Abstract
Based on its physical interactions with histone-modifying enzymes, the transcriptional corepressor Rcor1 has been implicated in the epigenetic regulation blood cell development. Previously, we have demonstrated that Rcor1 is essential for the maturation of definitive erythroid cells and fetal survival. To determine the functional role of Rcor1 in steady-state hematopoiesis in the adult, we used a conditional knockout approach. Here, we show that the loss of Rcor1 expression results in the rapid onset of severe anemia due to a complete, cell autonomous block in the maturation of committed erythroid progenitors. By contrast, both the frequency of megakaryocyte progenitors and their capacity to produce platelets were normal. Although the frequency of common lymphoid progenitors and T cells was not altered, B cells were significantly reduced and showed increased apoptosis. However, Rcor1-deficient bone marrow sustained normal levels of B-cells following transplantation, indicating a non-cell autonomous requirement for Rcor1 in B-cell survival. Evaluation of the myelomonocytic lineage revealed an absence of mature neutrophils and a significant increase in the absolute number of monocytic cells. Rcor1-deficient monocytes were less apoptotic and showed ~100-fold more colony-forming activity than their normal counterparts, but did not give rise to leukemia. Moreover, Rcor1−/− monocytes exhibited extensive, cytokine-dependent self-renewal and overexpressed genes associated with hematopoietic stem/progenitor cell expansion including Gata2, Meis1, and Hoxa9. Taken together, these data demonstrate that Rcor1 is essential for the normal differentiation of myeloerythroid progenitors and for appropriately regulating self-renewal activity in the monocyte lineage.
Keywords: Corepressor proteins, Epigenetics, Hematopoiesis, Erythropoiesis, Monocytes, Neutrophils
Introduction
Transcriptional corepressors are critical components of gene expression regulatory machinery. Their primary function is to serve as a platform to couple different histone modification activities that regulate gene expression. Through diverse transcription factor and cofactor interactions, tightly regulated gene expression patterns can be achieved by a limited number of proteins. Among the best characterized transcriptional corepressors is Rcor1 (CoREST) [1], a core component of a chromatin-modifying repressor complex that includes histone deacetylase 1/2 (HDAC1/2) [2–4] and the histone 3 lysine 4 (H3K4) demethylase Kdm1a (also known as LSD1) [5, 6]. HDAC1/2 removes acetyl groups from histone tails whereas Kdm1a removes monomethylation/dimethylation marks from H3K4 [7]. Although much attention has been focused on understanding the biochemical functions of Rcor1, its functional role in hematopoiesis has only recently been explored.
Rcor1 is expressed in hematopoietic stem cells, progenitor cells, and their differentiated progeny [8, 9]. Biochemical studies have identified several DNA binding transcription factors that physically interact with the Rcor1/Kdm1complex in different hematopoietic cell types. For example, Gfi1 and Gfi1b [10] have been shown to regulate key aspects of hematopoietic differentiation in vivo. The zinc finger protein Gfi1 is critical for granulocyte differentiation [11–13] and also regulates the production of common lymphoid progenitors as well as B-cell and T-cell differentiation [11, 12, 14–17]. The Gfi1 homolog, Gfi1b, is necessary for both erythroid and megakaryocytic differentiation [18, 19]. Although these interactions suggest potential roles for Rcor1 in multilineage differentiation, it is not yet known which hematopoietic cell lineages are functionally dependent on Rcor1 activity.
To directly assess Rcor1 function throughout the hematopoietic system and to bypass the embryonic lethality in the whole-animal Rcor1 knockout model [20], we generated an Mx1-Cre-driven Rcor1 knockout mouse. The loss of Rcor1 in adult hematopoietic cells leads to a complex phenotype that includes a complete block in erythroid and neutrophil differentiation but a sparing of the megakaryocyte lineage. These deficiencies are accompanied by an increase in monocytic cells that display abnormal self-renewal and reduced apoptosis.
Materials and Methods
Mice
Generation of Mx1-Cre; Rcor1flox/− mice (B6.CD45.2) and genotyping was performed as previously described [20]. Adult mice were injected every other day (three injections total) with ~175 μg of double stranded polyinosinic/polycytidylic acid (poly(I:C); 0.5 mg/ml, GE Healthcare Bio-Sciences, Pittsburgh, PA, http://www.gelifesciences.com). B6.CD45.1, B6.CD45.2/B6 Thy1.2 CD45.1 hybrids, or double congenic B6.CD45.1 Hbbd mice (8–12 weeks old) were used as transplant recipients. 1–3 × 106 control or Rcor1−/− unfractionated bone marrow (BM) was injected i.v. into irradiated (500–700 cGy) recipient mice. Sorted Lin−Sca1−c-kit+ (LSK) cells (1,500–2,500 cells) from untreated or poly(I:C) treated Mx1-Cre; Rcor1flox/− and control donors were cotransplanted with competitor BM (1–2 × 105 cells) from B6.CD45.1 or B6.CD45.1 EGFP+ (TgN(act-EGFP)OsbY01) donors into lethally irradiated recipient mice (1,000 cGy). Recipients were maintained on antibiotic-supplemented water [21]. All animal procedures were performed in accordance with the Institutional Animal Use and Care Committee at OHSU.
Flow Cytometry Analysis and Fluorescence-Activated Cell Sorting
Single-cell suspensions from BM, spleen, and peripheral blood were prepared as previously described [22]. Immunophenotyping of myeloerythroid progenitors [23], common myeloid progenitors (CMP), granulocyte-monocyte progenitors (GMP), megakaryocyte-erythroid progenitors (MEP) [24], and common lymphoid progenitors (CLP) [25] was performed on an LSRII, Canto, fluorescence-activated cell sorting (FACS) Calibur or Fortessa cytometer (BD, Franklin Lakes, NJ, http://www.bdbiosciences.com). Cell sorting was performed using a BD Influx or a BD Vantage sorter. Both dead cells (propidium iodine+) and doublets were excluded. Data were analyzed with FlowJo software (Ashland, OR, http://www.flowjo.com/). Antibodies (and clones) used in this study included: CD71 (R17217), Mac1 (M1/70), Gr1 (RB6–8C5), B220 (RA3–6B2), IgM (eB121–15F9), CD3 (145–2C11), and c-kit (2B8) (eBioscience, San Diego, CA, http://www.ebioscience.com); CD19 (1D3), CD4 (H29.19), CD5 (53–7.3), CD8 (53.6.7), CD16/32 (2.4G2), CD34 (RAM34), and CD115 (AFS98) (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com); TER119, Sca1 (D7), CD150 (TC15–12F12.2), CD105(MJ7/18), CD127/IL-7Rα (A7R34), and CD41 (MWReg30) (Biolegend, San Diego, CA, http://www.biolegend.com/). For apoptosis analysis, an Annexin-V Apoptosis Detection Kit I (BD Biosciences) was used with 7-AAD. For analysis of CMP/GMP/MEP, the lineage panel included B220, CD3, CD4, CD8, Gr1, Ter119, CD19, IgM, and CD127. For analysis of myeloerythroid progenitors, the lineage panel included B220, CD4, CD8, Mac1, and Gr1. To sort megakaryocyte progenitors (MkP), Ter119 was also included within this lineage mixture. For CLP analysis and LSK sorting, the lineage panel included B220, CD3, CD4, CD5, CD8, Mac1, Gr1, and Ter119.
Hemoglobin Electrophoresis
Performed as previously described [26].
