Summary
Lung transplantation can be lifesaving in end-stage cystic fibrosis (CF), but long-term survival is limited by chronic lung allograft dysfunction (CLAD). Persistent upper airway Pseudomonas aeruginosa (PsA) colonization can seed the allograft. While de novo PsA infection is associated with CLAD in non-CF recipients, this association is less clear for CF recipients experiencing PsA recolonization. Here, we evaluate host and pathogen contributions to this phenomenon. In the context of PsA infection, brushings from the airways of CF recipients demonstrate type 1 interferon gene suppression. Airway epithelial cell (AEC) cultures demonstrate similar findings in the absence of pathogens or immune cells, contrasting with the pre-transplant CF AEC phenotype. Type 1 interferon promoters are relatively hypermethylated in CF AECs. CF subjects in this cohort have more mucoid PsA, while non-CF PsA subjects have decreased microbiome α diversity. Peri-transplant protocols may benefit from consideration of this host and microbiome equilibrium.
Keywords: cystic fibrosis, lung transplantation, Pseudomonas aeruginosa, interferon, CLAD, allograft rejection, microbiome, transcriptome, epigenome, DNA methylation
Graphical Abstract
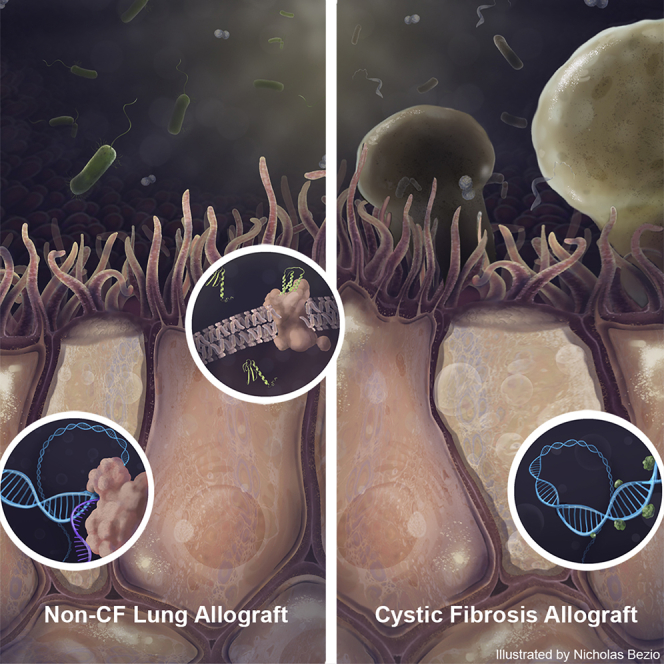
Highlights
Lung allograft Pseudomonas infection outcomes are better for recipients with CF
In CF, infected allograft airway cells demonstrate suppression of interferon genes
Differential DNA methylation may contribute to this distinct epithelial phenotype
Increased α diversity and mucoid forms characterize CF Pseudomonas infection
Cystic fibrosis (CF) lung transplant recipients tolerate Pseudomonas infection relatively well. Dugger et al. show that CF allograft airway cells have interferon gene suppression and promotor hypermethylation. Mucoid Pseudomonas is more common in CF, as was preserved α diversity. Thus, both host and microbiome differences appear to be associated with favorable outcomes in CF.
Introduction
Cystic fibrosis (CF), resulting from mutations in the CF transmembrane conductance regulator (CFTR), affects nearly 30,000 people in the United States alone.1 Because of its ubiquitous nature in human cells, CFTR mutation results in multiple organ dysfunctions, including pancreatic insufficiency, bowel obstruction, and sinopulmonary infections. However, it is chronic pulmonary infection, leading to bronchiectasis and respiratory failure, that limits the quality and quantity of life in most individuals with CF.2,3
From early life, CF individuals develop an airway microbiome that is distinct from healthy counterparts,4 and over time, the CF sinopulmonary microbiome is characterized by decreasing diversity and dominance of Staphylococcus aureus followed by Pseudomonas aeruginosa (PsA). Once established, PsA typically mutates to a mucoid form characterized by a protective alginate-containing matrix. This matrix sequesters PsA from host defenses and antibiotics, leading to progressive inflammation and end-stage lung failure.2,5
While lung transplantation can be a lifesaving option for CF and other end-stage lung diseases, chronic lung allograft dysfunction (CLAD) affects 50% of lung transplant recipients by 5 years post-transplant and is the major limitation to long-term survival.6 Post-transplant infections, including community-acquired respiratory viruses, fungi, PsA, and other bacteria, are important CLAD risk factors.7, 8, 9 PsA has been identified as a particularly important organism in this context.10 Because the host sinotracheal tract is not replaced during lung transplantation, recolonization with PsA is common in CF lung transplant recipients.11 Nonetheless, CF recipients do better following lung transplantation than non-CF recipients in many, although not all, reports.6,12, 13, 14, 15, 16 Given the potentially favorable outcomes for CF recipients, we hypothesized that host or microbiome adaptations in CF patients attenuate immune responses to PsA that would otherwise result in CLAD.
Results
PsA Differentially Influences Lung Allografts Based on CF Status
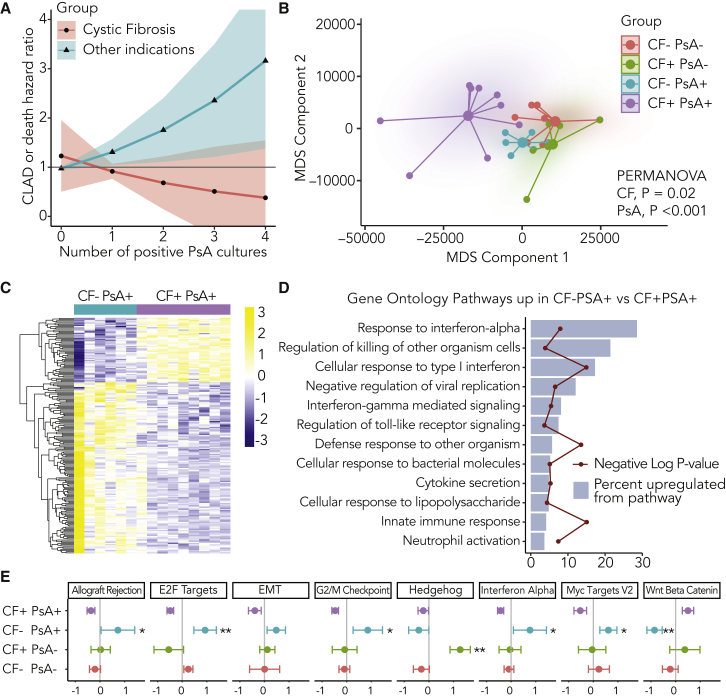
To understand the differential impact of PsA infection on lung transplant recipient outcomes with and without CF, we assessed time to CLAD or death in a single-center cohort of 397 lung transplant recipients (Figure S1; Table S1). We examined the interaction between CF status and the frequency of PsA+ bronchoalveolar lavage (BAL) culture events in a model, including recipient age, gender, and transplant indications other than CF. We found that CF status modified the association between PsA frequency and CLAD-free survival, as the CF-PsA interaction term was associated with a decreased risk of CLAD or death (hazard ratio [HR] 0.55, 95% confidence interval [CI] 0.30–0.99, p = 0.049). To explore this interaction finding in detail, we examined the risk of CLAD or death in age-adjusted models stratified by CF status (Figure 1A). In the subset of recipients with CF, the increasing frequency of PsA+ BAL cultures was not associated with a statistically significant difference in CLAD or death risk (HR 0.74, 95% CI 0.40–1.40, p = 0.36). By contrast, among non-CF recipients, each PsA+ culture was associated with a hazard ratio of 1.34 (95% CI 1.04–1.74, p = 0.025) for CLAD or death.
Figure 1.
Pseudomonas aeruginosa (PsA) Differentially Influences Lung Allografts Based upon CF Status
(A) Age-adjusted Cox proportional hazards models for CF+ (n = 34) and CF− (n = 362) subsets show CLAD or death hazard ratio (HR) as a function of the number of bronchoalveolar lavage cultures in which PsA was identified. PsA+ cultures were associated with increased CLAD or death risk for non-CF recipients (HR 1.34, 95% CI 1.04–1.74, p = 0.025), but not CF recipients (HR 0.75, 95% CI 0.40–1.40, p = 0.36). A Kaplan-Meier plot of subjects stratified by CF and ever-PsA+ status is shown in Figure S2A.
