Abstract
Acetate:succinate CoA transferase (ASCT) is a mitochondrial enzyme that catalyzes the production of acetate and succinyl-CoA, which is coupled to ATP production with succinyl-CoA synthetase (SCS) in a process called the ASCT/SCS cycle. This cycle has been studied in Trypanosoma brucei (T. brucei), a pathogen of African sleeping sickness, and is involved in (i) ATP and (ii) acetate production and proceeds independent of oxygen and an electrochemical gradient. Interestingly, knockout of ASCT in procyclic form (PCF) of T. brucei cause oligomycin A-hypersensitivity phenotype indicating that ASCT/SCS cycle complements the deficiency of ATP synthase activity. In bloodstream form (BSF) of T. brucei, ATP synthase works in reverse to maintain the electrochemical gradient by hydrolyzing ATP. However, no information has been available on the source of ATP, although ASCT/SCS cycle could be a potential candidate. Regarding mitochondrial acetate production, which is essential for fatty acid biosynthesis and growth of T. brucei, ASCT or acetyl-CoA hydrolase (ACH) are known to be its source. Despite the importance of this cycle, direct evidence of its function is lacking, and there are no comprehensive biochemical or structural biology studies reported so far. Here, we show that in vitro–reconstituted ASCT/SCS cycle is highly specific towards acetyl-CoA and has a higher kcat than that of yeast and bacterial ATP synthases. Our results provide the first biochemical basis for (i) rescue of ATP synthase-deficient phenotype by ASCT/SCS cycle in PCF and (ii) a potential source of ATP for the reverse reaction of ATP synthase in BSF.
Abbreviations: ASCT, acetate:succinate CoA transferase; SCS, succinyl-CoA synthetase; SCOT, succinyl-CoA:3-ketoacid CoA transferase; ACH, acetyl-CoA hydrolase; T. brucei, Trypanosoma brucei; SLPHOS, substrate-level phosphorylation; OXPHOS, oxidative phosphorylation; PCF, procyclic form; BSF, bloodstream form
Keywords: Mitochondria, Energy metabolism, Substrate-level phosphorylation, Acetate:succinate CoA transferase, Fatty acid biosynthesis, Trypanosoma brucei, Succinyl-CoA:3-ketoacid CoA transferase
Highlights
-
•
First biochemical and structural characterization of mitochondrial ASCT/SCS cycle
-
•
It is essential for mitochondrial acetate/ATP production and T. brucei BSF growth.
-
•
TbASCT/SCS cycle shows higher kcat than that of yeast and bacterial ATP synthases.
-
•
Detailed comparative biochemical analysis between ASCT and human SCOT
-
•
Active site residue and X-CoA binding site determined by site-directed mutagenesis
1. Introduction
Energy metabolism of parasites is quite diverse and essential for their survival in the host [1,2]. Human African trypanosomiasis (HAT) is a parasitic disease transmitted by the tsetse fly. HAT includes two forms: Trypanosoma brucei gambiense, which causes a slow-progressing form of the disease and is endemic in western and central Africa, and T. b. rhodesiense, which causes a rapidly progressing form endemic in eastern Africa [3]. Although five drugs are available for treating HAT, they are suboptimal in terms of safety and efficacy [4]. T. brucei also infects livestock animals such as cattle and sheep (Nagana), causing considerable economic losses in endemic countries [5,6]. Although efforts to control African trypanosomiasis have been ongoing for a century, the disease persists, and the outlook for eradication is poor [5,7,8].
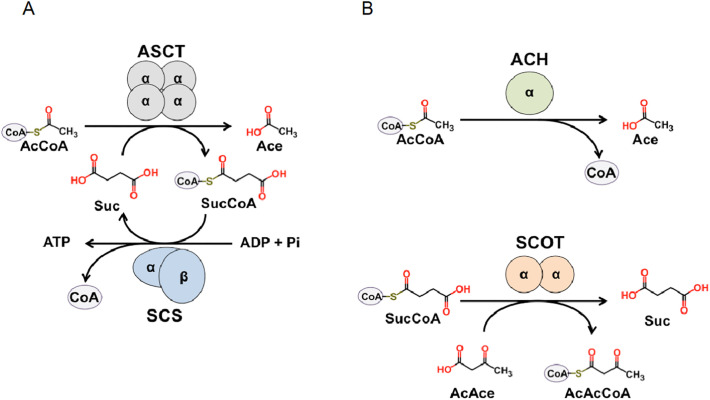
During its life cycle, T. brucei undergoes developmental changes including metabolic reprogramming dependent on the host environment (Fig. S1). When the bloodstream form (BSF) in mammalian host, which includes long slender (LS) and short stumpy (SS) forms, is ingested by the tsetse fly, the replicative LS form dies and the SS form differentiates into a procyclic form (PCF) in the midgut [9]. The parasite then migrates to the insect salivary glands, where it differentiates into infective metacyclic trypomastigote, which is injected into mammalian hosts during blood meals [9]. In the tsetse fly, the parasite switches from glucose to amino acid metabolism in order to adapt to its new environment. In the PCF in the insect vector, the end products of energy metabolism are, acetate, succinate, alanine, pyruvate and glycine [[10], [11], [12], [13]], whereas pyruvate followed by acetate and succinate are the end products in the LS and SS forms in mammalian host [[13], [14], [15]]. In both of the BSFs, enzymes associated with the mitochondrial electron transport chain (ETC) are downregulated [16,17], whereas cyanide-insensitive trypanosome alternative oxidase (TAO) is upregulated and functions as the sole terminal oxidase. TAO is required to re-oxidize NADH produced in the glycosomes via the glycerol-3-phosphate/dihydroxyacetone phosphate shuttle in order to maintain the glycolytic flux and ATP production [18]. Due to the downregulation of mitochondrial proton pump enzymes (complexes III and IV), BSFs maintain the electrochemical gradient by the reverse reaction of Fo/F1-ATP synthase hydrolyzing ATP, which is produced by substrate-level phosphorylation (SLPHOS) [16,17]. A potential candidate for ATP supply is the acetate:succinate CoA-transferase/succinyl-CoA synthetase (ASCT/SCS) cycle due to its ability to produce ATP independent of oxygen and an electrochemical gradient [19]. In this cycle, ASCT transfers the CoA moiety from acetyl-CoA to succinate, producing acetate and succinyl-CoA, which is used by SCS to produce ATP, CoA, and succinate [19] (Fig. 1A).
Fig. 1.
Schematic representation of the reactions catalyzed by (A) the ASCT/SCS cycle, (B) ACH and SCOT reaction. Abbreviations: AcCoA, acetyl-CoA; Ace, acetate; Suc, succinate; SucCoA, succinyl-CoA; AcAce, acetoacetate; AcAcCoA, acetoacetyl-CoA.
The ASCT/SCS cycle was initially investigated in T. brucei PCF, and is thought to have two functions: i) ATP and ii) acetate production [20,21]. The role of the ASCT/SCS cycle in ATP production was demonstrated indirectly by the hypersensitivity of TbASCT null mutants (~1000-fold) to oligomycin A (a specific Fo/F1-ATP synthase inhibitor) [22], indicating that ATP production by this cycle is essential for ATP synthesis when oxidative phosphorylation (OXPHOS) is impaired. In addition, SLPHOS observed in the presence of pyruvate and succinate was abolished upon RNAi knockdown of SCS β subunit [23], which further support the role of ATP production by ASCT/SCS cycle in PCF. From previous reports, SCS activity was detected in both BSF and PCF [24], and shown to be essential by RNAi knockdown [23,25]. Furthermore, the expression of TbASCT and SCS (α and β subunits) were also confirmed by proteome analyses in both BSF and PCF at levels comparable to other mitochondrial enzymes such as pyruvate dehydrogenase complex [26,27].
Regarding acetate production catalyzed by the ASCT/SCS cycle, glucose- and L-threonine-derived acetyl-CoA is converted into acetate by TbASCT. Mitochondrial acetate is also produced by acetyl-CoA hydrolase (TbACH) [10,22], which hydrolyzes acetyl-CoA to acetate and CoA (Fig. 1B). It is important to note that, in contrast to TbASCT, TbACH has no CoA transferase activity, as previously demonstrated [22]. Most of the acetate formed by TbASCT and TbACH is excreted as an end product, but a portion of it is used to supply de novo fatty acid biosynthesis in PCF trypanosomes [28] via a process termed the ‘acetate shuttle’, in which the acetate produced in mitochondria is transported to the cytosol, where it is re-converted to acetyl-CoA by ‘AMP-forming’ acetyl-CoA synthase (TbACS) at the expense of ATP [15,28]. To date, the acetate shuttle has been demonstrated only in trypanosomes and replaces the ubiquitous citrate shuttle, which does not operate in trypanosomatids [28]. This shuttling is essential for de novo biosynthesis of fatty acids, as RNAi silencing of TbACS inhibits the incorporation of acetate into fatty acids and is lethal for the PCF [28] and LS [15] form. Similarly, acetate production catalyzed by TbASCT and TbACH is essential, at least in the PCF, as shown by the lethal phenotype of the ∆ach/RNAiASCT double mutant [22].
The ASCT/SCS cycle has been identified in mitochondria and hydrogenosomes of microaerophilic organisms with an incomplete ETC (T. brucei, Fasciola hepatica, and Ascaris sp.) or that lack complex V (Trichomonas vaginalis, Tritrichomonas foetus, and Blastocystis sp.) [29]. In helminths such as Ascaris suum [1], Haemonchus contortus [30], and Echinococcus multilocularis [31], the adult stage live in hypoxic conditions and complexes III and IV are not functional [32]. In this case, complex I is the only active ETC enzyme with proton pump activity capable of maintaining the electrochemical gradient for OXPHOS, which is inefficient; thus, SLPHOS mediated by the ASCT/SCS cycle becomes significant.
