A common theme in antibody responses
In the fight against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), antibodies are a key tool, both as potential therapeutics and to guide vaccine development. Yuan et al. focused on finding shared antibody responses, in which multiple individuals develop antibodies against the same antigen using the same genetic elements and modes of recognition. The authors identified the immunoglobulin heavy-chain variable region 3-53 gene as the most frequently used among 294 antibodies that target the receptor-binding domain (RBD) of the viral spike protein. These antibodies have few somatic mutations, and crystal structures of two neutralizing antibodies bound to the RBD show that mostly germline-encoded residues are involved in binding. The minimal affinity maturation and high potency of these antibodies is promising for vaccine design.
Science, this issue p. 1119
IGHV3-53–encoded antibodies that display high-affinity germline binding to SARS-CoV-2 are a common response to COVID-19.
Abstract
Molecular understanding of neutralizing antibody responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could accelerate vaccine design and drug discovery. We analyzed 294 anti–SARS-CoV-2 antibodies and found that immunoglobulin G heavy-chain variable region 3-53 (IGHV3-53) is the most frequently used IGHV gene for targeting the receptor-binding domain (RBD) of the spike protein. Co-crystal structures of two IGHV3-53–neutralizing antibodies with RBD, with or without Fab CR3022, at 2.33- to 3.20-angstrom resolution revealed that the germline-encoded residues dominate recognition of the angiotensin I converting enzyme 2 (ACE2)–binding site. This binding mode limits the IGHV3-53 antibodies to short complementarity-determining region H3 loops but accommodates light-chain diversity. These IGHV3-53 antibodies show minimal affinity maturation and high potency, which is promising for vaccine design. Knowledge of these structural motifs and binding mode should facilitate the design of antigens that elicit this type of neutralizing response.
The ongoing coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in enormous global health and socioeconomic damage and requires urgent development of an effective vaccine (1). Although multiple vaccine candidates have entered clinical trials (2), the molecular features that contribute to an effective antibody response are not clear. Shared antibody responses to specific microbial pathogens have been found in which the same genetic elements and modes of recognition are observed in multiple individuals against a given antigen. Such responses to microbial pathogens have been observed against influenza (3), dengue (4), malaria (5), and HIV (6). Characterization of the molecular interactions between pathogens and cognate antigen can provide insight into how the immune repertoire is able to quickly respond to novel microbial pathogens and will facilitate the rational design of vaccines against them (7, 8).
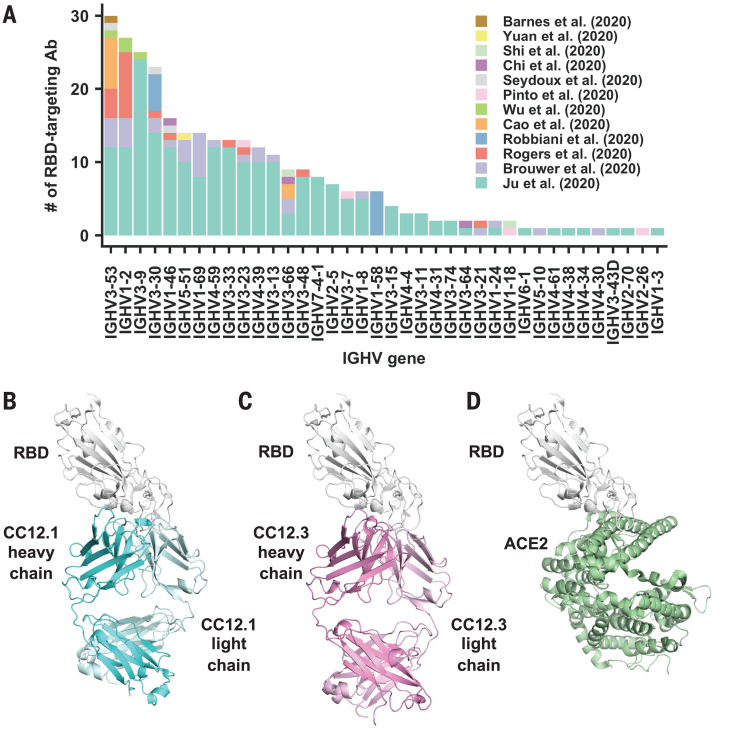
The spike (S) protein is the major surface antigen of SARS-CoV-2. The S protein uses its receptor-binding domain (RBD) to engage the host receptor angiotensin I converting enzyme 2 (ACE2) for viral entry (9–12). RBD-targeting antibodies could then neutralize SARS-CoV-2 by blocking ACE2 binding. A number of antibodies that target the RBD of SARS-CoV-2 have now been discovered (13–28). We compiled a list of 294 SARS-CoV-2 RBD-targeting antibodies for which information on immunoglobulin G heavy-chain variable (IGHV) gene usage is available (17–28) (table S1) and found that IGHV3-53 is the most frequently used IGHV gene among these antibodies (Fig. 1A), with 10% encoded by IGHV3-53, compared with 0.5 to 2.6% (mean 1.8%) in the repertoire of naïve healthy individuals (29, 30). IGHV3-53 antibodies were found in seven of 12 studies and in 17 of 32 COVID-19 patient samples (17–28, 31). These IGHV3-53 antibodies not only had lower somatic mutation rates but also were more potent compared with other germ lines in the cohort investigated here (27) (fig. S1). The prevalence of IGHV3-53 in the antibody response in SARS-CoV-2 patients has also been recognized in other antibody studies (20, 22, 27).
