Abstract
The purpose of this manuscript is to provide an overview of the technical specifications and architecture of the Cancer imaging Phenomics Toolkit (CaPTk www.cbica.upenn.edu/captk), a cross-platform, open-source, easy-to-use, and extensible software platform for analyzing 2D and 3D images, currently focusing on radiographic scans of brain, breast, and lung cancer. The primary aim of this platform is to enable swift and efficient translation of cutting-edge academic research into clinically useful tools relating to clinical quantification, analysis, predictive modeling, decision-making, and reporting workflow. CaPTk builds upon established open-source software toolkits, such as the Insight Toolkit (ITK) and OpenCV, to bring together advanced computational functionality. This functionality describes specialized, as well as general-purpose, image analysis algorithms developed during active multi-disciplinary collaborative research studies to address real clinical requirements. The target audience of CaPTk consists of both computational scientists and clinical experts. For the former it provides i) an efficient image viewer offering the ability of integrating new algorithms, and ii) a library of readily-available clinically-relevant algorithms, allowing batch-processing of multiple subjects. For the latter it facilitates the use of complex algorithms for clinically-relevant studies through a user-friendly interface, eliminating the prerequisite of a substantial computational background. CaPTk’s long-term goal is to provide widely-used technology to make use of advanced quantitative imaging analytics in cancer prediction, diagnosis and prognosis, leading toward a better understanding of the biological mechanisms of cancer development.
Keywords: CaPTk, Cancer, Imaging, Phenomics, Toolkit, Radiomics, Radiogenomics, Radiophenotype, Segmentation, Deep learning, Brain tumor, Glioma, Glioblastoma, Breast cancer, Lung cancer, ITCR
1. Introduction
The bane of computational medical imaging research has been its translation to the clinical setting. If novel research algorithms can be validated in ample and diverse data, then they have the potential to contribute to our mechanistic understanding of disease and hence substantially increase their value and impact in both the clinical and scientific community. However, to facilitate this potential translation a Graphical User Interface (GUI) is essential, supportive, and tangential to medical research. Designing and packaging such a GUI is a lengthy process and requires deep knowledge of technologies to which not all researchers are exposed. Providing a computational researcher an easy way to integrate their novel algorithms into a well-designed GUI would facilitate the use of such algorithms by clinical researchers, hence bringing the algorithm closer to clinical relevance.
Towards this effort, numerous open-source applications have been developed [1-4] with varying degrees of success in either the computational and/or clinical research communities. However, each of these had their own limitations. To address these limitations, we developed the Cancer imaging Phenomics Toolkit (CaPTk1) [5,6], which introduces a mechanism to integrate algorithms written in any programming language, while maintaining a lightweight viewing pipeline. CaPTk is a cross-platform (Windows, Linux, macOS), open-source, and extensible software platform with an intuitive GUI for analyzing both 2D and 3D images, and is currently focusing on radiographic scans of brain, breast, and lung cancer. CaPTk builds upon the integration of established open-source toolkits to bring together advanced computational functionality of specialized, as well as general-purpose, image analysis algorithms developed during active multidisciplinary collaborative research studies.
2. Tools Preceding CaPTk
Prior to beginning the development of CaPTk in 2015, we conducted a literature review of the current open-source applications developed by the community to address specific scientific needs such as Quantitative Image Phenomic (QIP) extraction and image annotation. While some of these tools (such as MIPAV [11] and MedINRIA [4]) could not be extended with customized applications, others (namely, 3D-Slicer [1] and MITK [2]) had complex software architecture that would be challenging for a limited team to extensively modify. Additional considerations in the decision to launch CaPTk as an independent toolkit included the substantial code base internal to the Center for Biomedical Image Computing and Analytics (CBICA) relating to existing software applications implemented in C++, existing research directions, software development expertise in C++, and licensing considerations. Furthermore, none of these applications could provide customized interactions for initializing seedpoints, as required by applications such as GLISTR [7] and GLISTRboost [8,9]. Below we refer to specific packages that preceded the development of CaPTk and we attempt to describe their advantages and disadvantages (at that time) compared to the requirements of CaPTk that led to its design and development.
