary Table 12.
| Difference in average log2 concentration (Remitters - Non-remitters) | p-value | |
|---|---|---|
| Glycocholic acid | 1.141 | 0.236 |
| Taurocholic acid | 0.069 | 0.749 |
| Glycochenodeoxycholic acid | 1.007 | 0.319 |
| Taurochenodeoxychoic acid | -0.494 | 0.704 |
| Cholic acid | -0.101 | 1.000 |
| Chenodeoxycholic acid | 0.086 | 0.912 |
| Deoxycholic acid | 1.753 | 0.053 |
| Lithocholic acid | 1.979 | 0.093 |
| Ursodeoxycholic acid | -2.763 | 0.043 |
| Acetate | -0.115 | 0.391 |
| Propionate | -0.347 | 0.571 |
| Butyrate | -0.228 | 0.583 |
| Valerate | 1.669 | 0.114 |
| Hexanoate | 2.476 | 0.050 |
Supplementary Table 13.
| Difference in average log2 concentration (Remitters - Non-remitters) | p-value | |
|---|---|---|
| N-acetylglycine | -0.891 | 0.03937 |
| N-acetylserine | -1.216 | 0.02142 |
| N-acetylthreonine | -2.193 | 0.01652 |
| N-methylalanine | 0.834 | 0.03926 |
| N-acetylglutamate | 1.020 | 0.02563 |
| pyroglutamine* | -1.212 | 0.04279 |
| 1-methylhistidine | -1.540 | 0.02128 |
| 3-methylhistidine | -2.245 | 0.01341 |
| imidazole propionate | -2.496 | 0.00315 |
| 4-imidazoleacetate | 1.537 | 0.00907 |
| 5-aminovalerate | -1.498 | 0.01343 |
| N-acetylphenylalanine | -1.205 | 0.00542 |
| N-methylphenylalanine | 1.838 | 0.01685 |
| phenyllactate (PLA) | -1.311 | 0.02799 |
| 4-hydroxyphenylacetate | -1.482 | 0.01624 |
| phenol sulfate | -3.136 | 0.02467 |
| kynurenine | -1.473 | 0.00109 |
| skatol | 1.681 | 0.03145 |
| indole | 1.529 | 0.02563 |
| N-acetylleucine | -1.435 | 0.00668 |
| alpha-hydroxyisocaproate | -1.860 | 0.03053 |
| 1-carboxyethylisoleucine | -0.949 | 0.04858 |
| ethylmalonate | 0.922 | 0.02142 |
| N-acetylvaline | -1.290 | 0.00200 |
| 1-carboxyethylvaline | -1.706 | 0.04029 |
| 2-hydroxy-3-methylvalerate | -1.522 | 0.02344 |
| 3-hydroxyisobutyrate | -1.191 | 0.03329 |
| 2,3-dihydroxy-5-methylthio-4-pentenoate (DMTPA)* | -1.078 | 0.04883 |
| hypotaurine | -2.656 | 0.00591 |
| taurine | -3.633 | 0.00282 |
| N-acetyltaurine | -4.291 | 0.00281 |
| norvaline | -1.735 | 0.01161 |
| dimethylarginine (ADMA + SDMA) | -2.119 | 0.00158 |
| N-acetylarginine | 1.297 | 0.01106 |
| N-acetylcitrulline | -1.143 | 0.01622 |
| creatine | -2.917 | 0.01783 |
| putrescine | -3.397 | 0.00148 |
| N-acetyl-isoputreanine* | -3.141 | 0.00176 |
| N1,N12-diacetylspermine | -2.093 | 0.01955 |
| N-acetylputrescine | -2.277 | 0.01343 |
| (N(1) + N(8))-acetylspermidine | -1.627 | 0.01002 |
| cysteinylglycine disulfide* | -1.037 | 0.04858 |
| 2-hydroxybutyrate/2-hydroxyisobutyrate | -1.982 | 0.00252 |
| histidylalanine | -1.844 | 0.01957 |
| tryptophylglycine | -1.639 | 0.02337 |
| valylglutamine | -1.175 | 0.04279 |
| valylglycine | -1.413 | 0.01624 |
| valylleucine | -1.389 | 0.02799 |
| lactate | -3.075 | 0.00282 |
| tricarballylate | 0.997 | 0.04643 |
| arachidonate (20:4n6) | -1.610 | 0.03939 |
| 3-methylglutarate/2-methylglutarate | 2.006 | 0.00602 |
| 3-carboxyadipate | 1.419 | 0.03939 |
| hexadecanedioate (C16) | 0.859 | 0.00315 |
| butyrylglycine (C4) | -1.148 | 0.03389 |
| palmitoylcarnitine (C16) | -2.363 | 0.02563 |
| stearoylcarnitine (C18) | -1.395 | 0.02799 |
| deoxycarnitine | -1.141 | 0.02344 |
| 2-hydroxynervonate* | 2.706 | 0.01781 |
| 3-hydroxydecanoate | -1.310 | 0.00194 |
| 3-hydroxylaurate | -0.977 | 0.00668 |
| 5-hydroxyhexanoate | 2.603 | 0.02281 |
| behenoyl ethanolamide (22:0)* | 1.574 | 0.01949 |