Fig. 1. Structures of two IGHV3-53 antibodies.
(A) The distribution of IGHV gene usage is shown for a total of 294 RBD-targeting antibodies (17–28). (B to D) Crystal structures of (B) CC12.1 in complex with SARS-CoV-2 RBD, (C) CC12.3 with SARS-CoV-2 RBD, and (D) human ACE2 with SARS-CoV-2 RBD (PDB 6M0J) (12).
To understand the molecular features that endow IGHV3-53 with favorable properties for RBD recognition, we determined the crystal structures of two IGHV3-53–neutralizing antibodies, CC12.1 and CC12.3, in complex with the SARS-CoV-2 RBD and with the cross-reactive Fab CR3022 to SARS-like CoVs (17). CC12.1 and CC12.3 were previously isolated from a SARS-CoV-2–infected patient and were shown to be specific for the RBD (27). CC12.1 and CC12.3 [median inhibitory concentration (IC50), ~20 ng/ml] were among the top four highly potent neutralizing antibodies in the panel of antibodies assayed against live replicating SARS-CoV-2 virus and pseudovirus (27). Although CC12.1 and CC12.3 are both encoded by IGHV3-53, CC12.1 uses IGHJ6, IGKV1-9, and IGKJ3, whereas CC12.3 uses IGHJ4, IGKV3-20, and IGKJ1. This variation in IGHJ, IGKV, and IGKJ usage indicates that CC12.1 and CC12.3 belong to different clonotypes but are encoded by a common IGHV3-53 germline gene (fig. S2). IgBlast analysis (32) showed that IGHV and IGKV of CC12.1 have acquired only four amino acid changes (somatic mutations) during affinity maturation from the original germline antibody sequence (fig. S2, A and B). Similarly, CC12.3 is also minimally somatically mutated, with three amino acid changes in IGHV and a single amino acid deletion in IGKV (fig. S2, A and C). The binding affinities (Kd) of the Fabs CC12.1 and CC12.3 to SARS-CoV-2 RBD are 17 and 14 nM, respectively (fig. S3). Moreover, competition experiments suggest that CC12.1 and CC12.3 bind to a similar epitope, which overlaps with the ACE2-binding site but not the CR3022 epitope (fig. S4).