Although ASCT activity has been described in T. brucei, F. hepatica, and T. vaginalis, the identified proteins shared no homology [20,21,33,34]. On the basis of amino acid sequence, ASCTs have been classified into three CoA-transferase subfamilies (IA, IB, and IC) [21]. Although ASCT activity has not been detected in mammals, TbASCT and human succinyl-CoA:3-ketoacid CoA-transferase (HsSCOT) are both annotated as subfamily IA CoA-transferases [21], exhibiting 52% amino acid sequence identity (Fig. S2). HsSCOT catalyzes the production of acetoacetyl-CoA and succinate from succinyl-CoA and acetoacetate (Fig. 1B), the rate-limiting step in ketone body catabolism [35]. Circulating acetoacetate produced by the liver is taken by extrahepatic cells, where acetoacetyl-CoA is produced and metabolized to acetyl-CoA to feed the tricarboxylic acid cycle for ATP synthesis [36]. Although HsSCOT and TbASCT are believed to have similar functions [21], neither biochemical evidence nor structural studies supporting this hypothesis have been reported so far. It has been previously demonstrated that a heterodimeric CoA transferase composed by ScoA and ScoB (WP_007410135.1 and WP_160182702.1, respectively) from Bacillus subtilis, which showed considerable high SCOT activity, exhibits 55% amino acid identity to corresponding N-terminal and C-terminal regions of homodimeric HsSCOT (Fig. S2), respectively [37]. Since TbASCT shares 52% amino acid identity to HsSCOT and currently annotated as SCOT, it is possible that TbASCT may also have SCOT activity and vise versa. Because ASCT and SCOT belong to different metabolic pathways, it is important to characterize and compare both enzymes.
Despite the essential roles of mitochondrial acetate production by TbASCT and TbACH in the PCF, their roles in BSFs have not been investigated. In addition, biochemical and structural biology studies of ASCT family enzymes have been hampered due to the difficulty to purify ASCT [20,33,34] in its active form. In this study, we report the first detailed biochemical features of purified TbASCT by using the new assay method, which is totally dependent on TbASCT substrates and enables to monitor the initial velocity of the reaction. Characterization of the in vitro reconstituted ASCT/SCS cycle and site-directed mutagenesis analyses provide insights into the catalytic mechanism of TbASCT and identification of the active site glutamate residue.
2. Material and methods
2.1. Knock-out of the ACH gene
Replacement of the acetyl-CoA hydrolase gene (ACH: Tb927.3.4260) with the phleomycin (BLE) and puromycin (PAC) resistance markers via homologous recombination was performed using DNA fragments containing the resistance marker gene flanked by the ACH UTR sequences, as described previously [22]. Briefly, the pGEMt plasmid was used to clone an HpaI DNA fragment containing the BLE and PAC resistance marker genes preceded by the ACH 5′-UTR fragment (537 or 492 bp) and followed by the ACH 3′-UTR fragment (521 or 485 bp) [22]. The ACH knock-out was generated in the 427 MITat1.2 13–90 (Hyg-Neo) monomorphic BSF cell line, which constitutively expresses the T7 RNA polymerase gene and the Tet repressor under control of a T7 RNA polymerase promoter for Tet-inducible expression [38]. Transfections were performed as previously reported, using the Nucleofector® system [39], and selection of drug-resistant clones, designated Δach (TetR-HYG T7RNAPOL-NEO Δach::BLE/Δach::PURO), was performed in HMI-9 medium containing hygromycin (5 μg/ml), neomycin (2.5 μg/ml), phleomycin (2.5 μg/ml), and puromycin (0.1 μg/ml). After transfection, the cells were re-suspended in 50 ml of conditioned medium and aliquoted into two 25-well titer plates; antibiotics were added the following day.
2.2. Inhibition of gene expression by RNAi
Inhibition of expression of the ASCT gene (Tb927.11.2690) by RNAi in the 427 MITat1.2 13–90 BSF strain was performed by expression of stem-loop “sense/anti-sense” RNA molecules of the targeted sequences introduced in the pHD1336 expression vector. Construction of the pLew-ASCT-SAS vector was as described previously [20]. The ASCT-SAS HindIII-BamHI cassette extracted from the pLew-ASCT-SAS plasmid was inserted into the HindIII-BamHI–digested pHD1336 vector. The resulting pHD-ASCT-SAS plasmid contained a sense and antisense version of the targeted gene fragment (572 bp fragment from nt positions 85 to 657), separated by a 64-bp fragment, under control of the PARP promoter linked to a prokaryotic Tet operator. The RNAiASCT and Δach/RNAiASCT cell lines were produced by transfecting (with the NotI-linearized pHD-ASCT-SAS plasmid) the 427 and Δach cell lines, respectively. Transfected cells were incubated in HMI-9 medium containing hygromycin (5 μg/ml), neomycin (2.5 μg/ml), and blasticidin (10 μg/ml) for selection of RNAiASCT cells or the same medium plus phleomycin (2.5 μg/ml) and puromycin (0.1 μg/ml) for selection of Δach/RNAiASCT cells. Induction of double-stranded RNA expression in the latter mutant was performed by addition of 10 μg/ml tetracycline.
2.3. Western blot analysis
Total protein extracts of wild-type or mutant T. brucei BSFs (5 × 106 cells) were size-fractionated by SDS-PAGE (10%) and immunoblotted onto Immobilon-P filters (Millipore) [40]. Immunodetection was performed as described elsewhere [40,41] using rabbit anti-ASCT (diluted 1:1000) [20], purified rabbit anti-ACH [22], and rabbit anti-GPDH (glycerol-3-phosphate dehydrogenase, EC 1.1.1.8; diluted 1:100) [42] as primary antibodies and anti-rabbit IgG conjugated to horseradish peroxidase (BioRad, 1:5000 dilution) as the secondary antibody. Blots were developed using SuperSignal® West Pico Chemiluminescent Substrate as described by the manufacturer (Thermo Scientific). Images were acquired and analyzed using a KODAK Image Station 4000 MM.
2.4. Bacterial strains, plasmids, and reagents for recombinant TbASCT and HsSCOT production
The TbASCT gene lacking the mitochondrial targeting signal (MTS; Δ1–9 residues) was amplified by PCR from genomic DNA (ILtat 1.4 strain [43]) and inserted into the pETSUMO vector (Thermo Fisher Scientific) by TA cloning [44] according to the manufacturer's protocol. The generated plasmid (pETSUMO/TbASCT) was used to transform chemically competent One Shot™ TOP10 (Thermo Fisher Scientific) Escherichia coli. In comparison to the TREU 927 strain, ASCT cloned from ILtat 1.4 had a single substitution of serine to asparagine at position 419, which is located at the protein surface. Next, BL21 star™ (DE3) (Thermo Fisher Scientific) was transformed with pETSUMO/TbASCT. The final expression host, BL21 star™ (DE3)pETSUMO/TbASCT, was stored in 20% (v/v) glycerol at −80 °C until use.
The plasmid used for HsSCOT expression was prepared similarly to that for TbASCT expression, with slight modifications. The HsSCOT (GI number: 4557817) gene lacking the MTS (Δ1–39 residues) and fused with a His10-SUMO tag at the N-terminus was codon optimized, amplified by PCR, and inserted into the pET101/D-TOPO vector (Thermo Fisher Scientific).
MagExtractor™-PCR & Gel Clean-up for gel extraction and PCR product purification kits, and MagExtractor™-plasmid for plasmid purification were purchased from TOYOBO (Japan). Ni-NTA affinity agarose was obtained from QIAGEN (Germany). SCS from prokaryote origin was obtained from Megazyme (Ireland). All other reagents were of the highest grade available and purchased from Sigma (USA), WAKO (Japan), or TCI (Japan).
2.5. Optimized large-scale purification of TbASCT and HsSCOT
A glycerol stock of E. coli BL21 star™ (DE3)pETSUMO/TbASCT was streaked onto an LB agar plate containing 50 μg/ml kanamycin and incubated overnight at 37 °C. The next day, a single colony was inoculated into 360 ml of Terrific Broth (TB) medium and cultured for 15 h. The pre-culture was inoculated (to a final OD600 of 1.0) into 6 Ultra Yield™ flasks (Thomson), each containing 600 ml of TB medium supplemented with 50 μg/ml kanamycin and 20 μM isopropyl β-D-thiogalactopyranoside (IPTG), and cultured for 12 h at 20 °C on a rotary shaker (200 rpm; BioShaker GBR-300; Taitec). The E. coli cells were harvested by centrifugation at 7000 ×g, 10 min, 4 °C and resuspended in 210 ml of lysis buffer (50 mM Tris-HCl pH 8.0, 5 mM imidazole, 20% [v/v] glycerol, 300 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 250 μM phenylmethylsulfonyl fluoride [PMSF]). Cells were broken by a single passage through a French Press (Ohtake) at 180 MPa and 4 °C. Unbroken cells and debris were removed by centrifugation at 30,000 ×g, 30 min, and 4 °C (R20A2 rotor, Hitachi), and the cytosolic fraction was obtained as the supernatant following ultracentrifugation at 200,000 ×g, 90 min, and 4 °C (45TI rotor, Hitachi). A pre-equilibrated 2-ml volume of Ni-NTA agarose (QIAGEN) was mixed with the cytosolic fraction overnight, loaded onto Econo-Pac® chromatography columns (Bio-Rad), and washed with 2 ml of lysis buffer. The column was consecutively washed with 5 ml of wash buffer A (50 mM Tris-HCl pH 8.0, 20 mM imidazole, 20% [v/v] glycerol, 10 mM ATP, 300 mM NaCl, 2.5 mM MgCl2, 250 μM PMSF) and 30 ml of wash buffer B (50 mM Tris-HCl pH 8.0, 20 mM imidazole, 20% [v/v] glycerol, 300 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 250 μM PMSF), and recombinant TbASCT was eluted with 10 volumes of elution buffer (50 mM Tris-HCl pH 8.0, 250 mM imidazole, 20% [v/v] glycerol, 300 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 250 μM PMSF), with collection of 1-ml fractions. Protein content in the fractions was measured at 280 nm using a Nanodrop 2000 (Thermo Fischer Scientific). Fractions exhibiting an absorbance >2.0 were pooled and concentrated to more than 10 mg/ml using a centrifugal filter device (Amicon Ultra-15, 50-kDa cutoff; Millipore). The concentrated fraction was diluted 10-fold with 50 mM Tris-HCl pH 8.0, 20% (v/v) glycerol, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA (protease buffer) to decrease the imidazole and NaCl concentrations. SUMO protease (Invitrogen) was added at a 1:20 protein ratio and incubated at 4 °C overnight. Pre-equilibrated Ni-NTA agarose (2 ml) was added to the mixture and mixed for 1 h. SUMO protease and the His6-SUMO tag were removed by loading the mixture onto an Econo-Pac® chromatography column. Pure TbASCT, collected as the flow-through, was concentrated to more than 10 mg/ml using a centrifugal filter device (Amicon Ultra-15, 50-kDa cutoff; Millipore), glycerol was added to a final concentration of 50% (v/v) and kept at −30 °C until use. The purification of TbASCT is summarized in Table S1.