| lignoceroyl ethanolamide (24:0)* | 1.846 | 0.01953 |
| choline | -1.154 | 0.01002 |
| phosphocholine | -3.211 | 0.00474 |
| trimethylamine N-oxide | -1.937 | 0.01343 |
| 1,2-dipalmitoyl-GPC (16:0/16:0) | -3.640 | 0.00027 |
| 1-palmitoyl-2-stearoyl-GPC (16:0/18:0) | -3.095 | 0.00068 |
| 1-palmitoyl-2-oleoyl-GPC (16:0/18:1) | -2.284 | 0.00542 |
| 1-palmitoyl-2-linoleoyl-GPC (16:0/18:2) | -1.721 | 0.03622 |
| 1-stearoyl-2-oleoyl-GPC (18:0/18:1) | -3.232 | 0.00096 |
| 1-palmitoyl-GPC (16:0) | -1.920 | 0.03327 |
| 1-stearoyl-GPC (18:0) | -3.063 | 0.00177 |
| 1-oleoyl-GPC (18:1) | -2.472 | 0.03690 |
| 1-linoleoyl-GPC (18:2) | -1.539 | 0.02333 |
| 1-stearoyl-GPE (18:0) | -2.492 | 0.00178 |
| 1-palmitoyl-GPS (16:0)* | -1.995 | 0.00223 |
| 1-stearoyl-GPS (18:0)* | -2.624 | 0.00224 |
| 1-stearoyl-GPG (18:0) | -1.655 | 0.03021 |
| 1-stearoyl-GPI (18:0) | -2.192 | 0.03939 |
| 1-(1-enyl-palmitoyl)-2-palmitoyl-GPC (P-16:0/16:0)* | -2.393 | 0.00279 |
| 1-(1-enyl-palmitoyl)-2-oleoyl-GPC (P-16:0/18:1)* | -1.407 | 0.04858 |
| 1-(1-enyl-palmitoyl)-GPE (P-16:0)* | -1.932 | 0.02142 |
| 1-(1-enyl-oleoyl)-GPE (P-18:1)* | -2.389 | 0.01811 |
| 1-(1-enyl-stearoyl)-GPE (P-18:0)* | -1.664 | 0.02563 |
| 1-dihomo-linoleoylglycerol (20:2) | -1.385 | 0.04719 |
| palmitoyl-myristoyl-glycerol (16:0/14:0) [2] | -0.924 | 0.02563 |
| palmitoyl-arachidonoyl-glycerol (16:0/20:4) [2]* | -2.743 | 0.00047 |
| stearoyl-arachidonoyl-glycerol (18:0/20:4) [2]* | -2.583 | 0.00914 |
| oleoyl-arachidonoyl-glycerol (18:1/20:4) [2]* | -2.406 | 0.01467 |
| linoleoyl-arachidonoyl-glycerol (18:2/20:4) [2]* | -2.780 | 0.01038 |
| glycosyl-N-palmitoyl-sphingosine (d18:1/16:0) | -1.553 | 0.03327 |
| glycosyl-N-tetracosadienoyl-sphingosine (d18:1/24:2)* | -1.687 | 0.03055 |
| lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) | -4.159 | 0.00002 |
| lactosyl-N-behenoyl-sphingosine (d18:1/22:0)* | -3.521 | 0.00015 |
| lactosyl-N-nervonoyl-sphingosine (d18:1/24:1)* | -4.576 | 0.00002 |
| palmitoyl sphingomyelin (d18:1/16:0) | -2.485 | 0.00252 |
| behenoyl sphingomyelin (d18:1/22:0)* | -2.144 | 0.00149 |
| tricosanoyl sphingomyelin (d18:1/23:0)* | -2.079 | 0.01311 |
| lignoceroyl sphingomyelin (d18:1/24:0) | -2.963 | 0.00162 |
| sphingomyelin (d18:2/16:0, d18:1/16:1)* | -2.696 | 0.00697 |
| sphingomyelin (d18:1/20:0, d16:1/22:0)* | -2.167 | 0.02077 |
| sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1)* | -2.107 | 0.01439 |
| sphingomyelin (d18:1/24:1, d18:2/24:0)* | -3.363 | 0.00133 |
| sphingomyelin (d18:2/24:1, d18:1/24:2)* | -4.014 | 0.00049 |
| coprostanol | 2.398 | 0.03348 |
| 4-cholesten-3-one | 1.682 | 0.03939 |
| androsterone sulfate | -2.137 | 0.03117 |
| tauro-beta-muricholate | -1.025 | 0.04858 |
| 6-oxolithocholate | 1.381 | 0.02341 |
| ursodeoxycholate sulfate (1) | -3.055 | 0.04633 |
| xanthosine | -1.258 | 0.03471 |
| adenine | 1.401 | 0.01343 |
| uridine-2',3'-cyclic monophosphate | -1.915 | 0.00162 |
| 3-ureidoisobutyrate | -2.958 | 0.00022 |
| 3-ureidopropionate | -2.075 | 0.00487 |
| beta-alanine | -1.429 | 0.01106 |
| cytidine 2',3'-cyclic monophosphate | -0.874 | 0.01311 |
| cytosine | 1.359 | 0.01612 |
| 2'-deoxycytidine | 1.666 | 0.02139 |
| 3-aminoisobutyrate | -2.148 | 0.00105 |
| methylphosphate | 0.899 | 0.04643 |
| (3'-5')-adenylylguanosine* | -1.455 | 0.00646 |
| (3'-5')-adenylyluridine | -1.329 | 0.04193 |
| (3'-5')-adenylyladenosine* | -1.526 | 0.01233 |
