SUMMARY
RNA polymerase II (RNA Pol II) is generally paused at promoter-proximal regions in most metazoans, and based on in vitro studies, this function has been attributed to the negative elongation factor (NELF). Here, we show that upon rapid depletion of NELF, RNA Pol II fails to be released into gene bodies, stopping instead around the +1 nucleosomal dyad-associated region. The transition to the 2nd pause region is independent of positive transcription elongation factor P-TEFb. During the heat shock response, RNA Pol II is rapidly released from pausing at heat shock-induced genes, while most genes are paused and transcriptionally downregulated. Both of these aspects of the heat shock response remain intact upon NELF loss. We find that NELF depletion results in global loss of cap-binding complex from chromatin without global reduction of nascent transcript 5’ cap stability. Thus, our studies implicate NELF functioning in early elongation complexes distinct from RNA Pol II pause-release.
Graphical Abstract

INTRODUCTION
In most metazoans, most genes exhibit RNA polymerase II (Pol II) pausing at promoter-proximal regions. Release of the paused Pol II into productive elongation state is thought to be a key regulatory step in response to environmental and developmental signals, while misregulation of Pol II pause-release is associated with oncogenesis such as pediatric leukemia and MYC-driven solid tumors (Chen et al., 2018). Using in vitro reconstitution assays of transcription and in vivo screens, many studies have identified protein complexes required for Pol II pausing, including negative elongation factor (NELF) (Wada et al., 1998), DRB sensitivity-inducible factor (DSIF) (Yamaguchi et al., 1999), Pol II-associated factor 1 (PAF1) (Chen et al., 2015a; 2017), and even the nucleosome: the basic structural unit of chromatin. Nucleosomes can serve as a molecular barrier for Pol II transcription (Kujirai et al., 2018; Lai and Pugh, 2017; Weber et al., 2014). Positive elongation factor b (P-TEFb), which is a complex of cyclin-dependent kinase 9 (CDK9) and Cyclin T, is required for release of Pol II from pausing (Marshall and Price, 1995). P-TEFb is catalytically activated by the super elongation complex (SEC) (Lin et al., 2010) or BRD4 (Jang et al., 2005; Yang et al., 2005) but inactivated by the HEXIM-7SK snRNP complex (Nguyen et al., 2001; Yang et al., 2001). How these factors and the nucleosome work together or separately to regulate promoter-proximal pausing of Pol II remains poorly understood.
5’ capping of mRNA-
Despite the pervasiveness of promoter-proximal pausing in metazoans and the abundant evidence that this is a highly regulated process, why this regulatory step is needed is still debated. Although having polymerase poised at the promoter waiting for an activation signal was thought to facilitate rapid transcriptional response (Nechaev and Adelman, 2011), rapid transcription can occur in the absence of promoter-proximal pausing (Lin et al., 2011). Pausing has also been proposed to facilitate synchronous gene activation in response to developmental cues (Boettiger and Levine, 2009; Lin et al., 2011). Another idea is that promoter-proximal pausing acts as a checkpoint for co-transcriptional processes such as to ensure mRNA capping (Sims et al., 2004). Eukaryotic mRNA contains a conserved 5’ m7G cap modification, thereby ensuring mRNA integrity partially through prevention of RNA degradation from its 5’-end (Ramanathan et al., 2016). Capping takes place co-transcriptionally upon ~20 nt RNA synthesis by Pol II in vitro and in vivo, indicating that 5’-end of nascent RNA can be capped when it emerges from the RNA exit tunnel of Pol II (Mandal et al., 2004; Martinez-Rucobo et al., 2015; Rasmussen and Lis, 1993). Pol II is paused at promoter-proximal regions after transcribing 20–120 nt RNA (Chen et al., 2018). These studies have suggested that the majority of nascent RNA that is associated with paused Pol II is capped. Decapping enzymes can be localized to promoters along with the 5’-3’ RNA exonuclease XRN2, which has been proposed to function as a torpedo for promoter-proximal termination (Brannan et al., 2012), suggesting that the 5’ cap modification can be reversible at promoters. The 5’ cap is further protected by the cap-binding complex (CBC) (Ramanathan et al., 2016). Interestingly, NELF has been shown to directly bind to the CBC and to regulate 3’-end processing for histone mRNAs (Narita et al., 2007; Schulze and Cusack, 2017). However, a connection between the CBC and promoter-proximal pausing has not been investigated.
Mechanism of pausing regulation by NELF in vitro and in vivo-
An inhibitory function of NELF for Pol II transcription has been extensively studied in vitro using reconstituted transcription systems. NELF was initially isolated from human nuclear extract as a complex that prevents Pol II transcription on a naked DNA template in vitro (Yamaguchi et al., 1999). Four distinct subunits NELF-A, NELF-B, NELF-C/D (alternative translation isoforms of the NELFCD gene), and NELF-E form the NELF complex, which is conserved among most metazoans, including human, mouse, and fly (Narita et al., 2003; Vos et al., 2016; Wu et al., 2005). The cryo-electron microscopy structure of Pol II with NELF and DSIF has further suggested how NELF “tentacles” could restrain Pol II mobility (Vos et al., 2018b).
NELF is proposed to be required for the establishment of pausing by Pol II for key biological events, including the heat shock response (Wu et al., 2003), estrogen signaling (Aiyar et al., 2004), IL-6 signaling (Aida et al., 2006), the inflammatory response (Adelman et al., 2009), FGF/ERK signaling (Williams et al., 2015), cell growth (Sun et al., 2011; Williams et al., 2015), and early embryonic development (Amleh et al., 2009; Williams et al., 2015), thus highlighting the physiological significance of NELF. However, reconciling in vitro studies of NELF with in vivo studies has been challenging. Unlike an essential role of NELF for pausing in vitro, NELF-depleted cells exhibited limited reduction of paused Pol II at promoters, with few genes upregulated and many genes downregulated at the mRNA level, and reduced Pol II occupancy in the gene body of many genes (Chen et al., 2015b; Core et al., 2012; Gilchrist et al., 2008; 2010; Muse et al., 2007; Rahl et al., 2010; Stadelmayer et al., 2014; Sun and Li, 2010; Sun et al., 2011; Williams et al., 2015; Yamamoto et al., 2014). These results have revealed a significant gap between our understanding of in vitro and in vivo pausing.
The recent emergence of rapid degradation strategies holds great promise for dissecting cellular mechanisms. Therefore, to examine NELF functions on Pol II pausing in human cells, we have established rapid NELF depletion in cells using the auxin-inducible degron (Natsume et al., 2016). We find that promoter-proximal pausing is largely intact in the absence of NELF, leading us to propose that NELF functions at a promoter-proximal step distinct from regulating the release of Pol II from pausing.
RESULTS
Establishment of a cellular system for acute depletion of NELF-
We employed the auxin-inducible degron (AID) system (Natsume et al., 2016) to rapidly deplete cells of NELF. The endogenous NELFC gene was tagged with the mini-AID cassette at its C-terminus in human DLD-1 cells that constitutively express the AID adapter protein OsTIR1 (Figure 1A, Methods). ~90% of NELF-C-AID protein disappeared within 1 h of auxin treatment in NELF-C-AID cells (Figure 1A). NELF-C protein was stable in auxin-treated parental cells (Figure 1A), confirming that NELF-C-AID is degraded in an AID-tag dependent manner. Notably, the NELF-E subunit was stable for up to 4 h of auxin treatment in NELF-C-AID cells, but it began to disappear at later timepoints (Figure 1A). This interdependences of NELF subunits for stability are consistent with previous studies (Narita et al., 2007; Sun et al., 2008; Williams et al., 2015). Long-term NELF-C-AID loss led to a severe growth defect (Figure 1B), which could lead to indirect effects on transcription. Therefore, we chose ≤ 4 h of auxin treatment for further experiments to ensure loss of NELF while minimizing indirect effects of NELF loss on Pol II transcription.
Figure 1. Acute NELF-C-AID degradation leads to loss of NELF complex on chromatin.

A. Schematic of auxin-inducible degradation of NELF-C-AID (upper panel). Western blots of whole cell extracts from NELF-C-AID or parental DLD-1 cells treated with auxin for the indicated times (lower panels). NELF-C is greatly depleted by 1 h. NELF-E and the DSIF subunit SPT5 are shown for comparison. Tubulin serves as a loading control.
B. Growth curves of untreated or auxin-treated NELF-C-AID cells (bottom panel). Relative cell number was measured by crystal violet staining. Mean ± SD, n = 3. Representative cell staining at 96 h is shown (top panel).
C. Genome browser track example of NELF-C, NELF-E, and SPT5 ChIP-seq in NELF-C-AID cells treated with or without auxin for 2 h. NELF-C loss leads to a concomitant loss of NELF-E on chromatin, while SPT5 occupancy is only partially reduced upon NELF-C loss.
D. ChIP-seq metaplot shows NELF-C, NELF-E, and SPT5 average occupancies in untreated or 2 h auxin-treated NELF-C-AID cells, centered on Pol II pause regions. N = 6,531 genes with PRO-seq signal in the control condition (see methods).
