Abstract
Context:
Ex-vivo microscopy encompasses a range of techniques to examine fresh or fixed tissue with microscopic resolution, eliminating the need to embed the tissue in paraffin or produce a glass slide. One such technique is light-sheet microscopy, which enables rapid 3D imaging. Our pathology-engineering collaboration has resulted in an open-top light-sheet (OTLS) microscope that is specifically tailored to the needs of pathology practice.
Objective:
To present an image atlas of OTLS images of prostate core needle biopsies.
Design:
Core needle biopsies (N=9) were obtained from fresh radical prostatectomy specimens. Each biopsy was fixed in formalin, dehydrated in ethanol, stained with TO-PRO3 and eosin, optically cleared, and imaged using OTLS microscopy. The biopsies were then processed, paraffin embedded, and sectioned. Hematoxylin and eosin (H&E)-staining and immunohistochemical (IHC) staining for cytokeratin 5 and cytokeratin 8 was performed.
Results:
Benign and neoplastic histologic structures showed high fidelity between OTLS and traditional light microscopy. OTLS microscopy had no discernible effect on H&E or IHC staining in this pilot study. The 3D histology information obtained using OTLS microscopy enabled new structural insights, including the observation of cribriform and well-formed gland morphologies within the same contiguous glandular structures, as well as the continuity of poorly formed glands with well-formed glands.
Conclusions:
3D OTLS microscopy images have a similar appearance to traditional H&E histology, with the added benefit of useful 3D structural information. Further studies are needed to continue to document the OTLS appearance of a wide range of tissues and to better understand 3D histologic structures.
Introduction:
Light-sheet microscopy is one of a family of emerging imaging techniques for slide-free, non-destructive ex-vivo microscopy (EVM). EVM gives pathologists the ability to examine fresh or fixed tissue with microscopic resolution, eliminating the need to embed the tissue in paraffin or produce a glass slide. EVM has potential to streamline laboratory workflow1, preserve valuable tissue for molecular assays2,3, and provide novel morphologic insights4.
Each EVM technique has its own strengths and weakness, reflecting tradeoffs between resolution, speed, cost, and ease-of-use. Light-sheet microscopy is a fluorescence microscopy technique with unparalleled ability to rapidly collect 3D microscopic information from intact specimens5. This ability has led to seminal discoveries in developmental biology and neuroscience research6–9. However, commercially available light-sheet microscopy systems are not practical for clinical specimens, in part due to elaborate specimen mounting procedures and constraints on specimen size and shape. To address this unmet need, our group built an open-top light-sheet (OTLS) microscope optimized for clinical specimens, as described in a previous publication10.
To demonstrate the potential impact of OTLS on the practice of pathology, we will present an image atlas of core needle biopsies from men with prostate cancer. Prostate cancer is the most common cancer in men, with greater than 160,000 new cases in the United States in 201711. Prostate cancer prognostication and treatment planning depend upon accurate histologic grading of prostate biopsies, which is entirely based on architectural features12,13. Because these architectural features are highly complex 3D structures, the prostate is an ideal organ to highlight the value of 3D microscopy using OTLS imaging. In this study, we present a range of benign and neoplastic structures using OTLS imaging with “pseudo-hematoxylin and eosin (H&E)” coloring in comparison to the corresponding H&E histology.
Materials and methods:
Case selection:
De-identified fresh prostate biopsies (9 samples from 2 patients) were obtained prospectively from radical prostatectomy specimens using an 18-gauge (approximately 1 mm inner diameter) needle biopsy device (Bard Max Core, Bard Biopsy). All patients were consented for tissue donation to the University of Washington Genitourinary Biorepository (study approved by the University of Washington Institutional Review Board, Seattle, WA).