2.1. Medical Imaging Interaction Toolkit (MITK)
One of the earliest works towards an open source extensible toolkit by the scientific community, MITK [2] had spearheaded the adoption of DICOM ingestion by the research community. Designed to be cross-platform, it had been extensively tested on various types of datasets, modalities and organ-systems and has arguably provided one of the best DICOM parsing engines in the community with a very nice mechanism for incorporating plugins that integrate natively into the application. Although MITK’s native plugin integration was not as user-/developer-friendly and required expert knowledge of MITK, it made incorporation of other tools such as XNAT [10] possible. However, we ended up not choosing MITK as an option for our solution as integrating an application written in a different programming language could not be done easily and any modifications of the graphical capabilities, such as custom interaction (e.g., GLISTR/GLISTRboost initializations) or quick GUI modification, was not possible without extensive background knowledge of toolkit’s internals.
2.2. Medical Image Processing, Analysis, and Visualization (MIPAV)
One of the earliest tools funded by the National Institutes of Health (NIH), the MIPAV [11] package was written to provide an easy-to-use user interface for image visualization and interaction. MIPAV is a cross-platform application, with a back-end written in Java. There were some performance issues with algorithms that required a high amount of computation capabilities and it was targeted for specific functionality and had no way to integrate third-party applications.
2.3. 3D Slicer
Perhaps the most popular open source tool for the imaging community, 3D Slicer incorporated a lot of functionality via its extension management system. It was relatively easy for a developer to write a Slicer extension since it provided out-of-the-box support for Python, which potentially resulted in performance issues. It was cross-platform and very well tested by the community and had extensive DICOM support. The user interface, however, was not very friendly for clinical researchers and any changes to either the interactive capabilities or the user interface could not be done easily. In addition, integrating an application written in a language that was not in C++ or Python was also not trivial.
2.4. MeVisLab
MeVisLab [3] was a semi-commercial tool made available by Fraunhofer MEVIS2 which was well tested by their industrial partners, had extensive DICOM support and was well-received by clinicians as well. However, it had limited capability of handling the number (5 in total) and size (2kB source) for extensions in the free version, a serious constraint if we were to use MeVisLab as the basis for providing multiple complex imaging applications to researchers. In addition, non-native extensions were not currently supported.
2.5. medInria
medInria [4] was an imaging platform that focused on algorithms related to image registration, diffusion image processing and tractography. Combined with extensive support for different types of DICOM image formats and its cross-platform capabilities, medInria had excellent annotation capabilities and was extensively used by clinical researchers by virtue of its easy-to-use interface. Nevertheless, because of its inability of assimilating extensions, it became a narrowly focused tool.
2.6. ITK-SNAP
ITK-SNAP [12] was one of the most widely used tools for segmentation and has been used to solve annotation problems in multiple organ systems. It was cross-platform and very well-tested. Nonetheless, because of its inability to support extensions and the architectural complexity of the visualization system, it was not a candidate to use as a base for CaPTk’s development.
3. CaPTk’s Infrastructure
The development of medical imaging tools that can be clinically relevant is driven by a specific medical need around which algorithms are developed, and involves understanding of a researcher’s/clinician’s workflow. As illustrated in Fig. 1, the planning begins with defining the clinical requirement of the task to be accomplished, followed by acquiring relevant sample datasets. Using these initial requirements and datasets, an algorithm is designed and then its usability is established using a series of refinements, including a robust visualization and interaction tool-chain.
Fig. 1.
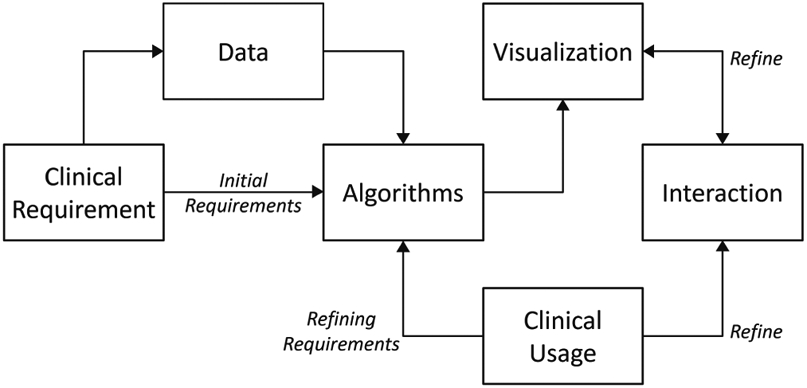
Data flow diagram for tool development planning. Addition of a GUI tremendously increases the usability of the algorithm. Feedback from clinical usage is used to refine the algorithm and rendering further.