In Vitro Colony-Forming Assays
For colony-forming unit (CFU) assays, cells were plated in duplicate or triplicate in 35 mm dishes in mouse methylcellulose complete medium (HSC007, R&D Systems, Minneapolis, MN, https://www.rndsystems.com) or in cytokine-free methylcellulose medium (M3234, Stem Cell Technologies, Vancouver, Canada, http://www.stemcell.com). Colonies were scored 7–10 days after plating. For serial replating assays, pooled day 8 colonies were washed in Dulbecco’s modified Eagle’s medium containing 10% serum and 1 × 104 cells were replated in fresh methylcellulose medium. For megakaryocyte (Mk) progenitor assays, 1 × 105 unfractionated BM were cultured in Mega-CultC (04950, Stem Cell Technologies) supplemented with mTPO (50 ng/ml) and mIL3 (10 ng/ml). Cultures were dehydrated, fixed, stained, and CFU-Mk were scored.
Morphological Analysis and Immunofluorescence Microscopy
BM touch preparations, peripheral blood, cytospun sorted cells, and colonies were stained with May-Grunwald and Giemsa stains. Tibias were fixed, decalcified, and cryopreserved as previously described [22] and vWF expression was detected with anti-vWF (A0082, Dako, http://www.dako.com), goat anti-rabbit cyanine 3, and 4′,6-diamidino-2-phenylindole. Imaging was performed with a Zeiss (Thornwood, NY, http://www.zeiss.com) Axiovert S-100, an AxioCam HRc camera and a Zeiss plan-neofluar ×20/0.50 lens, a Zeiss plan-neofluar ×40/1.30 lens, or a Zeiss plan-neofluar ×63/1.25 oil lens.
Quantitative RT-PCR
Sorted Mac1+Gr1lo cells from induced Mx1-Cre; Rcor1flox/− mice and controls treated with poly(I:C) and from primary recipients of Rcor1−/− and control BM were isolated by FACS. RNA was extracted using TRIzol and treated with DNase. Reverse transcription reactions were performed using Superscript III (Invitrogen, Grand Island, NY, http://www.lifetechnologies.com). Quantitative RT-PCR (qPCR) was performed with an Applied Biosystems PRISM 7900HT Fast Real-Time PCR system using SYBR green PCR master mix (Grand Island, NY, http://www.lifetechnologies.com). Relative abundance of each cDNA was determined according to the standard curve and normalized to 18S RNA levels. The primers used are listed below: 18Sfwd, 5′-CTCAACACGGGAAACCTCAC-3′; 18Srev, 5′-CGCTCCACCAACTAAGAACG-3′; Gata2fwd, 5′-CGCCTGTGGCCTCTACTACAA-3′; Gata2rev, 5′-TTTCTTGCTCTTCTTGGATTTGCT-3′; Hoxa9fwd, 5′-AACAATGCCGAGAATGAGAGC-3′; Hoxa9rev, 5′-CGAGTGGAGCGAGCATGTAG-3′; Meis1fwd, 5′-AAGATACAGGACTTACCATCCTTCA-3′; Meis1rev, 5′-GTCTATCATGGGCTGCACTATTCT-3′.
Statistical Analysis
Data were analyzed using Microsoft Excel or Prism 6. A two-tailed, unpaired Student’s t test was used and a p-value of <.05 was considered significant.
Results
Deletion of Rcor1 in Adult Hematopoietic Cells Produces a Lethal Anemia and Increases Myelomonocytic Cells
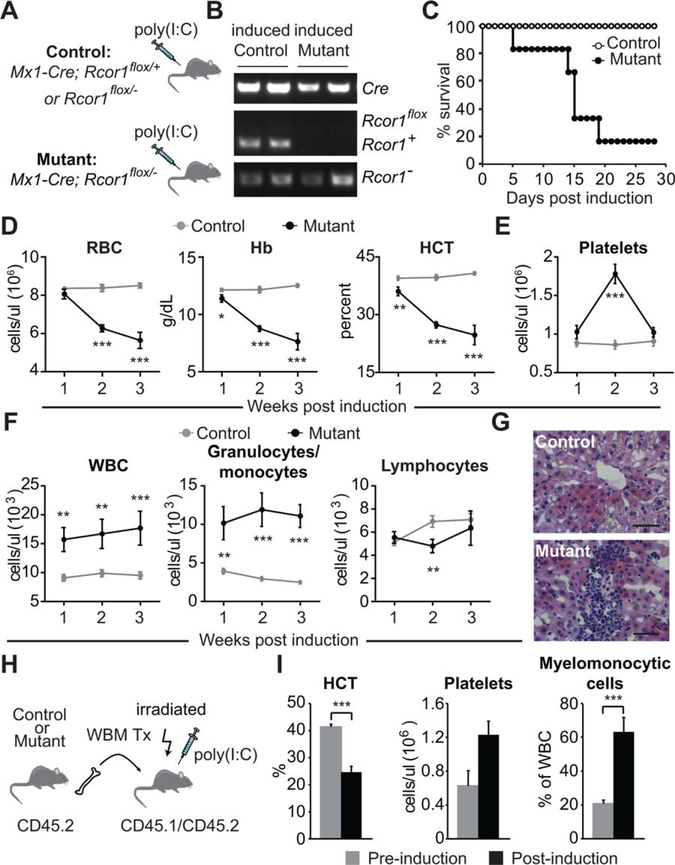
To assess Rcor1 function in steady-state, adult hematopoiesis, we generated mice carrying the Mx1-Cre transgene and a single functional Rcor1 allele in which exon 4 was flanked by loxP sites (Mx1-Cre; Rcor1flox/−). Administration of poly(I:C) was used to induce Cre expression in interferon-responsive cells [27], and 2 weeks following Cre induction, genotype analysis confirmed the absence of the Rcor1flox allele in the BM (Fig. 1A, 1B). Hereafter, we refer to these poly(I:C) treated, Rcor1-deficient mice as Rcor1−/−. Age-matched Rcor1flox/− and Mx1-Cre; Rcor1+/flox mice were used as controls.
Figure 1.
Loss of Rcor1 in adult mice causes a lethal anemia and the expansion of myelomonocytic cells. (A): Induction of Cre expression by injection of poly(I:C) in adult mice. (B): PCR genotype analysis of bone marrow (BM) cells 2 weeks after Cre induction. Only the Rcor1− allele was detected in the mutant BM. (C): Survival analysis after poly(I:C) treatment and Rcor1 deletion (nmutant = 6; ncontrol = 10). (D): Erythropoiesis in Rcor1−/− mice. Progressive reductions in circulating RBC number, serum Hb, and HCT were observed. (E): Rcor1-deficient mice had transiently elevated platelet counts after Cre induction. (F): Elevated circulating WBC in Rcor1−/− mice was the result of increased myelomonocytic cells. (G): Liver infiltration by myelomonocytic cells in Rcor1−/− mice. (H): Generation of BM-chimeric mice to exclusively delete Rcor1 from hematopoietic cells. (I): Peripheral blood analysis of BM-chimeric mice before and 2 weeks after Rcor1 deletion (n = 3). For (D–F), a minimum of seven mice for each genotype was tested at each time point. The average ± SEM is shown; *, p < .05; **, p < .01; ***, p < .001. Scale bar = 50 μm. Abbreviations: Hb, hemoglobin; HCT, hematocrit; RBC, red blood cells;WBC, white blood cells.