Staining protocol:
Samples were fixed in 10% neutral buffered formalin for 24 hours. After fixation, samples were washed in PBST (phosphate buffered saline with 0.2% Triton X-100). The samples were stained overnight in a solution of nuclear fluorescent dye TO-PRO3 Iodide at 1:1000 dilution in PBST with light shaking. The samples underwent a graded dehydration in solutions of 0%, 25%, 50%, 75%, and 100% ethanol and de-ionized water, 1 hour for each solution. Then, the samples were stained overnight in a 1:1000 dilution of stock Eosin solution (Surgipath, Leica) in 100% ethanol with light shaking. After staining, samples were immersed in ethyl cinnamate for 1 hour, followed by another 1 hour immersion with fresh ethyl cinnamate. Immersion in ethyl cinnamate renders biopsies transparent without altering the tissue structure in any noticeable way, a process known as optical clearing or chemical clarification (see review by Richardson and Lichtman14). The clearing method used here is similar to the iDISCO method15, but utilizes a much less toxic index-matching reagent (ethyl cinnamate) than the original iDISCO protocol. After clarification, the biopsies can be imaged in 3D, via OTLS microscopy, throughout their entire 1 mm diameter. All steps were performed at room temperature. The total dehydration, staining, and clarification time for each biopsy in this study was 36 hours. We have found that shorter protocols, requiring approximately 8 hours, produce similar OTLS images to those presented in this manuscript. The total imaging time for each biopsy was 30 minutes.
OTLS imaging:
Stained and cleared samples were placed on a custom OTLS system. Specimens were imaged using a 660 nm laser to excite nuclear dye TO-PRO3 and a 488 nm laser to excite eosin. The samples were gently compressed to reduce surface irregularities, then scanned at a speed of approximately 15 minutes per channel per biopsy. Each image channel was captured separately as 16-bit grayscale images, then false-colored using the algorithm described by Giacomelli et al. to mimic traditional H&E staining16. For more details about the staining, clearing, and imaging protocol, refer to our previous publication10. The approximate sampling pitch of the OTLS system used in this study was 0.75 microns per pixel (spatial resolution ~ 1.5 microns), similar to a whole-slide image scanned at 20x magnification.
Histopathological examination:
After OTLS imaging, the samples were processed and paraffin embedded. A single H&E-stained slide was prepared from each tissue sample (en face sectioning near the imaged tissue surface), then scanned at 20x magnification to produce a whole-slide image (Aperio, Leica Biosystems). Immunohistochemical (IHC) staining for cytokeratin 5 (CK5) and cytokeratin 8 (CK8) was performed to confirm the presence of carcinoma used the following protocol: incubation of sections in primary antibodies at 4°C overnight, followed by an appropriate biotinylated secondary antibody (1:150), ABC reagent (Vector Laboratories), and DAB (Invitrogen). A genitourinary pathology fellow and a board certified genitourinary pathologist reviewed each case.
Results:
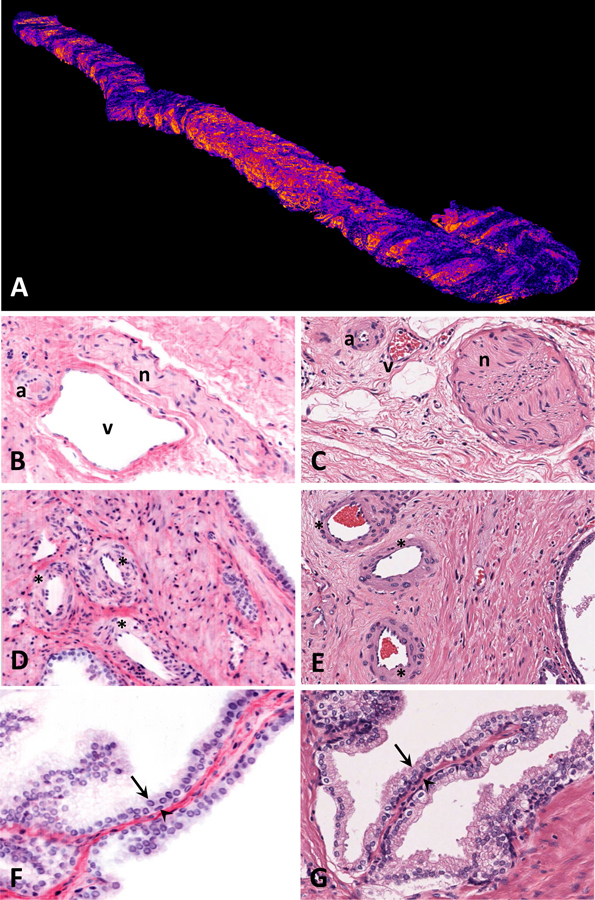
Prostate tissue is composed of glands with complex infoldings surrounded by fibromuscular stroma. Benign prostate glands are composed of a bilayered epithelium of basal cells and luminal epithelial cells. The prostate gland lacks a true capsule and is surrounded by tissue rich in vessels and nerves. Figures 1A–1G highlight these structures in images obtained using OTLS and traditional H&E-stained slides.