We determined four complex crystal structures, CC12.1/RBD, CC12.3/RBD, CC12.1/RBD/CR3022, and CC12.3/RBD/CR3022, at resolutions of 3.20, 2.33, 2.70, and 2.90 Å, respectively (table S2). CC12.1 and CC12.3 bind to the ACE2-binding site on SARS-CoV-2 RBD with an identical angle of approach (Fig. 1, B to D, and fig. S5). Another IGHV3-53 antibody, B38, the structure of which was determined recently (23), binds to the ACE2-binding site on SARS-CoV-2 RBD in a similar manner but with a Kd of 70.1 nM (fig. S6). Similar to the ACE2-binding site (11), the epitopes of these antibodies can only be accessed when the RBD is in the “up” conformation (fig. S7). Among 17 ACE2-binding residues on RBD, 15 and 16 are within the epitopes of CC12.1 and B38, respectively, and 11 are in the epitope of CC12.3 (Fig. 2, A to D). Many of the epitope residues are not conserved between SARS-CoV-2 and SARS-CoV (Fig. 2E), explaining their lack of cross-reactivity (27). The buried surface area (BSA) from the heavy-chain interaction is quite similar in CC12.1 (723 Å2), CC12.3 (698 Å2), and B38 (713 Å2). By contrast, the light-chain interaction is much smaller for CC12.3 (176 Å2) compared with CC12.1 (566 Å2) and B38 (495 Å2), consistent with different light-chain gene usage. Although both CC12.1 and B38 use IGKV1-9, CC12.3 uses IGKV3-20, which suggests that IGHV3-53 can pair with different light chains to target the ACE2-binding site of the SARS-CoV-2 RBD. CC12.1 (56% BSA from the heavy chain) binds the RBD with similar affinity to CC12.3 (80% BSA from the heavy chain) but with a slightly slower dissociation rate (fig. S3), which might be influenced by the different light chain and its greater contribution in CC12.1. Nevertheless, the light-chain identity seems not to be as critical as the heavy chain. In fact, among the RBD-targeting IGHV3-53 antibodies, nine different light chains are observed, although IGKV1-9 and IGKV3-20 are the most frequently found to date (fig. S8).
Fig. 2. Epitopes of IGHV3-53 antibodies.
(A to C) Epitopes of (A) CC12.1, (B) CC12.3, and (C) B38 (PDB 7BZ5) (23). Epitope residues contacting the heavy chain are shown in orange and those contacting the light chain are shown in yellow. CDR loops are labeled in the left panels; epitope residues are labeled in the right panels. For clarity, only representative epitope residues are labeled. Epitope residues that are also involved in ACE2 binding are shown in red. (D) ACE2-binding residues are shown in blue. ACE2 is shown in green in the left panel in a semitransparent cartoon representation. ACE2-binding residues are labeled in the right panel. A total of 17 residues were used for ACE2 binding (12), but only 15 are labeled here because the other two are at the back of the structure in this view and do not interact with the antibodies here. (E) Epitope residues for CC12.1, CC12.3, and B38 were identified by PISA (41) and annotated on the SARS-CoV-2 RBD sequence, which is aligned to the SARS-CoV RBD sequence with nonconserved residues highlighted. The 17 ACE2-binding residues were identified from a SARS-CoV-2 RBD–ACE2 complex structure as described previously (12).
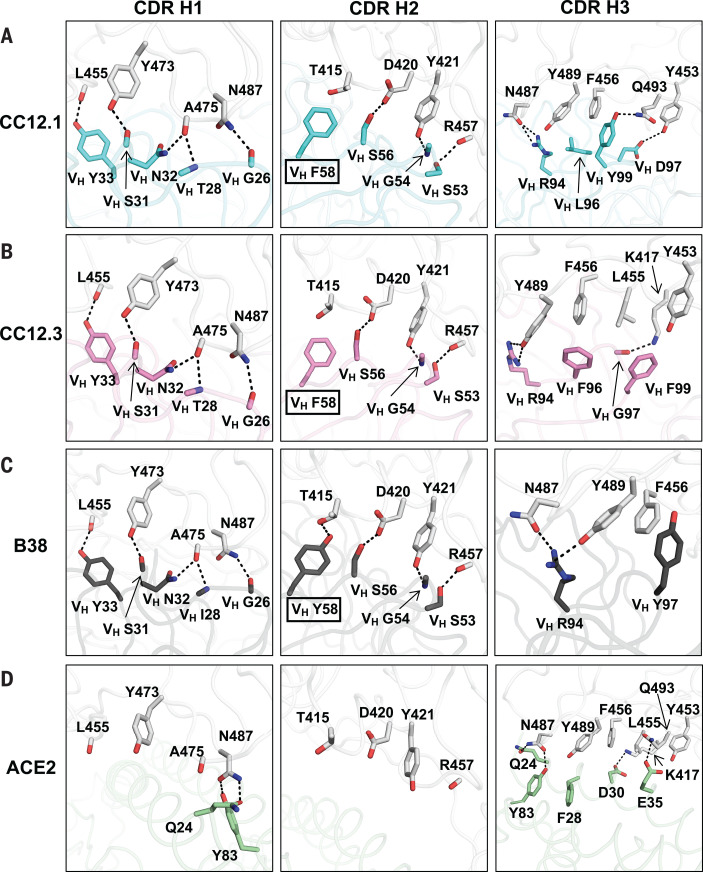