(B) Small airway brushings from CF and non-CF individuals with and without PsA were analyzed by RNA sequencing (RNA-seq; CF−PsA− n = 9, CF+PsA− n = 6, CF−PsA+ n = 6, CF+PsA+ n = 9). The multidimensional scaling plot (MDS) shows global changes in gene expression across the 4 groups. Global gene expression differences were identified by PERMANOVA across both CF (p = 0.02) and PsA (p < 0.001) strata.
(C) Heatmap analysis of differentially expressed genes between CF (purple, n = 9) and non-CF (blue, n = 6) recipients with positive PsA BAL cultures using an FDR-adjusted p value cutoff of 0.05. The yellow-purple shading indicates the relative expression scaled by row. In the context of PsA+ cultures, relative to non-CF individuals, CF recipients had a downregulation of 146 genes and an upregulation of 55 genes, detailed in Table S3.
(D) Gene Ontology pathway analysis was performed on genes upregulated in non-CF versus CF recipients with PsA with an FDR-adjusted p value cutoff of 0.05. The blue bars indicate the percentage of genes in the pathway that were upregulated, and the red line denotes the negative log of the p value for that pathway.
(E) Select MSigDB Hallmark gene expression pathways were compared across the 4 groups with CF+ PsA+ as the referent. Z scores for the mean and standard error of the sum of normalized counts for pathway genes in airway brushings are shown for each group (general linear models; ∗p < 0.05 and ∗∗p < 0.01).
During the study period, we prospectively collected airway brushes from lung transplant recipients with PsA+ cultures from 9 CF and 6 non-CF individuals (Figure S1). These were matched 1:1 with PsA− cases based on microbiologic culture results and time to post-transplant, and we performed next-generation RNA sequencing (RNA-seq) to evaluate the host transcriptome in these 30 unique lung transplant subjects. The subject characteristics for these groups were well matched, although the CF+PsA+ and CF−PsA− groups less commonly underwent bronchoscopy for surveillance, and subjects with CF were more likely to have had prior PsA+ airway cultures (Table S2).
We assessed the overall differences in the host transcriptome using the multidimensional scaling (MDS) plot (Figure 1B). Subjects with CF and PsA were distinct from the other groups primarily along MDS component 1. However, non-CF subjects with PsA overlapped with groups without PsA, suggesting that the interaction between CF and PsA was the dominant signal.
Next, we assessed which genes were differentially expressed in the PsA+ airway brush samples stratified by CF status. There were 55 genes upregulated and 146 genes downregulated in the CF+PsA+ airway samples versus CF−PsA+ referents (Figure 1C; Table S3). We performed Gene Ontology (GO) pathway analysis for these downregulated genes to characterize the nature of the attenuated transcripts in the CF subjects with PsA. Predominantly, pathways suppressed in CF subject airways with PsA were related to immune function (Figure 1D). The GO pathway with the largest difference in regulation was “response to interferon (IFN)-α” (6/21 genes, false discovery rate [FDR]-adjusted p = 5 × 10−6).
We also evaluated differential pathway responses using metagenes defined from curated gene sets.17, 18, 19 PsA culture positivity in the context of non-CF lung transplant recipients was associated with the increased expression of type 1 IFN genes, genes associated with allograft rejection, and multiple cell-cycle pathways, including E2F transcription factors, G2/M checkpoint inhibitors, and c-Myc gene targets (Figure 1E). Even though subjects in the CF+PSA− group demonstrated no evidence of acute cellular rejection by histopathology (Table S2), they had airway signatures of allograft rejection, including a signature of lymphocytic bronchitis (Figure S2B).20 By contrast PsA culture positivity in CF subjects was associated with the suppression of cell-cycle pathways and a trend toward the upregulation of Wnt-βcatenin signaling pathways. Hedgehog signaling, which may guard against airway fibrosis,21 was upregulated in CF subjects without PsA.
CF Airway Epithelial Phenotype Is Distinct before and after Transplantation
Although airway epithelial cells (AECs) are the predominant cell type in airway brushings, it is difficult to discern their specific contributions to gene expression changes from bulk RNA-seq of airway brushings. To understand the changes in AEC gene expression independent of direct stimulation from pathogens or infiltration of inflammatory cells, we cultured AECs from airway brushings under air-liquid interface (ALI) conditions. This approach also allowed comparison with pre-transplant (native) states in the form of AEC derived from CF lung explants or non-CF declined donor tissue.
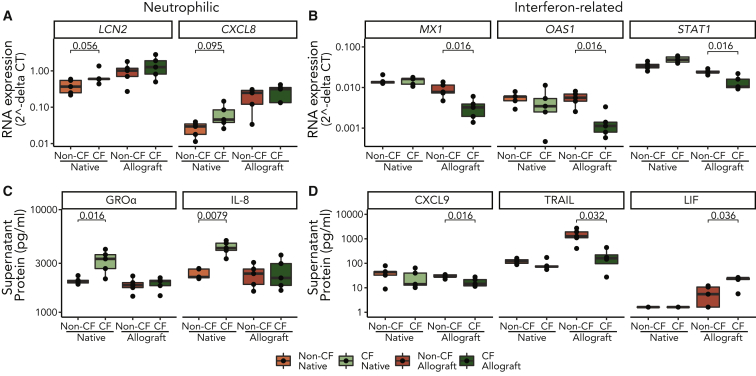
CXCL8 (human interleukin-8 [IL-8] gene) and LCN2 (antibacterial peptide gene) mRNA transcription trended toward higher expression in native CF AECs as compared to non-CF. In addition, CF AEC secreted significantly more IL-8 and growth-regulated oncogene-α (GRO-α) (neutrophilic chemokines) in culture supernatant versus non-CF AECs (Figure 2C). By contrast the IFN-related genes MX1, OAS1, and STAT1 were not differentially expressed pre-transplant (Figure 2B). We also stimulated cells with heat-killed PsA and found that both CXCL8 and LCN2 had greater upregulation in CF AECs as compared with non-CF AEC cultures (Figure S3A).
Figure 2.
CF Airway Epithelial Phenotype Is Distinct before and after Transplantation
Airway epithelial cells were cultured ex vivo either from native lung explants (n = 5 CF, n = 5 non-CF) or from airway brushings performed on allografts (n = 5 CF, n = 5 non-CF) 1 year post-transplant for subjects with and without CF.
(A and B) Neutrophilic transcripts LCN2 and CXCL8 (A) and interferon (IFN)-associated transcripts MX1, OAS1, and STAT1 (B), were quantified in cell lysates by PCR. The expression of mRNA is shown as 2−dCT values referenced to the housekeeping gene ACTB.
(C and D) Similarly, neutrophilic proteins IL-8 and GRO-α (C) and IFN-associated proteins CXCL-9, TRAIL, and LIF proteins (D) were measured in supernatant. LIF is negatively associated with IFN, while the CXCL-9 and TRAIL are reported to have a positive association. Box and whisker plots show median and range. (p values for the Wilcoxon rank-sum test are shown above each CF-to-non-CF comparison, except where values were >0.1 from pooled duplicates.)
We then compared AECs derived from airway brushings to lung allograft recipients with and without CF. The differences in neutrophilic protein secretion and antibacterial gene transcription pre-transplant were no longer present in CF allograft AEC (Figure 2A). Instead, we found relatively suppressed expression of IFN-related genes MX1, OAS1, and STAT1 in the CF allograft group (Figure 2B). Because these transcripts do not code for secreted proteins, we assayed supernatant for other IFN-related proteins (Figure 2D). CXCL-9 (monokine induced by IFN-γ) secretion and the IFN-dependent tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) secretion were attenuated in allograft epithelium from CF individuals.22 Leukemia inhibitory factor (LIF), a protein that counters the inflammatory STAT1 phosphorylation activity induced by IFNs,23, 24, 25 was found in greater abundance in the CF group.