Figure 1.
Open-top light-sheet microscopy (OTLS) images (from 3D pathology datasets) are labeled as panels A, B, D, and F, hematoxylin and eosin (H&E) images (from conventional slide-based histology), scanned at 20X magnification are labeled as panels C, E, and G. A, volumetric OTLS rendering of an entire prostate core needle biopsy. All other OTLS images are pseudo-colored to mimic H&E staining. B and C, neurovascular bundle with artery (a), vein (v), and nerve (n). D and E, benign prostatic glands, stroma, and vasculature; arterioles are designated with asterisks. F and G, benign prostatic glands with luminal epithelium (arrows) and basal epithelium (arrowheads).
Fig 1A is a volumetric rendering of the entire prostate core needle biopsy using OTLS imaging. OTLS obtains information from every micron of the tissue in all three dimensions, as opposed to a glass slide, which contains only a single 5 micron thick section of tissue. To find the best examples of each structure, we examined the OTLS prostate biopsy images in all three dimensions. Neurovascular bundles (Fig 1B and 1C) and arterioles (Fig 1D and 1E) have similar appearances in OTLS and H&E images. Benign basal and luminal cells can be identified using OTLS and H&E (Fig 1F and 1G).
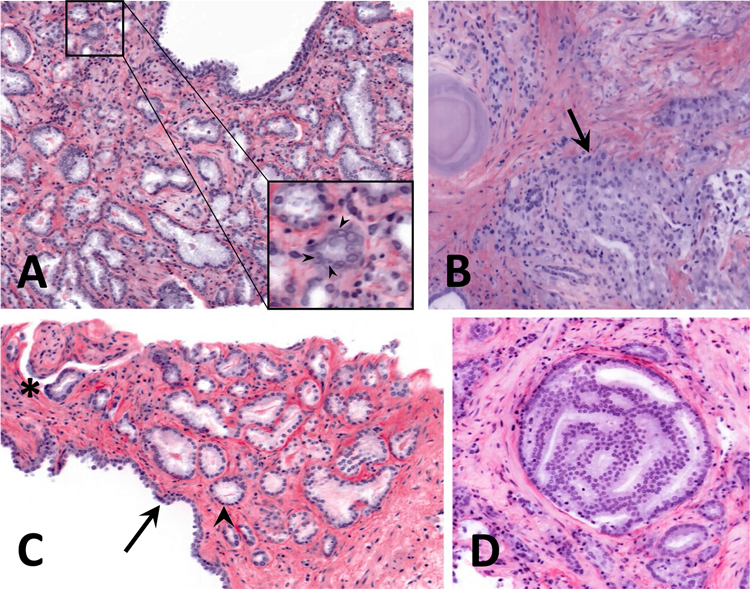
The prostate biopsies in this study contained examples of Gleason pattern (GP) 3 glands and all three variants of GP4 glands. It should be noted that there is high interobserver variability in diagnosing GP4, especially the fused and poorly formed gland variants17. Fig 2A shows an example of well-formed GP3 glands. The carcinoma glands are irregularly distributed and lack a basal cell layer, have abundant cytoplasm, and have basally located nuclei. The inset shows examples of readily identifiable nucleoli, which aid in diagnosing prostate carcinoma. Fig 2B shows an example of the fused gland variant of GP4, where several glands abut each other without intervening stroma. Fig 2C shows another example of well-formed GP3 glands adjacent to the epithelium of a benign gland. An asterisk marks an example of perineural invasion, a feature essentially pathognomonic for prostate carcinoma. An example of the cribriform variant of GP4, which has a sieve-like architecture, is shown in Fig 2D.