To understand why IGHV3-53 is elicited as a shared antibody response, the molecular interactions between the RBD and the heavy chains of CC12.1, CC12.3, and B38 were analyzed. The complementarity-determining regions (CDRs) H1 and H2 of these antibodies interact extensively with the RBD mainly through specific hydrogen bonds (Fig. 3, A and B). All residues on CDR H1 and H2 that hydrogen bond with the RBD are encoded by the germ line IGHV3-53 (fig. S2 and table S3). These interactions are almost identical among CC12.1, CC12.3, and B38, with the only difference at the variable region of immunoglobulin heavy chain (VH) residue 58. A somatic mutation VH Y58F in CC12.1 and CC12.3, but not in B38 (Fig. 3, A to C, boxed residues, and fig. S9), results in similar van der Waals interactions, with only a loss of a single hydrogen bond from the hydroxyl of the germ line Tyr in B38 to the RBD (Fig. 3C). None of these antibody interactions mimics ACE2 binding (Fig. 3D).
Fig. 3. Interactions between the RBD and heavy-chain CDR loops.
(A to C) Highly similar interaction modes between SARS-CoV-2 RBD and the antibody CDR H1 and H2 loops, but not the H3 loop, are observed for (A) CC12.1, (B) CC12.3, and (C) B38 (PDB 7BZ5) (23). The RBD is shown in white and antibody residues in cyan, pink, and dark gray, respectively. Oxygen atoms are shown in red and nitrogen atoms in blue. Hydrogen bonds are represented by dashed lines. (D) Interaction between ACE2 (green) and residues of the RBD (PDB 6M0J) (12) shown in (A) to (C).
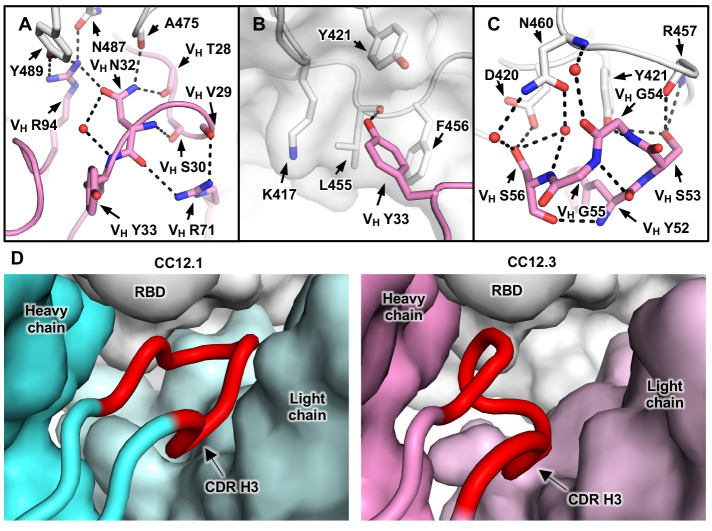
Our structural analysis reveals two key motifs in the IGHV3-53 germline sequence that are important for RBD binding: an NY motif at VH residues 32 and 33 in the CDR H1 and an SGGS motif at VH residues 53 to 56 in the CDR H2 (Fig. 3 and fig. S10). The side chain of VH N32 in the NY motif hydrogen bonds with the backbone carbonyl of A475 on the RBD, and this interaction is stabilized by an extensive network of hydrogen bonds with other antibody residues as well as a bound water molecule (Fig. 4A). VH N32 also hydrogen bonds with VH R94, which in turn hydrogen bonds with N487 and Y489 on the RBD (Fig. 4A). These polar contacts not only enhance the RBD-Fab interaction but also stabilize the CDR conformations with the surrounding residues (framework). VH Y33 in the NY motif inserts into a hydrophobic cage formed by RBD residues Y421, F456, and L455 and the aliphatic component of K417 (Fig. 4B). A hydrogen bond between VH Y33 and the carbonyl oxygen of L455 on the RBD further strengthens the interaction. The second key motif, SGGS, in CDR H2 forms an extensive hydrogen bond network with the RBD (Fig. 4C), including four hydrogen bonds that involve the hydroxyl side chains of VH S53 and VH S56 and four water-mediated hydrogen bonds to the backbone carbonyl of VH G54, the backbone amide of VH S56, and the side chain of VH S56. Along with VH Y52, the SGGS motif takes part in a type I beta turn, with a positive Φ angle for VH G55 at the end of the turn. In addition, the Cα of VH G54 is only 4 Å away from the RBD, indicating that side chains of other amino acids would clash with the RBD if they were present at this position.