3.1. Functionality
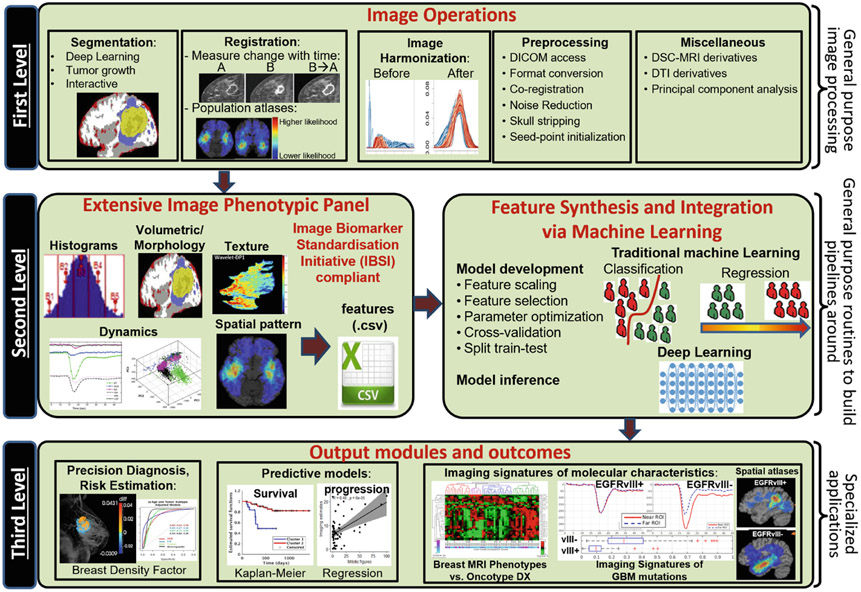
By keeping the data flow practices in mind (Fig. 1), CaPTk has been designed as a general-purpose tool spanning brain, breast, lung, and other cancers. It has a broad, three-level functionality as illustrated in Fig. 2. The first level provides basic image pre-processing tasks such as image input-output (currently supporting NIfTI [13] and DICOM file types), image registration [14], smoothing (denoising) [15], histogram-matching, and novel intensity harmonization for brain magnetic resonance imaging (MRI) scans (namely WhiteStripe [16]), among other algorithms. The second level comprises various general-purpose routines including extensive QIP feature extraction compliant with the Image Biomarker Standardization Initiative (IBSI) [17], feature selection, and a Machine Learning (ML) module. These routines are used within CaPTk for specialized tasks, but are also available to the community as general-purpose analysis steps that sites can use as the basis for pipelines customized for their data. In particular, this level targets extraction of numerous QIP features capturing different aspects of local, regional, and global visual and sub-visual imaging patterns, which, if integrated through the available ML module, can lead to the generation of predictive and diagnostic models. Finally, the third level of CaPTk focuses on specialized applications validated through existing scientific studies keeping reproducibility [18] as the cornerstone of development. These studies include: predictive models of potential tumor recurrence and patient overall survival [19-26], generating population atlases [27], automated extraction of white matter tracts [28], estimation of pseudo-progression for glioblastoma (GBM) [19], evaluating the Epidermal Growth Factor Receptor splice variant III (EGFRvIII) status in GBM [29,30], breast density estimation [31,32], estimation of tumor directionality & volumetric changes in 2 time-points [33], among others.
Fig. 2.
Diagram showcasing the three-level functionality of CaPTk.
CaPTk’s GUI was designed after multiple interactions with radiologists and other clinical collaborators, targeting single subject analysis primarily for clinical research. Along with the easy-to-use GUI, every specialized application within the CaPTk has an accompanying command-line interface (CLI) executable. These CLI applications allow the use of CaPTk functions as components of larger, more complicated pipelines and for efficient batch processing of large numbers of images.