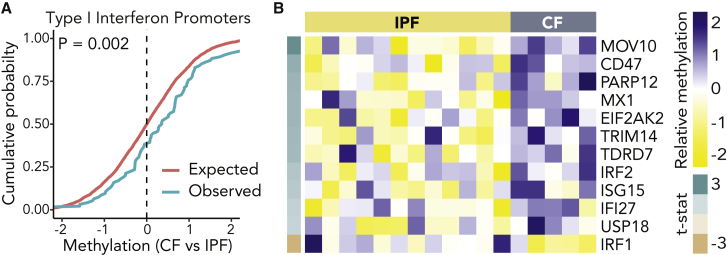
Type 1 IFN Promoters Are Hypermethylated in Allografts from Subjects with CF
To investigate possible epigenetic mechanisms for differential gene transcription by CF subjects before and after lung transplantation, we performed DNA methylation analysis. The proportion of methylated promotors within 50 Hallmark gene sets were compared between AECs derived from CF (n = 5) and non-CF (n = 12) allograft airway brushings (Table S4). Overall, we found a small global increase in promotor methylation in CF recipients (median β difference 4.5 × 10−4, p < 0.001). Across all 50 gene sets, there was a negative correlation between the degree of promotor methylation and the differences observed in metagene transcription from the RNA-seq data (ρ = −0.43, p = 0.002), which is consistent with the paradigm that promotor methylation may contribute to the observed transcriptional suppression observed in airways. Specifically, within the subset of genes in the type 1 IFN response pathway, there was increased promoter methylation when compared with the expected normal distribution (β difference 9.4 × 10−4, p = 0.002; Figure 3A). Within this type 1 IFN gene set, statistically significant differences in promotor methylation favored methylation in CF subjects, except for one gene (Figure 3B). While a random sampling of genes showed a distribution that matched the expected distribution, some other pathways were found to be hypermethylated in CF subjects, including G2/M and E2F cell-cycle pathways and allograft rejection genes, mirroring some differences observed from the gene expression patterns in non-CF recipients with PsA+ cultures (Figure S4).
Figure 3.
Type 1 IFN Promoters Are Hypermethylated in Allografts from Subjects with CF
(A) Kolmogorov-Smirnoff (K-S) plot for expected DNA methylation distribution in promoters of genes related to the type 1 IFN-related gene comparing allograft brush cells from CF (n = 5) and non-CF (IPF, n = 12) recipients. The pink line is the expected distribution and the green line is the observed distribution (K-S statistic p = 0.002).
(B) Heatmap of the 12 differentially methylated IFN-related gene promoters showing relative hypermethylation of 11 promoters in CF allograft cells.
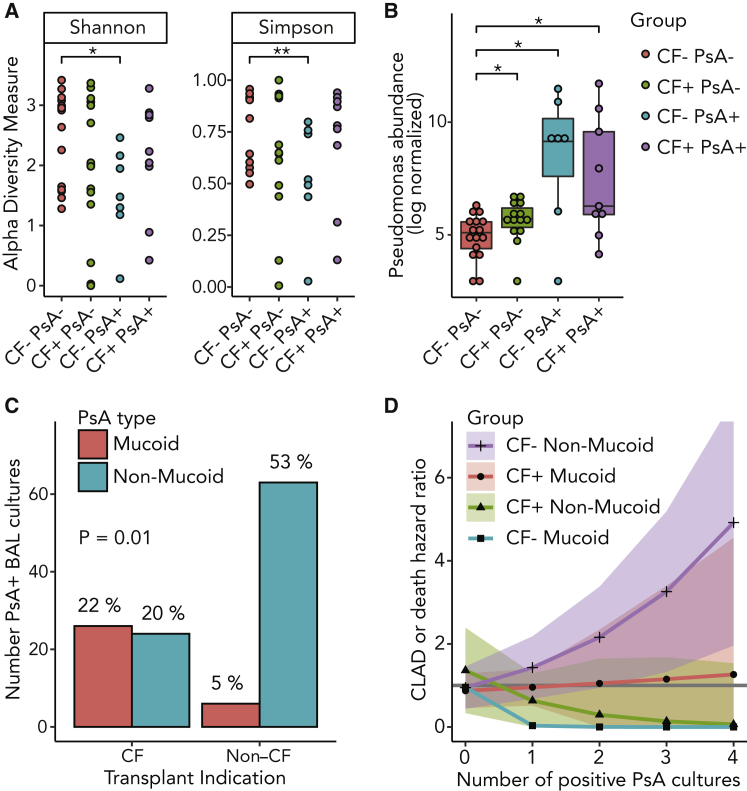
Non-mucoid PsA Is More Common in Non-CF Subjects and Is Associated with Increased CLAD or Death HR
To assess the contributions of PsA to the airway immune landscape, we compared the quantity and quality of PsA transcripts in airway brushings after transplantation. Even when PsA was not detected by culture, CF lung allograft recipients had greater abundance of PsA transcripts than non-CF recipients (p = 0.04; Figure 4B), and PsA groups had a higher normalized abundance of PsA transcripts (p = 2 × 10−13) than PsA culture-negative groups. In the context of PsA+ cultures, PsA was the dominant organism in 43% of non-CF and 33% of CF subjects (p = 1). However, we found a decrease in Shannon and Simpson measures of α diversity only in non-CF subjects with positive PsA cultures (Figure 4A). We also assessed β diversity across these four groups using UniFrac distances and by PERMANOVA, but we did not find a characteristic difference in the compositions of each group (Figure S3B).
Figure 4.
Non-CF Subjects with PsA Have Decreased α Diversity and Greater Prevalence of Non-mucoid PsA, Which Was Associated with an Increased Risk of CLAD or Death
(A) Shannon and Simpson α diversity metrics for transplant recipients stratified by CF and PsA culture status (CF−PsA− n = 9, CF+PsA− n = 6, CF−PsA+ n = 6, CF+PsA+ n = 9; Wilcoxon rank-sum; ∗p ≤ 0.05 and ∗∗p < 0.01).
(B) PsA abundance measured by qPCR and compared to culture status for CF and non-CF lung transplant recipients (Wilcoxon rank-sum; ∗p ≤ 0.05).
(C) Comparison of mucoid or non-mucoid PsA strains cultured from CF and non-CF transplant recipients (n = 119 PsA+ cultures, generalized estimating equations (GEE)-adjusted linear model, p = 0.01).
(D) Age-adjusted Cox proportional HR and 95% CI for CLAD or death based upon CF and mucoid PsA strata for increasing number of positive PsA cultures. HR = 1 (solid gray line).
Comparing all of the PsA+ BAL culture events, most isolates were non-mucoid strains from non-CF subjects (53%). Cumulatively, CF subjects had similar numbers of mucoid and non-mucoid strains, but mucoid PsA only accounted for 5% of the PsA cultures in the non-CF cohort (Figure 4C; p = 0.01).
We reexamined the association between the number of PsA+ BAL cultures and CLAD-free survival across CF strata for mucoid and non-mucoid PsA. As shown in Figure 4D, only non-mucoid PsA in non-CF individuals was associated with a statistically significant CLAD or death risk (HR 1.5, 95% CI 1.2–2.0, p = 0.006; p values for other comparisons ≥ 0.37).
To examine the potential impact of this mucoid dichotomy, we cultured clinical isolates of mucoid and non-mucoid PsA from CF and non-CF lung transplant recipients, respectively. Normal human airway epithelial cells, differentiated under ALI conditions, responded robustly to both strains of PsA in vitro, but there was a trend toward greater induction of CXCL8 by the non-mucoid PsA strain, while MX1 expression demonstrated a trend toward attenuation by the mucoid PsA strain (Figure S3C).
Discussion
While PsA infection was associated with decreased CLAD-free survival in non-CF lung transplant recipients, this association did not hold for CF lung transplant recipients. These findings are consistent with reports in some11,26 but not all lung transplant cohorts.27 However, comparisons between CF and non-CF recipients are limited by the confounding of subject characteristics, such as age and comorbidities. As an alternative approach, we assessed for potential host and microbiome mechanistic correlates of this protection. Brushes from the airway epithelium of lung transplant recipients demonstrated suppression of type 1 IFN-related genes unique to CF recipients in the context of PsA culture positivity. Cultured AEC demonstrated that these differences are present ex vivo in the absence of microbes or other cell types and are distinct from the native CF AEC phenotype. Furthermore, DNA methylation analysis showed that promotor methylation could explain some of these observed differences. At the same time, non-CF lung transplant recipients with PsA had decreased microbial α diversity and predominantly non-mucoid strains of PsA, for which increased flagellin expression and decreased antigen sequestration by alginate matrix could result in enhanced immunogenicity. The combination of attenuated host immune responsiveness and decreased antigenic stimulation by mucoid PsA may explain how CF patients can have favorable lung transplant outcomes despite PsA infection.