Fig. 4. Two IGHV3-53 germline-encoded motifs with a short CDR H3.
(A) Illustration of the extensive hydrogen bond network that involves VH N32 of the NY motif in CDR H1. (B) Hydrophobic cage interaction between the RBD and VH Y33 of the NY motif in CDR H1. (C) Hydrogen bond network that involves the SGGS motif in CDR H2. CC12.3 is shown here because its structure is at higher resolution than CC12.1. (D) CDR H3 length is constrained to fit in a relatively small pocket on the RBD surface. The heavy and light chains of CC12.1 (cyan) and the RBD (white) are shown in surface representation, with CDR H3 (red) highlighted in the diagram in the left panel. CC12.3 (pink) is shown in the right panel in the same representation.
The NY and SGGS motifs, important for RBD binding, are both encoded in the IGHV3-53 germline gene. In addition to IGHV3-53, only the closely related IGHV3-66 contains an NY motif in CDR H1 and an SGGS motif in CDR H2 (33). IGHV3-66 is commonly observed (19–22, 24, 26) in antibodies in SARS-CoV-2 patients (Fig. 1A) and is also well represented in the repertoire of healthy individuals (0.3 to 1.7% of total antibodies) (29). Overall, our structural analysis has identified two germline-encoded binding motifs that enable IGHV3-53 to target the SARS-CoV-2 RBD, with mutations apparently not required from affinity maturation.
Although the binding mode of CDR H1 and H2 to RBD is highly similar among CC12.1, CC12.3, and B38, the interaction of CDR H3 with the RBD varies (Fig. 3, A to C) because of differences in the CDR H3 sequences and conformations (fig. S1 and Fig. 4D). For example, whereas CDR H3 of CC12.1 interacts with RBD Y453 through a hydrogen bond, CDR H3 of CC12.3 and B38 do not form such a bond (Fig. 3, A to C). Similarly, because of the difference in light-chain gene usage, light-chain interactions with the RBD can vary substantially in IGHV3-53 antibodies (fig. S11). Overall, our structural analysis demonstrates that IGHV3-53 provides a versatile framework with which to target the ACE2-binding site in SARS-CoV-2 RBD.
An interesting feature of CC12.1 and CC12.3 is their relatively short CDR H3. Although the CDR H3 sequences of CC12.1 and CC12.3 differ, both are nine amino acids in length (Kabat numbering) (34). Antibody B38 has an even shorter CDR H3 of seven residues (23). The average CDR H3 length for human antibodies is ~13 (35), although very long CDR H3s (up to 30 residues or more) are found in many broadly neutralizing antibodies to HIV-1 (36). Longer CDR H3s cannot be accommodated in these IGHV3-53 antibodies because their epitopes are relatively flat, with only a small pocket to insert their CDR H3 loop (Fig. 4D). A similar conclusion was reached in another recent study, which also reported that SARS-CoV-2 RBD-targeting antibodies encoded by IGHV3-53 or IGHV3-66 tend to have a short CDR H3 (28, 37). Among the RBD-targeting antibodies reported recently (17–28), IGHV3-53 antibodies consistently have a significantly shorter CDR H3 compared with those encoded by other IGHV genes (P = 6e-8, Mann-Whitney U test) (fig. S12) and are also shorter than IGHV3-53–encoded antibodies in the naïve human antibody repertoire (30). Thus, a short CDR H3 length is one of the molecular features of the IGHV3-53–encoded antibody response to SARS-CoV-2 RBD, reminiscent of a below-average five-residue CDR L3 in IGHV1-2 antibodies to the receptor-binding site in HIV-1 Env gp120 (38). Nevertheless, a small subset of these IGHV3-53 antibodies have longer CDR H3s that warrant further investigation of their binding mode (fig. S12).