| (3'-5')-guanylyluridine | -1.957 | 0.00913 |
| gulonate* | -1.625 | 0.01696 |
| gamma-tocotrienol | 1.135 | 0.03053 |
| pterin | 1.024 | 0.02309 |
| L-urobilin | 2.768 | 0.03606 |
| beta-cryptoxanthin | 1.646 | 0.00541 |
| pyridoxamine | 1.250 | 0.01343 |
| 4-hydroxybenzoate | -2.095 | 0.01748 |
| p-cresol | 1.562 | 0.03618 |
| paraxanthine | 1.654 | 0.03792 |
| 1,3,7-trimethylurate | 1.972 | 0.04487 |
| 2-piperidinone | -1.884 | 0.04279 |
| ciliatine (2-aminoethylphosphonate) | 1.180 | 0.04564 |
| ergothioneine | -1.383 | 0.01394 |
| sitostanol | 1.983 | 0.00708 |
| glutamyl-meso-diaminopimelate | 1.661 | 0.00134 |
| N-methylpipecolate | 1.138 | 0.04279 |
| N-carbamoylglutamate | 1.129 | 0.02531 |
| sulfate* | -1.281 | 0.04643 |
| X - 12093 | 1.043 | 0.04990 |
| X - 12199 | 1.993 | 0.00984 |
| X - 13728 | 1.372 | 0.02904 |
| X - 14626 | -1.334 | 0.03568 |
| X - 14838 | 1.878 | 0.01478 |
| X - 15136 | 1.740 | 0.01957 |
| X - 16060 | 1.407 | 0.02584 |
| X - 17299 | -3.213 | 0.02128 |
| X - 17653 | 1.155 | 0.02892 |
| X - 17654 | 1.364 | 0.01333 |
| X - 17877 | -1.083 | 0.03568 |
| X - 18162 | -1.429 | 0.02842 |
| X - 20624 | -1.538 | 0.02346 |
| X - 21353 | -1.153 | 0.02309 |
| X - 21470 | -2.407 | 0.02121 |
| X - 21788 | -2.927 | 0.00723 |
| X - 21796 | 0.782 | 0.02563 |
| X - 22030 | 1.229 | 0.03716 |
| X - 22520 | -1.598 | 0.04858 |
| X - 23105 | 3.787 | 0.01313 |
| X - 23438 | 1.191 | 0.03935 |
| X - 23748 | 1.577 | 0.02318 |
| X - 23753 | 1.335 | 0.02631 |
| X - 23925 | 0.848 | 0.01883 |
| X - 24177 | -1.323 | 0.03939 |
| X - 24243 | -1.976 | 0.00351 |
| X - 24246 | -2.806 | 0.00437 |
| X - 24359 | -2.127 | 0.01323 |
| X - 24408 | -1.495 | 0.00906 |
| X - 24413 | -2.951 | 0.00542 |
| X - 24425 | 1.208 | 0.04643 |
| X - 24556 | 1.018 | 0.03053 |
| X - 24697 | 3.213 | 0.02281 |
| X - 24729 | -0.941 | 0.03327 |
| X - 24763 | 1.354 | 0.02977 |
| X - 24829 | 2.099 | 0.00702 |
| X - 24983 | -2.170 | 0.00703 |
| X - 25009 | -1.829 | 0.01193 |
| X - 25074 | -1.783 | 0.00513 |
| X - 25111 | 2.121 | 0.02329 |
| X - 25182 | -1.067 | 0.02142 |
References
- 1.Eckburg P.B., Bik E.M., Bernstein C.N., et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yatsunenko T., Rey F.E., Manary M.J., et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostic A.D., Xavier R.J., Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartor R.B., Wu G.D. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd-Price J., Arze C., Ananthakrishnan A.N., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plichta D.R., Graham D.B., Subramanian S., et al. Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host-microbiome relationships. Cell. 2019;178:1041–1056. doi: 10.1016/j.cell.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paramsothy S., Nielsen S., Kamm M.A., et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156:1440–1454.e2. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Kotlowski R., Bernstein C.N., Sepehri S., et al. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halfvarson J., Brislawn C.J., Lamendella R., et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan X.C., Tickle T.L., Sokol H., et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubin D.T., Ananthakrishnan A.N., Siegel C.A., et al. ACG Clinical Guideline: ulcerative colitis in adults. Am J Gastroenterol. 2019;114:384–413. doi: 10.14309/ajg.