Typically, 80% of the Rcor1−/− mice died within 2–3 weeks following poly(I:C) treatment (Fig. 1C), a time course consistent with acute BM failure. Analysis of the peripheral blood revealed a rapidly progressive anemia in Rcor1−/− mice suggestive of a near complete block in red cell production (Fig. 1D). By contrast, platelet counts increased transiently and then quickly returned to normal levels (Fig. 1E). The total number of circulating white blood cells was persistently elevated due exclusively to an increase in circulating myelomonocytic cells (Fig. 1F) and infiltrates of these cells were found in the liver (Fig. 1G).
As the Mx1 promoter has been reported to be active in BM stroma [28] it was important to determine whether the Rcor1−/− phenotype was due primarily to a hematopoietic cell-intrinsic defect. Chimeric mice in which Mx1-Cre; Rcor1flox/− cells contributed to >90% of hematopoietic cells were generated by the transplantation of unfractionated BM (Fig. 1H, 1I). Two weeks after Cre induction, these BM-restricted Rcor1−/− mice developed marked anemia, increased circulating myelomonocytic cells, and a transient increase in platelet counts that was indistinguishable from the non-BM restricted Rcor1−/− mice (Fig. 1D–1F). Together, these data demonstrate that Rcor1 expression is required in a hematopoietic cell-intrinsic manner to maintain the steady-state production of cells of both the myeloid and erythroid lineages.
Rcor1 Is Required for the Maturation of Committed Erythroid Progenitors
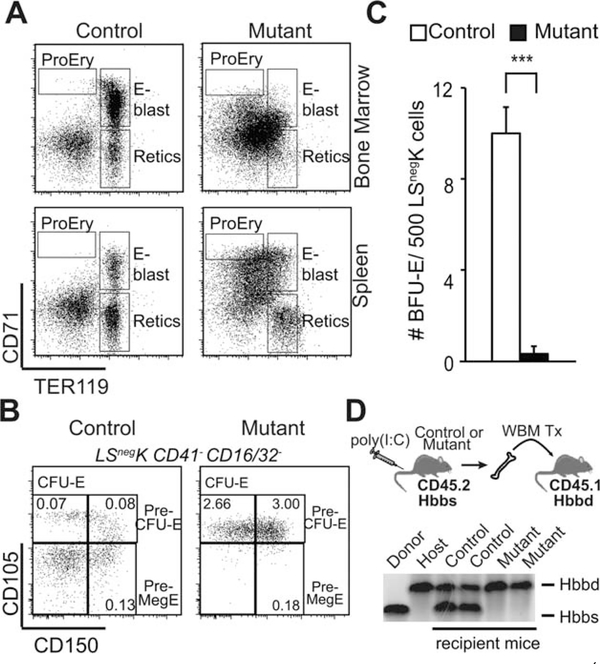
To identify the specific stages of adult erythroid cell maturation that were disrupted by the loss of Rcor1, progenitors expressing CD71 and Ter119 were evaluated [29]. These markers distinguish proerythroblasts (CD71hiTer119lo), maturing erythroblasts (CD71hi/intTer119hi), and reticulocytes (CD71−Ter119hi) in normal BM; however, this normal expression pattern was completely disrupted in Rcor1−/− BM and spleen (Fig. 2A). In order to resolve this, we identified phenotypically defined erythroid progenitors in the lineage-Sca1−Kit+ (LSnegK) myeloerythroid progenitor compartment as described by Weissman and colleagues [23]. This analysis revealed a normal frequency of bipotential megakaryocyte/erythroid progenitors (Pre-MegE) and a marked accumulation of Pre-CFU-E and CFU-E progenitors indicating that the loss of Rcor1 does not prevent erythroid progenitor specification (Fig. 2B). To functionally test the maturation of Rcor1−/− erythroid progenitors, LSnegK cells were plated in cytokine supplemented methylcellulose. Rcor1-deficient BM showed a 90% reduction in the formation of burst forming unit-erythroid (Fig. 2C) colonies comprised of maturing RBCs that develop from Pre-MegE and Pre-CFU-E. These findings indicate that Rcor1−/− deficiency blocks the terminal differentiation of erythroid-committed progenitor cells.
Figure 2.
Blocked erythroid lineage maturation in Rcor1−/− mice. (A): Flow cytometry analysis of developing red cells in the bone marrow (BM). Normal populations of maturing erythroid cells were not detected in mutant mice. (B): Flow cytometry analysis of BM progenitors by Pronk et al. [23]. Committed erythroid progenitors (CFU-E and Pre-CFU-E) were significantly expanded in Rcor1−/− mice (n = 3) relative to controls (n = 3), but bipotent erythroid and megakaryocyte progenitors (pre-MegE) were not. The frequency of each progenitor population in total BM is indicated. (C): Reduced erythroid colony activity in Rcor1−/− myeloerythroid progenitors (Lin−Sca1−c-kit+, LSnegK) (SEM is shown; ***, p < .001). Data from three independent experiments are shown. (D): Transplantation schema for assessing the RBC potential of Rcor1−/− BM cells in a wild-type hematopoietic microenvironment. Although PB donor cell analysis revealed a significant contribution of Rcor1−/− cells to circulating leukocytes in recipient mice 4 weeks post-transplant (not shown), hemoglobin derived from Rcor1−/− donor cells was below the level of detection. Abbreviations: BFU-E, burst forming unit-erythroid; CFU-E, colony-forming unit-erythroid; E-blast, erythroblasts; ProEry, proerythroblasts; Retics, reticulocytes.
To determine whether a normal hematopoietic microenvironment can attenuate this block in erythroid maturation, BM was isolated from CD45.2 Rcor1−/− mice with the Hbbd hemoglobin haplotype and transplanted into CD45.1 recipients that carry the Hbbs hemoglobin haplotype. Donor cell engraftment was monitored up to 9 weeks post-transplant, and all recipient mice appeared healthy with normal RBC indices (data not shown). Although >50% of circulating leukocytes were derived from Rcor1−/− donors, no Rcor1−/− donor-derived hemoglobin was detected in the peripheral blood (Fig. 2D). These data demonstrate a cell-autonomous requirement for Rcor1 during the maturation of committed erythroid progenitor cells into mature red blood cells.