Loss of NELF complex on chromatin upon NELF-C-AID depletion-
We carried out ChIP-seq experiments to test whether NELF-C-AID degradation reduces NELF-C-AID protein on chromatin. In the absence of auxin, ChIP-seq for NELF-C, NELF-E, and a DSIF subunit SPT5, showed that their peaks were colocalized at promoter regions (Figures 1C and S1A). This result indicates that the AID-tagged NELF-C protein is properly incorporated into the NELF-DSIF-Pol II pausing complex at promoters. When NELF-C-AID cells were treated with auxin for 2 h, NELF-C ChIP-seq peaks disappeared (Figure 1C). Interestingly, NELF-E ChIP-seq peaks simultaneously disappeared upon NELF-C-AID degradation (Figure 1C), despite NELF-E still being present in cells at this timepoint (Figure 1A). These results indicate that depletion of NELF-C-AID results in the loss of the NELF complex on chromatin. On the other hand, SPT5 ChIP-seq peaks largely remained upon NELF loss (Figure 1C), consistent with the NELF-independent binding of SPT5 to Pol II seen in vitro (Missra and Gilmour, 2010; Yamaguchi et al., 2002). Genome-wide analysis of ChIP-seq data showed global reduction of NELF-C-AID and NELF-E occupancy at Pol II pause regions upon auxin treatment, while SPT5 largely remained (Figure 1D). Together, our data indicate that rapid NELF-C-AID degradation leads to loss of the NELF complex from chromatin.
NELF loss reveals a 2-step Pol II pausing at promoters-
To precisely explore functions of NELF in Pol II transcription, we determined the genome-wide distribution of active Pol II with precision nuclear run-on sequencing (PRO-seq) (Kwak et al., 2013). PRO-seq track examples show that transcriptionally-active Pol II is paused at the promoter regions in the untreated condition (Figure 2A). Interestingly, the distribution of Pol II shows a modest shift downstream at 1 h and 4 h auxin treatment in NELF-C-AID cells (Figure 2A). To analyze this Pol II shift genome-wide, we defined the original pause region at each active gene: the nucleotide with maximum PRO-seq signal at the promoter-proximal region in untreated NELF-C-AID cells was defined as the location of the “1st pause region”, while the maximum in NELF-C-AID cells treated with auxin for 1h was defined as the location of the “2nd pause region” (see Methods). Metaplot analysis of PRO-seq signal centered on 1st pause regions showed that PRO-seq signal at 1st pause regions was decreased upon NELF loss, whereas increased PRO-seq peak corresponding to the 2nd pause region occurred at around 50 bp downstream of the 1st pause region upon NELF loss (Figure 2B). The histogram showed that the median distance between the 1st and 2nd pause regions is 49 bp and the majority (81.7%) of transcribing genes have a distance of no longer than 150 bp (Figure 2C). These results suggest that Pol II undergoes a “2-step” pausing and NELF loss results in transitioning of paused Pol II to 2nd pause regions.
Figure 2. NELF loss reveals a 2-step Pol II pausing at promoters.

A. Representative tracks of PRO-seq signal in NELF-C-AID cells treated with auxin for 0, 1, 4, 24 h at promoter regions of the SUV29H2, RAB8B, and FLCN genes. Scale bars, 50 bp.
B. Metaplot analysis shows PRO-seq mean coverage at pause regions in auxin-treated NELF-C-AID cells. N = 6,531.
C. The histogram shows base-pair distance between 1st and 2nd pause regions. Median is 49 bp change in pausing region. N = 6,531.
D. Heatmap analysis of PRO-seq and NELF-C ChIP-seq in NELF-C-AID cells treated with auxin as in A. PRO-seq signal is shown as log2 fold change relative to 0 h. NELF-C ChIP-seq signal at 0 h is shown. Gene regions (pause −5kb to pause +50 kb) are sorted by gene length with dashed lines indicating transcription end sites (top panel). Promoter regions (pause ± 0.5 kb) are sorted by NELF-C signal (bottom panel). N = 6,531.
E. Boxplots showing PRO-seq coverage (RPKM) at 1st pause region ± 5 bp, 2nd pause regions ± 5 bp, and gene bodies (1st pause region + 500 bp to + 100 kb or to transcription end sites). Genes with a low expression level (see Methods) are filtered out. N = 6,236. Whiskers, 1.5 times the interquartile range; boxes, the 25th and 75th percentiles. *P < 2.2 × 10−16 [Mann-Whitney U test]
To test whether NELF loss leads to release of paused Pol II to productive elongation, we generated heatmaps of PRO-seq log2 fold change at gene bodies. NELF-C ChIP-seq peaks overlapped 1st pause regions (Figure 2D), confirming that the NELF-Pol II pausing complex occupies 1st pause regions. Upon NELF loss at 1h and 4h auxin treatment, PRO-seq signal in gene bodies was uniformly and significantly decreased (Figures 2D and 2E). PRO-seq signal was significantly decreased at 1st pausing regions, and significantly increased at 2nd pause regions upon NELF loss (Figures 2D and 2E). These data suggest that the paused Pol II at 2nd pause regions is not able to enter productive elongation despite NELF loss.
Given this observation, we also examined the effects of long-term depletion of NELF. At 24 h auxin treatment, PRO-seq signal was significantly decreased at promoters and gene bodies. Given the discrepancy between the results with NELF loss within 1 to 4 hrs and 24 hrs, we propose that the global loss of PRO-seq signal throughout active genes after 24 hrs of NELF loss may be due to indirect effects of NELF loss, since we observed a severe growth defect at this timepoint. The effect of 24 hour NELF depletion with the AID system closely resembles what has been seen in studies depleting NELF with shRNA treatment over several days (Sun and Li, 2010).
NELF-E-AID depletion also showed a 2-step pausing-
To test the effects of depleting another NELF subunit, we generated NELF-E-AID cells. Western blotting showed that ≥ 80% of NELF-E-AID proteins were degraded within 1 h of auxin treatment (Figure S1B). ChIP-seq for NELF-C, NELF-E, and SPT5 in NELF-E-AID cells showed that NELF-E-AID degradation resulted in loss of NELF-C and NELF-E peaks, while SPT5 peaks were not reduced as much as NELF subunits (Figures S1C and S1D). Metaplot analysis of PRO-seq showed accumulation of PRO-seq signal around 50 bp downstream of 1st pause regions (Figure S2A). These results suggest that NELF-E-AID degradation leads to loss of the NELF complex on chromatin, and leads to Pol II accumulation at 2nd pause regions, consistent with results in NELF-C-AID cells. Importantly, parental DLD-1 cells did not display PRO-seq signal accumulation downstream of 1st pause regions upon auxin treatment (Figure S2B), excluding the possibility that this effect is due to an off-target effect of auxin treatment causing Pol II to accumulate at 2nd pause regions.
The 2nd pause regions seem to be associated with the +1 nucleosome-
We next examined whether the 2nd pause regions are associated with nucleosomes. First, we ordered heatmaps of PRO-seq signal by the distance to the 2nd pause region, and then aligned them to heatmaps of H3K4me3 ChIP-seq and MNase-seq signal (Figure 3A). MNase-seq heatmap centered on 1st pause regions showed nucleosome phasing, consistent with a recent study (Tome et al., 2018). Two patterns of nucleosome phasing were associated with distinct polymerase profiles: “Proximal 2nd pause” genes with ≤ 150 bp of distance to the 2nd pause region and “distal 2nd pause” genes with > 150 bp to the 2nd pause region. Proximal 2nd pause genes showed good +1, +2, and +3 nucleosome phasing, while distal 2nd pause genes showed relatively poor nucleosome phasing (Figures 3A and 3B).
Figure 3. The 2nd pause regions are associated with the +1 nucleosome.

A. Heatmaps of PRO-seq signal in auxin-treated NELF-C-AID cells shown alongside H3K4me3 ChIP-seq and MNase-seq signal in DLD-1 cells. Heatmaps are centered on promoter-proximal pause regions and are sorted by the distance to the 2nd pause region (pause ± 500 bp is shown). Log2 fold change of PRO-seq to 0 h is also shown. MNase-seq data from (Yamashita et al., 2011), accession number DRX000003. N = 6,531 (5,338 genes were classified as having a proximal 2nd pause region and 1,193 genes were classified as having a distal 2nd pause region).
B. Metaplot showing MNase-seq mean signal centered on the 1st pause region. N = 5,338 genes with a proximal 2nd pause region.
C. Metaplot of PRO-seq mean signal in auxin-treated NELF-C-AID cells, centered at the +1 nucleosomal dyad. The light green shading indicates the position of +1 nucleosome (+ 1 dyad ± 75 bp) as determined by MNase-seq. N = 1,843 genes that have a positioned +1 nucleosome among genes with a proximal 2nd pause (see methods).
We next examined how 1st pause regions and 2nd pause regions are distributed around the +1 nucleosome. We re-aligned the PRO-seq signal of proximal 2nd pause genes centered on the 1st nucleosomal dyad (Figure 3C, see methods). The peaks of 1st pause regions were observed around 60 bp upstream of the +1 dyad (Figure 3C), indicating that the Pol II-NELF complex is paused at the entrance of +1 nucleosome. Interestingly, upon NELF loss, PRO-seq peaks corresponding to the 2nd pause regions were observed at the region between the 1st pause regions and the +1 dyad (Figure 3C). This accumulation of PRO-seq signal up to the +1 dyad was also observed in auxin-treated NELF-E-AID cells (Figure S3A), but not observed in parental DLD-1 cells (Figure S3B). These results suggest that NELF loss results in transitioning of paused Pol II up to the +1 dyad. In distal 2nd pause genes, 2nd pause regions tend to be associated with the +2 or +3 nucleosome (Figure 3A). The ability to bypass the +1 dyad may be due to the lower stability of the +1 nucleosome at these genes (Figure 3B).
