Abstract
Bioaerosol samples were collected in an airborne infection isolation room, bathroom, and anteroom of a ventilated patient with coronavirus disease 2019. Twenty-eight samples were negative for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid, possibly due to the patient being on a closed-circuit ventilator or the efficiency of the air exchanges in the room.
Key Words: Airborne transmission, Hospital, Ventilator
Abbreviations: PPE, personal protective equipment; NIOSH, National Institute for Occupational Safety and Health; ICU, intensive care unit; PAPR, powered air-purifying respirator; HEPA, high-efficiency particulate air; HME, heat and moisture exchanger; RT-PCR, reverse transcription polymerase chain reaction; AIIR, airborne infection isolation room
Coronavirus disease 2019 (COVID-19) has now been detected in nearly every country in the world. The causative agent of COVID-19 has been identified as the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1 Research is now being conducted to learn more about the transmission of the virus and the best ways to protect the general population, as well as health care personnel working at the frontlines of disease management.
SARS-CoV-2 is believed to spread primarily through droplets or direct contact, with hospital-acquired transmission increasingly becoming a problem.2 Recent studies have shown that contact with contaminated surfaces is another possible method of transmission, with contamination of toilets and sinks being a priority in cleaning.3 , 4 However, regular hospital cleaning procedures appear to kill the virus, and viable virus in fecal samples is rarely identified.3 , 5 The virus may also be spread through aerosol-generating procedures, and it is possible that re-aerosolization of virus when health care workers remove personal protective equipment (PPE) increases exposure to the virus.6
With the number of cases and hospitalizations increasing, in order to protect health care personnel and uninfected patients in health care settings, it is important to understand the characteristics of aerosols containing SARS-CoV-2. While studies conducted during the pandemic have found aerosols containing virus within the health care setting, none have explicitly examined aerosols within the room of a patient on a ventilator.3 , 6 While ventilators are assumed to be closed systems, in practice, they can be disconnected during clinical care or while moving the patient for positioning or procedures.7 , 8 We sought to determine if aerosolized SARS-CoV-2 RNA could be found during the care of a ventilated patient who was being placed prone due to profound respiratory failure.
Methods
Bioaerosol sampling
Ten NIOSH BC 251 2-stage cyclone samplers were used.9 The NIOSH samplers separated particles into 3 size fractions, which are collected in a 15 mL centrifuge tube (>4 µm fraction), a 1.5 mL centrifuge tube (1-4 µm fraction) and on a filter cassette containing a 37-mm diameter, polytetrafluoroethylene filter with 2 µm pores (<1 µm fraction). Each sampler was connected with a 6.35-mm Tygon tubing to an air sampling pump (PCXR-4, SKC, Eighty Four, PA) at a 3.5 L/min flow rate.
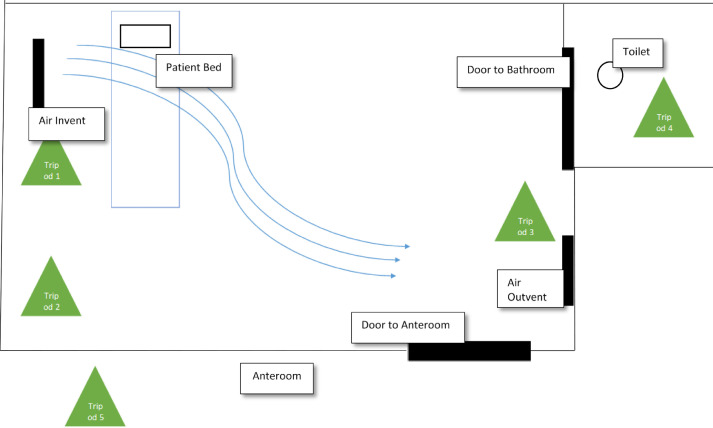
Two samplers were placed approximately 102 cm and 152 cm above the floor on each of 5 tripods. Three tripods were placed inside the patient room, 1 in the bathroom within the patient room, and 1 in the anteroom (Fig 1 ). The patient room samplers were placed next to the ventilator system (<2 ft away), in the corner along the same wall as the ventilator system, and by the anteroom door (<3 ft from the door). The anteroom sampler was placed <2 ft from the door to the patient room and the sampler in the bathroom was placed by the far wall (<2 ft from the toilet, <6 ft from the door). In an abundance of caution and to simplify disinfection, the sampler tripods were draped in plastic with holes cut out for the air intake and secured with tape. The pump cases were draped with a disposable underpad and secured to the tripods with tape.
Fig 1.
Location of the aerosol sampling within the patient room, patient bathroom, and anteroom.