Thrombopoiesis Is Not Rcor1 Dependent
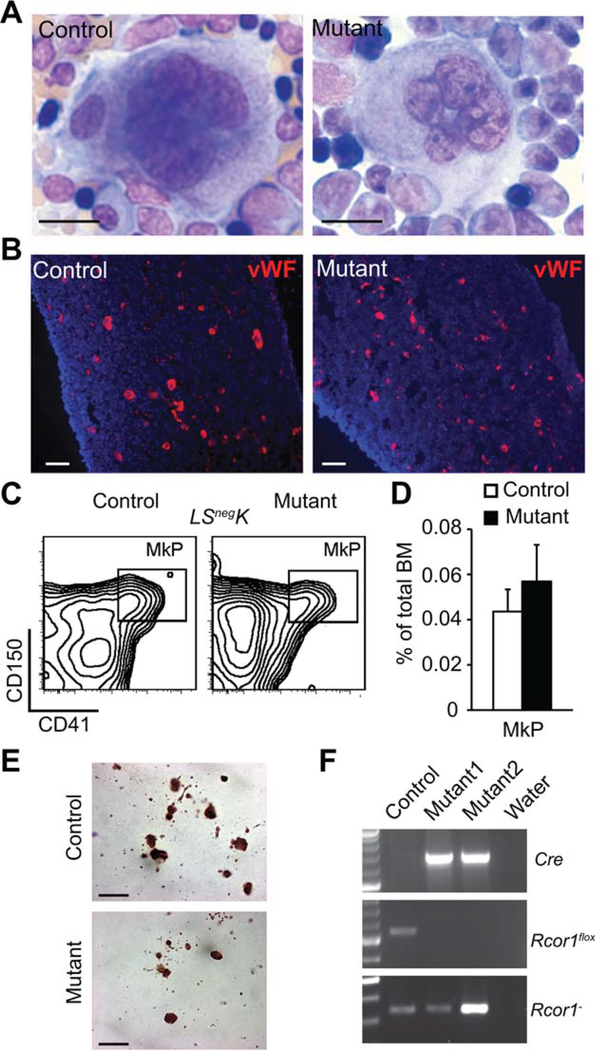
Although a previous in vitro study suggested that Rcor1 is essential for megakaryocytic maturation [10], platelet counts were not reduced following the deletion of Rcor1 in vivo (Fig. 1E). Megakaryocytes showed both a normal morphology and frequency in Rcor1−/− BM (Fig. 3A, 3B). The frequency of immunophenotypically defined megakaryocyte progenitors (MkP: Lin−, Scal-1−, c-kit+, CD150+, CD41+) [23] was indistinguishable from controls (Fig. 3C, 3D). In culture conditions supporting megakaryocyte differentiation from MkP, Rcor1−/− BM readily produced acetylcholinesterase+ (AchE+) megakaryocytic colonies (Fig. 3E). To exclude the possibility that Rcor1flox/− megakaryocytes persist after poly(I:C) treatment due to incomplete Cre-mediated recombination in vivo, MkP were sorted from BM (Fig. 3C) and genotyped. Only the Rcor1− allele was detected, indicating complete ablation of Rcor1 in progenitors of the megakaryocytic lineage (Fig. 3F). Together, these data indicate that Rcor1 is not essential for megakaryocytic progenitor specification, megakaryocyte maturation, or platelet production.
Figure 3.
Rcor1 is dispensable for thrombopoiesis. (A): May-Grunwald Giemsa stained bone marrow (BM) touch preparations revealed normal megakaryocyte morphology in Rcor1−/− mice. (B): vWF+ (red) megakaryocytes with a normal distribution in Rcor1−/− BM. Tibia sections were counterstained with 4′,6-diamidino-2-phenylindole (blue). (C): Flow cytometry analysis of MkP. (D): MkP frequency was not significantly altered in Rcor1-deficient BM (nmutant = 5, ncontrol = 3). (E): Typical acetylcholinesterase+ CFU-Mk derived from control and mutant BM cells. (F): The Rcor1flox allele was not detectable in sorted MkPs after Cre induction. Scale bars = (A–B) 20 μm; (E) 100 μm. Abbreviation: MkP, megakaryocyte progenitors.
Non-Cell Autonomous Suppression of B-Cell Survival in Rcor1-Deficient Mice
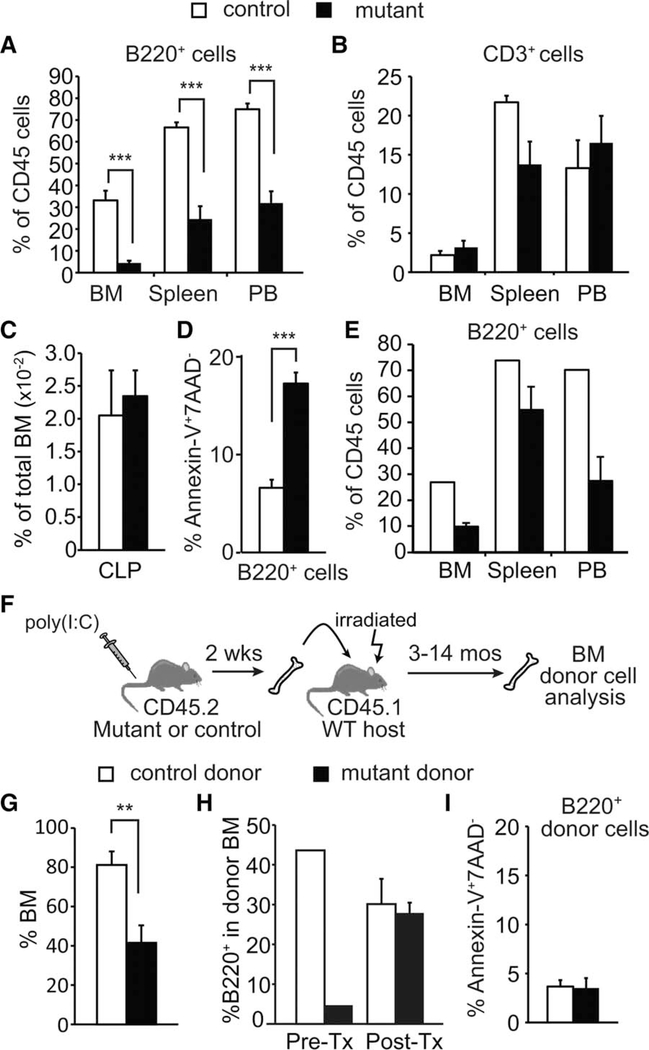
To determine whether Rcor1 is essential for normal lymphocyte production, we evaluated B220+ B cells and CD3+ T cells in Rcor1-deficient mice. Although B cells were reduced by greater than twofold in BM, spleen, and peripheral blood (PB), the number of T cells was not significantly altered (Fig. 4A, 4B). The frequency of CLP in the BM was not impacted by the loss of Rcor1 (Fig. 4C). However, apoptosis was increased 2.6-fold in Rcor1-deficient B cells compared to controls (Fig. 4D). Non-hematopoietic BM stromal cells have been shown to provide an important functional niche for B cells [30, 31]. To assess whether restricting the loss of Rcor1 to hematopoietic cells was sufficient to suppress B cells, we generated chimeric mice in which Mx1-Cre; Rcor1flox/− cells contributed to >90% of hematopoietic cells (Fig. 1H) and analyzed B cells 2 weeks after Cre induction (Fig. 4E). The loss of B cells observed in the hematopoietic-restricted knockout mice indicated a hematopoietic cell-intrinsic requirement for Rcor1 in supporting B-cell survival. Surprisingly, we found that in a transplant setting in which Rcor1-deficient donor cells comprise less than 50% of total BM cells (Fig. 4F, 4G), Rcor1−/− donor cells gave rise to B cells at the same frequency as control donor cells (Fig. 4H). Genotype analysis of peripheral blood isolated from host mice confirmed Rcor1 deletion in donor cells (data not shown). Consistent with normal B-cell levels in this setting, no difference in B-cell apoptosis was detected in Rcor1−/− B-cells post-transplant (Fig. 4I). These results indicate that the significant loss of B cells observed when Rcor1-deficient hematopoietic cells comprise nearly all BM is primarily due to a non-cell autonomous effect that is mediated by Rcor1−/− cells in other hematopoietic lineages.
Figure 4.