3.2. Architecture
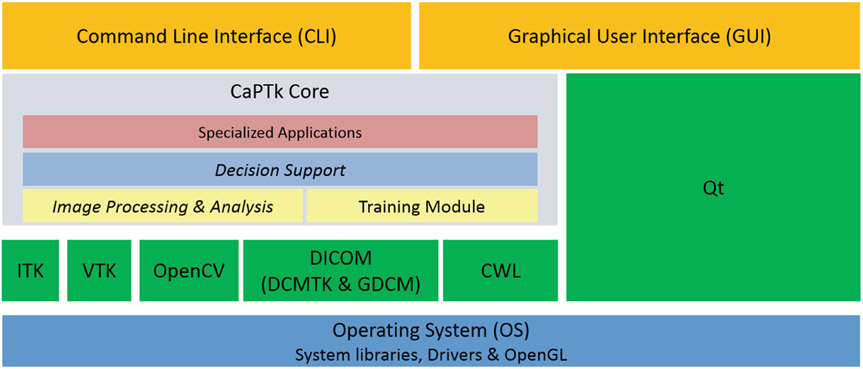
The architecture of CaPTk is depicted in Fig. 3. At the lowest level of the architecture (shown as blue in Fig. 3) is the operating system (OS) and its core components, such as system libraries (OpenGL, OpenMP) and hardware drivers. One level above (in green) are the libraries (e.g., ITK, VTK, OpenCV, Qt) that provide the lower level functionality of CaPTk. The next level consists of CaPTk core libraries constituting the algorithmic functionality of CaPTk, including all its three levels previously described and graphically shown in Fig. 2. At the very top (in orange), lie the GUI and the CLI components.
Fig. 3.
Detailed diagram of CaPTk’s architecture showing the various interdependencies between the components.
The Insight Toolkit (ITK) [34] has helped take research in the field of medical imaging to new heights by providing a set of common Application Programming Interfaces (APIs) and offered the ability for users to easily read, write, and perform computations with medical imaging datasets. ITK is used in CaPTk for tasks related to medical image registration (via the Greedy algorithm3) and segmentation, and ITK functions are components in new image analysis algorithms. The Visualization Toolkit (VTK) [35] has provided powerful functionalities for rendering and interaction of complex datasets. The Open Source Computer Vision Library (OpenCV) [36] has long fostered a healthy community of computer vision researchers and has established an industry-standard library for providing ML algorithms that can be used out-of-the-box, upon which CaPTk is building. For DICOM-related functionalities we leverage the DICOM Toolkit (DCMTK4), Grassroots DICOM (GDCM5), and have incorporated the DCMQI [37] tool for DICOM-Seg output. Qt6 is used for GUI related tasks and cross-platform features. We build upon these open source and well-validated tools for CaPTk’s development, with major emphasis towards being computationally efficient and fully cross-platform across the major desktop operating systems (i.e., Windows, Linux and macOS).
The architecture of CaPTk provides all source-level applications with efficient access to every imaging action and all common functions, treating imaging data as objects that can be passed between applications. This allows multiple applications access to common tasks (ie., pre-processing) without duplication of code or inefficient I/O. Simultaneously, the CaPTk design model that implements procedures written in C++ as both graphical and command-line applications also enables easy incorporation of stand-alone executables, such as ITK-SNAP7 and DeepMedic [38], written in any language.
The CaPTk core provides libraries for different specialized applications, such as those developed for brain, breast, and lung cancer. To ensure reproducibility of the algorithms as originally written, each of these specialized tools is designed as a monolithic application, and is independent of the other general-purpose applications.
3.3. Extending CaPTk
CaPTk can be easily extended with customized applications while leveraging all the graphical and algorithmic functionalities present in the application core. There are two ways for a developer to integrate their application into CaPTk8:
3.3.1. Application-Level Integration
This is the easiest way to call a custom application from CaPTk’s GUI. This can be done for an application written in any language as long as a self-contained executable can be created. Essentially, these applications are called in the same way as they would be executed interactively from the command line. Their dependencies can be provided through the OS platform, distributed as part of the CaPTk distribution (preferable), or can be downloaded from an external site during installation. Once the executable is created, integration in CaPTk requires only minor modification to the build script (CMake) and user interface code. An example of such an integration is the LIBRA application [31,32].
3.3.2. Source-Level Integration
This is the deeper level of application integration and is available if the application is written in C++, possibly incorporating ITK, VTK, and/or OpenCV. To ensure that a source-level integration happens for an application, it needs to be written as a CMake project, i.e., a CMakeLists.txt is essential to define the requirements, project structure, generated libraries and/or executable(s), and the corresponding install targets for CaPTk’s installation framework. The location of the entire project needs to be added to CaPTk’s build path via the appropriate CMake script files. The developer also has the option to add a customized interface for the application. Examples of this kind of integration are the Feature Extraction, Preprocessing and Utilities applications of CaPTk.