HsSCOT was purified by a similar protocol as that used for TbASCT purification, with minor modifications. Specifically, after induction, the cells were cultured for 16.5 h at 37 °C, the column was not washed with wash buffer containing ATP, and 50 ml of wash buffer containing 50 mM imidazole was used for the initial wash.
2.6. Protein quantification, denaturation, and BN-PAGE
Protein concentration was determined using a Bio-Rad Protein Assay kit with bovine serum albumin as the standard.
The cytosolic, flow-through, elution, and tag-free fractions from each purification step were subjected to discontinuous SDS-PAGE according to Laemmli [45]. The stacking and separating gels were 4% and 12% (w/v) acrylamide, respectively. Samples were mixed 1:3 (v/v) with 4 × SDS-PAGE loading buffer (240 mM Tris-HCl pH 6.8, 8% [w/v] SDS, 40% [v/v] glycerol, 0.04% [w/v] bromophenol blue, and 5% [v/v] β-mercaptoethanol) and heated for 5 min at 95 °C. Protein preparations were loaded onto the gel and run alongside broad-ranged Precision Plus Protein™ Standards (Bio-Rad). The electrophoresis was carried out at 25 mA for 120 min at room temperature, and the gel was stained with GelCode™ Blue Safe Protein Stain (Thermo Fisher Scientific).
BN-PAGE was performed as follows. Dilutions of purified TbASCT were prepared in 50 mM Tris-HCl pH 8.0, 8% (v/v) glycerol, 0.01% (w/v) n-Dodecyl-β-D-maltoside, 0.002% (w/v) G-250, and 25 mM NaCl and loaded onto a 4–16% (w/v) Bis-Tris gradient gel (Invitrogen) at final concentration of 1 μg to 1 ng for CBB staining and 0.1 μg to 0.01 ng for in-gel activity staining per lane, respectively. Anode and cathode buffers were prepared according to the manufacturer's protocol using 20 × running buffer (Invitrogen), with the cathode buffer supplemented with 0.002% (w/v) G-250. The gel was run at 4 °C under constant voltage of 150 V for 1 h and 250 V for 1 h.
2.7. In-gel ASCT activity stain
Free thiol groups from CoA can reduce nitroblue tetrazolium salt (NBT) in the presence of phenazine methosulfate (PMS) [46,47]. To assay TbASCT activity (in-gel ASCT-activity staining), the BN-PAGE gel was washed twice for 5 min each at room temperature with 50 mM Tris-HCl pH 7.2, 5 mM MgCl2, and 25 mM sodium phosphate. After washing, the BN-PAGE gel was incubated for 20 min in same buffer supplemented with 0.5 mM ADP, 0.5 mM acetyl-CoA, 2.5 mg/ml NBT, and 0.44 U/ml of SCS; thereafter, 50 mM succinate and 0.12 mg/ml PMS were added and incubated static in the dark at room temperature.
2.8. In vitro reconstitution of the TbASCT/SCS cycle
The in vitro reconstituted ASCT/SCS cycle was assayed by quantification of released CoA (Fig. 1A) using DTNB (Sigma). In this system, DTNB reacts with one molecule of CoA, forming TNB, which can be followed at 412 nm (ε412 = 14.0 mM−1 cm−1) [48]. The rate of CoA production was monitored using a V-660 spectrophotometer (JASCO) equipped with a water bath circulator (Taitec).
The dose-response range of TbASCT was determined at 30 °C. ASCT activity at different concentrations of purified TbASCT, ranging from 12.5 to 200 ng/ml, was assayed in 1 ml reaction mixture (100 mM Tris-HCl pH 7.0, 10 mM sodium phosphate, 2 mM MgCl2, 1 mM ADP, 0.1 mM DTNB, 0.5 mM acetyl-CoA, 0.44 U/ml SCS) using a black quartz cuvette. After addition of enzyme, the background was recorded for 5 min, and initial velocity was monitored after addition of succinate to a final concentration of 20 mM.
The optimal temperature for the TbASCT assay was also determined. ASCT activity was assayed in 100 mM Tris-HCl pH 6.8, 1 mM sodium phosphate, 2 mM MgCl2, 0.1% (v/v) Triton X-100, 1 mM ADP, 0.1 mM DTNB, 0.44 U/ml SCS, 0.5 mM acetyl-CoA, and 10 ng of purified TbASCT at 20, 25, 30, 35, 37, 40, and 45 °C in triplicate. The reaction was initiated by the addition of 10 mM succinate from a 1 M stock solution with pH adjusted to 7.0.
The optimal pH for TbASCT activity was determined using a 96-well plate (C96 Maxisorp Nunc-Immuno Plate, Thermo Scientific) and a SpectraMax® Paradigm® Multi-Mode Detection Platform (Molecular Devices) at 30 °C in triplicate. Background was recorded for 5 min at 412 nm in 195 μl of reaction mix per well (50 mM of each buffer with different pH, 10 mM sodium phosphate, 2 mM MgCl2, 1 mM ADP, 0.1 mM DTNB, 0.5 mM acetyl-CoA, 0.44 U/ml SCS, TbASCT 100 ng/ml), and the reaction was initiated by adding 5 μl of 400 mM succinate. ASCT activity was calculated using the end-point method after 10 min, and the highest activity (Tris-HCl pH 7.0) was set as 100%.
Routine TbASCT assays were conducted in 1.0 ml of reaction mixture (100 mM Tris-HCl pH 7.0, 10 mM sodium phosphate, 2 mM MgCl2, 1 mM ADP, 0.1 mM DTNB, 0.5 mM acetyl-CoA, 0.44 U/ml SCS) using a black quartz cuvette at 30 °C. After addition of enzyme, the background was recorded for 5 min, and the reaction was initiated by addition of succinate to a final concentration of 20 mM.
K m and V max values for succinate were determined by measuring the TbASCT activity at varying succinate concentrations ranging from 0.5 to 50 mM at a fixed acetyl-CoA concentration of 0.5 mM. Similarly, the kinetic parameters for acetyl-CoA were determined at concentrations ranging from 0.01 to 1 mM in the presence of 20 mM succinate.
The reaction mechanism was analyzed by varying the concentration of succinate (1, 2, 5, 10, or 20 mM) under a fixed acetyl-CoA concentration of 0.02, 0.05, or 0.1 mM and by varying the acetyl-CoA concentration (0.025, 0.05, 0.1, 0.25, or 0.5 mM) under a fixed succinate concentration of 5, 10, or 20 mM, with all samples in triplicate. All enzyme kinetic parameters were obtained by fitting the data via nonlinear regression analysis using GraphPad Prism 7 software.
2.9. CoA-donor specificity of TbASCT
The substrate specificity of TbASCT was determined in 96-well plates in a reaction mix containing 200 ng/ml TbASCT. CoA donors used in this study included acetyl-CoA, n-propionyl-CoA, butyril-CoA, 2-butenoyl-CoA, acetoacetyl-CoA, β-hydroxybutyryl-CoA, malonyl-CoA, hexanoyl-CoA, octanoyl-CoA, decanoyl-CoA, and stearoyl-CoA. Two microliters of each CoA donor (10 mM) was transferred to the plate and mixed with 193 μl of reaction mix. After recording the background for 5 min, the reaction was initiated by adding 5 μl of 800 mM succinate. Each condition was assayed in quadruplicate; the ASCT activity in the presence of acetyl-CoA was set to 100%, and relative activity of TbASCT under each condition was then calculated.
2.10. Biochemical characterization of HsSCOT
HsSCOT was characterized similarly to TbASCT, as described above. Briefly, the dose-response range of HsSCOT was assayed in 1 ml of reaction mixture containing 100 mM Tris-HCl pH 8.6, 15 mM MgCl2, and 1 mM succinyl-CoA in the presence of 58–1860 ng of purified HsSCOT at 37 °C. To start the reaction, 7.5 mM acetoacetate was added, and the increase in absorbance at 310 nm, which indicates the formation of acetoacetyl-CoA (ε310 = 7.8 mM−1 cm−1) [49], was monitored. Next, HsSCOT activity was assayed under different pH and temperature conditions to optimize the assay. The kinetic parameters (K m and V max) for acetoacetate and succinyl-CoA were determined under optimal conditions (232 ng purified HsSCOT, 100 mM Tris-HCl pH 8.6, 15 mM MgCl2 and 1 mM succinyl-CoA at 37 °C).
2.11. Site-directed mutagenesis
TbASCT mutants were obtained by quick change (QIAGEN) according to the supplier's protocol using pETSUMO/TbASCT as the template. After the desired plasmid was obtained, it was used to transform BL21 star™ (DE3) cells. Each mutant was expressed and purified as described for wild-type TbASCT.
2.12. Crystallization of TbASCT
Crystallization conditions were screened using the sitting-drop vapor diffusion method with reservoir solutions supplied in commercially available screening kits (Crystal Screen, Crystal Screen 2, and PEG-ION 1, 2, and Grid Screen PEG6000 from Hampton Research; and Wizard Classic 1, 2, and Wizard Cryo 1, 2, from Molecular Dimensions). A droplet made by mixing 0.5 μl of purified TbASCT (5, 10 mg/ml) with an equal volume of reservoir solution was equilibrated against 100 μl of the reservoir solution at 293 K. After the initial crystallization, the crystals of TbASCT were generated under two conditions (PEG-ION Screen No. 5 and PEG-ION 2 Screen No. 48); crystallization conditions were optimized by fine-tuning the pH and precipitant concentration using the hanging drop vapor diffusion method at 293 K. The best crystals were obtained using a reservoir solution containing 0.05 M HEPES-NaOH buffer pH 7.4, 18% (w/v) PEG3350, 0.35 M CaCl2 with 10 mg/ml TbASCT dissolved in 20 mM Tris-HCl pH 8.5, 100 mM NaCl, and 10% (v/v) glycerol. Before being flash-cooled in liquid nitrogen, TbASCT crystals were soaked in reservoir solution supplemented with 20% (v/v) glycerol as a cryoprotectant.