Figure 2.
Open-top light-sheet microscopy (OTLS) images of prostate carcinoma exhibiting various Gleason patterns. A, well-formed Gleason pattern 3 glands. The inset shows a zoom-in to highlight nucleoli (arrowheads). B, example of fused Gleason pattern 4 glands, indicated by an arrow. C, benign prostatic glands (arrow) adjacent to well-formed Gleason pattern 3 glands (arrowhead points to one example). The asterisk in the upper left corner highlights an area of perineural invasion. D, example of a cribriform Gleason pattern 4 gland.
Because OTLS imaging is non-destructive, it is possible to examine the effect of OTLS imaging on subsequent H&E and IHC staining. In comparison to the OTLS image (Fig 3A), the H&E image (Fig 3B) exhibits more basophilic cytoplasm and more deeply eosinophilic stromal tissue. Eosinophilic crystalloids, often seen in prostate carcinoma, are present in the lumens of some of the carcinoma glands in both the OTLS and H&E images. IHC staining for CK5 shows the presence of basal cells around benign glands and absence of basal cells around the carcinoma glands (Fig 3C). CK8 IHC staining highlights all luminal epithelial cells in benign and carcinoma glands (Fig 3D). In these images (Fig 3B–3D), OTLS imaging did not have any effects on downstream H&E or IHC staining of the biopsy that would alter the diagnostic interpretation.
Figure 3.
Open-top light-sheet microscopy (OTLS), hematoxylin and eosin (H&E), and immunohistochemistry (IHC) images of a region of prostate carcinoma. H&E and IHC slides were scanned at 20X magnification and the images were cropped to simulate a 5X field of view. A, an OTLS image showing benign prostatic glands (arrow) adjacent to well-formed Gleason pattern 3 glands (arrowhead points to one example). B, an H&E stained slide from the same region as the OTLS image, showing benign prostatic glands (arrow) adjacent to well-formed Gleason pattern 3 glands (arrowhead points to one example). C, a cytokeratin 5 (CK5) immunostain highlights basal cells. The benign glands have a basal cell layer and the carcinoma glands lack a basal cell layer. D, a cytokeratin 8 (CK8) immunostain highlights luminal epithelial cells in both benign and carcinoma glands.