3.4. Utilizing CaPTk in Custom Pipelines
The Common Workflow Language (CWL) [39] is an open standard to enable easy description of an executable for analysis workflows and tools in a way that ensures portability and scalability across a wide range of software and hardware environments, ranging from different operating systems to personal machines and High Performance Computing (HPC) environments. CWL has been conceived to meet the needs of high-throughput data-intensive scientific areas, such as Bioinformatics, Medical Imaging, Astronomy, High Energy Physics, and ML.
CaPTk leverages CWL as a means of relaying compact, human- and machine-readable descriptions of applications within the software suite. These descriptions, either in memory or as text files, are used to validate the optional and required inputs to an application. Each native application is specified in the CWL grammar in it’s abstract form that includes all possible parameters, and CWL is further used to pass the actual arguments used in applying an analysis routine to a specific set of data. These CWL descriptions can improve validity of results and enhance data provenance for end-users by recording all details about data and options used to generate results from any CWL pipeline in a compact, reproducible, verifiable format.
We have contributed the C++ implementation of the CWL parser that is used within CaPTk’s CLI9 to the CWL development community. This CWL parser is an easy and portable tool for developers to structure and write a CLI application (Fig. 4). Our contribution should i) allow the easy construction of pipelines without requiring any additional scripting, and ii) provide the ability to run both local and cloud applications. The reading and writing of a CWL definition file is embedded within the Command Line Parser itself to reduce the effort of writing pipelines.
Fig. 4.

The principle of CaPTk’s CWL CLI parser.
3.5. Online Accessibility
CBICA’s Image Processing Portal (IPP10) can be used by anyone to run resource-intensive applications of CaPTk on CBICA’s HPC servers at no charge. CaPTk can be used to initialize the inputs that are required for all applications that are semi-automated, such as GLISTR [7] and GLISTRboost [8]. In addition, applications that may be impractical to run on a personal machine due to their long-running nature can be run remotely on IPP.
3.6. Code Maintenance
To ensure that good practices in open science principles are followed during the CaPTk development, the source code and issue tracker for CaPTk are maintained publicly on GitHub11. The entire history of the source code can be found on the repository. A stable version of CaPTk is compiled and released twice every year, and it includes the source code, as well as self-contained installers for multiple platforms.
CaPTk also follows the software development best practices [40] and employs Continuous Integration and Continuous Deployment (CI/CD) via GitHub and Azure DevOps12 as mechanisms to encourage a rapid development cycle while incorporating work from multiple developers. Any code contributions, either from the CaPTk development team or external users, are merged into the master only after successful completion of CI/CD checks and a code review13. Each push into the master GitHub code repository for CaPTk automatically produces a new binary installer for each supported OS that gives users immediate access to the bleeding edge development of CaPTk.
4. Example of General Purpose Applications
4.1. Quantitative Image Phenomic (QIP) Feature Panel
Building around the functionalities provided by ITK [34] and MITK [2], we have made the Feature Panel available in CaPTk as generic as possible, while at the same time maintaining safeguards to ensure clinical validity of the extracted features. For example, all computations are performed in the physical space of the acquired scan instead of the image space. Options to control the Quantization Extent (i.e., whether it should happen for the entire image or only in the annotated region of interest), the Quantization Type (e.g., Fixed Bin Number, Fixed Bin Size or Equal width [17]), resampling rate, and interpolator type, are all defined in the physical space and accessible to the user to alter as needed for a particular study. The user also has the capability to perform lattice-based feature computations [41,42] for both 2D and 3D images.
There are two broad types of features getting extracted by CaPTk (detailed mathematical formulations can be found at: cbica.github.io/CaPTk/tr_FeatureExtraction.html):
- First Order Features:
- Intensity-based: Minimum, maximum, mean, standard deviation, variance, skewness and kurtosis.
- Histogram-based: Bin frequency & probability, intensity values at the 5th and 95th quantiles, statistics on a per-bin level.
- Volume-based: Number of pixels (for 2D images) or voxels (for 3D images) and their respective area or volume.
- Morphology-based: Various measures of the region of interest such as Elongation, Perimeter, Roundness, Eccentricity, Ellipse Diameter in 2D and 3D, Equivalent Spherical Radius.
- Second Order Features:
- Texture Features: Grey Level Co-occurrence Matrix, Grey Level Run-Length Matrix, Grey Level Size-Zone Matrix, Neighborhood Grey-Tone Difference Matrix are part of this section [17].
- Advanced Features: Includes but is not limited to Local Binary Patterns, frequency domain features, e.g., Gabor and power spectrum [41].