Non-cell autonomous suppression of B-cell survival in Rcor1-deficient mice. (A, B): Deletion of Rcor1 leads to a decreased number of B220+ B cells in bone marrow, spleen, and peripheral blood, but does not significantly impact CD3+ T cells (nmutant = 6, ncontrol = 5). (C): Normal frequency of CLP in Rcor1-deficient BM (nmutant = 5, ncontrol = 4). (D): Increased apoptosis in BM B cells following Rcor1 deletion (nmutant = 4, ncontrol = 4). (E): Transplantation of Mx1-Cre;Rcor1−/flox BM into WT hosts followed by Cre induction (Fig. 1H for details) shows a reduced frequency of B cells in the BM, spleen, and PB after 2 weeks (nmutant cell recipients = 3, ncontrol cell recipients = 2). (F): BM transplant schema. Donor WBM or sorted Lin−Sca1+c-kit+ cells were transplanted into sublethally irradiated WT hosts. (G): Percent donor cell contribution in BM after transplant (nmutant cell recipients = 10, ncontrol cell recipients = 10). (H): Representative donor cell contribution to B cells. Donor BM was analyzed for B-cell content prior to transplant (Pre-Tx) and 10 months post-transplant (Post-Tx). B cells derived from Rcor1−/− donor cells corrected to control levels following engraftment into WT hosts. (I): Apoptosis is not increased in Rcor1−/− donor B220+ B cells post-transplant. Pooled data from three donors for each genotype are shown (nmutant cell recipients = 5, ncontrol cell recipients = 4). SEM is shown; **, p < .01; ***, p < .001. Abbreviations: BM, bone marrow; CLP, common lymphoid progenitors; PB, peripheral blood; WBM, whole bone marrow.
Rcor1 Deficiency Leads to an Absence of Mature Neutrophils and an Increase in Monocytic Cells
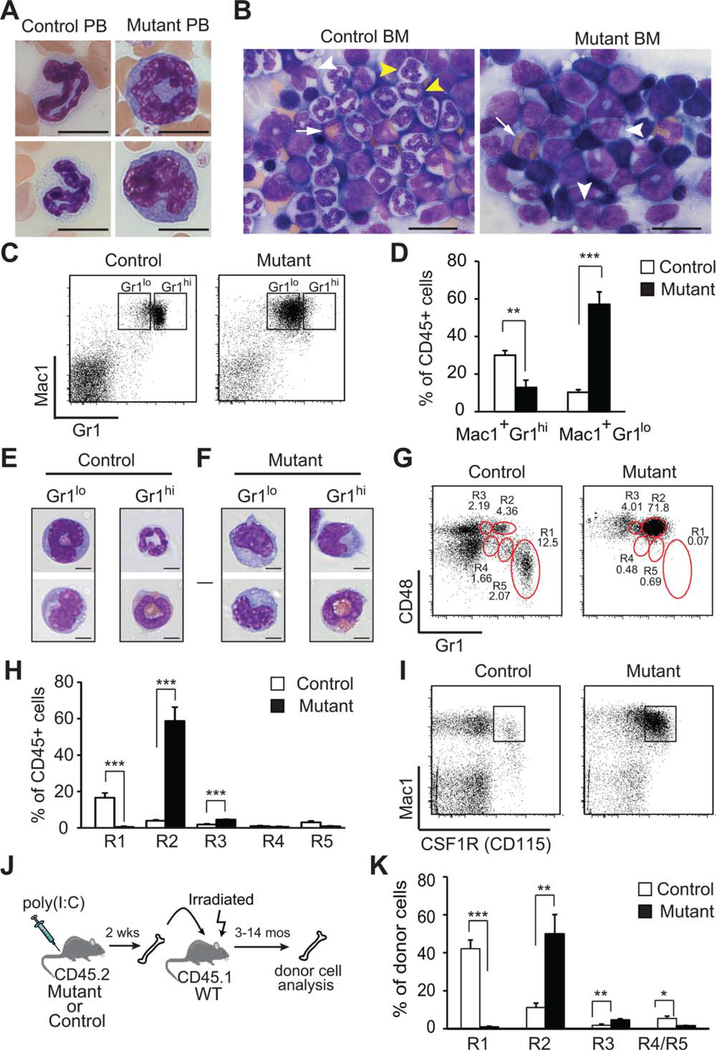
Evaluation of leukocytes in the blood and BM, the Rcor1−/− mice revealed the presence of many immature myelomonocytic cells and an almost complete absence of maturing/mature neutrophils (Fig. 5A, 5B). Cells expressing the Mac1+Gr1hi neutrophil phenotype were markedly decreased within the BM, spleen, and blood, and those Mac1+Gr1hi cells that were detected were either eosinophils or monocytes (Fig. 5C–5F and Supporting Information Fig. S1A). More than 50% of the Rcor1−/− BM cells had a Mac1+Gr1lo phenotype compared to 10% in control mice (Fig. 5C, 5D). Both the expansion of Mac1+Grlo cells and loss of Mac1+Gr1hi cells were also observed in Rcor1−/− BM chimeric mice indicating this phenotype is due to a hematopoietic cell-autonomous defect (Fig. 1H and Supporting Information Fig. S1B). Normally, the Mac1+Gr1lo cell population is primarily comprised of maturing granulocytes, with a few monocytes (Fig. 5E) [32–34]. By contrast, the Mac1+Gr1lo cell population from Rcor1−/− BM was almost exclusively monocytes, with only a few immature granulocytes (Fig. 5F). Five functionally and developmentally distinct subpopulations of myelomonocytic cells (R1-R5) can be resolved by distinct patterns of CD48 and Gr-1 expression [34]. In the Rcor1−/− mice, although the maturing granulocytic cell population (R1) was essentially absent, granulocyte-committed precursor populations (R4, R5) were detected at a normal frequency, suggesting that neutrophil differentiation but not specification is Rcor1 dependent (Fig. 5G, 5H). The frequency of monocytic cells (R2) and mixed potential cells (R3) was increased 15-fold and 2.3-fold, respectively. Supporting this finding of expanded monopoiesis, most of the Mac1+ cells in the Rcor1-deficient mice also coexpressed CD115, the CSF-1 receptor (Fig. 5I). Furthermore, Rcor1−/− cells displayed the identical myelomonocytic differentiation defects after transfer into wild-type hosts (Fig. 5J, 5K and Supporting Information Fig. S1C, S1D). Although in some mice the Rcor1−/− myelomonocytic cells slowly increased over time, progression to leukemia was not observed for up to 12 months.
Figure 5.
Rcor1-deficiency produces a loss of neutrophils and increased numbers of monocytes. May-Grunwald Giemsa stained (A) blood smears and (B) BM touch preparations. Maturing and mature neutrophils in PB and the BM (yellow arrow heads) were absent from Rcor1−/− mice, whereas monocytes (white arrow head) and eosinophils (white arrow) were present. (C): Mac1 and Gr1 expression on bone marrow cells. (D): A significant reduction in the proportion of Mac1+Gr1hi cells and a significant increase in the proportion of Mac1+Gr1lo cells were observed in Rcor1-deficient BM (nmutant = 9, ncontrol = 9). (E, F): Morphology of sorted myelomonocytic cells from control (E) and Rcor1-deficient (F) BM. Most Rcor1−/− cells were monocytes. (G): Phenotype of myelomonocytic cells mice based on CD48 and Gr1 expression patterns. (H): Although Rcor1-deficient mice lacked mature granulocytes (R1), they retained granulocytic precursor cells (R4, R5) and had significantly increased monocytic (R2) and bipotential (R3) cell populations relative to control mice, (nmutant = 4, ncontrol = 4). (I): Consistent with their monocytic phenotype, most Rcor1−/− Mac1+ cells coexpressed CSF1R. (J-K) Rcor1−/− donor cells maintained their abnormal monocytic phenotype after transplant into wild-type hosts (nmutant = 5, ncontrol = 5). SEM is shown; **, p < .01; ***, p < .001. Scale bars = 10 μm (A), 20 μm (B), and 5 μm (E, F). Abbreviations: BM, bone marrow; PB, peripheral blood.