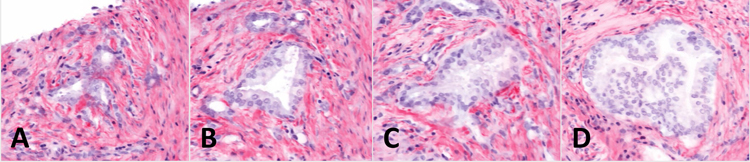
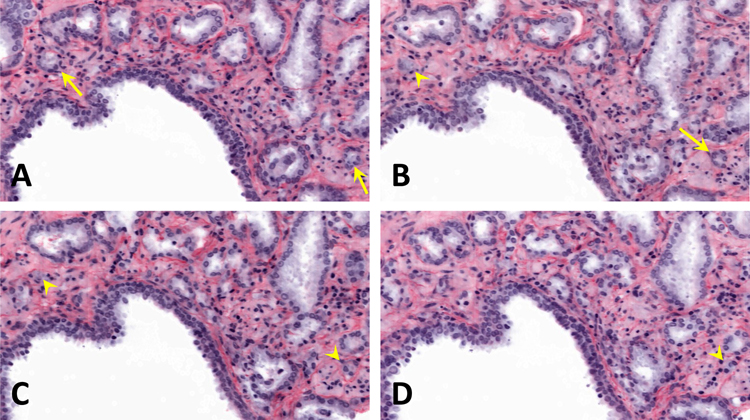
To show the potential for 3D histology to provide new structural insights, we traced individual carcinoma glands through the depth of the biopsy. In one example, a region of carcinoma exhibits three well-formed GP3 glands (Fig 4A). At 40 and 80 microns deeper than the cross section shown in Fig 4A, the carcinoma region shows just one well-formed gland (Fig 4B and 4C). At 120 microns deeper than the cross section shown in Fig 4A, the same carcinoma region exhibits a cribriform GP4 gland (Fig 4D). These three different morphologies form one contiguous structure when traced micron-by-micron throughout the depth of the biopsy. In another example, tracing poorly formed glands through the depth of the biopsy shows that they exhibit well-formed gland morphology at deeper depths within the biopsy (Fig 5A–D). In our experience using OTLS, we have seen that all poorly formed glands are contiguous with well-formed glands when examined in 3D.
Figure 4.
Open-top light-sheet microscopy (OTLS) images of a carcinoma gland that changes morphology and grade with depth. Each sequential image shows an optical section 40 microns deeper than the previous image. A, three separate well-formed Gleason pattern 3 glands. B and C, a single well-formed Gleason pattern 3 gland. D, a single cribriform Gleason pattern 4 gland.
Figure 5.
Open-top light-sheet microscopy (OTLS) images of carcinoma glands that change morphology with depth. Arrows point to well-formed Gleason pattern 3 glands and arrowheads point to poorly formed Gleason pattern 4 glands. Each sequential image shows an optical section 25 microns deeper than the previous image. A, the arrows point to two well-formed glands. B, one gland is poorly formed and the other gland is well-formed. C, both glands are poorly formed at this depth. D, one gland is no longer present and the other gland is poorly formed.
Discussion:
We have shown examples of OTLS images from a range of prostate biopsies that showcase diverse histomorphologies. Small molecule fast-acting nuclear (TO-PRO3) and cytoplasmic (eosin) fluorescent dyes, along with false-coloring algorithms, were used in this study to demonstrate the advantages and limitations of thick tissue fluorescent staining and OTLS microscopy. Pseudo-H&E images, in which our two channel fluorescence images are false colored to mimic the color palette of H&E staining, have the substantial advantage of familiarity to the practicing pathologist. Only minimal training is required to recognize instances where pseudo-H&E images may deviate from conventional H&E images, e.g. where the cytoplasm of carcinoma glands appears to be more strongly stained by hematoxylin than by TO-PRO3 in the pseudo-H&E images. With the custom OTLS device used here, we can image intact prostate biopsies at resolutions similar to what is achieved with a 20x objective on a standard trans-illumination brightfield light microscope. Features of benign prostatic tissue and carcinoma were readily identified using our OTLS microscopy approach. Perhaps the highest impact application of OTLS to the practice of pathology is non-destructive 3D microscopy of entire, intact biopsies. We chose prostate core-needle biopsies to demonstrate this clinical application because of the complex 3D morphology of prostate carcinoma. Assessment of prostate carcinoma gland morphology is the sole criterion for Gleason grading, which plays a major role in clinical cancer care guidelines. For example, a region containing three well-formed carcinoma glands is traced through the depth of a biopsy in Figure 4, in which the same contiguous gland structure exhibits a cribriform morphology at a depth that is 120 microns deeper than the original imaging plane. This difference in morphology in the same contiguous gland could influence a clinician’s recommendation for active surveillance vs. definitive treatment (surgery and/or radiation)12. Other studies using 3D microscopy have shown anecdotal examples of similarly impactful observations in bladder18 and colon cancer19. In the future, more robust studies are needed with 3D microscopy to improve our understanding of cancer biology and its clinical correlations.