Following a 6-hour sampling period, collected samples from each sampler were processed as follows: 1) 1,000 µL viral transport medium was added to the 15 mL tube, the tube was vortexed, inverted and vortexed, then frozen at −80°C, 2) 400 µL viral transport medium was added to the 1.5 mL tube, the tube was vortexed, inverted and vortexed and frozen at −80°C, 3) sterile forceps were used to remove the filter from its cassette and place it in a 15 mL tube, 1,000 µL of viral transport medium was added to wet the entire filter, and the tube was vortexed and stored at −80°C. RNA extraction occurred on the m2000 (Abbott Molecular, Abbott Park, IL) with 600 µL of input sample volume, and sample elute of 50 uL. The extracted samples underwent real-time polymerase chain reaction (RT-PCR) for selected gene regions of the SARS-CoV-2 virus nucleocapsid (N1, N2, N3) and human RNase P gene10 using a laboratory developed protocol adapted from the protocol published by the Centers for Disease Control and Prevention.11 Following RNA extraction, a 20 µL reaction was set up containing 5 µL of sample RNA, 8.5 µL of nuclease-free water, 1.5 µL of combined primer/probe mix and 5µL of TaqPath 1-Step RT-qPCR Master Mix (ThermoFisher, Waltham, MA). Thermal cycling was performed at 25°C for 2 minutes followed by 50°C for 15 minutes, followed by an initial denaturation at 95°C for 2 minutes, followed by 45 cycles of amplification at 95°C for 3 seconds and 55.0°C for 30 seconds. A previously characterized SARS-CoV-2 sample was tested concurrently as a positive control. Fluorescence growth curves which cross the threshold line within 40 cycles (<40 Ct) were considered positive.
Patient room
Sampling took place in an ICU-level airborne infection isolation room (AIIR) in the Serious Communicable Diseases Unit at Emory University Hospital. The AIIR has negative air pressure relative to the anteroom (0.016-0.018 in of w.c.) with 20 air exchanges per hour, laminar flow across the patient bed, and HEPA filtration. The adjoining anteroom has negative air pressure relative to the hallway.
Patient
The patient was confirmed PCR positive for COVID-19 the day before sampling took place by both nasopharyngeal and oropharyngeal swabs. The patient's legal authorized representative provided informed consent under a protocol approved by the Emory IRB. During sampling, the patient was mechanically ventilated on a Hamilton ventilator, with HEPA end exhaust and in-line heat and moisture exchanger, via an 8.0 endotracheal tube with in-line, closed suction device. Between 2 and 5 multidisciplinary health care workers, including advanced practice providers, an attending physician, ICU nurses, and a respiratory therapist, were present in the patient room or anteroom during the entire 6-hour sampling period. With 5 health care personnel present in the patient room, the patient was re-positioned from prone to supine. However, the patient failed to tolerate supination, with decreasing oxygen saturation by SpO2 monitoring and increased peak pressures on the ventilator, and therefore required re-proning. During this time, there was one brief disconnection of the proximal and distal part of the ventilator hose.
Results
In total, 30 samples were collected, 3 from each sampler: one 15 mL centrifuge tube, one 1.5 mL centrifuge tube, and an additional 15 mL centrifuge tube containing the removed 37-mm filter. Two samples were unable to be used for testing: a 15 mL tube from the “Ventilator” tripod and a filter from the “Corner” tripod which fell on the floor. Of the 28 tested samples, 4 were positive for RP primer, indicating presence of human nucleic acid material. These included 2 samples from the patient room “Corner” 102 cm-high sampler, one sample from the room “Door”102 cm-high sampler and one sample from the “Bathroom” 152-high cm sampler (Table 1 ). None of the 28 samples tested were positive for SARS-CoV-2 nucleic acid.
Table 1.
Location and sampling time in patient room, bathroom, and anteroom
| Tripod designation | Tripod location | Tripod placement (start time) | Tripod removal (end time) | Sampling time (minutes) | SARS-CoV-2 detected (+/total) | Human RNase P gene detected (+/total) |
|---|---|---|---|---|---|---|
| T1 “Vent” | Patient room by ventilator system | 10:45 | 16:15 | 330 | 0/5 | 0/5 |
| T2 “Corner” | Patient room corner along ventilator wall | 10:45 | 16:30 | 345 | 0/5 | 2/5 |
| T3 “Door” | Patient room by anteroom door | 10:45 | 16:45 | 360 | 0/6 | 1/6 |
| T4 “Bathroom” | Patient room bathroom by toilet | 10:45 | 16:50 | 365 | 0/6 | 1/6 |
| T5 “Anteroom” | Anteroom by patient room door | 10:45 | 16:00 | 315 | 0/6 | 0/6 |
Discussion
In order to protect health care personnel with appropriate PPE, there has been considerable discussion on what constitutes an aerosol-generating procedure. Our preliminary study shows that SARS-CoV-2 aerosols were not detected in an AIIR with a COVID-19 positive patient mechanically ventilated on a closed-suction device. This was particularly important because this patient was substantially manipulated through pronation and supination in the bed by many health care personnel during the 6 hours of sampling. His respiratory status was so poor that these aggressive measures were attempted to keep the patient able to be oxygenated. The health care workers were wearing powered air purifying respirators (PAPRs) as well as disposable gowns, 2 sets of gloves, and booties, in accordance with hospital policy. All PPE except for the PAPR were doffed in the patient room, with the PAPR being doffed in the anteroom.