Type 1 IFNs play key roles in infection, autoimmunity, and allograft rejection.28 In a cohort of kidney transplant recipients, type 1 IFN gene expression was increased in the context of antibody-mediated rejection.29 Poly(I:C), a viral pathogen-associated molecular pattern mimic, can break tolerance and induce obliterative airway disease in a murine heterotopic tracheal transplant model, while knockout of the type 1 IFN receptor abrogated this effect.30 In a mouse model of intestinal graft-versus-host disease, knockout of IFN-α and -β receptors protected from intestinal injury.31 An in vitro infection model using HeLa cells showed the induction of type 1 IFN responses that lead to an increase in self-peptide presentation by major histocompatibility complex (MHC) class I complexes,32 potentially linking it to deleterious autoimmune or alloimmune consequences if extrapolated to the context of transplantation. Consistent with the results presented here, PsA was shown to attenuate IFN responses to rhinovirus in CF AECs, but not in normal human bronchial cells.33 Finally, the inhibition of type 1 IFN signaling attenuated dermal fibrosis in a systemic sclerosis model, linking inflammation and fibrosis via type 1 IFN signaling.34 Thus, the inhibition of alloimmune responses or direct protection from fibrosis could be a link between the downregulation of type 1 IFN signaling in CF lung transplant recipients and the relative protection from CLAD in the context of PsA exposure.
Methylation of promoter region DNA can sterically inhibit RNA transcription. We identified multiple pathways for which promotors were relatively hypermethylated in AECs from CF lung transplant recipients, including the IFN-α gene set. Such inhibition could result from chronic stimulation, as has been previously observed.35 Alternatively, bacteria can influence the transcriptional landscape of a cell by various epigenetic mechanisms, including methylation, histone acetylation, and altering the activity of chromatin-binding proteins that may confer an evolutionary advantage.36 For example, Listeria monocytogenes secretes a virulence factor that influences the chromatin repressor BAHD1 and, subsequently, transcription of IFN-stimulated genes.37 Even extracellular bacteria such as PsA have been shown to be capable of altering the host epigenome by interfering with histone phosphorylation and acetylation.38 While we were not able to assay histone modification in this cohort, this would be another plausible mechanism for how AEC could adopt a hyporesponsive phenotype. It is also possible that downregulation of type I IFN signaling may reflect recipient immune exhaustion from chronic colonization or CF-specific differences in lymphocyte function.39
A key finding is that native and allograft AECs maintain the phenotype obtained in vivo when they are grown in culture without direct microbial or circulating (i.e., classical immune cells) host factors. In general, primary airway cells maintain their phenotype for at least four passages ex vivo.40,41 Our own unpublished data show that inflammatory responses due to microbial stimulation are maintained for at least two passages ex vivo.
In our cohort, non-CF individuals with PsA also had reduced microbial α diversity. Low microbial diversity can result from the overgrowth of a dominant pathogen and is typically associated with true infection (as opposed to colonization) and worse clinical outcomes. Pre-transplant, lower diversity is associated with the development of chronic infection over time in CF patients.5 However, in this study, CF patients with PsA had preserved α diversity. Also, mucoid PsA was common in CF lung transplant recipients, while non-CF recipients had mostly non-mucoid PsA. CF patients are often infected with non-mucoid PsA early in life and transition from non-mucoid to mucoid strains by their early teens.2 Conversely, mucoid PsA lung infections are highly unusual in non-CF individuals. Notably, PsA abundance was greater in CF patients, even when not detected by culture. The presence of mucoid PsA before and after transplant suggests recolonization, likely from the sinotracheal tract. Clinically, mucoid strains are typically accompanied by higher morbidity and mortality as compared to non-mucoid phenotypes.42 However, mucoid PsA variants have decreased infectivity, resulting from the loss of immunogenic virulence factors and development of an exopolysaccharide matrix that facilitates biofilm formation and immune evasion.43, 44, 45 When comparing mucoid to non-mucoid PsA strains in vitro, we did not observe a statistically significant difference in AEC immune responses. However, we used heat-killed PsA, which may have denatured virulence factors or counteracted the effect of the alginate matrix, diminishing the apparent differences between strains.
Conclusion
We observed a distinct epithelial immunophenotype after lung transplantation in CF patients, characterized by type 1 IFN pathway suppression, and possibly secondary to DNA hypermethylation. This immune suppression may be beneficial for CF recipient allografts in the presence of PsA. Future studies of PsA clinical management should thus consider the potential detrimental effects of eradicating a mucoid PsA-dominated microbiome.
Limitations of Study
These data were generated at a single center and thus may not be generalizable to other centers where patient characteristics and management protocols differ. Also, while we attempted to control for age in our analysis statistical adjustments, comparisons between CF and non-CF allograft recipients are limited by the lack of overlap in age between the two groups. Other transcriptomic pathways may be evident with larger sample numbers and/or greater read depth. Our in vitro data exclude many cell types from the allograft milieu, and co-culture with classical immune cells could reveal other interesting and nuanced interactions that we have not reported here. Last, our epigenetic analysis only included DNA methylation and other mechanisms, like histone acetylation should be considered in future analyses.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Heat-Killed P. aeruginosa laboratory strain PAO1 | InvivoGen | Cat#tlrl-hkpa |
| Mucoid P. aeruginosa clinical strain | UCSF Microbiology | Acc#F24698 |
| P. aeruginosa clinical strain | UCSF Microbiology | Acc#S59951 |
| Biological Samples | ||
| Airway explant tissue | UCSF Department of Pathology | IRB 10-02253 |
| Airway brush samples | UCSF Dep. of Medicine | IRB 13-10738 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Rho kinase inhibitor, Y-27632 | Selleck Chem | Cat# S1049 |
| QIAzol | QIAGEN | Cat#79306 |
| TRIzol | Invitrogen | Cat#15596026 |
| Trypsin-EDTA 0.25% | ThermoFisher | Cat#25200056 |
| Protease from Streptomyces griseus Type XIV | Millipore Sigma | Cat#P5147 |
| Tryptic Soy Broth | BD-Bacto | Cat#211825 |
| Critical Commercial Assays | ||
| miRNeasy Mini Kit | QIAGEN | Cat#217004 |
| SuperScript cDNA Synthesis Kit | Invitrogen | Cat#11904018 |
| Taqman Gene Expression Master Mix | Applied Biosystems | Cat#4369542 |
| Human Cytokine/Chemokine Array 65-Plex Panel | Eve Technologies | Cat#HD65 |
| NEBNext RNA First Strand Synthesis Module | New England BioLabs | Cat#E7525 |
| NEBNext Ultra® Second Strand Synthesis Module | New England BioLabs | Cat#E7550 |
| NEBNext Ultra® II DNA Library Prep Kit for Illumina | New England BioLabs | Cat#E7645 |
| GenElute Mammalian Genomic DNA Miniprep Kit | Millipore Sigma | Cat#G1N70 |
| QIAshredder | QIAGEN | Cat#217084 |
| Human Cytokine/Chemokine Array | Eve Technologies | Cat#HD71 |
| Human Adipokine Array 5-Plex | Eve Technologies | Cat#HDADK5 |
| Infinium MethylationEpic Array | Illumina | Cat#WG-317-1002 |
| Deposited Data | ||
| Airway Epithelial Transcriptome in Lung Transplant Recipients with Cystic Fibrosis and/or Pseudomonas | Mendeley Data | https://doi.org/10.17632/89nzfyvkvc.1 |
| Experimental Models: Cell Lines | ||
| Primary cell cultures derived from explanted tissue | See Biological Samples above | n/a |
| Experimental Models: Organisms/Strains | ||
| Pseudomonas aeruginosa | See Bacterial and Virus Strains above | n/a |
| Oligonucleotides | ||
| Cxcl8 Hs00174103_m1 | ThermoFisher | Cat#4331182 |
| Lcn2 Hs01008571_m1 | ThermoFisher | Cat#4331182 |
| Mx1 Hs00895608_m1 | ThermoFisher | Cat#4331182 |
| Oas1 Hs00973637_m1 | ThermoFisher | Cat#4331182 |
| Actb Hs01060665_g1 | ThermoFisher | Cat#4331182 |
| Stat1 Hs01013996_m1 | ThermoFisher | Cat#4331182 |
| Other | ||
| ALI Transwell® Inserts | Corning Life Sciences | Cat#3460 |
Resource Availability
Lead Contact
Requests for information or resource sharing can be directed to the Lead Contact, John Greenland (John.Greenland@ucsf.edu).