All features are in conformance with the Image Biomarker Standardisation Initiative (IBSI) [17], unless otherwise indicated within the documentation of CaPTk. Most implementations are taken from ITK [34] or MITK [2] with additions made to ensure conformance when the physical coordinate space rather than the image space, which is one of the major sources of variation in between CaPTk and comparative packages, such as PyRadiomics [43]. Other sources of variation include the type of binning/quantization used and the extent on which it is applied. The user also has the option of performing batch processing with multiple subjects14.
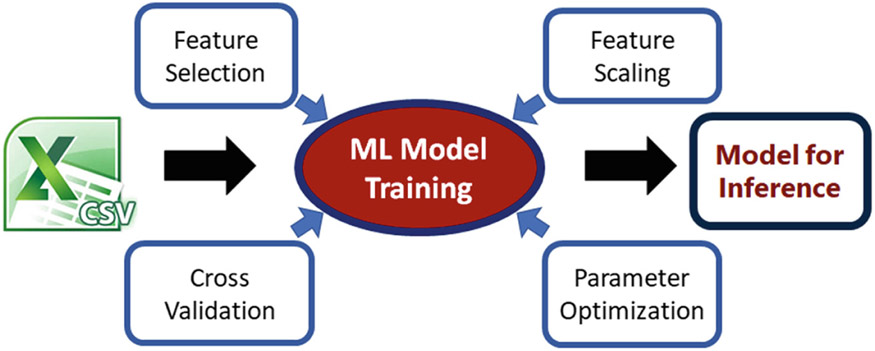
4.2. ML Training Module
We have based our machine learning module on OpenCV’s ML back-end15 and exposed the utility of Support Vector Machines (SVM) using Linear and Radial Basis Function kernels to the user, offering a grid search optimization for c and g. Additional kernels and algorithms will also be offered later. In addition, there is the capability to perform k-fold cross validation and to split the training and testing sets by percentages (Fig. 5).
Fig. 5.
The principle of CaPTk’s machine learning module.
5. Future Directions
We developed CaPTk to address a gap at the time in the existing available tools and to make our preexisting advanced computational studies available to clinicians and imaging researchers in a tool that did not require a computational expert. The growing number of users of CaPTk and the reach of specialized applications within the suite validates our initial decision to produce a software package with interface features designed to make the specialized algorithms accessible in a way that we could not have accomplished with toolkits available when CaPTk was initiated. However, feedback from this wider set of researchers and clinicians has exposed some limitations in the current version of CaPTk that we intend to address in its next major revision, i.e., CaPTk v.2.0.
Importantly, the variability and inconsistency among imaging headers has turned out to be a serious challenge in medical imaging. CaPTk is currently able to handle NIfTI file format and has limited support for handling DICOM files (which is currently based on DCMTK16 and GDCM17) and does not support network retrieval from DICOM-compliant Picture Archiving and Communication Systems (PACS18). Over the course of its evaluation by clinicians, it has become evident that seamless integration with PACS is a necessary element for CaPTk’s future.
Furthermore, a graphical interface for cohort selection and a batch processing interface, combined with the use of CWL to provide data provenence for each step in a pipeline, would also be essential to the construction of replicable research studies. In order to achieve this goal, integration with a data management, archival, and distribution system such as XNAT [10] is essential.
Finally, integration with state-of-the-art tools that rely on server-side communication such as the Deep Learning based NVIDIA-Clara annotation engine19, cannot be easily accomplished with CaPTk in its current state.
We have recently surveyed the state of other image processing applications and toolkits and conducted extensive outreach to prospective collaborators as we begin to design the architecture for the next generation of the Cancer imaging Phenomics Toolkit. While we continue to support and develop the current implementation of CaPTk, plans for the 2.0 branch include support (via CWL) for web- and cloud-based services and leveraging the current and updated functionality of MITK [2] and/or Slicer Core in the future.
6. Conclusions
We developed the CaPTk to provide a library for advanced computational analytics based on existing published multi-disciplinary studies, to meet the need at the time in the field of medical image processing applications for a suite that could be easy extended with components using any programming language. We consider medical image analysis to be much more than just a collection of algorithms; to ensure success of a tool, having the capability to visualize and interact with the original and processed image datasets is equally important. Building upon the powerful segmentation and registration framework provided by ITK and the rendering and interaction capabilities (particularly for 3D datasets) provided by VTK, we have built a light-weight application, CaPTk, that can visualize and interact with different datasets and also provide a solid annotation pipeline, thereby making it a powerful tool for use by clinical researchers. By enabling easy integration of computation algorithms regardless of their programming language, we have further ensured that computational researchers have a quick path for potential translation to the clinic.