Pol II CTD phosphorylation states are stable upon NELF loss-
The phosphorylation states of the Pol II C-terminal domain (CTD) have been shown to regulate Pol II throughout the transcription cycle by a series of cyclin-dependent kinases, including initiation when CDK7 phosphorylates the CTD on serine 5, and promoter-proximal pause-release, which is associated with serine 2 phosphorylation by Cdk9-CyclinT (P-TEFb) (Chen et al., 2018). Accordingly, phospho-Ser5 is enriched at promoters, while phospho-Ser2 occupancy increases along gene bodies. We tested whether NELF loss affects CTD phosphorylation states. Metaplot analysis of ChIP-seq for phospho-Ser2 and phospho-Ser5 of Pol II CTD showed that both phosphorylation levels at promoters were maintained upon NELF loss. Notably, these phosphorylation peaks shifted slightly downstream of pause regions (Figure 4A), corresponding to the transition of paused Pol II to 2nd pause regions. These results suggest that NELF loss does not result in alteration of CTD phosphorylation states and therefore, the transition of paused Pol II to the 2nd pause region can occur without changes in CTD phosphorylation.
Figure 4. P-TEFb-independent transition to 2nd pause region.
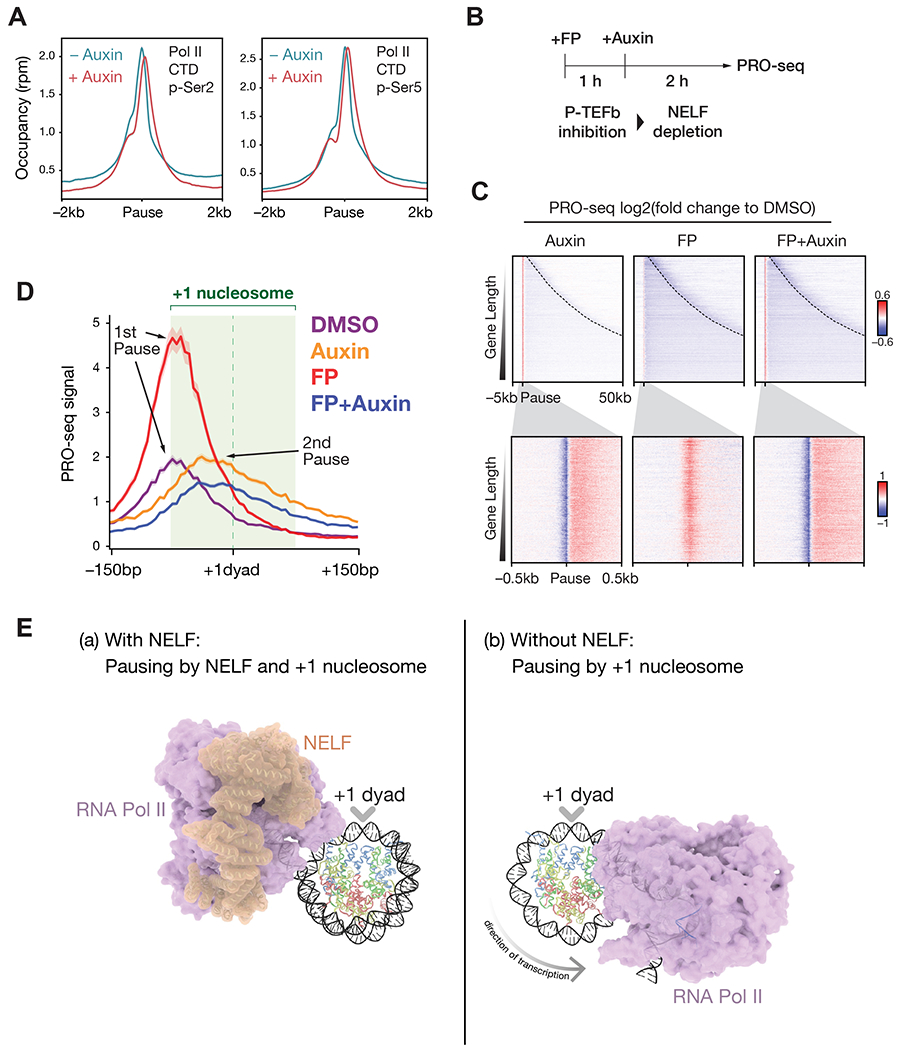
A. Metaplots of Pol II CTD phospho-Ser2 or phospho-Ser5 ChIP-seq signal centered on pause regions in NELF-C-AID cells treated with or without auxin for 2h. N = 6,531.
B. Schematic showing the experimental design used for C and D. NELF-C-AID cells are treated with the P-TEFb inhibitor flavopiridol (FP) for 1 h followed by auxin treatment for 2 h to deplete NELF on chromatin.
C. Heatmaps of log2 fold changes of PRO-seq signal compared to the DMSO control are shown. Dashed lines in upper panels indicate transcription end sites. Rows are ordered by gene length. N = 6,531.
D. Metaplot of PRO-seq mean signal in NELF-C-AID cells treated with flavopiridol and auxin, centered at the +1 nucleosomal dyad. The light green shading indicates the position of +1 nucleosome (+ 1 dyad ± 75 bp) as determined by MNase-seq. N = 1,843 genes that have a positioned +1 nucleosome among genes with a proximal 2nd pause (see methods).
E. Proposed mechanism of Pol II pausing at promoters: (a) NELF and the +1 nucleosome cooperatively regulate pausing. NELF physically inhibits Pol II transcription while the +1 nucleosome (or perhaps factors associated with the +1 nucleosome) could serve as an additional mechanism of pausing. (b) NELF depletion allows Pol II to advance up to the +1 dyad in a P-TEFb-independent manner. P-TEFb activity is required for Pol II to traverse the nucleosome and proceed into productive elongation. Structures of Pol II transcribing through nucleosomes are adapted from (Kujirai et al., 2018). Structure of NELF-Pol II complex is adapted from (Vos et al., 2018b).
The transition of paused Pol II to the +1 dyad is independent of P-TEFb activity-
Release from promoter-proximal pausing relies on the activity of the Cdk9-Cyclin T complex known as P-TEFb and may involve phosphorylation of DSIF and NELF in addition to phospho-Ser2 of the Pol II CTD (Chen et al., 2018). Given that NELF loss does not affect phospho-Ser2 levels, we examined whether the transition of paused Pol II to 2nd pause regions depends on P-TEFb activity. We treated NELF-C-AID cells with the P-TEFb inhibitor flavopiridol (FP), followed by auxin treatment to deplete NELF (Figure 4B). Heatmaps of PRO-seq log2 fold change showed that FP treatment greatly induced pausing at 1st pause regions when NELF was present (Figure 4C). Interestingly, depleting NELF with auxin treatment canceled the FP-induced 1st region of pausing, indicating that transition of paused Pol II upon NELF loss is independent of P-TEFb (Figure 4C). To examine 2nd pause positions at the +1 nucleosome, we generated PRO-seq metaplots centered on the +1 dyad as described for Figure 3C. The metaplot analysis showed that FP-induced pausing occurred at the entrance of the +1 nucleosome, while PRO-seq peaks in FP + auxin condition was observed at the region between 1st pause regions and the +1 dyad, similar to PRO-seq peaks in the auxin-alone condition (Figure 4D). Since there was no increase in Pol II occupancy at the 2nd pause region after FP treatment in the absence of NELF, this suggests that NELF is generally needed for the establishment of pausing. To exclude potential off-target effects of flavopiridol, we also tested the effects of NVP-2, a highly selective P-TEFb inhibitor with sub-nanomolar potency (Olson et al., 2018). Consistent with our results with flavopiridol treatment, NVP-2-induced pausing occurred at the 1st pause region in the presence of NELF, and shifted to the 2nd pause region upon NELF depletion (Figures S4A–S4C). These data demonstrate that Pol II can be paused at the entrance of the +1 nucleosome in a NELF-dependent manner (Figure 4E (a)) and that Pol II can resume transcription toward the +1 dyad upon NELF loss without P-TEFb activity (Figure 4E (b)) (please see Discussion).
Functional role of acute loss of NELF in the heat shock response-
In mammalian cells, the heat shock response has been demonstrated to result in the upregulation of a small subset of genes through release from the paused state, while downregulating the majority of genes through increased promoter-proximal pausing (Chen et al., 2017; Mahat et al., 2016a). To test whether acute loss of NELF is required for the heat shock response, we treated NELF-C-AID cells with auxin for 2 h, followed by heat shock treatment at 42 °C for 1 h (Figure 5A). We defined heat shock-upregulated genes and heat shock-downregulated genes based on PRO-seq density at gene bodies (please see Methods).
Figure 5. The heat shock response is robust in the absence of NELF.

A. Schematic showing the experimental design. NELF-C-AID cells are treated with auxin for 2 h followed by heat shock at 42 °C for 1 h before performing PRO-seq.
B. Heatmaps of PRO-seq signal for heat shock-upregulated genes. Gene regions (pause −5kb to pause +50 kb) are sorted by gene length with dashed lines indicating transcription end regions (top panels). Promoter regions (pause ± 0.5 kb) are sorted by NELF-C signals (bottom panels). N = 258.