Materials Availability
There are restrictions to the availability of our bacterial strains due to the lack of an external centralized repository for its distribution and our need to maintain the stock. We are glad to share clinical Pseudomonas strains with reasonable compensation by requestor for its processing and shipping, and we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Primary airway cells from this study are unavailable because they were consumed by experiments for this study.
Data and Code Availability
The RNASeq and DNA methylation datasets generated during this study are available at Mendeley Data under https://doi.org/10.17632/89nzfyvkvc.1. Transcriptomic count data are provided in GEO data matrix format. Raw DNA methylation data are provided as in EPIC report format. Analytic code is available from the corresponding author upon request.
Experimental Models and Subject Details
Human Subjects
We included lung transplant recipients from an ongoing longitudinal cohort (Figure S1, detailed summary demographic data in Table S1) at the University of California San Francisco. Lung transplant candidates were approached for study enrollment at the time of listing or as soon as possible following lung transplant. All subjects provided informed consent for medical record review, research airway brushing during bronchoscopy, or both (UCSF Department of Medicine IRB 13-10738). Post-transplant immunosuppression followed institutional protocols, with basiliximab and methylprednisolone induction. During the first 3 months, prednisone was dosed at 20 mg daily and tacrolimus dosed to 10 to 14 ng/mL trough concentrations. For month 3-6, prednisone was tapered to 0.2 mg/kg daily and tacrolimus targeted 10 to 12 ng/mL. Prednisone was then tapered to 0.1 mg/kg by 12 months and tacrolimus adjusted to 8 to 10 ng/mL. Mycophenolate mofetil targeting 2 g daily in divided doses was started post-operatively. Immunosuppression was tailored to address side effects.46
All subjects who consented for records review and underwent bronchoscopy with BAL were included in our longitudinal cohort to assess CLAD-free survival. Subjects undergoing for-cause (e.g., symptomatic) or surveillance bronchoscopy (scheduled at 2, 4, 8, 12, 26, 52, and 78 weeks post-transplant) were eligible for an additional research airway sampling after post-transplant week 6, at the bronchoscopy provider’s discretion. For example, in cases with acute symptoms or clinical instability, providers may have elected to forgo research brushing to ensure adequate time to obtain transbronchial biopsies. From this longitudinal brushing cohort, we selected nested case-control cohorts for transcriptome and metagenome comparisons, and DNA methylation array. Study enrollment is depicted in Figure S1).
CLAD or Death Hazard Assessment
All consenting lung allograft recipients with pulmonary function test (PFT) during the study period, including those only with consent for medical records review, were included in this analysis. CLAD cases were determined based on irreversible decline of > 20% in FEV1.47 Two investigators then reviewed PFT data for each CLAD case and excluded cases with diagnostic uncertainty or alternative causes for FEV1 decline. Death was determined from UCSF and UNOS records. PsA positive cultures were abstracted from medical records. Mucoid status was determined by clinical labs based on visual inspection. Demographic summary, including age and gender information, is displayed in Table S1.
Transcriptome & Metagenome Analysis
All available airway brushes from lung transplant recipients with CF (with or without PsA) and non-CF (with PsA) were included. Controls (non-CF subjects without PsA) were matched based on microbiologic culture results and time-post transplant. When multiple brushing samples were available per subject, only the first was included. Age, gender and other demographic data are included in Table S2.
DNA Methylation Analysis
All available one-year brushings that yielded epithelial cell cultures from subjects with CF were included. Controls, also from one-year brushings with cell cultures, were selected from subjects with IPF and matched based upon donor-age in 1:2 ratio. Demographic information, including age and gender are available in Table S4.
Primary Cell Cultures
At the time of necropsy (non-diseased lungs) or lung transplant (CF affected lungs), airways were isolated from deidentified subjects under UCSF Department of Pathology IRB 10-02253 and processed as described below. Other than lung health status, no other identifying information was available from the tissue repository.
Pseudomonas aeruginosa Strains
A retrospective medical records search was performed to find laboratory-confirmed P. aeruginosa (PsA) lung infections among the UCSF lung transplant longitudinal cohort with cryopreserved and banked bacteria. We identified 1 mucoid PsA from a CF lung transplant recipient (UCSF Microbiology Laboratory accession #F24698) and 1 non-mucoid PsA from a non-CF recipient (accession #S59951). Live bacteria were cultured then heat-killed as described below.
Method Details
Airway Brushing
Following bronchoalveolar lavage and prior to transbronchial biopsies for clinical purposes, airway brushing was performed using ConMed #129 2-mm brushes in distal airways under fluoroscopic guidance. Airway brushes were immediately placed in 700 ul Qiazol and frozen at −20°C for up to 24 hours prior to vortexing and removal of brush. Qiazol with lysed airway cells was then passed through a QIAshredder prior to long-term storage at −80°C.
RNA-seq Host Transcriptome and Microbiome
RNA was extracted from airway brushes using the QIAGEN miRNeasy Mini Kit following manufacturer’s instructions and then reverse transcribed with NEBNext RNA First Strand Synthesis and Ultra® Second Strand Synthesis Modules. Library construction was performed with NEBNext Ultra® II Library Prep Kit according to described methods48 on an Agilent Bravo liquid handling instrument. Depletion of abundant sequences by hybridization (DASH) was employed to selectively deplete human mitochondrial cDNA, thus enriching for both microbial and human protein coding transcripts.49 The final RNA-seq libraries underwent 150 nucleotide paired-end Illumina sequencing on a Novaseq 6000.
Clinical Pseudomonas Strain Culture and Quantification
UCSF Microbiology Department supplied blood agar plates with Pseudomonas aeruginosa: One mucoid and one non-mucoid strain each harvested from the airways of separate lung transplant recipients (CF and non-CF individuals respectively). One colony was taken from each dish and placed into 20ml of tryptic soy broth and grown overnight in 37°C and 250rpm shaking incubator. Spectrophotometry was used to quantify organism numbers per ml of culture media. Bacteria were centrifuged and broth discarded. Pellet was washed with sterile PBS and centrifuged again, supernatant discarded. Finally, the cell pellet was resuspended to 1010 bacteria/ml of PBS. Aliquots of 1ml were heat-killed at 65°C for 45 minutes. Heat-killing was verified by streaking 1μl of cells onto blood agar and incubating in 37°C for 3 days checking for growth every 24hr. Based on preliminary experiments to generate an optimal IL-8 signal, heat-killed bacteria were diluted to 6⋅108/ml in sterile culture media for epithelial stimulation experiments.
Airway Epithelial Cell Culture
Airway epithelial cell culture techniques were based upon published techniques.50 For pre-transplantation in vitro experiments: Segments of human bronchus were harvested from 5 explanted CF lungs and 5 explanted non-CF lungs. Bronchi were washed 3 times in cold PBS with antibiotics and 5mM dithiothreitol to digest mucus, rinsed in PBS with antibiotics, and then submerged in 0.1% protease in PBS with antibiotics overnight at 4°C. The following day, protease was aspirated, 5% FBS in DMEM added, and the container was shaken vigorously to loosen epithelial cells from the airway scaffold tissue. The remaining bronchial tissue was removed, cells were centrifuged at 300xg and 4°C for 6 minutes and supernatant discarded. Cell pellet was resuspended in 0.05% trypsin-EDTA and pipetted repeatedly until the cell suspension was uniformly cloudy and no visible cell clumps remained. The suspension was neutralized with 5% FBS in DMEM, flirted through 100μm cell strainer and centrifuged again. The supernatant was discarded and pellet resuspended in 5% FBS in DMEM and cell concentration counted. Cells (5⋅105/cm2) resuspended in 5% FBS in DMEM were plated onto 6-mm, 0.4-μm cell culture Transwell® inserts coated with human placental collagen (20 μg/cm2), and 5% FBS in DMEM was added to the basal compartment. Cultures were placed in 37°C, 5% CO2 incubator. The next day, media was removed from both compartments, cells were washed in PBS, and ALI-media (see components table below) was added to the basal compartment only. Media was changed every 2-3 days until experimentation at day 28.