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 13.Paramsothy S., Kamm M.A., Kaakoush N.O., et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 14.Costello S.P., Hughes P.A., Waters O., et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–164. doi: 10.1001/jama.2018.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moayyedi P., Surette M.G., Kim P.T., et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102–109.e6. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Carlson P.E., Jr. Regulatory considerations for fecal microbiota transplantation products. Cell Host Microbe. 2020;27:173–175. doi: 10.1016/j.chom.2020.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Colwell R.K., Coddington J.A. Estimating terrestrial biodiversity through extrapolation. Philos Trans R Soc Lond B Biol Sci. 1994;345:101–118. doi: 10.1098/rstb.1994.0091. [DOI] [PubMed] [Google Scholar]
- 18.Parks D.H., Beiko R.G. Measures of phylogenetic differentiation provide robust and complementary insights into microbial communities. ISME J. 2013;7:173–183. doi: 10.1038/ismej.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd-Price J., Mahurkar A., Rahnavard G., et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–666. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown E.M., Ke X., Hitchcock D., et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe. 2019;25:668–680.e7. doi: 10.1016/j.chom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franzosa E.A., Sirota-Madi A., Avila-Pacheco J., et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sofia M.A., Ciorba M.A., Meckel K., et al. Tryptophan metabolism through the kynurenine pathway is associated with endoscopic inflammation in ulcerative colitis. Inflamm Bowel Dis. 2018;24:1471–1480. doi: 10.1093/ibd/izy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feagan B.G., Sandborn W.J., D'Haens G., et al. The role of centralized reading of endoscopy in a randomized controlled trial of mesalamine for ulcerative colitis. Gastroenterology. 2013;145:149–157.e2. doi: 10.1053/j.gastro.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 24.Jairath V., Zou G., Parker C.E., et al. Systematic review and meta-analysis: placebo rates in induction and maintenance trials of ulcerative colitis. J Crohns Colitis. 2016;10:607–618. doi: 10.1093/ecco-jcc/jjw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.US Food and Drug Administration . US Food and Drug Administration; Washington, DC: 2016. Ulcerative Colitis: Clinical Trial Endpoints Guidance for Industry. [Google Scholar]
- 26.Horn H.S. The ecology of secondary succession. Annu Rev Ecol Syst. 1974;5:25–37. [Google Scholar]
- 27.Walter J., Maldonado-Gomez M.X., Martinez I. To engraft or not to engraft: an ecological framework for gut microbiome modulation with live microbes. Curr Opin Biotechnol. 2018;49:129–139. doi: 10.1016/j.copbio.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dethlefsen L., Huse S., Sogin M.L., et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seekatz A.M., Young V.B. Clostridium difficile and the microbiota. J Clin Invest. 2014;124:4182–4189. doi: 10.1172/JCI72336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGovern B.H., Ford C.B., Henn M.R., et al. SER-109, an investigational microbiome drug to reduce recurrence after Clostridioides difficile infection: lessons learned from a phase 2 Trial. Clin Infect Dis. 2020 Apr 7 doi: 10.1093/cid/ciaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopetuso L.R., Scaldaferri F., Petito V., et al. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrett W.S., Gallini C.A., Yatsunenko T., et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coyte K.Z., Schluter J., Foster K.R. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 34.Johnson K.V., Burnet P.W. Microbiome: should we diversify from diversity? Gut Microbes. 2016;7:455–458. doi: 10.1080/19490976.2016.1241933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCormick B.A. Bacterial-induced hepoxilin A3 secretion as a pro-inflammatory mediator. FEBS J. 2007;274:3513–3518. doi: 10.1111/j.1742-4658.2007.05911.x. [DOI] [PubMed] [Google Scholar]
- 36.Mumy K.L., Bien J.D., Pazos M.A., et al. Distinct isoforms of phospholipase A2 mediate the ability of Salmonella enterica serotype typhimurium and Shigella flexneri to induce the transepithelial migration of neutrophils. Infect Immun. 2008;76:3614–3627. doi: 10.1128/IAI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.US Food and Drug Administration . US Food and Drug Administration; Washington, DC: 2019. Important Safety Alert Regarding Use of Fecal Microbiota for Transplantation and Risk of Serious Adverse Reactions Due to Transmission of Multi-Drug Resistant Organisms. [Google Scholar]
- 38.US Food and Drug Administration . US Food and Drug Administration; Washington, DC: 2020. Safety Alert Regarding Use of Fecal Microbiota for Transplantation and Risk of Serious Adverse Events Likely Due to Transmission of Pathogenic Organisms. [Google Scholar]
- 39.Wilcox M.H., McGovern B.H., Hecht G.A. The efficacy and safety of fecal microbiota transplant for recurrent Clostridium difficile infection: current understanding and gap analysis. Open Forum Infect Dis. 2020;7:ofaa114. doi: 10.1093/ofid/ofaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y., Guo C., Tang L., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blaser M.J. Fecal microbiota transplantation for dysbiosis—predictable risks. N Engl J Med. 2019;381:2064–2066. doi: 10.1056/NEJMe1913807. [DOI] [PubMed] [Google Scholar]
- 42.Rubin D.T. Curbing our enthusiasm for fecal transplantation in ulcerative colitis. Am J Gastroenterol. 2013;108:1631–1633. doi: 10.1038/ajg.2013.279. [DOI] [PubMed] [Google Scholar]
- 43.Tomkovich S., Dejea C.M., Winglee K., et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest. 2019;130:1699–1712. doi: 10.1172/JCI124196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauss J., Kaplan G.G., Beck P.L., et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 45.Thorpe C.M., Kane A.V., Chang J., et al. Enhanced preservation of the human intestinal microbiota by ridinilazole, a novel Clostridium difficile-targeting antibacterial, compared to vancomycin. PLoS One. 2018;13 doi: 10.1371/journal.pone.0199810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kump P., Wurm P., Grochenig H.P., et al. The taxonomic composition of the donor intestinal microbiota is a major factor influencing the efficacy of faecal microbiota transplantation in therapy refractory ulcerative colitis. Aliment Pharmacol Ther. 2018;47:67–77. doi: 10.1111/apt.14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segata N., Faust K., Izard J., et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truong D.T., Franzosa E.A., Tickle T.L., et al. MetaPhIAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]