C. Boxplot analysis of PRO-seq coverage (RPKM) in gene bodies (pause region + 500 bp to + 100 kb or to transcription end sites) in heat shock-upregulated genes. N = 258. Whiskers, 1.5 times the interquartile range; boxes, the 25th and 75th percentiles. n.s., P > 0.06. [Mann-Whitney U test]
D. Heatmaps of PRO-seq signal for heat shock-downregulated genes shown as in B. N = 5334.
E. Boxplot analysis of PRO-seq coverage (RPKM) for heat shock-downregulated genes shown as in C. N = 5,334. *P < 0.01 [Mann-Whitney U test]
F. Metaplot of PRO-seq mean signal in NELF-C-AID cells treated with heat shock and auxin, centered at the +1 nucleosomal dyad. The light green shading indicates the position of the +1 nucleosome (+ 1 dyad ± 75 bp) as determined by MNase-seq. N = 1,490 genes that have a positioned +1 nucleosome among genes with a proximal 2nd pause in heat shock-downregulated genes.
PRO-seq tracks demonstrates that transcriptional upregulation upon heat shock appeared normal in the absence of NELF (Figure S5A). Genome-wide analysis of heat shock-upregulated genes showed that PRO-seq density in gene bodies at 42 °C in the absence of NELF is not significantly different from what is seen in the presence of NELF (Figures 5B and 5C). These results suggest that transcriptional induction upon heat shock is robust in the absence of NELF. For heat shock-downregulated genes, PRO-seq tracks showed that transcriptional downregulation upon heat shock also appeared relatively normal in the absence of NELF (Figure S5B). PRO-seq density was reduced in gene bodies at 42 °C, with even slightly lower PRO-seq signal, in the absence of NELF (Figures 5D and 5E).
Heat shock in the presence of NELF resulted in increased PRO-seq signal at 1st pause regions (Figure 5D), confirming that heat shock induces canonical pausing (Chen et al., 2017; Mahat et al., 2016a). Surprisingly, in the absence of NELF, PRO-seq signal at the 2nd pause regions were increased upon heat shock, while PRO-seq signal at the 1st pause regions was not increased. Metaplot analysis of PRO-seq showed that PRO-seq signal at the entrance of +1 nucleosome was increased upon heat shock in the presence of NELF, while PRO-seq signal around the +1 dyad were increased upon heat shock in the absence of NELF (Figure 5F). These results demonstrate that both gene upregulation and downregulation upon heat shock are intact in the absence of NELF, and that nucleosome-associated pausing can serve as a complementary step for pausing in heat shock-downregulated genes.
NELF is required for recruitment of the cap-binding complex to promoters-
Next, we addressed the significance of NELF-dependent pausing for nascent RNA stability. Cap-binding complex (CBC) has been shown to directly bind to NELF (Narita et al., 2007; Schulze and Cusack, 2017). The binding affinity between CBC and NELF can be increased 8-fold by the 5’ cap analog m7GTP (Schulze and Cusack, 2017), suggesting that CBC could be efficiently targeted to the NELF-Pol II complex upon completion of the capping reaction. However, the molecular function of NELF-CBC binding at promoters has not been determined. We first characterized genome-wide occupancies of CBP80, a subunit of CBC. ChIP-seq peak analysis showed that most CBP80 peaks highly overlap NELF-C peaks (Figure 6A). Annotation of ChIP-seq peaks demonstrated that CBP80 and NELF co-occupy promoters, while NELF peaks without CBP80 peaks are enriched in intron/intergenic regions (Figure S6A). Scatter plot of NELF-C peak intensity versus CBP80 peak intensity at promoters showed significant positive correlation between them (Figure 6B, Spearman correlation coefficient R2 = 0.77). These results demonstrate a genome-wide association between CBP80 and NELF at promoters.
Figure 6. NELF is required for recruitment of cap-binding complex.

A. Venn diagram showing overlap of peaks between NELF-C and CBP80 ChIP-seq.
B. Scatter plot of CBP80 coverage versus NELF-C coverage at each promoter. R2 = 0.81 [Spearman’s rank correlation coefficient]
C. Western blots for CBP80 using whole-cell extracts of NELF-C-AID cells treated with or without auxin for 2 h.
D. ChIP-seq track examples for CBP80 and NELF-C in NELF-C-AID cells treated as in C.
E. Metaplots of CBP80 ChIP-seq signal at pause regions in NELF-C-AID cells treated as in C. N = 6,531.
The CBP80 protein level was stable upon NELF loss as assayed by western blotting (Figure 6C), but CBP80 ChIP-seq in auxin-treated NELF-C-AID cells showed that most of the CBP80 peaks disappeared upon NELF loss (Figures 6D). Metaplot analysis of CBP80 ChIP-seq signal demonstrated substantial reduction of CBP80 at pause regions upon NELF loss (Figure 6E). Heatmap analysis showed that CBP80 occupies the 1st pause regions with Pol II, while the residual CBP80 in NELF-depleted cells colocalized with Pol 2 at 2nd pause regions (Figure S6B). This suggests that although a large fraction of CBP80 is dissociated from chromatin upon NELF loss, CBP80 may be weakly recruited to Pol II paused at the +1 dyad.
NELF loss does not result in global loss of nascent transcript capping-
Cap-binding proteins protect 5’ capping against decapping enzymes (Gonatopoulos-Pournatzis and Cowling, 2014). Given that NELF loss impairs CBC recruitment, we considered the possibility that 5’ capping might be unstable. Therefore, we performed PRO-cap (Kwak et al., 2013) to map the 5’-end of capped nascent transcripts in the presence and absence of NELF. We identified ~90,000 transcription start sites (TSS) (de novo TSS called), with ~15,000 significantly increased PRO-cap signal (Cap Up) and ~6,000 with decreased PRO-cap signal (Cap Down) in the absence of NELF (Figure 7A). Metaplot analysis of PRO-cap and PRO-seq signal showed that Cap Up TSS were associated with Pol II maintaining pausing but transitioning from 1st to 2nd pause regions (Figure 7B). In contrast, Cap Down TSS were characterized by decreased PRO-seq signal (Figure 7C). The precise location of TSS identified by PRO-cap were unchanged after NELF depletion (Figure S7A), suggesting that the observed transitioning of Pol II from the 1st to 2nd pause region was not due to changes in TSS usage.
Figure 7. Analysis of nascent transcript capping in the absence of NELF.

A. Differential expression analysis of PRO-cap data in NELF-C-AID cells in untreated versus 2h auxin treated condition. Each dot indicates TSS identified from PRO-cap data (N = 96,565). Red dots indicate TSS with increased PRO-cap signals (N = 16,593, Padj < 0.05), and blue dots indicate TSS with decreased PRO-cap signals (N = 6,125, Padj < 0.05).
B. Metaplots of PRO-cap (left panel) and PRO-seq (right panel) at Cap Up TSS in NELF-C-AID cells.
C. Metaplots at Cap Down TSS as in B.
D. Heatmaps of NELF-C, CBP80 and MNase-seq in NELF-C-AID cells. Upper panels show Cap Up TSS clusters (N = 9,614), and lower panels show Cap Down TSS cluters (N = 5,336). Rows are sorted by length of TSS clusters.
E. Model of NELF function distinct from regulating release from promoter-proximal pausing. Please see the discussion for the details. Structures of CBC with the NELF-E C-terminal tail are adapted from (Schulze and Cusack, 2017).
Both Cap Up and Cap Down TSS showed similar decreases in NELF and CBP80 occupancy after NELF depletion (Figure 7D). However, Cap Up and Cap Down promoters showed different nucleosome profiles, with Cap Up TSS exhibiting better nucleosome phasing (Figure 7D). Since NELF loss leads to loss of protection of the cap by the CBC, we looked at the occupancy of decapping factor DCP2 and 5’-3’ exonuclease XRN2, which had previously been implicated in promoter-proximal early termination of Pol II (Brannan et al., 2012; Davidson et al., 2012) . ChIP-seq shows a pronounced increase of XRN2 and DCP2 at some Cap Down genes (Figure S7B). However, metaplot analysis shows similar increases of XRN2 and DCP2 at Cap Up and Cap Down TSS upon NELF depletion, suggesting that additional contextual information, such as promoter and/or chromatin structure could underlie the differential stability of Cap Up and Cap down TSS.
DISCUSSION
The function of the NELF-CBC interaction-
Using rapid depletion of NELF, we have dissected in vivo functions of NELF in human cells. NELF loss substantially reduces the CBC, alters cap stability, and increases occupancies of XRN2 and DCP2 at TSS. Here, we propose that NELF has primary roles in modulating cap stability through the CBC recruitment, which sets up a regulatory mechanism for promoter-proximal early termination of Pol II (Figure 7E, please see below).
Following NELF depletion, we found decreased PRO-cap signal at a subset of TSS that also exhibited decreased PRO-seq signal, rather than the typical transitioning to a 2nd pause region (Figure 7, Cap Down TSS). This raises the possibility that these transcripts were susceptible to early termination: at these TSS, loss of CBC, and increased DCP2, may promote the removal of unprotected 5’ cap, and increased XRN2 may stimulate 5’-3’ exonucleolytic degradation, which could lead to early termination of Pol II by the “torpedo” mechanism (Proudfoot, 2016). Since XRN2 coimmunoprecipitates with DCP2 (Brannan et al., 2012), their recruitment upon NELF loss may be cooperatively regulated.