Acknowledgments.
CaPTk is primarily funded by the Informatics Technology for Cancer Research (ITCR)20 program of the National Cancer Institute (NCI) of the NIH, under award number U24CA189523, as well as partly supported by the NIH under award numbers NINDS:R01NS042645, NCATS:UL1TR001878, and by the Institute for Translational Medicine and Therapeutics (ITMAT) of the University of Pennsylvania. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH, or the ITMAT of the UPenn.
Footnotes
References
- 1.Kikinis R, Pieper SD, Vosburgh KG: 3D slicer: a platform for subject-specific image analysis, visualization, and clinical support In: Jolesz FA (ed.) Intraoperative Imaging and Image-Guided Therapy, pp. 277–289. Springer, New York: (2014). 10.1007/978-1-4614-7657-3_19 [DOI] [Google Scholar]
- 2.Wolf I, et al. : The medical imaging interaction toolkit. Med. Image Anal 9(6), 594–604 (2005) [DOI] [PubMed] [Google Scholar]
- 3.Link F, Kuhagen S, Boskamp T, Rexilius J, Dachwitz S, Peitgen H: A flexible research and development platform for medical image processing and visualization In: Proceeding Radiology Society of North America (RSNA), Chicago: (2004) [Google Scholar]
- 4.Toussaint N, Souplet J-C, Fillard P: MedINRIA: medical image navigation and research tool by INRIA (2007) [Google Scholar]
- 5.Davatzikos C, et al. : Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging 5(1), 011018 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rathore S, et al. : Brain cancer imaging phenomics toolkit (brain-CaPTk): an interactive platform for quantitative analysis of glioblastoma In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 133–145. Springer, Cham: (2018). 10.1007/978-3-319-75238-9_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gooya A, et al. : GLISTR: glioma image segmentation and registration. IEEE Trans. Med. Imaging 31(10), 1941–1954 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakas S, et al. : GLISTRboost: combining multimodal MRI segmentation, registration, and biophysical tumor growth modeling with gradient boosting machines for glioma segmentation In: Crimi A, Menze B, Maier O, Reyes M, Handels H (eds.) BrainLes 2015. LNCS, vol. 9556, pp. 144–155. Springer, Cham: (2016). 10.1007/978-3-319-30858-6_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng K, et al. : Segmentation of gliomas in pre-operative and post-operative multimodal magnetic resonance imaging volumes based on a hybrid generative-discriminative framework In: Crimi A, Menze B, Maier O, Reyes M, Winzeck S, Handels H (eds.) BrainLes 2016. LNCS, vol. 10154, pp. 184–194. Springer, Cham: (2016). 10.1007/978-3-319-55524-9_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL: The extensible neuroimaging archive toolkit. Neuroinformatics 5(1), 11–33 (2007). 10.1385/NI:5:1:11 [DOI] [PubMed] [Google Scholar]
- 11.McAuliffe MJ, et al. : Medical image processing, analysis and visualization in clinical research In: Proceedings 14th IEEE Symposium on Computer-Based Medical Systems, CBMS 2001, Bethesda, MD, USA, pp. 381–386 (2001). https://ieeexplore.ieee.org/document/941749 [Google Scholar]
- 12.Yushkevich PA, et al. : User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3), 1116–1128 (2006) [DOI] [PubMed] [Google Scholar]
- 13.Cox R, et al. : A (sort of) new image data format standard: NIfTI-1: we 150. Neuroimage 22 (2004). https://www.scienceopen.com/document?vid=6873e18e-a308-4d49-b4aa-8b7f291c613c [Google Scholar]
- 14.Yushkevich PA, Pluta J, Wang H, Wisse LE, Das S, Wolk D: Fast automatic segmentation of hippocampal subfields and medial temporal lobe subregions in 3 tesla and 7 tesla T2-weighted MRI. Alzheimer’s Dement. J. Alzheimer’s Assoc 12(7), P126–P127 (2016) [Google Scholar]
- 15.Smith SM, Brady JM: Susanâ”a new approach to low level image processing. Int. J. Comput. Vis 23(1), 45–78 (1997) [Google Scholar]
- 16.Shinohara RT, et al. : Statistical normalization techniques for magnetic resonance imaging. NeuroImage: Clin 6, 9–19 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zwanenburg A, Leger S, Vallières M, Löck S: Image biomarker standardisation initiative. arXiv preprint arXiv:1612.07003 (2016) [Google Scholar]
- 18.Wilkinson MD, et al. : The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akbari H, Bakas S, Martinez-Lage M, et al. : Quantitative radiomics and machine learning to distinguish true progression from pseudoprogression in patients with GBM. In: 56th Annual Meeting of the American Society for Neuroradiology, Vancouver, BC, Canada: (2018) [Google Scholar]