2.13. Determination of the TbASCT structure
X-ray diffraction data were collected using the synchrotron radiation at Spring-8 (beamline BL44XU, Japan). X-ray diffraction data set at 2.01-Å resolution were obtained after processing and scaling using XDS and XSCALE [50], respectively. The structure of TbASCT was solved according to the molecular replacement method with the MOLREP [51] program using the structure of HsSCOT (PDB code 3DLX; 52% amino-acid sequence identity with TbASCT) as the initial model and refined using the REFMAC5 [52] and COOT [53] programs. Data collection and refinement statistics are summarized in Supplementary Table 4. All figures of protein structures were prepared using the graphics program PyMol (http://www.pymol.org/). The atomic coordinates and structural factors were deposited in the Protein Data Bank under accession code 6LP1.
3. Results
3.1. Growth phenotype of Δach,RNAiASCT and Δach/RNAiASCT in BSF
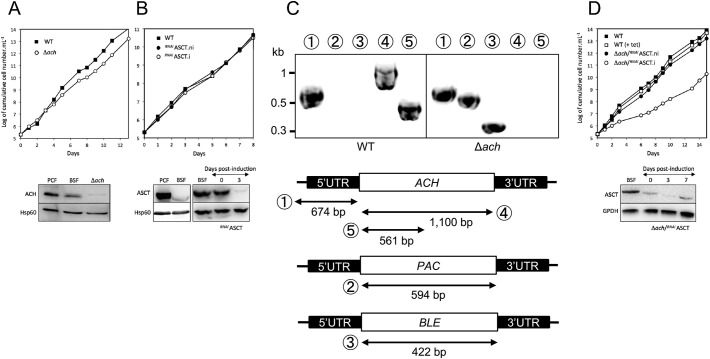
In this study, we investigated the roles of TbACH and TbASCT in the monomorphic 427 BSF strain, in which differentiation into the SS form is defective, leading to an almost exclusive composition of LS cells in vitro. Western blot analyses of the BSF show that both TbACH and TbASCT were expressed in the T. brucei wild type 427 strain, although at a much lower level for TbASCT compared to the PCF (Fig. 2A and B, bottom panels). This was consistent with the 6-fold higher level of TbASCT expression in PCF trypanosomes compared to LS BSFs quantified in a previous proteome study [54]. Similar result was shown for TbACH in other studies [26,27].
Fig. 2.
Analysis of TbASCT and TbACH mutant cell lines. Growth curves of the (A) ∆ach and (B) RNAiASCT mutants incubated in the presence (.i, ○) or absence (.ni, ●) of 10 μg/ml tetracycline. Bottom panel show the Western blot analyses of the PCF, parental BSF trypanosomes, and mutant cell lines using immune serum indicated in the left margin. Integration of targeting vector into the ACH locus in the ∆ach cell line. (C) The top panel shows results of PCR analysis of genomic DNA isolated from the parental 427 MITat1.2 13–90 BSF strain (WT) and ∆ach cell lines. The lower panel shows PCR fragments (①–⑤) amplified using primers based on sequences that flank the 5′UTR used to target the ACH gene for disruption (black boxes), internal sequences from ACH, and puromycin resistance (PAC) or phleomycin (BLE) resistance genes. As expected, using primers derived from the ACH gene and drug resistance genes, PCR amplification was only observed for the parental 427 (WT) and ∆ach cell lines, respectively. (D) Growth curves of the ∆ach/RNAiASCT mutants incubated in the presence (.i, ○) or absence (.ni, ●) of 10 μg/ml tetracycline. Bottom panel show the Western blot analyses of the parental BSF trypanosomes, and ∆ach/RNAiASCT mutant cell line using immune serum indicated in the left margin. Growth of the parental cell line was not affected by addition of 10 μg/ml tetracycline. Cells were maintained in the exponential growth phase (between 105 and 2 × 106 cells/ml), and cumulative cell numbers reflect normalization for dilution during cultivation. Induction (.i) refers to addition of 10 μg/ml tetracycline.
To study the role of TbACH and TbASCT in the BSF, we have prepared mutant cell lines of either ACH knock-out, ASCT knockdown, and its combination. We generated the TbACH knock-out mutant (Δach::BLE/Δach::PAC, abbreviated as Δach) by replacing both ACH alleles with phleomycin (BLE) and puromycin (PAC) resistance markers in the 427 BSF background. Deletion of both ACH alleles in the selected Δach cell line was confirmed by PCR (Fig. 2C) and Western blotting (Fig. 2A) using immunopurified anti-ACH antibodies raised against recombinant histidine-tagged TbACH [22]. TbASCT expression was down-regulated by RNAi by introducing the pHD-ASCT-SAS plasmid in the same parental BSF cell line to generate the RNAiASCT mutant. Neither the Δach nor RNAiASCT mutants exhibited significant change in growth phenotype (Fig. 2A and B, top panels).
To generate the Δach/RNAiASCT double mutant, the pHD-ASCT-SAS construct was introduced in the Δach cell line. After several transfection attempts, a single cell line was obtained, although all of the transfections of the parental 427 cells with the same plasmid preparation (pHD-ASCT-SAS) each produced approximately a dozen cell lines. Three days after induction by addition of tetracycline, the Δach/RNAiASCT.i double mutant exhibited a strong reduction in growth rate, correlated with reduced TbASCT expression indicating that mitochondrial acetate production is essential in BSF as well as PCF [22].
3.2. Expression and purification of recombinant TbASCT and HsSCOT
Recombinant His6-SUMO TbASCT was successfully expressed in E. coli BL21 star™ (DE3) cells and purified to homogeneity. Under optimal conditions, bacterial growth reached an OD600 of 10, from which 48 g of wet cell pellet per 3.6-L culture of TB medium was obtained. Finally, after adding SUMO protease, 26 mg of tag-free TbASCT was purified with a yield of 18% and specific ASCT activity of 119 μmol/min/mg protein, corresponding to a 20-fold increase compared to the cytosolic fraction (Table S1). Under similar conditions, we purified 0.485 mg of tag-free HsSCOT with a 6% yield and specific SCOT activity of 16.6 μmol/min/mg protein, representing a 325-fold increase compared to the cytosolic fraction (Table S2). According to our protocol, both enzymes were purified to homogeneity as judged by SDS-PAGE (Fig. S3).
3.3. The ASCT/SCS cycle coupled assay system
As previously reported assay method of ASCT is not suitable for the detailed kinetic analysis of the enzyme, we established a new system. In the ASCT/SCS cycle, one molecule of acetyl-CoA is consumed by TbASCT, and equimolar amounts of ATP and CoA are produced by SCS (Fig. 1A) [55]. We developed a new coupled enzyme assay for ASCT based on this cycle and the reaction of 5,5′-Dithiobis(2-nitrobenzoic acid) (DTNB) with the thiol group of CoA to produce one molecule of 2-nitro-5-thiobenzoic acid (TNB), which can be followed colorimetrically.
To validate the in vitro reconstituted ASCT/SCS cycle assay, the dependence on each assay component was investigated (Fig. S4A). When TbASCT, acetyl-CoA, or succinate was removed, no activity was detected. However, when either SCS, ADP, phosphate or MgCl2 was removed, a minimal amount of activity remained, ranging from 3.76 to 14.7 μmol/min/mg protein, which was consistent with the non-enzymatic degradation of succinyl-CoA produced by TbASCT in the assay system (equivalent to 15.5 μmol/min/mg protein with 0.1 mM succinyl-CoA).
3.4. Biochemical characterization of TbASCT
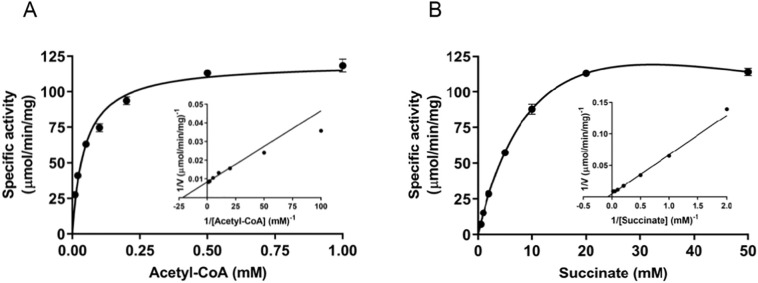
Purified recombinant TbASCT was biochemically characterized using the in vitro reconstituted ASCT/SCS cycle assay. TbASCT activity exhibited a linear dose response over the range of 6.25 to 200 ng/ml in the assay mixture, with an R 2 value of 0.981 (Fig. S4B). As such, subsequent assays were conducted using 100 ng/ml TbASCT. The optimum temperature and pH for the reaction were determined to be 30 °C and 7.0, respectively (Fig. S4C, D). The K m values for acetyl-CoA and succinate were 0.046 mM and 13.8 mM, respectively (Fig. 3A, B and Table 1 ). At a high concentration of succinate, substrate inhibition was clearly observed, with a K i of 75.7 mM (Fig. 3B). Considering substrate inhibition, the V max of succinate was calculated as 221 μmol/min/mg (k cat = 191 s−1) according to the following equation:
Fig. 3.
Biochemical characterization of TbASCT. Kinetic parameters of TbASCT were determined by varying the concentration of (A) acetyl-CoA under a fixed succinate concentration of 20 mM or (B) succinate under a fixed acetyl-CoA concentration of 0.5 mM, and the Lineweaver-Burk plot is shown in the inset. Error bars are ± SD (n = 3). At high succinate concentrations, substrate inhibition was observed, with a Ki of 75.7 mM.
Table 1.
Biochemical parameters of WT and E319A mutant TbASCT.
| Acetyl-CoA |
Succinate |
|||
|---|---|---|---|---|
| Km (mM) | kcat (s−1) | Km (mM) | kcat (s−1) | |
| WT | 0.046 ± 0.007 | 104 ± 3.91 | 13.8 ± 0.474 | 191 ± 4.25 |
| E319A | N.D. | N.D. | N.D. | N.D. |
Values represent the average ± SE (n = 3) calculated using GraphPad Prism 7. N.D. indicates non-detectable level.