However, despite a general increase in DCP2 and XRN2 occupancy after NELF depletion, our PRO-cap analysis showed no corresponding global loss of capping at the majority of TSS following short periods of NELF depletion. Interestingly, Cap Down TSS showed a distinct nucleosome profile from TSS with a 2nd pause region (Cap Up TSS), although we are currently unable to determine whether this difference is a cause or consequence of these genes preferentially requiring NELF. It is also not clear why the 5’ cap remains stable at many TSS even though XRN2 and DCP2 recruitment is increased. Future studies will be needed to better understand the relationship between 5’ cap protection and Pol II pausing.
Function of nucleosome-associated pausing in the heat shock response-
We have found that both the upregulation and downregulation of gene expression during the heat shock response are robust in the absence of NELF in human cells (Figure 7). NELF has previously been shown to be dispensable for heat shock-induced gene upregulation in Drosophila cells (Ghosh et al., 2011), which is consistent with our findings. It has also been previously shown that gene downregulation upon heat shock is associated with pausing (Aprile-Garcia et al., 2019; Chen et al., 2017; Mahat et al., 2016a). We found that upon heat shock, Pol II is paused at 1st pause regions for the downregulated genes, consistent with these studies. Importantly, our results have demonstrated that Pol II is also paused at the 2nd pause regions in the absence of NELF (Figure 7), indicating that pausing can be NELF-independent with the canonical region of pausing being dependent on NELF pausing.
Pol II pausing associated with the +1 nucleosome-
Although the +1 nucleosome is a widely conserved structure of eukaryotic promoters (Lai and Pugh, 2017), whether the +1 nucleosome is required for promoter-proximal pausing in metazoans has not been answered. Cryo-electron microscopy structure studies have shown that transcribing Pol II can be paused at superhelical locations SHL(−6), SHL(−5), SHL(−2), and SHL(−1) of the nucleosome (Kujirai et al., 2018), and that the nucleosome can be accommodated on the NELF-Pol II-DSIF pausing complex (Farnung et al., 2018). Consistent with these studies, our data suggested that Pol II is paused at −60 bp upstream of the +1 dyad, which corresponds to SHL(−6), in a NELF-dependent manner. In Drosophila, Pol II pausing regions have been shown to be associated with the −60 bp site of the promoter-proximal nucleosome (Weber et al., 2014) or even upstream of the +1 nucleosome (Kwak et al., 2013). Therefore, the NELF-Pol II complex paused around SHL(−6) may be a primary feature of promoter-proximal pausing among metazoans. Future structural analyses could uncover how NELF determines 1st pause regions for Pol II in the context of chromatin.
We found that in the absence of NELF, Pol II is found at 2nd pause regions, which are associated with regions between 1st pause regions and the +1 dyad. These pause regions may be SHL(−5), SHL(−2), and SHL(−1) of the 1st nucleosome. The elongation factors Elf1 and DSIF have been shown to cooperatively prevent Pol II pausing at SHL(−6), SHL(−5), SHL(−2) (Ehara et al., 2019). Recruitment of these elongation factors to Pol II may facilitate effective release of paused Pol II into productive elongation. Of course, it still remains possible that the 2nd pause region is associated with unknown factors other than the nucleosome. Future in vivo studies will determine the mechanistic details for the 2nd pausing of Pol II upon NELF loss.
Reconciling in vitro and in vivo roles of NELF-
We find that acute loss of NELF does not release Pol II from promoter-proximal pausing in living cells (Figure 2). Rather, Pol II is found at the +1 dyad-associated regions in the absence of NELF. This means that NELF’s function in the CBC recruitment can be functionally distinct from regulation of Pol II pause-release. Consistent with this in vivo finding is that the C-terminal “tentacle” domain of the NELF-E subunit, which has an RNA recognition motif (RRM) and CBC binding domain, is not required for pausing in vitro (Missra and Gilmour, 2010; Vos et al., 2018b). In vivo, the NELF-E RRM may capture nascent RNA to facilitate its binding to CBC.
The transition of paused Pol II to 2nd pause regions in the absence of NELF occurs in a P-TEFb-independent manner (Figure 4), further demonstrating that NELF functions at a step distinct from pause-release. We reconcile the in vitro requirement of NELF for P-TEFb-dependent pause-release with our in vivo findings as follows. In an initial in vitro transcription system, NELF, when bound to Pol II with DSIF, was shown to possess an intrinsic ability to inhibit Pol II and this could be relieved by P-TEFb (Yamaguchi et al., 1999). However, when a minimal transcription system with recombinant proteins was used, P-TEFb alone was unable to relieve this Pol II inhibition (Vos et al., 2018a; Wada et al., 2000), suggesting that P-TEFb phosphorylation of NELF/DSIF/Pol II CTD is not sufficient for Pol II release in vitro. In cells, inhibition of Pol II by NELF and DSIF at the entrance of the +1 nucleosome could serve to allow time for NELF to recruit the CBC. CBC binding could in turn help activate P-TEFb (Lenasi et al., 2011). Since P-TEFb phosphorylation of NELF/DSIF/Pol II CTD was not sufficient for release from pausing in vitro, we propose that P-TEFb-dependent phosphorylation of unknown factors could be required for Pol II to traverse the +1 nucleosome into productive elongation.
The broad presence of NELF across metazoans raises the possibility that it is part of an added layer of transcriptional regulation needed for these more complex genomes and developmental programs. Recently, another metazoan complex, Integrator has been shown to mediate early termination of gene expression through its intrinsic endonuclease activity (Tatomer et al., 2019) (Elrod et al., 2019). Therefore, multiple early termination mechanisms could be used to achieve a greater diversity of gene regulatory mechanisms especially during critical differentiation decisions. Future studies, including expanding the use of acute depletion techniques, can be used to further explore and deconvolve the numerous gene regulatory mechanisms as they occur in their natural chromatin environment in vivo.
STAR*Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Ali Shilatifard (ASH@Northwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
The endogenous NELF-C or NELF-E genes were homozygously tagged with mini-AID tag (Natsume et al., 2016) at its C-terminus in DLD-1 cells that constitutively express the OsTIR1 gene (Holland et al., 2012), as described previously (Chen et al., 2017). Briefly, co-transfection of 2 donor plasmids (Neo and Hyg) and 1 Cas9 plasmid were performed using lipofectamine 3000 (TheremoFisher) (please see below for the details of plasmids). Clones that show resistance for 250 μg/ml Geneticin (ThermoFisher) and 100 μg/ml Hygromycin B (ThermoFisher) were picked up and genotype for each clone was validated by PCR. OsTIR1-expressing DLD-1 cells were grown in DMEM (Corning) supplemented with 10% FBS (Corning), 1X GlutaMAX (Gibco), 2 μg/ml puromycin (Gibco), 100 U/ml Penicillin and 100 μg/ml Streptomycin (Gibco). For the growth assay in Figure 1B, cells were fixed by 4% formaldehyde in PBS and were stained by crystal violet solution (Sigma), followed by OD590 measurement of crystal violet dissolved in 10% acetic acid.
Plasmids, Peptides and Chemicals
For homozygous mAID tagging to the endogenous NELF-C gene, donor plasmids YNP37 (NELF-C-AID_Neo) and YNP38 (NELF-C-AID_Hyg) were prepared using pMK286 and pMK287 (Natsume et al., 2016). Cas9 plasmid YNP39 (gRNA: CTGCAAATCTAACTTCATCA) was prepared using pX330 (Cong et al., 2013). Similarly, for NELF-E gene tagging, donor plasmid YNP47 (NELF-E-AID_Neo) and YNP48 (NELF-E-AID_Hyg) and Cas9 plasmid YNP41 (gRNA: CTACAGTGATGACGTCTACA) were prepared. Compounds for cell treatment: 1 M stock solution of Auxin (3-indole-acetic acid sodium salt, abcam) was prepared in H2O and added in culture at 500 μM. 1 mM stock solution of Flavopiridol (Cayman) was prepared in DMSO and added in culture at 1 μM. 1 mM stock solution of NVP-2 (MedChemExpress) was prepared in DMSO and added in culture at 250 nM.
METHOD DETAILS
Heat Shock Induction
Heat shock of mammalian cells was performed using ~70-80% confluent DLD-1 cells by adding pre-heated (42 °C) conditioned media collected from identically growing cells (Mahat et al., 2016a). The heat shock cells were incubated at 42 °C for 1 hour. After washing with PBS, the heat shock and non-heat shock DLD-1 cells were processed for PRO-seq as described below.