Rcor1 Deficiency Leads to the Accumulation of Abnormal Monocytic Cells
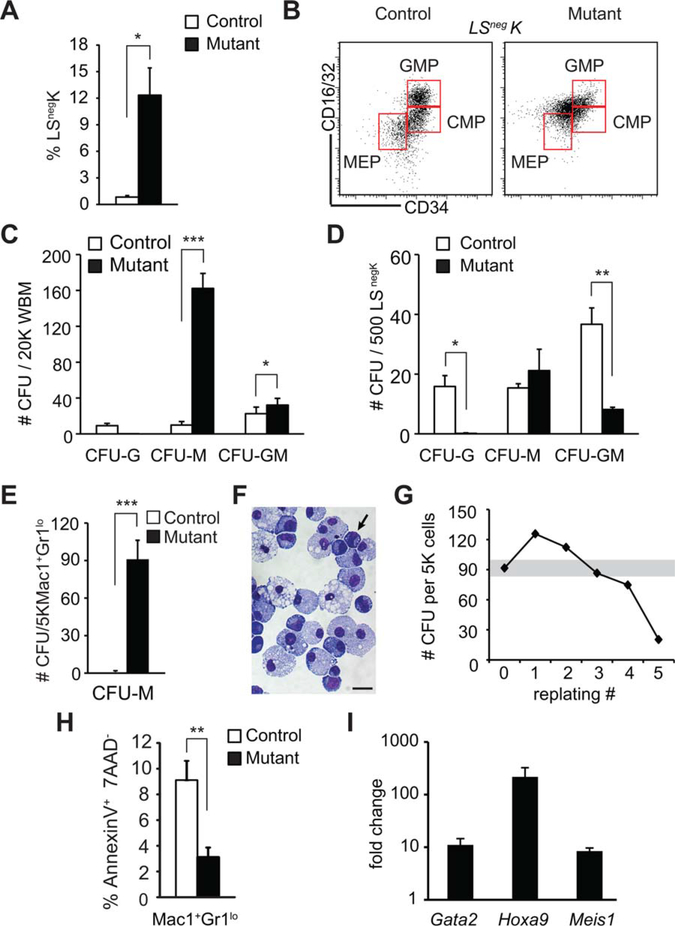
To further evaluate monocytic cell expansion in the Rcor1−/− mice, the frequency of both phenotypically and functionally defined myeloid progenitor cells was assessed. Although the number of the LSnegK myeloerythroid progenitors was increased 14-fold in the Rcor1−/− BM (Fig. 6A), distinct populations of the downstream CMP and GMP could not be easily identified due to aberrant patterns of cell surface marker expression (Fig. 6B). Using the phenotypic analysis described by Pronk et al. [23] as an alternative approach, we found that some myeloid progenitors inappropriately expressed CD105 and that pre-GM (GMP precursors) were reduced by 66%, whereas GMPs were increased by ~60% (Supporting Information Fig. S2A–S2D).
Figure 6.
Enhanced self-renewal activity in Rcor1-deficient monocytic cells. (A): Expansion of Lin−Sca1−ckit+ (LSnegK) myeloerythroid progenitor cells in Rcor1−/− bone marrow (BM) (nmutant = 5, ncontrol = 4). (B): Aberrant CMP/GMP/MEP progenitor [24] fluorescence-activated cell sorting profile in mutant mice. (C, D): Myelomonocytic progenitor colony-forming activity in: (C) unfractionated BM; (D) sorted LSnegK cells; (E) sorted Mac1+Gr1lo cells. Pooled results from three independent experiments are shown. (F): May-Grunwald Giemsa stained cells from pooled Rcor1−/− colonies shown in (E). Immature cells similar to the Mac1+Gr1lo cells directly isolated from mutant mice persist in culture (arrow). (G): CFU activity following serial replating every 8 days. Mac1+Gr1lo cells from Rcor1−/− mutants produced stable levels of CFU-activity through four of five serial replatings. (H): Decreased apoptosis in Mac1+Gr1lo cells in Rcor1−/− BM (nmutant = 5, ncontrol = 4). (I): Gata2, Hoxa9, and Meis1 are derepressed in Rcor1-deficient Mac1+Gr1lo cells. Pooled results from three to five qRT-PCR analyses are shown. SEM is indicated, *, p < .05; **, p < .01; ***, p < .001. Scale bar = 10 μm. Abbreviations: CFU, colony-forming unit; CMP, common myeloid progenitors; GMP, granulocyte-monocyte progenitors; MEP, megakaryocyte-erythroid progenitors.
To functionally assess myeloid cell progenitor activity in Rcor1−/− mice, the myeloid colony-forming activity of both unfractionated BM and sorted LSnegK cells was tested. Rcor1−/− BM formed significantly more myeloid colonies than controls on a per cell basis, indicative of increased progenitor activity (Fig. 6C). Specifically, colony-forming activity for monocytes/macrophages (CFU-M) was 16-fold higher in Rcor1−/− BM. Although mutant LSnegK cells were also CFU-M biased, they formed 50% fewer myeloid CFU than their normal counter parts (Fig. 6D), produced no detectable granulocytic colonies (CFU-G) and showed a 66% decrease in granulocyte/monocyte/macrophage colonies (CFU-GM). Thus, the majority of the total myelomonocytic CFU activity in the Rcor1−/− BM must have arisen from cells outside of the LSnegK progenitor cell phenotype (Fig. 6A).
As the Mac1+Gr1lo myeloid cells comprise a large proportion of the Lin+ BM cells in Rcor1−/− mice, we directly tested their functional activity. As expected, control Mac1+Gr1lo cells had minimal CFU potential, forming <1 colony per 5,000 cells. By contrast, Rcor1−/− Mac1+Gr1lo cells generated a mean of 92 CFU-M colonies per 5,000 cells (Fig. 6E).Morphological analysis of the colonies derived from these mutant Mac1+Gr1lo cells confirmed their monocytic identity and also revealed that these cells were capable of differentiating into macrophages (Fig. 6F). Remarkably, the Mac1+Gr1lo cells from Rcor1-deficient mice also possessed extensive serial replating activity and continually produced CFU-M for up to five serial passages in culture (Fig. 6G). Importantly, this robust CFU-M activity was dependent on the presence of cytokines, indicating these monocytic cells were not able to proliferate autonomously. Finally, analysis of apoptosis in Mac1+Gr1lo BM cells directly isolated from Rcor1−/− mice revealed a 60% reduction in the frequency of apoptotic cells (Fig. 6H). Together, these data indicate that the Mac1+Gr1lo cell subset in the Rcor1−/− mice is cytokine-dependent, monocyte lineage cells that are resistant to apoptosis, and possess extensive, abnormal self-renewal activity.
Transcription Factors that Regulate Stem/Progenitor Cell Proliferation Are Derepressed in Rcor1-Deficient Monocytic Cells
As we previously demonstrated that in fetal liver erythroid progenitors, Rcor1 suppresses hematopoietic stem/progenitor cell gene expression signatures [20], we evaluated the expression levels of Gata2, Meis1, and Hoxa9 in Rcor1-deficient Mac1+Gr1lo cells. These transcription factors are known regulators of hematopoietic stem cell proliferation [35–38] and are normally expressed at high levels in hematopoietic stem cells relative to more differentiated myelomonocytic cells [39–42]. Moreover, their activity has been implicated in myeloid leukemia [43–46], and expression of Gata2 and Hoxa9 has been demonstrated to be elevated in patients with myelodysplastic syndrome (MDS) [47–50]. qPCR revealed large increases in the transcript levels (8- to >200-fold) of all three genes in Rcor1-deficient Mac1+Gr1lo cells relative to controls (Fig. 6I). Thus, Gata2, Hoxa9, and/or Meis1 derepression may enhance the proliferative capacity and self-renewal activity of Rcor1-deficient monocytic cells. Future studies will elucidate whether Rcor1 directly regulates their expression in myeloid cells.