OTLS is one of a family of emerging slide-free thick-tissue fluorescence microscopy techniques that includes confocal microscopy20, multi-photon microscopy21,22, microscopy with UV surface excitation (MUSE)23, and structured illumination24. All of these techniques image tissue by using exogenously applied fluorescent dyes or endogenous contrast mechanisms. The differences between the techniques reflect trade-offs between resolution, depth of focus, imaging speed, and cost. For instance, multi-photon microscopy has high resolution and imaging depth, but requires extensive imaging time, up to 24 hours for a single core needle biopsy21. In contrast, MUSE is inexpensive with remarkably quick imaging speed, making it an attractive technique for rapid fresh tissue imaging, but cannot image below a tissue surface due to the use of UV wavelengths for illumination25. Selection of the optimal technique depends on both the clinical application and the needs/resources of the pathology laboratory. OTLS strikes a balance between resolution, speed of imaging, depth, and cost, making the technology well-suited for 3D microscopy of biopsy specimens.
There are multiple limitations of light-sheet microscopy that prevent its immediate adoption to the clinic: (1) Commercially available light-sheet microscopy systems are designed for small transparent specimens with complex tissue mounting requirements, (2) there is a learning curve for pathologists to interpret new imaging artifacts, (3) the staining and imaging procedure has unknown effects on downstream laboratory processes, and (4) the terabyte-sized imaging datasets generated by these microscopes pose many informatics challenges. All of these limitations are surmountable in the near-term. In response to limitation (1), we have designed an OTLS system that enables rapid imaging of large, irregularly shaped specimens without elaborate tissue-mounting requirements. Detailed validation studies, further development of clarification techniques26,27,28, and shared image repositories will address limitations (2) and (3). Finally, the use of cloud-based parallelized compute clusters, rapid 3D reconstruction algorithms29, fast internet connections, and machine learning algorithms18, can mitigate many of the informatics challenges created by current limitations (4).
In summary, clinical adoption of 3D pathology using OTLS microscopy has the potential to improve the quality of patient care. In the near term, OTLS can serve as a complement rather than an alternative to traditional light microscopy. Continued documentation of the appearance of a wide range of tissues imaged with 3D OTLS microscopes, as well as clinical studies to document the added value of 3D OTLS microscopy, are essential to accelerate clinical adoption30.
References
- 1.Jung W, Boppart SA. Optical coherence tomography for rapid tissue screening and directed histological sectioning. Stud Health Technol Inform 2013;185:109–128. [PubMed] [Google Scholar]
- 2.Forest F, Cinotti E, Yvorel V, et al. Ex vivo confocal microscopy imaging to identify tumor tissue on freshly removed brain sample. J Neurooncol 2015;124(2):157–164. [DOI] [PubMed] [Google Scholar]
- 3.Georges J, Zehri A, Carlson E, et al. Label-free microscopic assessment of glioblastoma biopsy specimens prior to biobanking. Neurosurg Focus 2014;36(2):E8. [DOI] [PubMed] [Google Scholar]
- 4.Jain M, Robinson BD, Wu B, Khani F, Mukherjee S. Exploring Multiphoton Microscopy as a Novel Tool to Differentiate Chromophobe Renal Cell Carcinoma From Oncocytoma in Fixed Tissue Sections. Arch Pathol Lab Med 2018;142(3):383–390. [DOI] [PubMed] [Google Scholar]
- 5.Keller PJ, Dodt HU. Light sheet microscopy of living or cleared specimens. Curr Opin Neurobiol 2012;22(1):138–143. [DOI] [PubMed] [Google Scholar]
- 6.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004;305(5686):1007–1009. [DOI] [PubMed] [Google Scholar]
- 7.Dodt HU, Leischner U, Schierloh A, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 2007;4(4):331–336. [DOI] [PubMed] [Google Scholar]