PRO-seq Library Preparation and Alignment
PRO-seq was performed according to the previously published protocol (Mahat et al., 2016b) with minor modifications. 10 million DLD-1 nuclei were mixed with 0.5 million spike-in Drosophila S2 cell nuclei. Nuclear run-on assays were performed with 25 μM Biotin-11-ATP/UTP/CTP/GTP (PerkinElmer) for 3 min at 30 °C. RNA fragmentation was performed with 0.2 M NaOH for 10 min on ice. Biotin-labeled RNA was purified by streptavidin beads M-280 (Thermo). The 5’ cap removal and triphosphate repair were performed with RppH (NEB), 5’ hydroxyl repair was then performed with PNK (NEB). 5’ and 3’ adapter ligation was performed with T4 RNA ligase I (NEB). Reverse transcription was performed with SuperScript III (ThermoFisher). cDNA was amplified using Phusion Hot Start II DNA polymerase (ThermoFisher). DNA Libraries were size selected by PippinHT (Sage) or AMpure XP (Beckman Coulter) and sequenced on a NextSeq 500 (Illumina). Low-quality bases and adapters from 3’ ends of reads were removed using cutadapt 1.14 (Martin, 2011) requiring a read length of 16–36 bp. Reads derived from ribosomal RNA were filtered out by mapping reads on human and fly ribosomal DNA. The remaining reads were aligned using bowtie 2.2.6 with --very-sensitive option (Langmead and Salzberg, 2012), to a concatenated genome comprised of human hg19 and fly dm3 assemblies. The 5’ ends of aligned reads with MAPQ ≥ 30 were taken by bedtools genomecov 2.25.0 (Quinlan and Hall, 2010) with -strand and −5 options, and strands of the 5’ ends of reads were then swapped. Read counts were normalized to total reads aligned to the spike-in genome.
PRO-seq Data Analysis
Protein coding transcripts were chosen from Ensembl version 75 database by R 3.3.3, according to the previously described protocol (Liang et al., 2018) with minor modifications. Briefly, we selected RefSeq-validated transcripts with TSSs that overlapping NELF-C ChIP-seq peaks. We filtered out transcripts that are ≤ 2 kb long and ≤ 2 kb away from the nearest genes. Next, we chose transcripts that have ≥ 2 rpm total PRO-seq coverage at the region between TSS − 100 bp and TSS + 300 bp, using 3 independent PRO-seq data in untreated NELF-C-AID cells. In these selected transcripts, the nucleotide position with maximum PRO-seq coverage in the region between TSS − 100 bp and TSS + 300 bp was assigned to the 1st pause region. Transcripts that have ≤ 0.2 rpm coverage at the 1st pause region nucleotide were excluded. The resulting transcripts with assigned the 1st pause regions were used for further analyses. The nucleotide positions with maximum PRO-seq coverage in 1h auxin-treated NELF-C-AID cells at the region from +10 to +500 bp downstream of the 1st pause region was assigned to the 2nd pause region. For Heat shock experiments, transcripts with ≥ 1.5-fold increase or decrease in PRO-seq coverage at gene bodies (the 1st pause regions +500 to +100kb or to transcription termination sites) upon heat shock were assigned to HS-upregulated or HS-downregulated genes, respectively. Heatmaps and metaplots of PRO-seq signal was generated by deepTools 3.0.0/3.1.1/3.1.2 computeMatrix, plotHeatmap, and plotProfile (Ramírez et al., 2016). Log2 fold change of PRO-seq signal at nucleotide resolution was calculated by deepTools 3.0.0/3.1.1/3.1.2 bigwigCompare with pseudocount 0.25 rpm.
ChIP-seq Library Preparation
ChIP-seq was performed as described previously (Liang et al., 2018) with minor modification. Briefly, 20-50 million cells were crosslinked with 1% paraformaldehyde (Electron Microscopy Sciences) in PBS for 10 min at r.t. and were quenched with 0.2 M glycine for 5 min. Spike-in mouse embryonic fibroblast fixed cells were added to samples as described previously (Orlando et al., 2014) NELF-C, NELF-E, SPT5, Pol II CTD Ser2P, Pol II CTD Ser5P, XRN2, DCP2 ChIP-seq in NELF-C-AID cells. Chromatin was sonicated with the Covaris E220 for 4 min with 10% duty cycle, 140 peak intensity power, 200 cycles per burst. Immunoprecipitations were carried out overnight with antibodies as indicated (see antibodies section) and Protein A/G-agarose beads (Santa Cruz) or Dynabeads Protein G (Invitrogen). Protein degradation by 400 μg/ml Protease K (roche) and reverse crosslinking reaction were performed at 65 °C overnight. Immunoprecipitated DNA was purified by chloroform extraction and ethanol precipitation or using PCR purification kit (QIAGEN). DNA libraries were prepared by the HTP Library Preparation Kit for Illumina (KAPA) and sequenced on the NextSeq 500 or NovaSeq 6000 (Illumina).
ChIP-seq Alignment and Data Analysis
Single-end raw reads trimmed by Trimmomatic 0.33 (Bolger et al., 2014) were aligned to a concatenated genome comprised of human hg19 and mouse mm9 assemblies, using bowtie 2.2.6 (Langmead and Salzberg, 2012) with --sensitive option. The aligned reads with MAPQ ≥ 30 were extended to 150 bp and read counts were normalized to total reads aligned to spike-in genome. Metaplots of ChIP-seq signal were generated using deepTools 3.0.0/3.1.1/3.1.2 computeMatrix, plotHeatmap, plotProfile (Ramírez et al., 2016). Peaks were called using MACS 2.1.0 (Zhang et al., 2008) with a q-value cutoff of 0.05. Venn diagrams were generated using intervene 0.6.4 (Khan and Mathelier, 2017) with bedtools 2.25.0. For the correlation plot in Figure 6B, reads at each pause site ± 2 kb were counted using featureCounts 2.0.0 (Liao et al., 2014) then read counts were visualized in R 3.3.3. Peak annotation for NELF-C and CBP80 ChIP-seq in Figure S6A was performed using HOMER 4.10 (Heinz et al., 2010). Merged heatmap in Figure S6B was prepared by stacking heatmaps with reduced opacity. For identification of XRN2 peaks around TSS for metaplots in Figure S7C and S7D, XRN2 peaks were called in auxin-treated NELF-C-AID cells, then XRN2 peaks overlapping Cap Up TSS ± 1 kb or Cap Down TSS ± 1 kb were selected using bedtools window 2.25.0 (Quinlan and Hall, 2010).
MNase-seq Data Analysis
MNase-seq single-end raw reads in DLD-1 cells (Yamashita et al., 2011) (SRA ID:DRX000003) were used to determine nucleosome positions. Low quality bases were trimmed from the 3’ end and 16–36 bp reads were taken by Trimmomatic 0.33 (Bolger et al., 2014). Processed reads were aligned to the human hg19 genome with bowtie 1.1.2 (Langmead et al., 2009) with options -m 1 -v 2. The resulting BAM files were sorted and converted to BED files. The BED files were processed by improved nucleosome-positioning algorithm iNPS 1.2.2 (Chen et al., 2014) to determine nucleosome positions. For annotation of the +1 nucleosome, the closest nucleosome that was found downstream of the pause region for each gene and that was annotated as MainPeak by iNPS was chosen. Note that nucleosomes that did not overlap the regions of H3K4me3 ChIP-seq peaks ± 75 bp were excluded to discard false-positive nucleosome positions. The resulting nucleosomes are referred to as the +1 nucleosome.
PRO-cap Library Preparation and Read Alignment
PRO-cap libraries were prepared as described previously (Kwak et al., 2013; Mahat et al., 2016b) with minor modifications. NELF-C-AID cells were treated with or without 500 μM auxin for 2 h, followed by nuclear extraction. ~10 million NELF-C-AID nuclei with 0.5 million spike-in drosophila S2 nuclei were subjected to nuclear run-on reaction. Uncapped RNA with 5’-monophosphate was degraded using Terminator 5’-phosphate dependent exonuclease (Lucigen). 5’ triphosphate and monophosphate from uncapped RNA were removed using Quick CIP (NEB). 5’ cap repair was performed with RppH (NEB). For adapter ligation, PRO-seq adapter sequences were used instead of PRO-cap adapters. Libraries were then sequenced by NextSeq 500 in paired-end sequencing mode. Raw reads were processed as described for PRO-seq above, requiring a read length of 18–36 bp. Processed paired-end reads were aligned to a concatenated genome comprised of hg19 and dm3 assemblies using bowtie2 (Langmead and Salzberg, 2012) with options (--very-sensitive -X 1000 –no-mixed –no-discordant –no-unal) as described previously (Tome et al., 2018). The 5’ ends of aligned R2 reads with MAPQ ≥ 40 were taken by bedtools genomecov 2.25.0 (Quinlan and Hall, 2010) with -strand and −5 options. Read counts were normalized to total reads aligned to the spike-in genome.
PRO-cap Data Analysis
de novo transcription start sites were identified from PRO-cap data using HOMER 4.11 with findcsRNATSS option (Duttke et al., 2019; Heinz et al., 2010). TSS identified in untreated and auxin-treated samples were merged then defined as TSS regions. Raw PRO-cap reads were counted using HOMER 4.11 annotatePeaks. Read counts were normalized to spike-in counts using RUVseq with RUVg option (Risso et al., 2014) in R 3.3.3. Differential expression analysis was performed using DESeq2 (Love et al., 2014) and visualized in R 3.3.3. TSS with significant gain of PRO-cap signal upon NELF loss (adjusted p-value < 0.05) was defined as Cap Up TSS, while TSS with significant loss of PRO-cap signal upon NELF loss (adjusted p-value < 0.05) was defined as Cap Down TSS. Metaplots of PRO-cap and PRO-seq at Cap Up TSS or Cap Down TSS were generated using deepTools 3.1.1. For heatmaps of ChIP-seq and MNase-seq in Figure 7D, TSS were merged with gap of no longer than 600 bp to obtain TSS clusters. Heatmaps were then generated at Cap Up TSS clusters and Cap Down TSS clusters. To measure distances of PRO-cap signals between untreated and 2h auxin-treated NELF-C-AID cells, each position of TSS in the untreated sample was compared with the position of the nearest TSS in the treated sample in a strand-specific manner using HOMER 4.11 annotatePeaks.