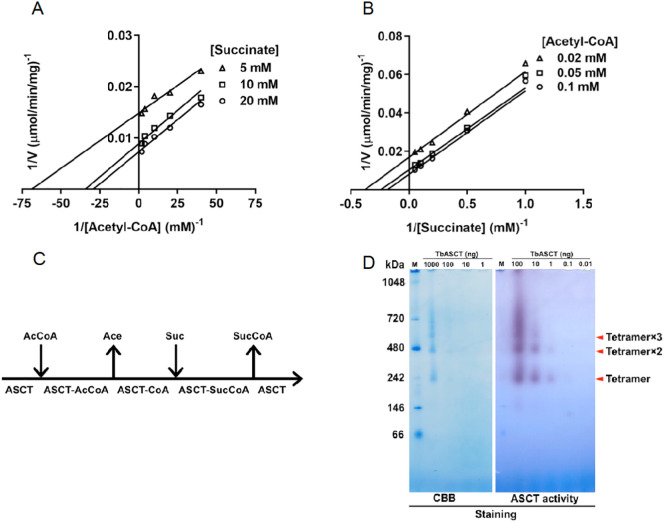
Next, the TbASCT reaction mechanism was examined. The reaction of TbASCT requires two substrates (acetyl-CoA and succinate). When the enzyme activity was determined by varying the concentration of one substrate under a fixed concentration of the second substrate and vice versa, double reciprocal plots of both cases exhibited parallel lines (Fig. 4A, B), indicating that TbASCT follows a ping-pong bi-bi catalytic mechanism (Fig. 4C) [56].
Fig. 4.
Reaction mechanism of TbASCT. The double reciprocal plot of varying the concentration of (A) acetyl-CoA under a set of fixed succinate concentrations of 5, 10, and 20 mM and varying the concentration of (B) succinate under a set of fixed acetyl-CoA concentrations of 0.02, 0.05, and 0.1 mM. (C) Schematic representation of ping-pong bi-bi mechanism catalyzed by TbASCT. (D) BN-PAGE and In-gel ASCT activity staining of TbASCT, M: Protein marker. Major bands identified from both staining are shown in red arrows with calculated molecular weight of 225, 414 and 564 kDa, which corresponds to one, two or three tetramers of TbASCT, respectively.
3.5. Blue native (BN)-PAGE and in-gel ASCT-activity staining
To investigate the oligomeric state of TbASCT in solution, BN-PAGE and in-gel ASCT activity staining were performed (Fig. 4D). Two bands corresponding to 225 and 414 kDa were detected by CBB staining (100 ng TbASCT/lane), while a third band at 564 kDa could be observed by ASCT activity staining (10 ng TbASCT/lane), which are consistent with calculated molecular weight of one, two and three tetramers of TbASCT, respectively (Fig. 4D, red arrows). At lower concentration of TbASCT, the 225 kDa band become predominant indicating that in solution TbASCT is a tetramer. At high concentration, smearing above 225 kDa band was observed for both staining methods, highlighting the sticky nature of TbASCT. Surprisingly, in-gel ASCT activity staining showed 1000 times more sensitivity compared with CBB staining (Fig. 4D) and can be used to detect TbASCT as low as 0.1 ng/lane.
3.6. Specificity of CoA donors to the TbASCT reaction
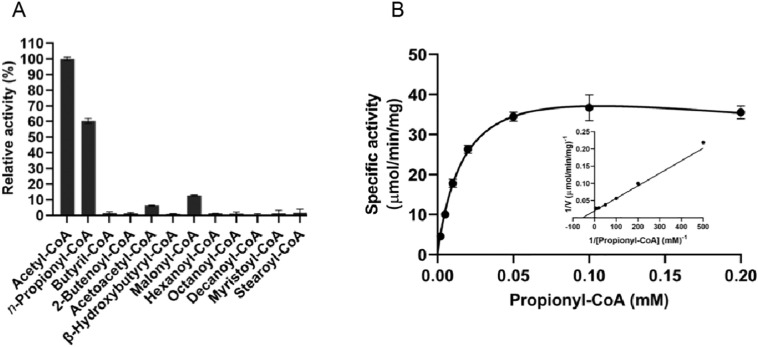
Although acetyl-CoA is known to be a substrate for TbASCT, the specificity of CoA donors has not been demonstrated. Hence, we examined the ability of TbASCT to utilize different CoA donors (Fig. 5A) and found that in the presence of n-propionyl-CoA, acetoacetyl-CoA, or malonyl-CoA, the activity remaining relative to acetyl-CoA was 60%, 6%, and 13%, respectively. Other CoA donors tested in this study exhibited negligible CoA-transferase activity (Fig. 5A). As TbASCT exhibited significantly high CoA-transferase activity with n-propionyl-CoA, we determined the kinetic parameters and found K m, V max, k cat values of 0.018 mM, 49.7 μmol/min/mg, and 42.9 s−1, respectively. At n-propionyl-CoA concentrations above 0.1 mM, CoA-transferase activity was inhibited, with a K i value of 0.633 mM (Fig. 5B).
Fig. 5.
CoA donor specificity of TbASCT. (A) TbASCT shows activity in the presence of n-propionyl-CoA, acetoacetyl-CoA, and malonyl-CoA of 60%, 6%, and 13% relative to acetyl-CoA (100%), respectively. Error bars are ± SD (n = 4). (B) Biochemical characterization of propionyl-CoA:succinate CoA transferase activity of TbASCT. Lineweaver-Burk plot is shown in the inset. Error bars are ± SD (n = 3). The Km, Vmax, kcat, and Ki values for propionyl-CoA were 0.018 ± 0.002 mM, 49.7 ± 2.75 μmol/min/mg, 42.9 ± 2.38 s−1, and 0.633 ± 0.175 mM, respectively. Kinetic parameters described above represent the average ± SE (n = 3) calculated using GraphPad Prism 7.
3.7. Biochemical characterization of HsSCOT, ASCT/SCOT reaction specificity and site-directed mutagenesis of TbASCT
Because TbASCT shares 52% amino acid sequence identity with HsSCOT, and TbASCT had SCOT activity (Table S5), we also examined whether HsSCOT exhibits ASCT activity. Recombinant HsSCOT was purified, and after removal of the His10-SUMO tag, a single 52-kDa band corresponding to HsSCOT was detected by SDS-PAGE (Fig. S3B). HsSCOT activity exhibited a linear dose response over the concentration range of 58 to 1860 ng/ml in the assay mixture, with an R 2 value of 0.998 (Fig. S5A); therefore, subsequent assays were conducted using 232 ng/ml. The optimum temperature and pH for this reaction were determined to be 37 °C and 8.6, respectively (Fig. S5B, C). Under optimal conditions, purified recombinant HsSCOT followed classical Michaelis-Menten kinetics, with K m, V max, and k cat values for acetoacetate and succinyl-CoA of 0.765 mM, 121 μmol/min/mg, and 105 s−1 and 0.971 mM, 219 μmol/min/mg, and 190 s−1, respectively (Table S3). Acetoacetate exhibited substrate inhibition at high concentrations, with a K i value of 285 mM (Fig. S5D), which is extremely high and under physiological condition [57], inhibition by acetoacetate should be insignificant. The ASCT and SCOT activities of TbASCT and HsSCOT were also analyzed and compared, with the activity assayed under optimal conditions. The SCOT activity of WT TbASCT was 0.038 μmol/min/mg, whereas HsSCOT did not exhibit detectable ASCT activity. Thus, TbASCT exhibited 3000-fold greater ASCT than SCOT activities.
3.8. Crystal structure of TbASCT
To compare detailed structural difference between ASCT and SCOT, the crystal structure of TbASCT was investigated. The ligand-free TbASCT crystal (Fig. S6D and S7A) was solved by molecular replacement using the structure of HsSCOT (Fig. S6C and S7B, PDB entry 3DLX) and Sus scrofa SCOT (SsSCOT, Fig. S6A, PDB entry 3OXO) as search models and refined to 2.01-Å resolution. Four TbASCT protomers were identified in the crystallographic asymmetric unit (chains A-D; Fig. S8A). Each TbASCT protomer associated to form a tight homotetramer, consistent with the oligomerization state revealed by BN-PAGE and in-gel ASCT-activity staining (Fig. 4D). Each protomer of TbASCT was shown to have a similar structure, as indicated by root-mean-square (RMS) deviations of 0.73–0.97 Å as calculated for 463 superimposed Cα positions between the chains. The RMS deviation of monomers within TbASCT (Chain A) and HsSCOT (Chain A) was 1.06 Å between the two structures for 456 Cα atoms (Fig. S8D). Succinyl-CoA is a product and a substrate of ASCT and SCOT, respectively, which is likely to bind to common substrate/product tunnel and reacts with the active site glutamate residue. In the crystal structure of SsSCOT complexed with CoA [58] (Fig. S6B), CoA was found forming thioester bond with the active site glutamate residue (E344) in the SENG motif conserved in family IA ASCTs and SCOTs. In the crystal structure of TbASCT, E319 corresponding to active site glutamate residue in the SENG motif could be clearly identified (Fig. S6D) as predicted previously [21]. Consistently, ASCT and SCOT activities of TbASCT were abolished in the E319A mutant (Table S5), even with 10-fold more protein than used in the WT assay (Table 1).
4. Discussion
The results of this study demonstrate an essential role of mitochondrial acetate production in the T. brucei BSF energy metabolism and provide the first biochemical characterization of an ASCT family enzyme. The gene encoding T. brucei ASCT was identified in 2004 [20] followed by ACH [22], it has been shown to play an essential role in mitochondrial acetate production. The acetate production catalyzed by ASCT is coupled to the generation of ATP via the ASCT/SCS cycle [22]. Although the RNAiASCT.i mutant BSF lacks a growth phenotype (Fig. 2), it has been previously demonstrated in PCF that the knockdown of F1β subunit of Fo/F1-ATP synthase is lethal in Δasct background, indicating that TbASCT and ATP synthase are both responsible for ATP production in mitochondria. Since in BSF, the ATP synthase functions to maintain the electrochemical gradient by pumping protons using ATP (reverse reaction) [16,17], the TbASCT/SCS cycle can be the main or the only source of mitochondrial ATP.