For post-transplantation in vitro experiments: Airway epithelial cells were obtained from 5 CF recipients and 5 non-CF recipients by sterile brushing during a surveillance bronchoscopy (asymptomatic monitoring) 12 months after transplantation procedure. The cells were agitated from the brush into sterile ALI-media and centrifuged as above. Supernatant is discarded and cell pellet resuspended in 0.25% tryspin-EDTA for 5-10 minutes at room temperature. Three volumes of DMEM or ALI with 5% FBS added to quench trypsin and centrifuged again. Supernatant discarded and cell pellet resuspended in ALI with 10 μM rho kinase inhibitor (promoting growth from very small numbers of cells) and plated onto a collagen-coated culture dish (size and volume depending on number of cells). Cells were grown in 37°C, 5%CO2 incubator and media changed every 2-3 days until about 80% confluent. Cells were washed once with 2-5 mL of PBS then incubated with 0.25% trypsin-EDTA in 37°C for 5 minutes. Cells and trypsin were removed into a conical tube and 3 volumes of DMEM with 5% FBS used to quench trypsin. Cells were centrifuged and supernatant discarded. Cell pellet was resuspended in ALI-media, counted, and plated as in cells from explanted tissue. Remaining methods match the explanted tissue protocol above.
| ALI Component | Concentration |
|---|---|
| LHC Basal Medium | 50% of medium |
| DMEM high glucose with pyruvate | 50% of medium |
| Glutamine | 2mM |
| Bovine serum albumin | 0.5mg/ml |
| Bovine pituitary extract | 0.24mg/ml |
| Insulin | 5μg/ml |
| Transferrin | 10μg/ml |
| Hydrocortisone | 0.1μM |
| Triiodothyronine | 0.01μM |
| Epinephrine | 2.7μM |
| Recombinant human EGF | 0.5ng/ml |
| Retinoic acid | 0.05μM |
| Phosphorylethanolamine | 0.5μM |
| Ethanolamine | 0.5μM |
| Zinc sulfate | 3μM |
| Penicillin, Streptomycin | 100U/ml, 100μg/ml |
| Selenium | 0.03μM |
| Manganese | 0.001μM |
| Silicone | 0.5μM |
| Molybdenum | 0.001μM |
| Vanadium | 0.005μM |
| Nickel sulfate | 0.001μM |
| Tin | 0.0005μM |
| Ferrous sulfate | 1.5μM |
| Magnesium chloride | 0.6mM |
| Calcium chloride | 0.11mM |
ALI Cell Bacterial Exposures
Once epithelial cells were grown in ALI conditions for 28 days, selected wells were exposed to heat-killed P. aeruginosa (HKPA): PAO1, clinical mucoid strain, or clinical non-mucoid strain (see Key Resources Table). Apical surfaces were washed with 100μl of warm PBS and then the PBS was discarded. ALI media was used to dilute HKPA strains to 6⋅108/ml. For controls, plain ALI-media was used. Control or a HKPA dilution were applied to cultures, 100μl in apical compartment and 600μl in basal compartment. Cultures were incubated for 24hr then supernatants from apical and basal compartments collected and immediately frozen in −80°C. Cells were harvested with TRIzol reagent and then stored at −80°C until RNA extraction.
Supernatant Protein Analysis
At the end of experiments, 100μl of basal media was used to wash the apical compartment of each well then pooled back with the basal supernatant. Supernatants were frozen immediately and stored at −80°C until analysis. Samples were shipped overnight on dry ice to Eve Technologies for analysis by BioPlex 200 HD71 multiplex immunoassay after determination of sample dilution requirements on a single sample.
RT-PCR
Cells were harvested at the end of experiments with TRIzol. Total RNA was prepared using the miRNeasy Mini kit according to manufacturer’s protocols. RNA concentration was determined by NanoDrop spectrophotometer. Using 500ng of RNA, cDNA was synthesized using SuperScript First-Strand Synthesis System following manufacturer’s protocol. RT-PCR was performed on QuantStudio 7 Flex using TaqMan reagents and commercially available primer-probes (see Oligonucleotides in the Key Resources Table) with an input of 1ul of cDNA per reaction. Cycle thresholds (Ct) for transcripts of interest were normalized to Actb housekeeping gene (dCt) within each sample and expression shown as 2-dCt.
Quantification and Statistical Analysis
CLAD or death hazard assessment
We examined the interaction between CF as a transplant indication and the frequency of PsA-positive BAL culture events, adjusted for recipient age, gender, and transplant indications other than CF using Cox proportional hazards models in the “survival” package.51 Cumulative exposure to PsA as a function of time from transplant was used as a time-dependent covariate. The interaction term was PsA exposure ∗ CF status. To explore this interaction finding in detail, we examined risk of CLAD or death in age-adjusted models stratified by CF status, and examined mucoid and non-mucoid PsA separately. Results were visualized using the “predict.coxph” function with risk score type with 95% confidence intervals derived as 1.96 ∗ standard error.
Host transcriptome analysis
RNA sequences were aligned to the human genome (NCBI GRC h38) using STAR,52 and coding RNAs were grouped by ENTREZ symbol. Kruskal’s non-metric multidimensional scaling plots were created using the “MASS” library. Global gene expression differences between groups were calculated by PERMANOVA using the “vegan” library. Differential gene expression and normalization was performed using “DESeq2,” using a false discovery rate-adjusted p value cutoff of 0.05. A heatmap of differentially expressed genes in PsA+ brushes was created using “pheatmap.” Pathway analysis was performed using the Gene Ontology Biologic Process pathways through “goanna.” Similar pathways were grouped for readability. Metagenes scores were calculated using MSigDB Hallmark Gene set collections as the z-score derived from sum of variance stabilizing transformation-normalized gene counts by subtracting the mean and dividing by the standard deviation. Differences in metagene score were determined by generalized linear modeling.
Airway epithelial cell gene expression and protein quantification
We compared quantitative PCR and protein concentrations between CF and non-CF samples using two-tailed Wilcoxon rank-sum tests, without adjustment for multiple comparisons.
DNA methylation analysis
Airway epithelial cells were cultured to confluence for a single passage prior to DNA extraction by GenElute Mammalian Genomic DNA Miniprep Kit. DNA methylation was determined by Illumina Infinium MethylationEPIC BeadChip Kit. Preprocessing was performed using RnBeads.53 Differential promotor methylation was determined between CF and IPF AEC using unpaired Student’s t test. Comparisons between observed and expected t-statistic distributions within Hallmark gene sets were determined by Kolmogorov-Smirnov tests.
Pathogen Detection Bioinformatics
Detection of host transcripts and airway microbes leveraged the open-source ID-Seq pipeline54 that incorporated subtractive alignment of the human genome (NCBI GRC h38) using STAR,52 quality filtering using PRICESeqfilter and removal of additional non-fungal eukaryotes, cloning vectors and phiX phage using Bowtie2.55 The identities of the remaining microbial reads were determined by querying the NCBI nucleotide (NT) and non-redundant (NR) protein databases using GSNAP-L and RAPSEARCH2, respectively. Microbial alignments were aggregated to the genus-level and only those microbes aligning to both NT and NR databases were utilized in subsequent downstream pathogen versus commensal analyses. Pathogen counts were normalized using DESeq2. MDS and PERMANOVA were performed as above to assess beta-diversity. Alpha-diversity was compared using Simpson and Shannon metrics using the “physeq” package.56
Association between CF and mucoid status was determined by generalized estimated equation-adjusted logistic regression model (GEE) with Mucoid status as the dependent variable and subject identifier as the cluster variable.
All statistical analyses were performed in R version 3.6.0.
Acknowledgments
This work was supported by Cystic Fibrosis Foundation Therapeutics grant GREENL16XX0 (to J.R.G.), the UCSF Cystic Fibrosis Foundation Research Development grant (to W.F.), Veterans Affairs Office of Research and Development awards CX001034 and CX002011 (to J.R.G.), National Heart, Lung, and Blood Institute (NHLBI) R01HL151552 (to J.R.G.), NHLBI K23HL138461 (to C.L.), and National Institute of Diabetes and Digestive and Kidney Diseases P30DK072517 (to W.F.). The graphical abstract was illustrated by Nicholas Bezio (Bezio Studios, Colchester, VT). The authors would like to acknowledge Steve Miller from University of California, San Francisco (UCSF) Microbiology for help obtaining clinical PsA strains.
Author Contributions
D.T.D., M.F., C.L., E.G., and J.R.G. designed the experiments. D.T.D., M.F., L.Z., S.C., and L.S. performed the experiments. W.F. provided the primary cells for the cultures and consulted on experimental design. S.R.H., J.P.S., L.E.L., J.A.G., R.J.S., J.K., and M.E.K. obtained and provided the clinical samples for the RNA-seq and culture data. D.T.D., M.F., C.L., and J.R.G. performed the data analysis. D.T.D. and J.R.G. wrote the manuscript. All of the authors participated in the manuscript revisions.