Discussion
This study provides the first in vivo functional analysis of the corepressor Rcor1 in adult hematopoiesis and identifies its critical roles in myeloerythroid lineage differentiation. The Rcor1/Kdm1a complex was previously shown to be a cofactor for Gfi1 or Gfi1b mediated transcriptional repression in hematopoietic cell lines [10]. A comparison of hematopoietic defects in our Rcor1 knockout models with those reported for Kdm1a, Gfi1, and Gfi1b knockout mice provides additional insights into the biological relevance of these biochemical interactions. For example, if Rcor1 works together with Kdm1a, Gfi1, or Gfi1b to regulate a specific hematopoietic lineage in vivo, then it would be predicted that the loss of any component of this complex would cause identical lineage-specific defects. Indeed, the erythroid defects observed in the Rcor1−/− mice largely phenocopy those observed in both Kdm1a-deficient and Gfi1b-deficient mice [19, 51]. Likewise, the ablation of Kdm1a, Gfi1, or Rcor1 causes severe neutropenia in adult mice [11, 12, 51]. Thus, all available data support previous reports that Rcor1/Kdm1a/Gfi1b and Rcor1/Kdm1a/Gfi1 complexes regulate erythroid and granulocytic maturation, respectively.
Importantly, hematopoietic defects associated with Rcor1-, Kdma1-, Gfi1-, or Gfi1b-deficiency in vivo demonstrate several functional differences. One of the most discordant phenotypes is within the megakaryocyte lineage. While loss of Kdm1a or Gfi1b produces severe reduction in platelets counts [19, 51], the loss of Rcor1 causes a transient increase in platelets. Moreover, Gfi1b−/− cells derived from either fetal liver or adult BM cannot form AchE+ megakaryocytic colonies in culture [18, 19], whereas Rcor1−/− BM readily produces AchE+ CFU-Mk colonies. A second marked phenotypic difference among Rcor1, Kdm1a, and Gfi1 mutant mice is within the lymphoid progenitors. Whereas loss of Kdma1 or Gfi1 impairs the formation of CLP [51, 52], a normal CLP frequency was found in Rcor1-deficient mice. The maintenance of thrombopoiesis and CLP production in Rcor1-deficient mice might reflect functional redundancy among Rcor family members [53]; however, our findings also raise the possibility that Gfi1b, Gfi1, and Kdm1a may normally use a cofactor other than Rcor1 to maintain these two lineages.
Despite a striking loss of B cells in the Rcor1-deficient mice, our transplant studies demonstrate that this phenotype is attributable to a non-cell autonomous mechanism. Suppression of lymphopoiesis can be associated with emergency granulopoiesis [54]. Given that loss of Rcor1 throughout the BM leads to a severe neutropenia, we suspect that the ensuing stress response contributes to B-cell loss. Thus, in a transplant setting where Rcor1−/− cells do not contribute to the majority of BM cells (Fig. 4G), we speculate that because the remaining host cells produce normal levels of neutrophils, Rcor1−/− B cells are not suppressed. Future studies will determine whether there is a cell-autonomous requirement for Rcor1 in B- and T-cell maturation and/or function.
Interestingly, the adult Rcor1 knockout mice exhibited a subset of phenotypes consistent with several clinical features associated with MDS and myeloproliferative disorders [55, 56]. Specifically, the differentiation of two major erythromyelocytic lineages, erythrocytes and neutrophils, was completely blocked following Rcor1 deletion and occurred concomitantly with monocytosis and monocytic liver infiltration. These features meet the World Health Organization criteria for chronic myelomonocytic leukemia (CMML) [57, 58]. Although we have not observed transformation to leukemia in Rcor1−/− BM recipient mice for more than a year following transplant, more time may be required to develop the necessary secondary mutations. The CMML-like disease in mice lacking Rcor1 expression in BM raises the possibility that disruption of RCOR1 expression and/or function may also contribute to CMML pathology in humans. RCOR1 mutations or copy number alterations have not yet been reported in CMML or myeloid malignancies. Given that in the mouse model, Rcor1 haploinsufficiency does not affect normal hematopoiesis, we anticipate that a complete or near complete loss of RCOR1 expression is likely required to elicit phenotypic alterations in humans. Consistent with this possibility, RCOR1 loss was recently reported in a subset of diffuse large B-cell lymphoma patients and is associated with a prognostically significant gene expression signature [59].
Conclusions
We have shown that the maturation of both erythroid cells and neutrophils is critically dependent on the transcription corepressor Rcor1. By contrast, monocytic cell survival and self-renewal are dramatically enhanced in the absence of Rcor1. These findings provide new insights into the complexity of the transcription regulatory networks that regulate normal hematopoiesis. As Rcor1−/− mice develop a neutropenia and monocytosis, it will be important to determine whether RCOR1 contributes to pathogenesis in a wide spectrum of human myeloid disorders.
Supplementary Material
Significance Statement.
In vitro interactions between histone modifying enzymes and the transcriptional co-repressor Rcor1 suggest it has an important role in the epigenetic regulation blood cell development. This study provides the first in vivo evidence that Rcor1 is essential for the normal production of mature red blood cells and neutrophils but not platelets. Rcor1-deficient monocytes are increased in number, have an enhanced capacity to self-renew, but remain cytokine dependent and do not give rise to leukemia. Rcor1 loss also leads to the aberrant expression of transcription factors that normally promote the replication of hematopoietic stem/progenitor cells. These findings demonstrate that Rcor1 is essential for red cell and neutrophil maturation while restricting the self-renewal potential of monocytes.