Visualization of Protein Structures
Atomic models of the RNAPII-nucleosome complexes in which Pol II is paused at SHL(−5) (PDB ID: 6A5P) or SHL (−1) (PDB ID: 6A5T) were adapted from (Kujirai et al., 2018). The structure of NELF-Pol II complex (PDB ID: 6GML) was adapted from (Vos et al., 2018b). The structure of CBC with NELF-E C-terminal tail and m7GTP (PDB ID: 5OOB) was adapted from (Schulze and Cusack, 2017).
QUANTIFICATION AND STATISTICAL ANALYSES
We have performed at least 2 biological replicates for all studies and analyses of any of the replicates results in the same conclusions as the other replicates. n indicates the number of biologically independent samples, and N indicates the number of genes. Mann-Whitney U test for Figure 2E, 5C, 5E was performed in R 3.3.3. Adjusted p value for Figure 7A was calculated using DESeq2 (Love et al., 2014) in R 3.3.3.
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw and processed ChIP-seq, PRO-seq and PRO-cap data reported in this paper is available at GEO: GSE144786.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TH1L (D5G6W) Rabbit mAb (NELFC/D) | Cell Signaling | Cell Signaling Technology Cat# 12265, RRID:AB_2797862 |
| Recombinant Anti-NELFe antibody [EPR11600] | Abcam | Abcam Cat# ab170104, RRID:AB_2827280 |
| Purified Mouse Anti-DSIF Clone 17/DSIF (SPT5) |
BD Biosciences | BD Biosciences Cat# 611107, RRID:AB_398420 |
| Anti-RNA polymerase II subunit B1 (phospho CTD Ser-2), clone 3E10 | Millipore | Millipore Cat# 04-1571, RRID:AB_11212363 |
| Anti-RNA polymerase II subunit B1 (phospho-CTD Ser-5), clone 3E8 | Millipore | Millipore Cat# 04-1572, RRID:AB_10615822 |
| NCBP1/CBP80 Antibody | Bethyl | Bethyl Cat# A301-793A, RRID:AB_1211224 |
| XRN2 antibody | Bethyl | Bethyl Cat# A301-103A, RRID:AB_2218876 |
| DCP2 antibody | Bethyl | Bethyl Cat# A302-597A, RRID:AB_10555903 |
| HSP 90alpha/beta (F-8) antibody | Santa Cruz Biotechnology | Santa Cruz Biotechnology Cat# sc-13119, RRID:AB_675659 |
| Anti-Tubulin, beta E7 | DSHB | DSHB Cat# E7, RRID:AB_528499 |
| Rabbit anti-H3K4me3 serum | Shilatifard Laboratory | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Flavopiridol | Cayman | Cat# 10009197 |
| 3-indole-acetic acid sodium salt | Abcam | Cat# ab146403 |
| NVP-2 | MedChemExpress | Cat# HY-12214A |
| Paraformaldehyde (Electron Microscopy Sciences) | Fisher Scientific | Cat# 50-980-487 |
| Biotin-11-ATP | PerkinElmer | Cat# NEL544001EA |
| Biotin-11-CTP | PerkinElmer | Cat# NEL542001EA |
| Biotin-11-GTP | PerkinElmer | Cat# NEL545001EA |
| Biotin-11-UTP | PerkinElmer | Cat# NEL543001EA |
| RNA 5’ Pyrophosphohydrolase (RppH) | NEB | Cat# M0356S |
| PNK | NEB | Cat# M0201L |
| T4 RNA ligase I | NEB | Cat# M0204L |
| Phusion Hot Start II DNA polymerase | ThermoFisher | Cat# F549S |
| Terminator 5’-triphosphate dependent exonuclease | Lucigen | Cat# TER51020 |
| Quick CIP | NEB | Cat# M0525S |
| Proteinase K | Roche | Cat# 3115828001 |
| Dynabeads Protein G | ThermoFisher | Cat# 10003D |
| Protein A/G PLUS-Agarose | Santa Cruz Biotechnology | Cat# sc-2003 |
| Dynabeads Streptavidin M-280 | ThermoFisher | Cat# 11205D |
| 2% Agarose, PippinHT, 100-600 bp.10/pkg. | Sage science | Cat# HTC2010 |
| Critical Commercial Assays | ||
| HTP Library Preparation Kit | KAPA Biosciences | Cat# KK8234 |
| Agencourt AMPure XP | Beckman Coulter | Cat# A63882 |
| Deposited Data | ||
| Raw and processed data | This paper | GEO: GSE144786 |
| Human reference genome GRCh37/hg19 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/human |
| Drosophila reference genome BDGP Release 5/dm3 | Genome Reference Consortium | |
| Ensembl version 75 | http://www.ensembl.org/ | |
| RNAPII-nucleosome structure SHL(−5) | (Kujirai et al., 2018) | PDB: 6A5P |
| RNAPII-nucleosome structure SHL (−1) | (Kujirai et al., 2018) | PDB: 6A5T |
| Pol II-NELF structure | {Vos:2018cp} | PDB: 6GML |
| Complex of human nuclear cap-binding complex with m7GTP and NELFe c-terminal peptide | (Schulze and Cusack, 2017) | PDB: 5OOB |
| DLD1_normoxia_nucleosome | (Yamashita et al., 2011) | https://www.ncbi.nlm.nih.gov/sra/DRX000003 |
| Experimental Models: Cell Lines | ||
| DLD-1 OsTIR1 | (Holland et al., 2012) | N/A |
| NELF-C-AID DLD-1 OsTIR1 #7-10B | This paper | N/A |
| NELF-E-AID DLD-1 OsTIR1 #20-1B | This paper | N/A |
| Mouse embryonic fibroblasts | Stem cell technology | cat# 00325 |
| S2-DGRC | DGRC | FlyBase: FBtc0000006 |
| Recombinant DNA | ||
| YNP37 (Donor plasmid: NELF-C-AID_Neo) | This paper | N/A |
| YNP38 (Donor plasmid: NELF-C-AID_Hyg) | This paper | N/A |
| YNP39 (Cas9 plasmid for NELF-C, gRNA: CTGCAAATCTAACTTCATCA) | This paper | N/A |
| YNP41 (Cas9 for NELF-E, gRNA: CTACAGTGATGACGTCTACA) | This paper | N/A |
| YNP47 (Donor plasmid: NELF-E-AID_Neo) | This paper | N/A |
| YNP48 (Donor plasmid: NELF-E-AID_Hyg) | This paper | N/A |
| Software and Algorithms | ||
| Bowtie 2.2.6 | (Langmead and Salzberg, 2012) | |
| BEDTools 2.25.0 | (Quinlan and Hall, 2010) | |
| R 3.3.3 | https://www.r-project.org/ | |
| deepTools 3.0.0/3.1.1/3.1.2 | (Ramírez et al., 2016) | |
| Bowtie 1.1.2 | (Langmead et al., 2009) | |
| iNPS 1.2.2 | (Chen et al., 2014) | |
| HOMER 4.10/4.11 | (Duttke et al., 2019; Heinz et al., 2010) | |
| RUVseq | (Risso et al., 2014) | |
| DESeq2 | (Love et al., 2014) | |
| Trimmomatic 0.33 | (Bolger et al., 2014) | |
| cutadapt 1.14 | (Martin, 2011) | |
| intervene 0.6.4 | (Khan and Mathelier, 2017) | |
| featureCounts 2.0.0 | (Liao et al., 2014) | |
Highlights.