Nevertheless, the growth inhibition observed in the Δach/RNAiASCT.i double mutant generated in this study demonstrated that mitochondrial acetate production is indeed essential not only in PCF [22], but also in BSF. It is noteworthy that the Δach/RNAiASCT cell line obtained in this study exhibited an important leakage (i.e., reduced TbASCT expression even in the absence of tetracycline) (Day 0 post-induction in Fig. 2D). TbASCT was rapidly re-expressed (Day 7 in Fig. 2D) with restoration of normal growth at 7 days post-induction. Re-expression of RNAi-targeted genes upon induction is commonly observed in the BSF as well as PCF when the targeted gene is essential for their growth [59,60]. The quite early reversion of growth, correlated with TbASCT re-expression supports an essential role for ASCT-ACH genes in the BSF. Thus, collectively, these data demonstrate that although TbASCT and TbACH catalyze distinct reactions, they share a common role in terms of acetate production to feed the de novo fatty acid biosynthesis [15,28], act redundantly, and are indispensable for growth of BSF trypanosomes as previously observed for the PCF [22].
Biochemical and structural biology studies of ASCT have thus far been hampered due to a difficulty to express and purify the recombinant enzyme because of its low expression level and solubility and high degree of instability [20,33,34]. Expression of recombinant TbASCT fused with a His6-SUMO tag at the N-terminus in this study is therefore critical for increasing the expression level as well as solubility of the enzyme [[61], [62], [63], [64]], and the addition of glycerol [18,64] in all purification steps is necessary to stabilize the enzymatic activity.
Previously, a method to assay ASCT using [1-14C]acetyl-CoA was reported [19,33,34]. However, such radioisotope (RI) methods require specialized facilities and the purification of released [1-14C]acetate using anion-exchange chromatography, which is not suitable for steady-state kinetic studies and high-throughput screening of inhibitors. Hence, a new assay method was developed based on real-time quantification of free CoA released by the in vitro reconstituted ASCT/SCS cycle. This assay is totally dependent on TbASCT substrates, which enables monitoring of the initial velocity, thus allowing us to perform the first detailed biochemical characterization of TbASCT. The affinity (K m) values determined for acetyl-CoA (0.046 mM) and succinate (13.8 mM) were similar to reported values determined using a RI method with partially purified ASCT from L. mexicana promastigotes (0.26 mM for acetyl-CoA and 6.9 mM for succinate) [19]. This result suggests that the trypanosomatid ASCT/SCS cycle functions under high concentrations of succinate, which is a major end-product of metabolism in the T. brucei PCF [[10], [11], [12], [13]] and BSF [14]. In T. brucei PCF, succinate is produced by glycosomal and mitochondrial NADH-fumarate reductases (NADH-FRDg and NADH-FRDm1, respectively), from which mitochondrial NADH-FRDm1 is responsible for up to 44% of total succinate excreted [65]. Although succinate production from glucose metabolism is reduced by 92% in the PCF RNAiFRDg/m1 double mutant [65], the cytosolic dihydroorotate dehydrogenase, which used fumarate as the electron acceptor, may also significantly contributes to succinate production [66,67]. Thus, succinate is expected to be present at high concentrations in T. brucei mitochondrion. Under optimal conditions, the ASCT/SCS cycle exhibits a turnover (k cat) of 191 s−1. Considering a stoichiometry of 1 mol of ATP produced per mole of CoA detected, the k cat of the TbASCT/SCS cycle is higher than the reported values for purified Fo/F1-ATP synthase from several organisms, including Bacillus PS3 (16 s−1) [68], E. coli (27 s−1) [69], and Saccharomyces cerevisiae (120 s−1) [70], clearly demonstrating that the ASCT/SCS cycle produces ATP with high efficiency. The Fo/F1-ATP synthase is a multi-subunit mega-complex enzyme, with the simplest form of E. coli composed of 8 subunits, α3β3γδϵ (Fo) and ab2c10 (F1) [71]. Compared to the reported ATP synthases, the ASCT/SCS cycle is encoded by a total of three genes (for ASCT α4 and SCS αβ subunits) and does not interact each other. The most prominent advantage of the ASCT/SCS cycle is the ability to synthesize ATP and acetate independently of the presence of oxygen, an electrochemical gradient, and OXPHOS, hence, this cycle fuels and boosts ATP and acetate production in the parasite's mitochondria. Such an observation is consistent with the fact that the ASCT/SCS cycle is conserved in the hydrogenosomes and mitochondria of many anaerobic protists and helminths [21], in which ATP synthase is not conserved (hydrogenosomes) or exhibits limited activity due to low environmental oxygen concentrations.
Despite the high amino acid identity between TbASCT and HsSCOT, our comprehensive biochemical analysis clearly demonstrates that TbASCT have low SCOT activity and that HsSCOT does not have ASCT activity, in accordance with the lack of experimental evidence of β-oxidation in T. brucei [72]. Moreover, we investigated the reaction mechanism of TbASCT and found that TbASCT employs a ping-pong bi-bi mechanism of catalysis, similar to other family IA CoA transferases, including SCOTs [21]. We also analyzed the substrate specificity in terms of CoA donor and found that TbASCT can utilize n-propionyl-CoA with an affinity higher than that for acetyl-CoA, with similar catalytic efficiency (k cat/K m): 2375 and 2267 s−1 mM−1, respectively. Interestingly, F. hepatica excretes acetate and propionate as end products of glucose catabolism [73], and it was demonstrated that F. hepatica ASCT utilizes both propionyl-CoA and acetyl-CoA as CoA donors [33]. It is important to note that T. brucei does not accumulate propionate [[10], [11], [12], [13], [14]]; consistent with these reports, n-propionyl-CoA was not detected in a previous metabolome analysis [74]. Taken together, our results indicate that acetyl-CoA is the physiological and the only CoA donor for TbASCT in vivo.
The catalytic glutamate residue located within the SENG motif was identified in SsSCOT by site-directed mutagenesis [75] and analysis of the crystal structure in complex with CoA (PDB: 3OXO) [58]. Although TbASCT exhibited extremely low SCOT activity (0.038 μmol/min/mg), the enzyme has 52% identity with SsSCOT and the SENG motif is conserved. In contrast to homodimeric mammalian SCOTs, the TbASCT forms a tight homotetramer as revealed by the crystal structure obtained in this study (Fig. S8) and by BN-PAGE followed by CBB and activity staining (Fig. 4D). The RMS deviation of monomers within TbASCT (Chain A) and SsSCOT (Chain E, PDB: 3OXO) was 1.50 Å between the two structures for 454 Cα atoms. In the crystal structure of SsSCOT, CoA was found covalently bound to E344, which corresponds to E319 of TbASCT. In addition, the carbonyl group of M423 located within the X-CoA tunnel from SsSCOT, which is not conserved in TbASCT and is replaced by P398, is found making hydrogen bond with the amine group of β-mercaptoethylamine of bound CoA. As the ASCT activity of the E319A and P398A mutants were completely abolished (Table. S4 and S5), we confirmed that E319 is the catalytic glutamate residue and P398 is located in the predicted X-CoA tunnel from TbASCT.
In the structure of SsSCOT bound to CoA, a second tunnel, adjacent to CoA binding site, is found with potential to bind the CoA acceptor (Fig. S6). In the second tunnel, several residues conserved in mammalian SCOTs (L327, L350, Y353), which are not conserved in trypanosomatid ASCTs (T302, M325, F328) are identified (Fig. S7). However, TbASCT mutants targeting T302, M325 and F328 showed no drastic changes in ASCT or SCOT activities (Table S4 and S5) and thus, we conclude that the second tunnel is not involved in catalysis. As a crystal structure of ligand free TbASCT was obtained (PDB: 6LP1), study on the binding site of the CoA acceptor by solving the crystal structure of TbASCT in complex with acetate or succinate is ongoing.
5. Conclusion
In conclusion, we demonstrated that mitochondrial acetate production is essential for growth of the BSF of trypanosomes. Previously, we have reported the remarkable effect of combination of ascofuranone and glycerol targeting TAO and glycerol kinase, respectively, for trypanocidal activity against BSF [76]. Considering the growth phenotype of Δach/RNAiASCT.i double mutant described in this study, a dual inhibitor targeting TbASCT and TbACH can be a suitable drug candidate to combat African trypanosomes due to their common function in terms of acetate production, although they catalyze distinct reaction. Furthermore, the absence of the ACH gene in the T. cruzi (American trypanosome) genome suggests that ASCT is the sole source of mitochondrial acetate and may be a potential drug target against Chagas disease. Because of the high degree of amino acid identity, the kinetic characterization of TbASCT reported here might facilitate the development of high-throughput screening systems and development of useful tools for structure-based design of ASCT-specific inhibitors targeting African and American trypanosomiasis.
Data availability
The Table S4 and S5 include mutant analyses data. The ligand-free structure of TbASCT with PDB accession code 6LP1 is described in METHODS. Source data for Fig. S6 and 7 can be found in Table S6.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank all of the staff members of beamline BL44XU (SPring-8) and BL-17A (KEK-PF) for assistance with X-ray diffraction data collection. The synchrotron radiation experiments were performed at BL-17A of the Photon Factory (proposal nos. 2013G055, 2013G107, 2015G510, and 2017G022) and at BL44XU of SPring-8 (proposal nos. 2016A6635, 2017A6722, 2017B6722, 2018A6820, and 2018B6820). The synchrotron beamline BL44XU at SPring-8 was used under the Cooperative Research Program of the Institute for Protein Research, Osaka University.
Funding source
This work was supported in part by Infectious Disease Control from the Science and Technology Research Partnership for Sustainable Development (SATREPS, nos. 10000284 to K.K. and 14425718 to D.K.I.); a Grant-in-aid for the Bilateral Joint Research Project (no. 16035611-000722 to K.K.); a Grant-in-aid for Scientific Research on Priority Areas 18073004 (to K.K.); a Creative Scientific Research Grant 18GS0314 (to K.K.) from the Japan Society for the Promotion of Science; a Grant-in-aid from the Program for the Promotion of Basic and Applied Research for Innovations in Bio-Oriented Industry (BRAIN) (no. 26020A to S.H. and K.K.); Grants-in-aid for Scientific Research (B) 16K19114 to D.K.I and (C) 23570131 and 26234567 to T.S.; and The Leading Initiative for Excellent Young Researchers (LEADER) 16811362 to D.K.I. and (B) 19H03436 to K.K. and D.K.I. and (C) 19K07523 to D.K.I. from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT). This work was also supported by the Japanese Initiative for Progress of Research on Infectious Diseases for Global Epidemics (JP18fm0208027 to D.K.I. and T.S.) from the Japan Agency for Medical Research and Development (AMED); a Grant-in-aid for research on emerging and re-emerging infectious diseases from the Japanese Ministry of Health and Welfare (to K.K.); and the Strategic Japanese-French Cooperative Program 11102218 (to K.K.). This work was also supported by the Centre National de la Recherche Scientifique (CNRS), the Université de Bordeaux, the Agence Nationale de la Recherche (ANR) through grants ACETOTRYP of the ANR-BLANC-2010 call and GLYCONOV of the “Générique” call and the Laboratoire d'Excellence (LabEx) ParaFrap ANR-11-LABX-0024 to F.B.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbabio.2020.148283.