- 20.Akbari H, et al. : Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery 78(4), 572–580 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macyszyn L, et al. : Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-oncology 18(3), 417–425 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akbari H, et al. : Pattern analysis of dynamic susceptibility contrast-enhanced MR imaging demonstrates peritumoral tissue heterogeneity. Radiology 273(2), 502–510 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rathore S, et al. : Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning. J. Med. Imaging 5(2), 021219 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akbari H, et al. : Survival prediction in glioblastoma patients using multi-parametric MRI biomarkers and machine learning methods. In: ASNR, Chicago, IL: (2015) [Google Scholar]
- 25.Rathore S, Bakas S, Akbari H, Shukla G, Rozycki M, Davatzikos C: Deriving stable multi-parametric MRI radiomic signatures in the presence of inter-scanner variations: survival prediction of glioblastoma via imaging pattern analysis and machine learning techniques In: Medical Imaging 2018: Computer-Aided Diagnosis, vol. 10575, p. 1057509 International Society for Optics and Photonics; (2018) [Google Scholar]
- 26.Li H, Galperin-Aizenberg M, Pryma D, Simone CB II, Fan Y: Unsupervised machine learning of radiomic features for predicting treatment response and overall survival of early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy. Radiother. Oncol 129(2), 218–226 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilello M, et al. : Population-based MRI atlases of spatial distribution are specific to patient and tumor characteristics in glioblastoma. NeuroImage: Clin. 12, 34–40 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tunç B, et al. : Individualized map of white matter pathways: connectivity-based paradigm for neurosurgical planning. Neurosurgery 79(4), 568–577 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakas S, et al. : In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index. Clin. Cancer Res 23(16), 4724–4734 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akbari H, et al. : In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro-oncology 20(8), 1068–1079 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller BM, et al. : Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c-means clustering and support vector machine segmentation. Med. Phys 39(8), 4903–4917 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller BM, Kontos D: Preliminary evaluation of the publicly available laboratory for breast radiodensity assessment (LIBRA) software tool. Breast Cancer Res. 17, 117 (2015). 10.1186/s13058-015-0626-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweitzer M, et al. : SCDT-37. Modulation of convection enhanced delivery (CED) distribution using focused ultrasound (FUS). Neuro-Oncology 19(Suppl 6), vi272 (2017) [Google Scholar]
- 34.Yoo TS, et al. : Engineering and algorithm design for an image processing API: a technical report on ITK-the insight toolkit. Stud. Health Technol. Inform 85, 586–592 (2002) [PubMed] [Google Scholar]
- 35.Schroeder WJ, Lorensen B, Martin K: The visualization toolkit: an object-oriented approach to 3D graphics. Kitware (2004) [Google Scholar]
- 36.Bradski G: The OpenCV Library. Dr. Dobb’s J. Softw. Tools (2000). https://github.com/opencv/opencv/wiki/CiteOpenCV [Google Scholar]
- 37.Herz C, et al. : DCMQI: an open source library for standardized communication of quantitative image analysis results using DICOM. Cancer Res. 77(21), e87–e90 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamnitsas K, et al. : Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal 36, 61–78 (2017) [DOI] [PubMed] [Google Scholar]
- 39.Amstutz P, et al. : Common workflow language, v1.0 (2016) [Google Scholar]
- 40.Knuth DE: Computer programming as an art. Commun. ACM 17(12), 667–673 (1974) [Google Scholar]
- 41.Gastounioti A, et al. : Breast parenchymal patterns in processed versus raw digital mammograms: a large population study toward assessing differences in quantitative measures across image representations. Med. Phys 43(11), 5862–5877 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, et al. : Parenchymal texture analysis in digital mammography: a fully automated pipeline for breast cancer risk assessment. Med. Phys 42(7), 4149–4160 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Griethuysen JJ, et al. : Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77(21), e104–e107 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]