Declarations of Interests
The authors declare no competing interests.
Published: July 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100055.
Supplemental Information
A positive-fold change indicates upregulated in CF+ versus CF− referents. (Related to Figure 1)
References
- 1.Stephenson A.L., Sykes J., Stanojevic S., Quon B.S., Marshall B.C., Petren K., Ostrenga J., Fink A.K., Elbert A., Goss C.H. Survival Comparison of Patients With Cystic Fibrosis in Canada and the United States: A Population-Based Cohort Study. Ann. Intern. Med. 2017;166:537–546. doi: 10.7326/M16-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malhotra S., Hayes D., Jr., Wozniak D.J. Cystic Fibrosis and Pseudomonas aeruginosa: the Host-Microbe Interface. Clin. Microbiol. Rev. 2019;32:e00138-18. doi: 10.1128/CMR.00138-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chmiel J.F., Aksamit T.R., Chotirmall S.H., Dasenbrook E.C., Elborn J.S., LiPuma J.J., Ranganathan S.C., Waters V.J., Ratjen F.A. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, gram-negative bacteria, and multiple infections. Ann. Am. Thorac. Soc. 2014;11:1120–1129. doi: 10.1513/AnnalsATS.201402-050AS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salsgiver E.L., Fink A.K., Knapp E.A., LiPuma J.J., Olivier K.N., Marshall B.C., Saiman L. Changing Epidemiology of the Respiratory Bacteriology of Patients With Cystic Fibrosis. Chest. 2016;149:390–400. doi: 10.1378/chest.15-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pritt B., O’Brien L., Winn W. Mucoid Pseudomonas in cystic fibrosis. Am. J. Clin. Pathol. 2007;128:32–34. doi: 10.1309/KJRPC7DD5TR9NTDM. [DOI] [PubMed] [Google Scholar]
- 6.Chambers D.C., Yusen R.D., Cherikh W.S., Goldfarb S.B., Kucheryavaya A.Y., Khusch K., Levvey B.J., Lund L.H., Meiser B., Rossano J.W., Stehlik J., International Society for Heart and Lung Transplantation The Registry of the International Society for Heart and Lung Transplantation: Thirty-Fourth Adult Lung And Heart-Lung Transplantation Report-2017; Focus Theme: Allograft Ischemic Time. J. Heart Lung Transplant. 2017;36:1047–1059. doi: 10.1016/j.healun.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 7.Walter S., Gudowius P., Bosshammer J., Römling U., Weissbrodt H., Schürmann W., von der Hardt H., Tümmler B. Epidemiology of chronic Pseudomonas aeruginosa infections in the airways of lung transplant recipients with cystic fibrosis. Thorax. 1997;52:318–321. doi: 10.1136/thx.52.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaparro C., Maurer J., Gutierrez C., Krajden M., Chan C., Winton T., Keshavjee S., Scavuzzo M., Tullis E., Hutcheon M., Kesten S. Infection with Burkholderia cepacia in cystic fibrosis: outcome following lung transplantation. Am. J. Respir. Crit. Care Med. 2001;163:43–48. doi: 10.1164/ajrccm.163.1.9811076. [DOI] [PubMed] [Google Scholar]
- 9.Allyn P.R., Duffy E.L., Humphries R.M., Injean P., Weigt S.S., Saggar R., Shino M.Y., Lynch J.P., 3rd, Ardehali A., Kubak B. Graft Loss and CLAD-Onset Is Hastened by Viral Pneumonia After Lung Transplantation. Transplantation. 2016;100:2424–2431. doi: 10.1097/TP.0000000000001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregson A.L. Infectious Triggers of Chronic Lung Allograft Dysfunction. Curr. Infect. Dis. Rep. 2016;18:21. doi: 10.1007/s11908-016-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritchard J., Thakrar M.V., Somayaji R., Surette M.G., Rabin H.R., Helmersen D., Lien D., Purighalla S., Waddell B., Parkins M.D. Epidemic Pseudomonas aeruginosa infection in patients with cystic fibrosis is not a risk factor for poor clinical outcomes following lung transplantation. J. Cyst. Fibros. 2016;15:392–399. doi: 10.1016/j.jcf.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Lynch J.P., 3rd, Sayah D.M., Belperio J.A., Weigt S.S. Lung transplantation for cystic fibrosis: results, indications, complications, and controversies. Semin. Respir. Crit. Care Med. 2015;36:299–320. doi: 10.1055/s-0035-1547347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samano M.N., Pêgo-Fernandes P.M., Fonseca Ribeiro A.K., Turaça K., Abdalla L.G., Fernandes L.M., Correia A.T., Jatene F.B. Lung transplantation in patients with cystic fibrosis. Transplant. Proc. 2013;45:1137–1141. doi: 10.1016/j.transproceed.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Singer J.P., Katz P.P., Soong A., Shrestha P., Huang D., Ho J., Mindo M., Greenland J.R., Hays S.R., Golden J. Effect of Lung Transplantation on Health-Related Quality of Life in the Era of the Lung Allocation Score: A U.S. Prospective Cohort Study. Am. J. Transplant. 2017;17:1334–1345. doi: 10.1111/ajt.14081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vos R., Vanaudenaerde B.M., Geudens N., Dupont L.J., Van Raemdonck D.E., Verleden G.M. Pseudomonal airway colonisation: risk factor for bronchiolitis obliterans syndrome after lung transplantation? Eur. Respir. J. 2008;31:1037–1045. doi: 10.1183/09031936.00128607. [DOI] [PubMed] [Google Scholar]
- 16.Botha P., Archer L., Anderson R.L., Lordan J., Dark J.H., Corris P.A., Gould K., Fisher A.J. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation. 2008;85:771–774. doi: 10.1097/TP.0b013e31816651de. [DOI] [PubMed] [Google Scholar]
- 17.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhakta N.R., Christenson S.A., Nerella S., Solberg O.D., Nguyen C.P., Choy D.F., Jung K.L., Garudadri S., Bonser L.R., Pollack J.L. IFN-stimulated Gene Expression, Type 2 Inflammation, and Endoplasmic Reticulum Stress in Asthma. Am. J. Respir. Crit. Care Med. 2018;197:313–324. doi: 10.1164/rccm.201706-1070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen K., Campfield B.T., Wenzel S.E., McAleer J.P., Kreindler J.L., Kurland G., Gopal R., Wang T., Chen W., Eddens T. Antiinflammatory effects of bromodomain and extraterminal domain inhibition in cystic fibrosis lung inflammation. JCI Insight. 2016;1:e87168. doi: 10.1172/jci.insight.87168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenland J.R., Wang P., Brotman J.J., Ahuja R., Chong T.A., Kleinhenz M.E., Leard L.E., Golden J.A., Hays S.R., Kukreja J. Gene signatures common to allograft rejection are associated with lymphocytic bronchitis. Clin. Transplant. 2019;33:e13515. doi: 10.1111/ctr.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng T., Frank D.B., Kadzik R.S., Morley M.P., Rathi K.S., Wang T., Zhou S., Cheng L., Lu M.M., Morrisey E.E. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature. 2015;526:578–582. doi: 10.1038/nature14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peteranderl C., Herold S. The Impact of the Interferon/TNF-Related Apoptosis-Inducing Ligand Signaling Axis on Disease Progression in Respiratory Viral Infection and Beyond. Front. Immunol. 2017;8:313. doi: 10.3389/fimmu.2017.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascual-García M., Bonfill-Teixidor E., Planas-Rigol E., Rubio-Perez C., Iurlaro R., Arias A., Cuartas I., Sala-Hojman A., Escudero L., Martínez-Ricarte F. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8+ T cell tumor-infiltration impairing anti-PD1 therapy. Nat. Commun. 2019;10:2416. doi: 10.1038/s41467-019-10369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dallagi A., Girouard J., Hamelin-Morrissette J., Dadzie R., Laurent L., Vaillancourt C., Lafond J., Carrier C., Reyes-Moreno C. The activating effect of IFN-γ on monocytes/macrophages is regulated by the LIF-trophoblast-IL-10 axis via Stat1 inhibition and Stat3 activation. Cell. Mol. Immunol. 2015;12:326–341. doi: 10.1038/cmi.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regis G., Pensa S., Boselli D., Novelli F., Poli V. Ups and downs: the STAT1:STAT3 seesaw of Interferon and gp130 receptor signalling. Semin. Cell Dev. Biol. 2008;19:351–359. doi: 10.1016/j.semcdb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Burkett A., Vandemheen K.L., Giesbrecht-Lewis T., Ramotar K., Ferris W., Chan F., Doucette S., Fergusson D., Aaron S.D. Persistency of Pseudomonas aeruginosa in sputum cultures and clinical outcomes in adult patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:1603–1610. doi: 10.1007/s10096-011-1483-8. [DOI] [PubMed] [Google Scholar]