Appendix A. Supplementary data
Supplementary material
References
- 1.Kita K., Hirawake H., Miyadera H., Amino H., Takeo S. Role of complex II in anaerobic respiration of the parasite mitochondria from Ascaris suum and Plasmodium falciparum. Biochim. Biophys. Acta Bioenerg. 2002;1553(1–2):123–139. doi: 10.1016/s0005-2728(01)00237-7. [DOI] [PubMed] [Google Scholar]
- 2.Harada S., Inaoka D.K., Ohmori J., Kita K. Diversity of parasite complex II. Biochim. Biophys. Acta Bioenerg. 2013;1827(5):658–667. doi: 10.1016/j.bbabio.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Buscher P., Cecchi G., Jamonneau V., Priotto G. Human African trypanosomiasis. Lancet. 2017;390(10110):2397–2409. doi: 10.1016/S0140-6736(17)31510-6. [DOI] [PubMed] [Google Scholar]
- 4.Baker C.H., Welburn S.C. The long wait for a new drug for human African Trypanosomiasis. Trends Parasitol. 2018;34(10):818–827. doi: 10.1016/j.pt.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Yaro M., Munyard K.A., Stear M.J., Groth D.M. Combatting African animal Trypanosomiasis (AAT) in livestock: the potential role of trypanotolerance. Vet. Parasitol. 2016;225:43–52. doi: 10.1016/j.vetpar.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Shaw A.P. Assessing the economics of animal trypanosomosis in Africa--history and current perspectives. Onderstepoort J Vet Res. 2009;76(1):27–32. doi: 10.4102/ojvr.v76i1.57. [DOI] [PubMed] [Google Scholar]
- 7.Nakayima J., Nakao R., Alhassan A., Mahama C., Afakye K., Sugimoto C. Molecular epidemiological studies on animal trypanosomiases in Ghana. Parasit. Vectors. 2012;5:217. doi: 10.1186/1756-3305-5-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aksoy S., Buscher P., Lehane M., Solano P., Van Den Abbeele J. Human African trypanosomiasis control: achievements and challenges. PLoS Negl. Trop. Dis. 2017;11(4) doi: 10.1371/journal.pntd.0005454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S.H., Stephens J.L., Englund P.T. A fatty-acid synthesis mechanism specialized for parasitism. Nat Rev Microbiol. 2007;5(4):287–297. doi: 10.1038/nrmicro1617. [DOI] [PubMed] [Google Scholar]
- 10.Millerioux Y., Ebikeme C., Biran M., Morand P., Bouyssou G., Vincent I.M., Mazet M., Riviere L., Franconi J.M., Burchmore R.J., Moreau P., Barrett M.P., Bringaud F. The threonine degradation pathway of the Trypanosoma brucei procyclic form: the main carbon source for lipid biosynthesis is under metabolic control. Mol. Microbiol. 2013;90(1):114–129. doi: 10.1111/mmi.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Weelden S.W., Fast B., Vogt A., van der Meer P., Saas J., van Hellemond J.J., Tielens A.G., Boshart M. Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J. Biol. Chem. 2003;278(15):12854–12863. doi: 10.1074/jbc.M213190200. [DOI] [PubMed] [Google Scholar]
- 12.Coustou V., Biran M., Breton M., Guegan F., Riviere L., Plazolles N., Nolan D., Barrett M.P., Franconi J.M., Bringaud F. Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J. Biol. Chem. 2008;283(24):16342–16354. doi: 10.1074/jbc.M709592200. [DOI] [PubMed] [Google Scholar]
- 13.Smith T.K., Bringaud F., Nolan D.P., Figueiredo L.M. Metabolic reprogramming during the Trypanosoma brucei life cycle. F1000Res. 2017;6 doi: 10.12688/f1000research.10342.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Grinsven K.W., Van Den Abbeele J., Van den Bossche P., van Hellemond J.J., Tielens A.G. Adaptations in the glucose metabolism of procyclic Trypanosoma brucei isolates from tsetse flies and during differentiation of bloodstream forms. Eukaryot. Cell. 2009;8(8):1307–1311. doi: 10.1128/EC.00091-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazet M., Morand P., Biran M., Bouyssou G., Courtois P., Daulouede S., Millerioux Y., Franconi J.M., Vincendeau P., Moreau P., Bringaud F. Revisiting the central metabolism of the bloodstream forms of Trypanosoma brucei: production of acetate in the mitochondrion is essential for parasite viability. PLoS Negl. Trop. Dis. 2013;7(12) doi: 10.1371/journal.pntd.0002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnaufer A., Clark-Walker G.D., Steinberg A.G., Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24(23):4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown S.V., Hosking P., Li J., Williams N. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot. Cell. 2006;5(1):45–53. doi: 10.1128/EC.5.1.45-53.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiba T., Kido Y., Sakamoto K., Inaoka D.K., Tsuge C., Tatsumi R., Takahashi G., Balogun E.O., Nara T., Aoki T., Honma T., Tanaka A., Inoue M., Matsuoka S., Saimoto H., Moore A.L., Harada S., Kita K. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc. Natl. Acad. Sci. U. S. A. 2013;110(12):4580–4585. doi: 10.1073/pnas.1218386110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Hellemond J.J., Opperdoes F.R., Tielens A.G. Trypanosomatidae produce acetate via a mitochondrial acetate:succinate CoA transferase. Proc. Natl. Acad. Sci. U. S. A. 1998;95(6):3036–3041. doi: 10.1073/pnas.95.6.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riviere L., van Weelden S.W., Glass P., Vegh P., Coustou V., Biran M., van Hellemond J.J., Bringaud F., Tielens A.G., Boshart M. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J Biol Chem. 2004;279(44):45337–45346. doi: 10.1074/jbc.M407513200. [DOI] [PubMed] [Google Scholar]
- 21.Tielens A.G., van Grinsven K.W., Henze K., van Hellemond J.J., Martin W. Acetate formation in the energy metabolism of parasitic helminths and protists. Int. J. Parasitol. 2010;40(4):387–397. doi: 10.1016/j.ijpara.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Millerioux Y., Morand P., Biran M., Mazet M., Moreau P., Wargnies M., Ebikeme C., Deramchia K., Gales L., Portais J.C., Boshart M., Franconi J.M., Bringaud F. ATP synthesis-coupled and -uncoupled acetate production from acetyl-CoA by mitochondrial acetate:succinate CoA-transferase and acetyl-CoA thioesterase in Trypanosoma. J. Biol. Chem. 2012;287(21):17186–17197. doi: 10.1074/jbc.M112.355404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochud-Allemann N., Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 2002;277(36):32849–32854. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins T.M., Eisenthal R., Weitzman P.D. Two distinct succinate thiokinases in both bloodstream and procyclic forms of Trypanosoma brucei. Biochem. Biophys. Res. Commun. 1988;151(1):257–261. doi: 10.1016/0006-291x(88)90587-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Cui J., Nilsson D., Gunasekera K., Chanfon A., Song X., Wang H., Xu Y., Ochsenreiter T. The Trypanosoma brucei MitoCarta and its regulation and splicing pattern during development. Nucleic Acids Res. 2010;38(21):7378–7387. doi: 10.1093/nar/gkq618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butter F., Bucerius F., Michel M., Cicova Z., Mann M., Janzen C.J. Comparative proteomics of two life cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite’s host adaptation machinery. Mol. Cell. Proteomics. 2013;12(1):172–179. doi: 10.1074/mcp.M112.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dejung M., Subota I., Bucerius F., Dindar G., Freiwald A., Engstler M., Boshart M., Butter F., Janzen C.J. Quantitative proteomics uncovers novel factors involved in developmental differentiation of Trypanosoma brucei. PLoS Pathog. 2016;12(2) doi: 10.1371/journal.ppat.1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riviere L., Moreau P., Allmann S., Hahn M., Biran M., Plazolles N., Franconi J.M., Boshart M., Bringaud F. Acetate produced in the mitochondrion is the essential precursor for lipid biosynthesis in procyclic trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 2009;106(31):12694–12699. doi: 10.1073/pnas.0903355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller M., Mentel M., van Hellemond J.J., Henze K., Woehle C., Gould S.B., Yu R.Y., van der Giezen M., Tielens A.G., Martin W.F. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 2012;76(2):444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omura S., Miyadera H., Ui H., Shiomi K., Yamaguchi Y., Masuma R., Nagamitsu T., Takano D., Sunazuka T., Harder A., Kolbl H., Namikoshi M., Miyoshi H., Sakamoto K., Kita K. An anthelmintic compound, nafuredin, shows selective inhibition of complex I in helminth mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2001;98(1):60–62. doi: 10.1073/pnas.011524698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumoto J., Sakamoto K., Shinjyo N., Kido Y., Yamamoto N., Yagi K., Miyoshi H., Nonaka N., Katakura K., Kita K., Oku Y. Anaerobic NADH-fumarate reductase system is predominant in the respiratory chain of Echinococcus multilocularis, providing a novel target for the chemotherapy of alveolar echinococcosis. Antimicrob. Agents Chemother. 2008;52(1):164–170. doi: 10.1128/AAC.00378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inaoka D.K., Shiba T., Sato D., Balogun E.O., Sasaki T., Nagahama M., Oda M., Matsuoka S., Ohmori J., Honma T., Inoue M., Kita K., Harada S. Structural insights into the molecular design of flutolanil derivatives targeted for fumarate respiration of parasite mitochondria. Int. J. Mol. Sci. 2015;16(7):15287–15308. doi: 10.3390/ijms160715287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Grinsven K.W., van Hellemond J.J., Tielens A.G. Acetate:succinate CoA-transferase in the anaerobic mitochondria of Fasciola hepatica. Mol. Biochem. Parasitol. 2009;164(1):74–79. doi: 10.1016/j.molbiopara.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 34.van Grinsven K.W., Rosnowsky S., van Weelden S.W., Putz S., van der Giezen M., Martin W., van Hellemond J.J., Tielens A.G., Henze K. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J. Biol. Chem. 2008;283(3):1411–1418. doi: 10.1074/jbc.M702528200. [DOI] [PubMed] [Google Scholar]