- 8.Jährling N, Becker K, Schönbauer C, Schnorrer F, Dodt HU. Three-dimensional reconstruction and segmentation of intact Drosophila by ultramicroscopy. Front Syst Neurosci 2010;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holekamp TF, Turaga D, Holy TE. Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron 2008;57(5):661–672. [DOI] [PubMed] [Google Scholar]
- 10.Glaser AK, Reder NP, Chen Y, et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat Biomed Eng 2017;1(7):0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 12.Carroll PR, Parsons JK, Andriole G, et al. NCCN Guidelines Insights: Prostate Cancer Early Detection, Version 2.2016. J Natl Compr Canc Netw 2016;14(5):509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol 2016;40(2):244–252. [DOI] [PubMed] [Google Scholar]
- 14.Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell 2015;162(2):246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renier N, Wu Z, Simon DJ, et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014;159(4):896–910. [DOI] [PubMed] [Google Scholar]
- 16.Giacomelli MG, Husvogt L, Vardeh H, et al. Virtual Hematoxylin and Eosin Transillumination Microscopy Using Epi-Fluorescence Imaging. PLoS One 2016;11(8):e0159337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egevad L, Ahmad AS, Algaba F, et al. Standardization of Gleason grading among 337 European pathologists. Histopathology 2013;62(2):247–256. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka N, Kanatani S, Tomer R, et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat Biomed Eng 2017;1(10):796–806. [DOI] [PubMed] [Google Scholar]
- 19.Bronsert P, Enderle - Ammour K, Bader M, et al. “Cancer cell invasion and EMT marker expression: a three - dimensional study of the human cancer–host interface.” J Pathol. 2014;234(3):410–422. [DOI] [PubMed] [Google Scholar]
- 20.Abeytunge S, Li Y, Larson B, et al. Confocal microscopy with strip mosaicing for rapid imaging over large areas of excised tissue. J Biomed Opt 2013;18(6):61227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson E, Levene MJ, Torres R. Multiphoton microscopy with clearing for three dimensional histology of kidney biopsies. Biomed Opt Express 2016;7(8):3089–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao YK, Shen D, Sheikine Y, et al. Assessment of breast pathologies using nonlinear microscopy. Proc Natl Acad Sci U S A 2014;111(43):15304–15309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fereidouni F, Harmany ZT, Tian M, et al. Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat Biomed Engin 2017;1(12):957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Tulman DB, Sholl AB, et al. Gigapixel surface imaging of radical prostatectomy specimens for comprehensive detection of cancer-positive surgical margins using structured illumination microscopy. Sci Rep 2016;6:27419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshitake T, Giacomelli MG, Quintana LM, et al. Rapid histopathological imaging of skin and breast cancer surgical specimens using immersion microscopy with ultraviolet surface excitation. Sci Rep 2018;8(1):4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan C, Cai R, Quacquarelli FP, et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods 2016;13(10):859–867. [DOI] [PubMed] [Google Scholar]
- 27.Li W, Germain RN, Gerner MY. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc Natl Acad Sci U S A 2017;114(35):E7321–E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park Y-G, Sohn C, Chen R, et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat Biotechnol 2018; 10.1038/nbt.4281. [DOI] [PMC free article] [PubMed]
- 29.Hörl D, Rojas Rusak F, Preusser F, et al. BigStitcher: Reconstructing high-resolution image datasets of cleared and expanded samples. bioRxiv 2018; 10.1101/343954. [DOI] [PubMed]
- 30.Uhlén P, Tanaka N. Improved Pathological Examination of Tumors with 3D Light-Sheet Microscopy. Trends Cancer 2018;4(5):337–41. [DOI] [PubMed] [Google Scholar]