Some studies have detected low amounts of SARS-CoV-2 RNA in aerosol samples collected in health care settings where patients were breathing and coughing into the environment while others have not.3 , 6 , 12 , 13 Our study was performed in an AIIR with laminar flow and a high air exchange rate with the patient on a ventilator with a HEPA filter. These early findings, based upon a single sampling event, suggest that the amount of airborne SARS-CoV-2 was very low.
However, it is important to note the limitations of our study. First, we cannot comment on the possibility of virus transmission in this setting, because this study only measured aerosols in the air and did not examine other potential pathways for virus transmission. Further, the sampling may have been too dilute and the amount of RNA may have been below the limit of detection of 10 viral copies/mL. Second, aerosol sampling was conducted for only 1 patient during one 6-hour sampling period. Third, previous studies have shown that the filtration efficiency of ventilator exhaust filters varies widely, and thus these results cannot be assumed to apply to all ventilators and exhaust filters.14 Finally, although the patient was positive for COVID-19 in the upper respiratory tract and was diagnosed with acute respiratory distress syndrome, and therefore likely positive in the lower respiratory tract based on recent evidence,13 we cannot confirm that the patient was actively shedding virus on the day of sampling. Given we cannot understand the full extent of possible environmental contamination in the patient room with aerosol samples alone, surface sampling studies will be conducted along with any aerosol sampling in the future.
Footnotes
Funding: This work was supported in part by the Center for AIDS Research (P30 AI050409). Supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR002378. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
Conflicts of interest: None to report.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh P-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ong SWX, Tan YK, Chia PY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323:E1–E3. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC. Coronavirus disease 2019 (COVID-19)—transmission. 2020. Available at:https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/how-covid-spreads.html. Accessed May 8, 2020.
- 5.WHO. Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19). 2020. Available at:https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf. Accessed May 20, 2020.
- 6.Liu Y, Ning Z, Chen Y, et al. Aerodynamic characteristics and RNA concentration of SARS-CoV-2 aerosol in Wuhan hospitals during COVID-19 outbreak. Microbiology. 2020;582:557–560. [Google Scholar]
- 7.Cobley M, Atkins M, Jones PL. Environmental contamination during tracheal suction: a comparison of disposable conventional catheters with a multiple-use closed system device. Anaesthesia. 1991;46:957–961. doi: 10.1111/j.1365-2044.1991.tb09858.x. [DOI] [PubMed] [Google Scholar]
- 8.Gorman LJ, Sanai L, Notman AW, Grant IS, Masterton RG. Cross infection in an intensive care unit by Klebsiella pneumoniae from ventilator condensate. J Hosp Infect. 1993;23:27–34. doi: 10.1016/0195-6701(93)90127-l. [DOI] [PubMed] [Google Scholar]
- 9.Blachere FM, Lindsley WG, Pearce TA, et al. Measurement of airborne influenza virus in a hospital emergency department. Clin Infect Dis. 2009;48:438–440. doi: 10.1086/596478. [DOI] [PubMed] [Google Scholar]
- 10.Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC/NCIRD/DVD. Real-time RT-PCR panel for detection 2019-novel coronavirus. 2020: 12. Available at:https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-for-detection-instructions.pdf. Accessed May 15, 2020.
- 12.Santarpia JL, Rivera DN, Herrera V, et al. Transmission potential of SARS-CoV-2 in viral shedding observed at the University of Nebraska Medical Center. Infect Dis. 2020;10:1–8. [Google Scholar]
- 13.Chia PY, Coleman KK, Tan YK, et al. Detection of air and surface contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in hospital rooms of infected patients. Infect Dis. 2020;11:2800. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkes A. Preventing the transmission of pathogenic microbes during anesthesia. Expert Rev Med Devices. 2005;2:319–326. doi: 10.1586/17434440.2.3.319. [DOI] [PubMed] [Google Scholar]