- 35.Kayer M.A. Disorders of ketone production and utilization. Mol. Genet. Metab. 2006;87(4):281–283. doi: 10.1016/j.ymgme.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Fukao T., Lopaschuk G.D., Mitchell G.A. Pathways and control of ketone body metabolism: on the fringe of lipid biochemistry. Prostaglandins Leukot Essent Fatty Acids. 2004;70(3):243–251. doi: 10.1016/j.plefa.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Stols L., Zhou M., Eschenfeldt W.H., Millard C.S., Abdullah J., Collart F.R., Kim Y., Donnelly M.I. New vectors for co-expression of proteins: structure of Bacillus subtilis ScoAB obtained by high-throughput protocols. Protein Expr. Purif. 2007;53(2):396–403. doi: 10.1016/j.pep.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Wirtz E., Leal S., Ochatt C., Cross G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99(1):89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 39.Burkard G., Fragoso C.M., Roditi I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;153(2):220–223. doi: 10.1016/j.molbiopara.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Harlow E.L.D. Cold Spring Harbor Laboratory Press; New York: 1988. Antibodies: A Laboratory Manual. [Google Scholar]
- 41.Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 42.Denise H., Giroud C., Barrett M.P., Baltz T. Affinity chromatography using trypanocidal arsenical drugs identifies a specific interaction between glycerol-3-phosphate dehydrogenase from Trypanosoma brucei and Cymelarsan. Eur. J. Biochem. 1999;259(1–2):339–346. doi: 10.1046/j.1432-1327.1999.00048.x. [DOI] [PubMed] [Google Scholar]
- 43.Miller E.N., Turner M.J. Analysis of antigenic types appearing in first relapse populations of clones of Trypanosoma brucei. Parasitology. 1981;82(1):63–80. doi: 10.1017/s0031182000041871. [DOI] [PubMed] [Google Scholar]
- 44.Zhou M.Y., Gomez-Sanchez C.E. Universal TA cloning. Curr Issues Mol Biol. 2000;2(1):1–7. [PubMed] [Google Scholar]
- 45.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Rothe G.M. Series; Springer Lab manual Springer, Berlin: 1994. Electrophoresis of Enzymes: Laboratory Methods. [Google Scholar]
- 47.Manchenko G.P. CRC press; Boca Raton: 2002. Handbook of Detection of Enzymes on Electrophoretic Gels. [Google Scholar]
- 48.Watanabe T., Kitabatake N., Doi E. Method for the accurate measurement of freezing-induced denaturation of ovalbumin with 5,5′-dithiobis-(2-nitrobenzoic acid) Biosci. Biotechnol. Biochem. 1994;58:359–362. [Google Scholar]
- 49.Howard J.B., Zieske L., Clarkson J., Rathe L. Mechanism-based fragmentation of coenzyme A transferase. Comparison of alpha 2-macroglobulin and coenzyme A transferase thiol ester reactions. J Biol Chem. 1986;261(1):60–65. [PubMed] [Google Scholar]
- 50.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A. Vagin, A. Teplyakov, MOLREP: an Automated Program for Molecular Replacement, J Appl Cryst (30) (1997) 1022–1025.
- 52.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Gunasekera K., Wuthrich D., Braga-Lagache S., Heller M., Ochsenreiter T. Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics. 2012;13:556. doi: 10.1186/1471-2164-13-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunger-Glaser I., Linder M., Seebeck T. Histidine-phosphorylation of succinyl CoA synthetase from Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;100(1):43–52. doi: 10.1016/s0166-6851(99)00025-0. [DOI] [PubMed] [Google Scholar]
- 56.Segel I.H. John Wiley & Sons; New York: 1975. Enzyme Kinetics. [Google Scholar]
- 57.Wentz A.E., d’Avignon D.A., Weber M.L., Cotter D.G., Doherty J.M., Kerns R., Nagarajan R., Reddy N., Sambandam N., Crawford P.A. Adaptation of myocardial substrate metabolism to a ketogenic nutrient environment. J. Biol. Chem. 2010;285(32):24447–24456. doi: 10.1074/jbc.M110.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fraser M.E., Hayakawa K., Brown W.D. Catalytic role of the conformational change in succinyl-CoA:3-oxoacid CoA transferase on binding CoA. Biochemistry. 2010;49(48):10319–10328. doi: 10.1021/bi100659s. [DOI] [PubMed] [Google Scholar]
- 59.Tyc J., Klingbeil M.M., Lukes J. Mitochondrial heat shock protein machinery hsp70/hsp40 is indispensable for proper mitochondrial DNA maintenance and replication. mBio. 2015;6(1) doi: 10.1128/mBio.02425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klein C., Terrao M., Clayton C. The role of the zinc finger protein ZC3H32 in bloodstream-form Trypanosoma brucei. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malakhov M.P., Mattern M.R., Malakhova O.A., Drinker M., Weeks S.D., Butt T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genom. 2004;5(1–2):75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 62.Zuo X., Mattern M.R., Tan R., Li S., Hall J., Sterner D.E., Shoo J., Tran H., Lim P., Sarafianos S.G., Kazi L., Navas-Martin S., Weiss S.R., Butt T.R. Expression and purification of SARS coronavirus proteins using SUMO-fusions. Protein Expr. Purif. 2005;42(1):100–110. doi: 10.1016/j.pep.2005.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marblestone J.G., Edavettal S.C., Lim Y., Lim P., Zuo X., Butt T.R. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15(1):182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X., Inaoka D.K., Shiba T., Balogun E.O., Allmann S., Watanabe Y.I., Boshart M., Kita K., Harada S. Expression, purification, and crystallization of type 1 isocitrate dehydrogenase from Trypanosoma brucei brucei. Protein Expr. Purif. 2017;138:56–62. doi: 10.1016/j.pep.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 65.Coustou V., Besteiro S., Riviere L., Biran M., Biteau N., Franconi J.M., Boshart M., Baltz T., Bringaud F. A mitochondrial NADH-dependent fumarate reductase involved in the production of succinate excreted by procyclic Trypanosoma brucei. J. Biol. Chem. 2005;280(17):16559–16570. doi: 10.1074/jbc.M500343200. [DOI] [PubMed] [Google Scholar]
- 66.Takashima E., Inaoka D.K., Osanai A., Nara T., Odaka M., Aoki T., Inaka K., Harada S., Kita K. Characterization of the dihydroorotate dehydrogenase as a soluble fumarate reductase in Trypanosoma cruzi. Mol. Biochem. Parasitol. 2002;122(2):189–200. doi: 10.1016/s0166-6851(02)00100-7. [DOI] [PubMed] [Google Scholar]
- 67.Arakaki T.L., Buckner F.S., Gillespie J.R., Malmquist N.A., Phillips M.A., Kalyuzhniy O., Luft J.R., Detitta G.T., Verlinde C.L., Van Voorhis W.C., Hol W.G., Merritt E.A. Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies. Mol. Microbiol. 2008;68(1):37–50. doi: 10.1111/j.1365-2958.2008.06131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soga N., Kinosita K., Jr., Yoshida M., Suzuki T. Efficient ATP synthesis by thermophilic Bacillus FoF1-ATP synthase. FEBS J. 2011;278(15):2647–2654. doi: 10.1111/j.1742-4658.2011.08191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fischer S., Etzold C., Turina P., Deckers-Hebestreit G., Altendorf K., Graber P. ATP synthesis catalyzed by the ATP synthase of Escherichia coli reconstituted into liposomes. Eur. J. Biochem. 1994;225(1):167–172. doi: 10.1111/j.1432-1033.1994.00167.x. [DOI] [PubMed] [Google Scholar]
- 70.Forster K., Turina P., Drepper F., Haehnel W., Fischer S., Graber P., Petersen J. Proton transport coupled ATP synthesis by the purified yeast H+ -ATP synthase in proteoliposomes. Biochim. Biophys. Acta Bioenerg. 2010;1797(11):1828–1837. doi: 10.1016/j.bbabio.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 71.Okuno D., Iino R., Noji H. Rotation and structure of FoF1-ATP synthase. J. Biochem. 2011;149(6):655–664. doi: 10.1093/jb/mvr049. [DOI] [PubMed] [Google Scholar]
- 72.Allmann S., Mazet M., Ziebart N., Bouyssou G., Fouillen L., Dupuy J.W., Bonneu M., Moreau P., Bringaud F., Boshart M. Triacylglycerol storage in lipid droplets in procyclic Trypanosoma brucei. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0114628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mansour T.E. Studies on the carbohydrate metabolism of the liver fluke Fasciola hepatica. Biochim. Biophys. Acta. 1959;34:456–464. doi: 10.1016/0006-3002(59)90298-7. [DOI] [PubMed] [Google Scholar]
- 74.Creek D.J., Mazet M., Achcar F., Anderson J., Kim D.H., Kamour R., Morand P., Millerioux Y., Biran M., Kerkhoven E.J., Chokkathukalam A., Weidt S.K., Burgess K.E., Breitling R., Watson D.G., Bringaud F., Barrett M.P. Probing the metabolic network in bloodstream-form Trypanosoma brucei using untargeted metabolomics with stable isotope labelled glucose. PLoS Pathog. 2015;11(3) doi: 10.1371/journal.ppat.1004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rochet J.C., Bridger W.A. Identification of glutamate 344 as the catalytic residue in the active site of pig heart CoA transferase. Protein Sci. 1994;3(6):975–981. doi: 10.1002/pro.5560030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balogun E.O., Inaoka D.K., Shiba T., Tsuge C., May B., Sato T., Kido Y., Nara T., Aoki T., Honma T., Tanaka A., Inoue M., Matsuoka S., Michels P.A.M., Watanabe Y.I., Moore A.L., Harada S., Kita K. Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase of Trypanosoma brucei brucei. FASEB J. 2019;33(11):13002–13013. doi: 10.1096/fj.201901342R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The Table S4 and S5 include mutant analyses data. The ligand-free structure of TbASCT with PDB accession code 6LP1 is described in METHODS. Source data for Fig. S6 and 7 can be found in Table S6.