Acute NELF depletion reveals a 2-step pausing of Pol II at promoters
The 1st to 2nd pausing transition is independent of P-TEFb/SEC activity
The heat shock response remains intact in the absence of NELF
NELF recruits the cap binding complex with modest effects on 5’ cap stability
ACKNOWLEDGMENTS
We thank the Shilatifard lab members, D. Reinberg, D. Taatjes, A. Stark, E. Bartom, K. Eagen, J. Lis, J. Conaway for helpful discussion and P. Ozark for advice for the data analysis. We thank M. Kanemaki, A. Holland, I. Cheeseman, D. Foltz for providing reagents. We thank N. Ethen for providing protein structure-based illustration and graphical abstract. Y.A. was supported by the JSPS Research Fellowship for Young Scientists and the Uehara Memorial Foundation Research Fellowship. E.R.S. was supported by the National Institutes of Health grant R50CA211428. R.S. was supported by funding from University of Miami Miller School of Medicine, Sylvester Comprehensive Cancer Center and grants R01 GM078455 and DP1 CA228041 from the National Institute of Health. Studies in the Shilatifard laboratory related to transcription elongation are funded by the National Institutes of Health grant R01CA214035.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, and Rogatsky I (2009). Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proceedings of the National Academy of Sciences 106, 18207–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, and Handa H (2006). Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Molecular and Cellular Biology 26, 6094–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar SE, Sun J-L, Blair AL, Moskaluk CA, Lu Y-Z, Ye Q-N, Yamaguchi Y, Mukherjee A, Ren D-M, Handa H, et al. (2004). Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes & Development 18, 2134–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amleh A, Nair SJ, Sun J, Sutherland A, Hasty P, and Li R (2009). Mouse Cofactor of BRCA1 (Cobra1) Is Required for Early Embryogenesis. PLoS ONE 4, e5034–e5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprile-Garcia F, Tomar P, Hummel B, Khavaran A, and Sawarkar R (2019). Nascent-protein ubiquitination is required for heat shock–induced gene downregulation in human cells. Nat. Struct. Mol. Biol 26, 1–16. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, and Levine M (2009). Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. (2012). mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Molecular Cell 46, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Smith ER, and Shilatifard A (2018). Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 19, 464–478. [DOI] [PubMed] [Google Scholar]
- Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, Smith ER, Shiekhattar R, and Shilatifard A (2015a). PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell 162, 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Xie P, Collings CK, Cao K, Aoi Y, Marshall SA, Rendleman EJ, Ugarenko M, Ozark PA, Zhang A, et al. (2017). PAF1 regulation of promoter-proximal pause release via enhancer activation. Science 357, 1294–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Gao X, and Shilatifard A (2015b). Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes & Development 29, 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Liu Y, Zhu S, Green CD, Wei G, and Han J-DJ (2014). Improved nucleosome-positioning algorithm iNPS for accurate nucleosome positioning from sequencing data. Nature Communications 5, 4909. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Gilchrist DA, Fargo DC, Kwak H, Adelman K, and Lis JT (2012). Defining the status of RNA polymerase at promoters. Cell Rep 2, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttke SH, Chang MW, Heinz S, and Benner C (2019). Identification and dynamic quantification of regulatory elements using total RNA. Genome Res. 29, 1836–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, and Sekine S-I (2019). Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science 363, 744–747. [DOI] [PubMed] [Google Scholar]
- Elrod ND, Henriques T, Huang K-L, Tatomer DC, Wilusz JE, Wagner EJ, and Adelman K (2019). The Integrator complex terminates promoter-proximal transcription at protein-coding genes. bioRxiv 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnung L, Vos SM, and Cramer P (2018). Structure of transcribing RNA polymerase II-nucleosome complex. Nature Communications 9, 5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SKB, Missra A, and Gilmour DS (2011). Negative elongation factor accelerates the rate at which heat shock genes are shut off by facilitating dissociation of heat shock factor. Molecular and Cellular Biology 31, 4232–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SKB, Collins JB, Li L, Gilmour DS, and Adelman K (2008). NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & Development 22, 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Santos, dos G., Fargo DC, Xie B, Gao Y, Li L, and Adelman K (2010). Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, and Cowling VH (2014). Cap-binding complex (CBC). Biochem. J 457, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, and Cleveland DW (2012). Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proceedings of the National Academy of Sciences 109, E3350–E3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, and Ozato K (2005). The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular Cell 19, 523–534. [DOI] [PubMed] [Google Scholar]
- Khan A, and Mathelier A (2017). Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18, 287–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujirai T, Ehara H, Fujino Y, Shirouzu M, Sekine S-I, and Kurumizaka H (2018). Structural basis of the nucleosome transition during RNA polymerase II passage. Science 362, 595–598. [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, and Lis JT (2013). Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WKM, and Pugh BF (2017). Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol 18, 548–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Meth 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T, Peterlin BM, and Barboric M (2011). Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb). Journal of Biological Chemistry 286, 22758–22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Smith ER, Aoi Y, Stoltz KL, Katagi H, Woodfin AR, Rendleman EJ, Marshall SA, Murray DC, Wang L, et al. (2018). Targeting Processive Transcription Elongation via SEC Disruption for MYC-Induced Cancer Therapy. Cell 175, 766–779.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, and Shilatifard A (2011). Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes & Development 25, 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, and Shilatifard A (2010). AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Molecular Cell 37, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, and Lis JT (2016a). Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Molecular Cell 62, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, and Lis JT (2016b). Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat Protoc 11, 1455–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, and Reinberg D (2004). Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.a 101, 7572–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, and Price DH (1995). Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem 270, 12335–12338. [DOI] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from highthroughput sequencing reads. EMBnet 17, 10–12. [Google Scholar]
- Martinez-Rucobo FW, Kohler R, van de Waterbeemd M, Heck AJR, Hemann M, Herzog F, Stark H, and Cramer P (2015). Molecular Basis of Transcription-Coupled Pre-mRNA Capping. Molecular Cell 58, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Missra A, and Gilmour DS (2010). Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proceedings of the National Academy of Sciences 107, 11301–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, and Adelman K (2007). RNA polymerase is poised for activation across the genome. Nat. Genet 39, 1507–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim D-K, Hasegawa J, Omori M, Inukai N, et al. (2003). Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Molecular and Cellular Biology 23, 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TMC, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, and Handa H (2007). NELF interacts with CBC and participates in 3’ end processing of replication-dependent histone mRNAs. Molecular Cell 26, 349–365. [DOI] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, and Kanemaki MT (2016). Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep 15, 210–218. [DOI] [PubMed] [Google Scholar]
- Nechaev S, and Adelman K (2011). Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 1809, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, and Bensaude O (2001). 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414, 322–325. [DOI] [PubMed] [Google Scholar]
- Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, et al. (2018). Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nature Chemical Biology 14, 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Chen MW, Brown VE, Brown VE, Solanki S, Solanki S, Choi YJ, Choi YJ, Olson ER, et al. (2014). Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9, 1163–1170. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ (2016). Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352, aad9926–aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, and Young RA (2010). c-Myc Regulates Transcriptional Pause Release. Cell 141, 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Robb GB, and Chan S-H (2016). mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, and Lis JT (1993). In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. U.S.a 90, 7923–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, and Dudoit S (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol 32, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze WM, and Cusack S (2017). Structural basis for mutually exclusive co-transcriptional nuclear cap-binding complexes with either NELF-E or ARS2. Nature Communications 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Belotserkovskaya R, and Reinberg D (2004). Elongation by RNA polymerase II: the short and long of it. Genes & Development 18, 2437–2468. [DOI] [PubMed] [Google Scholar]
- Stadelmayer B, Micas G, Gamot A, Martin P, Malirat N, Koval S, Raffel R, Sobhian B, Severac D, Rialle S, et al. (2014). Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nature Communications 5, 5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, and Li R (2010). Human negative elongation factor activates transcription and regulates alternative transcription initiation. Journal of Biological Chemistry 285, 6443–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Pan H, Lei C, Yuan B, Nair SJ, April C, Parameswaran B, Klotzle B, Fan J-B, Ruan J, et al. (2011). Genetic and genomic analyses of RNA polymerase II-pausing factor in regulation of mammalian transcription and cell growth. Journal of Biological Chemistry 286, 36248–36257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Watkins G, Blair AL, Moskaluk C, Ghosh S, Jiang WG, and Li R (2008). Deregulation of cofactor of BRCA1 expression in breast cancer cells. J. Cell. Biochem 103, 1798–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatomer DC, Elrod ND, Liang D, Xiao M-S, Jiang JZ, Jonathan M, Huang K-L, Wagner EJ, Cherry S, and Wilusz JE (2019). The Integrator complex cleaves nascent mRNAs to attenuate transcription. Genes & Development 33, 1525–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome JM, Tippens ND, and Lis JT (2018). Single-molecule nascent RNA sequencing identifies regulatory domain architecture at promoters and enhancers. Nat. Genet 50, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, and Cramer P (2018a). Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560, 607–612. [DOI] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Urlaub H, and Cramer P (2018b). Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Pöllmann D, Caizzi L, Hofmann KB, Rombaut P, Zimniak T, Herzog F, and Cramer P (2016). Architecture and RNA binding of the human negative elongation factor. Elife 5, e14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Orphanides G, Hasegawa J, Kim DK, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, et al. (2000). FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Molecular Cell 5, 1067–1072. [DOI] [PubMed] [Google Scholar]
- Wada T, Wada T, Takagi T, Takagi T, Yamaguchi Y, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, et al. (1998). DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Development 12, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, and Henikoff S (2014). Nucleosomes Are Context-Specific, H2A.Z-Modulated Barriers to RNA Polymerase. Molecular Cell 53, 819–830. [DOI] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, and Adelman K (2015). Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Molecular Cell 58, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-H, Lee C, Fan R, Smith MJ, Yamaguchi Y, Handa H, and Gilmour DS (2005). Molecular characterization of Drosophila NELF. Nucleic Acids Res. 33, 1269–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-H, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, and Gilmour D (2003). NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & Development 17, 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Yamaguchi Y, Takagi T, Takagi T, Wada T, Wada T, Yano K, Yano K, Furuya A, Furuya A, et al. (1999). NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Inukai N, Narita T, Wada T, and Handa H (2002). Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Molecular and Cellular Biology 22, 2918–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto J, Hagiwara Y, Chiba K, Isobe T, Narita T, Handa H, and Yamaguchi Y (2014). DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nature Communications 5, 4263. [DOI] [PubMed] [Google Scholar]
- Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, Tanaka Y, Hashimoto SI, Sugano S, Nakai K, and Suzuki Y (2011). Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res. 21, 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, and Zhou Q (2001). The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414, 317–322. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, and Zhou Q (2005). Recruitment of P-TEFb for Stimulation of Transcriptional Elongation by the Bromodomain Protein Brd4. Molecular Cell 19, 535–545. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw and processed ChIP-seq, PRO-seq and PRO-cap data reported in this paper is available at GEO: GSE144786.