Acknowledgments
We thank Carly Hernandez, Kim Hamlin, Hyunjung Lee, Andrea Ansari, and Travis Polston for technical support. This work was supported by NIH grants to W.H.F. (HL069133 and S10RR027376) and G.M. (NS22518). H.Y. was supported by an American Heart Association predoctoral fellowship. G.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Andres ME, Burger C, Peral-Rubio MJ et al. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA 1999;96:9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphrey GW, Wang Y, Russanova VR et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem 2001;276:6817–6824. [DOI] [PubMed] [Google Scholar]
- 3.You A, Tong JK, Grozinger CM et al. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc Natl Acad Sci USA 2001;98:1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hakimi MA, Bochar DA, Chenoweth J et al. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA 2002;99:7420–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang M, Gocke CB, Luo X et al. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell 2006;23:377–387. [DOI] [PubMed] [Google Scholar]
- 6.Lee MG, Wynder C, Cooch N et al. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437:432–435. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Lan F, Matson C et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119: 941–953. [DOI] [PubMed] [Google Scholar]
- 8.Lattin J, Zidar DA, Schroder K et al. G-protein-coupled receptor expression, function, and signaling in macrophages. J Leukoc Biol 2007;82:16–32. [DOI] [PubMed] [Google Scholar]
- 9.Wu C, Orozco C, Boyer J et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 2009;10:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saleque S, Kim J, Rooke HM et al. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell 2007; 27:562–572. [DOI] [PubMed] [Google Scholar]
- 11.Karsunky H, Zeng H, Schmidt T et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet 2002;30:295–300. [DOI] [PubMed] [Google Scholar]
- 12.Hock H, Hamblen MJ, Rooke HM et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 2003;18:109–120. [DOI] [PubMed] [Google Scholar]
- 13.Phelan JD, Shroyer NF, Cook T et al. Gfi1-cells and circuits: Unraveling transcriptional networks of development and disease. Curr Opin Hematol 2010;17:300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: Key regulators of hematopoiesis. Leukemia 2010;24:1834–1843. [DOI] [PubMed] [Google Scholar]
- 15.Moroy T, Khandanpour C. Growth factor independence 1 (Gfi1) as a regulator of lymphocyte development and activation. Semin Immunol 2011;23:368–378. [DOI] [PubMed] [Google Scholar]
- 16.Doan LL, Kitay MK, Yu Q et al. Growth factor independence-1B expression leads to defects in T cell activation, IL-7 receptor alpha expression, and T cell lineage commitment. J Immunol 2003;170:2356–2366. [DOI] [PubMed] [Google Scholar]
- 17.Yucel R, Karsunky H, Klein-Hitpass L et al. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med 2003;197: 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saleque S, Cameron S, Orkin SH. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev 2002;16:301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foudi A, Kramer DJ, Qin J et al. Distinct, strict requirements for Gfi-1b in adult bone marrow red cell and platelet generation. J Exp Med 2014;211:909–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao H, Goldman DC, Nechiporuk T et al. Corepressor Rcor1 is essential for murine erythropoiesis. Blood 2014;123:3175–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Bailey AS, Jiang S et al. Endothelial cells mediate the regeneration of hematopoietic stem cells. Stem Cell Res 2010;4:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldman DC, Bailey AS, Pfaffle DL et al. BMP4 regulates the hematopoietic stem cell niche. Blood 2009;114:4393–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pronk CJ, Rossi DJ, Mansson R et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 2007; 1:428–442. [DOI] [PubMed] [Google Scholar]
- 24.Akashi K, Traver D, Miyamoto T et al. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000;404:193–197. [DOI] [PubMed] [Google Scholar]
- 25.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 1997; 91:661–672. [DOI] [PubMed] [Google Scholar]
- 26.Baumann CI, Bailey AS, Li W et al. PECAM-1 is expressed on hematopoietic stem cells throughout ontogeny and identifies a population of erythroid progenitors. Blood 2004;104:1010–1016. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn R, Schwenk F, Aguet M et al. Inducible gene targeting in mice. Science 1995;269:1427–1429. [DOI] [PubMed] [Google Scholar]
- 28.Park D, Spencer JA, Koh BI et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 2012;10: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Socolovsky M, Nam H, Fleming MD et al. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood 2001;98:3261–3273. [DOI] [PubMed] [Google Scholar]
- 30.Day RB, Bhattacharya D, Nagasawa T et al. Granulocyte colony-stimulating factor reprograms bone marrow stromal cells to actively suppress B lymphopoiesis in mice. Blood 2015;125:3114–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokoyoda K, Egawa T, Sugiyama T et al. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity 2004;20:707–718. [DOI] [PubMed] [Google Scholar]
- 32.Hestdal K, Ruscetti FW, Ihle JN et al. Characterization and regulation of RB6–8C5 antigen expression on murine bone marrow cells. J Immunol 1991;147:22–28. [PubMed] [Google Scholar]
- 33.Walkley CR, Yuan YD, Chandraratna RA et al. Retinoic acid receptor antagonism in vivo expands the numbers of precursor cells during granulopoiesis. Leukemia 2002;16:1763–1772. [DOI] [PubMed] [Google Scholar]
- 34.Vassen L, Duhrsen U, Kosan C et al. Growth factor independence 1 (Gfi1) regulates cell-fate decision of a bipotential granulocytic-monocytic precursor defined by expression of Gfi1 and CD48. Am J Blood Res 2012;2:228–242. [PMC free article] [PubMed] [Google Scholar]
- 35.Cai M, Langer EM, Gill JG et al. Dual actions of Meis1 inhibit erythroid progenitor development and sustain general hematopoietic cell proliferation. Blood 2012;120:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence HJ, Christensen J, Fong S et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 2005;106:3988–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling KW, Ottersbach K, van Hamburg JP et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med 2004;200:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigues NP, Boyd AS, Fugazza C et al. GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood 2008;112: 4862–4873. [DOI] [PubMed] [Google Scholar]
- 39.Chen W, Kumar AR, Hudson WA et al. Malignant transformation initiated by Mll-AF9: Gene dosage and critical target cells. Cancer Cell 2008;13:432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pineault N, Helgason CD, Lawrence HJ et al. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol 2002;30:49–57. [DOI] [PubMed] [Google Scholar]
- 41.Tsai FY, Keller G, Kuo FC et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994;371: 221–226. [DOI] [PubMed] [Google Scholar]
- 42.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 1997;89:3636–3643. [PubMed] [Google Scholar]
- 43.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene 2007;26:6766–6776. [DOI] [PubMed] [Google Scholar]
- 44.Argiropoulos B, Yung E, Humphries RK. Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes Dev 2007;21:2845–2849. [DOI] [PubMed] [Google Scholar]
- 45.Eklund E The role of Hox proteins in leukemogenesis: Insights into key regulatory events in hematopoiesis. Crit Rev Oncog 2011;16:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vicente C, Conchillo A, Garcia-Sanchez MA et al. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol 2012;82:1–17. [DOI] [PubMed] [Google Scholar]
- 47.Bresolin S, Trentin L, Zecca M et al. Gene expression signatures of pediatric myelodysplastic syndromes are associated with risk of evolution into acute myeloid leukemia. Leukemia 2012;26:1717–1719. [DOI] [PubMed] [Google Scholar]
- 48.Fadilah SA, Cheong SK, Roslan H et al. GATA-1 and GATA-2 gene expression is related to the severity of dysplasia in myelodysplastic syndrome. Leukemia 2002;16: 1563–1565. [DOI] [PubMed] [Google Scholar]
- 49.Heinrichs S, Berman JN, Ortiz TM et al. CD34+ cell selection is required to assess HOXA9 expression levels in patients with myelodysplastic syndrome. Br J Haematol 2005;130:83–86. [DOI] [PubMed] [Google Scholar]
- 50.Inoue D, Kitaura J, Togami K et al. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J Clin Invest 2013;123:4627–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kerenyi MA, Shao Z, Hsu YJ et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife 2013;2: e00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng H, Yucel R, Kosan C et al. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 2004;23:4116–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang P, Wang Y, Chen J et al. RCOR2 is a subunit of the LSD1 complex that regulates ESC property and substitutes for SOX2 in reprogramming somatic cells to pluripotency. Stem Cells 2011;29:791–801. [DOI] [PubMed] [Google Scholar]
- 54.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol 2014;14:302–314. [DOI] [PubMed] [Google Scholar]
- 55.Beachy SH, Aplan PD. Mouse models of myelodysplastic syndromes. Hematol Oncol Clin North Am 2010;24:361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vardiman JW, Thiele J, Arber DA et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood 2009;114:937–951. [DOI] [PubMed] [Google Scholar]
- 57.Patnaik MM, Parikh SA, Hanson CA et al. Chronic myelomonocytic leukaemia: A concise clinical and pathophysiological review. Br J Haematol 2014;165:273–286. [DOI] [PubMed] [Google Scholar]
- 58.Swerdlow SH, International Agency for Research on Cancer, World Health Organization. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer, 2008:439. [Google Scholar]
- 59.Chan FC, Telenius A, Healy S et al. An RCOR1 loss-associated gene expression signature identifies a prognostically significant DLBCL subgroup. Blood 2015;125:959–966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.