- 27.Vital D., Hofer M., Benden C., Holzmann D., Boehler A. Impact of sinus surgery on pseudomonal airway colonization, bronchiolitis obliterans syndrome and survival in cystic fibrosis lung transplant recipients. Respiration. 2013;86:25–31. doi: 10.1159/000339627. [DOI] [PubMed] [Google Scholar]
- 28.Assadiasl S., Shahi A., Salehi S., Afzali S., Amirzargar A. Interferon regulatory factors: where to stand in transplantation. Transpl. Immunol. 2018;51:76–80. doi: 10.1016/j.trim.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Rascio F., Pontrelli P., Accetturo M., Oranger A., Gigante M., Castellano G., Gigante M., Zito A., Zaza G., Lupo A. A type I interferon signature characterizes chronic antibody-mediated rejection in kidney transplantation. J. Pathol. 2015;237:72–84. doi: 10.1002/path.4553. [DOI] [PubMed] [Google Scholar]
- 30.Miller H.L., Shah P.D., Orens J.B., McDyer J.F. Prevention of airway allograft tolerance by polyinosinic:polycytidylic acid requires type I interferon responsiveness for mouse airway obliteration. J. Heart Lung Transplant. 2013;32:914–924. doi: 10.1016/j.healun.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamarthée B., Malard F., Gamonet C., Bossard C., Couturier M., Renauld J.C., Mohty M., Saas P., Gaugler B. Donor interleukin-22 and host type I interferon signaling pathway participate in intestinal graft-versus-host disease via STAT1 activation and CXCL10. Mucosal Immunol. 2016;9:309–321. doi: 10.1038/mi.2015.61. [DOI] [PubMed] [Google Scholar]
- 32.Spencer C.T., Bezbradica J.S., Ramos M.G., Arico C.D., Conant S.B., Gilchuk P., Gray J.J., Zheng M., Niu X., Hildebrand W. Viral infection causes a shift in the self peptide repertoire presented by human MHC class I molecules. Proteomics Clin. Appl. 2015;9:1035–1052. doi: 10.1002/prca.201500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chattoraj S.S., Ganesan S., Faris A., Comstock A., Lee W.M., Sajjan U.S. Pseudomonas aeruginosa suppresses interferon response to rhinovirus infection in cystic fibrosis but not in normal bronchial epithelial cells. Infect. Immun. 2011;79:4131–4145. doi: 10.1128/IAI.05120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu M., Skaug B., Bi X., Mills T., Salazar G., Zhou X., Reveille J., Agarwal S.K., Blackburn M.R., Mayes M.D., Assassi S. Interferon regulatory factor 7 (IRF7) represents a link between inflammation and fibrosis in the pathogenesis of systemic sclerosis. Ann. Rheum. Dis. 2019;78:1583–1591. doi: 10.1136/annrheumdis-2019-215208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merino A., Zhang B., Dougherty P., Luo X., Wang J., Blazar B.R., Miller J.S., Cichocki F. Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J. Clin. Invest. 2019;129:3770–3785. doi: 10.1172/JCI125916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bierne H., Hamon M., Cossart P. Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lebreton A., Lakisic G., Job V., Fritsch L., Tham T.N., Camejo A., Matteï P.J., Regnault B., Nahori M.A., Cabanes D. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science. 2011;331:1319–1321. doi: 10.1126/science.1200120. [DOI] [PubMed] [Google Scholar]
- 38.Dortet L., Lombardi C., Cretin F., Dessen A., Filloux A. Pore-forming activity of the Pseudomonas aeruginosa type III secretion system translocon alters the host epigenome. Nat. Microbiol. 2018;3:378–386. doi: 10.1038/s41564-018-0109-7. [DOI] [PubMed] [Google Scholar]
- 39.Duan Y., Guangqiang L., Miaomiao X., Zhinan Y. CFTR, which not only serves as a TCR signaling molecule but also function as an anion channel, dual-negatively regulates IFN gamma production and tumor immunity in gamma delta T cells. J. Immunol. 2019;2(1, Suppl. 1) 60.10. [Google Scholar]
- 40.Rayner R.E., Makena P., Prasad G.L., Cormet-Boyaka E. Optimization of Normal Human Bronchial Epithelial (NHBE) Cell 3D Cultures for In Vitro Lung Model Studies. Sci. Rep. 2019;9:500. doi: 10.1038/s41598-018-36735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiemstra P.S., McCray P.B., Jr., Bals R. The innate immune function of airway epithelial cells in inflammatory lung disease. Eur. Respir. J. 2015;45:1150–1162. doi: 10.1183/09031936.00141514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., Kosorok M.R., Farrell P.M., Laxova A., West S.E., Green C.G., Collins J., Rock M.J., Splaingard M.L. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA. 2005;293:581–588. doi: 10.1001/jama.293.5.581. [DOI] [PubMed] [Google Scholar]
- 43.Gellatly S.L., Hancock R.E. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 2013;67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 44.Lorè N.I., Cigana C., De Fino I., Riva C., Juhas M., Schwager S., Eberl L., Bragonzi A. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLOS ONE. 2012;7:e35648. doi: 10.1371/journal.pone.0035648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfgang M.C., Jyot J., Goodman A.L., Ramphal R., Lory S. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc. Natl. Acad. Sci. USA. 2004;101:6664–6668. doi: 10.1073/pnas.0307553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greenland J.R., Chong T., Wang A.S., Martinez E., Shrestha P., Kukreja J., Hays S.R., Golden J.A., Singer J.P., Tang Q. Suppressed calcineurin-dependent gene expression identifies lung allograft recipients at increased risk of infection. Am. J. Transplant. 2018;18:2043–2049. doi: 10.1111/ajt.14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verleden G.M., Glanville A.R., Lease E.D., Fisher A.J., Calabrese F., Corris P.A., Ensor C.R., Gottlieb J., Hachem R.R., Lama V. Chronic lung allograft dysfunction: definition, diagnostic criteria, and approaches to treatment-a consensus report from the Pulmonary Council of the ISHLT. J. Heart Lung Transplant. 2019;38:493–503. doi: 10.1016/j.healun.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Langelier C., Kalantar K.L., Moazed F., Wilson M.R., Crawford E.D., Deiss T., Belzer A., Bolourchi S., Caldera S., Fung M. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl. Acad. Sci. USA. 2018;115:E12353–E12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu W., Crawford E.D., O’Donovan B.D., Wilson M.R., Chow E.D., Retallack H., DeRisi J.L. Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:41. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fulcher M.L., Gabriel S., Burns K.A., Yankaskas J.R., Randell S.H. Well-Differentiated Human Airway Epithelial Cell Cultures. In: Picot J., editor. Series Volume 107. Humana Press; 2005. pp. 183–206. (Human Cell Culture Protocols: Methods in Molecular Medicine). [DOI] [PubMed] [Google Scholar]
- 51.Therneau, T.M. (2020). A Package for Survival Analysis in R. https://cran.r-project.org/web/packages/survival/vignettes/survival.pdf.
- 52.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Müller F., Scherer M., Assenov Y., Lutsik P., Walter J., Lengauer T., Bock C. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol. 2019;20:55. doi: 10.1186/s13059-019-1664-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramesh A., Nakielny S., Hsu J., Kyohere M., Byaruhanga O., de Bourcy C., Egger R., Dimitrov B., Juan Y.F., Sheu J. Metagenomic next-generation sequencing of samples from pediatric febrile illness in Tororo, Uganda. PLOS ONE. 2019;14:e0218318. doi: 10.1371/journal.pone.0218318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A positive-fold change indicates upregulated in CF+ versus CF− referents. (Related to Figure 1)
Data Availability Statement
The RNASeq and DNA methylation datasets generated during this study are available at Mendeley Data under https://doi.org/10.17632/89nzfyvkvc.1. Transcriptomic count data are provided in GEO data matrix format. Raw DNA methylation data are provided as in EPIC report format. Analytic code is available from the corresponding author upon request.