In addition to IGHV3-53, several other IGHV genes, such as IGHV1-2, IGHV3-9, and IGHV3-30, are also more frequently observed than other germ lines in SARS-CoV-2 RBD-targeting antibodies (Fig. 1A). Future work will investigate the molecular mechanisms of these IGHV responses to SARS-CoV-2, as well as whether other germline gene segments, including IGHD and the light chain, contribute in recurring motifs to the SARS-CoV-2 antibody response. The characterization of these IGHV3-53 antibodies to SARS-CoV-2 is a promising starting point for rational vaccine design (39), given that limited to no affinity maturation is required to achieve a highly potent neutralizing antibody response to the RBD. Because IGHV3-53 is found at a reasonable frequency in healthy individuals (29, 30), this particular antibody response could be commonly elicited during vaccination (40).
Acknowledgments
We thank R. Stanfield for assistance in data collection, B. Briney for naïve antibody germline analysis, and the staff of Stanford Synchrotron Radiation Laboratory (SSRL) Beamline 12-1 for assistance. Funding: This work was supported by the NIH (grant no. K99 AI139445 to N.C.W.), the Bill and Melinda Gates Foundation (grant no. OPP1170236 to I.A.W. and D.R.B.), NIH NIAID CHAVD (grant no. UM1 AI44462 to I.A.W., D.S., and D.R.B.), and the IAVI Neutralizing Antibody Center. Use of the SSRL, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). Author contributions: M.Y., H.L., N.C.W., F.Z., D.H., T.F.R., E.L., D.S, J.G.J., D.R.B., and I.A.W. conceived and designed the study. F.Z., D.H., T.F.R., E.L., D.S., J.G.J., and D.R.B. isolated the antibodies and provided clones and sequences. M.Y., H.L., N.C.W., C.-C.D.L., W.Y., and Y.H. expressed and purified the proteins. M.Y. and C.-C.D.L. performed biolayer interferometry binding assays. M.Y., H.L., N.C.W., X.Z., and H.T. performed the crystallization and x-ray data collection. M.Y. and X.Z. determined and refined the x-ray structures. M.Y., H.L., N.C.W., C.-C.D.L., and X.Z. analyzed the data. M.Y., H.L., N.C.W., and I.A.W. wrote the manuscript, and all authors reviewed and/or edited the manuscript. Competing interests: D.R.B., D.H., J.G.J., E.L., T.F.R., D.S., and F.Z. are listed as inventors on pending patent applications describing the SARS-CoV-2 antibodies originally published in Rogers et al. (27). Data and materials availability: X-ray coordinates and structure factors are deposited in the RCSB Protein Data Bank under identification nos. 6XC2, 6XC3, 6XC4, and 6XC7. Antibody plasmids are available from D.R.B. under a material transfer agreement from The Scripps Research Institute. Other materials are available from I.A.W. on request. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/369/6507/1119/suppl/DC1
Materials and Methods
Figs. S1 to S12
Tables S1 to S3
MDAR Reproducibility Checklist
References and Notes
- 1.Kissler S. M., Tedijanto C., Goldstein E., Grad Y. H., Lipsitch M., Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 368, 860–868 (2020). 10.1126/science.abb5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lurie N., Saville M., Hatchett R., Halton J., Developing COVID-19 vaccines at pandemic speed. N. Engl. J. Med. 382, 1969–1973 (2020). 10.1056/NEJMp2005630 [DOI] [PubMed] [Google Scholar]
- 3.Andrews S. F., McDermott A. B., Shaping a universally broad antibody response to influenza amidst a variable immunoglobulin landscape. Curr. Opin. Immunol. 53, 96–101 (2018). 10.1016/j.coi.2018.04.009 [DOI] [PubMed] [Google Scholar]
- 4.Parameswaran P., Liu Y., Roskin K. M., Jackson K. K. L., Dixit V. P., Lee J.-Y., Artiles K. L., Zompi S., Vargas M. J., Simen B. B., Hanczaruk B., McGowan K. R., Tariq M. A., Pourmand N., Koller D., Balmaseda A., Boyd S. D., Harris E., Fire A. Z., Convergent antibody signatures in human dengue. Cell Host Microbe 13, 691–700 (2013). 10.1016/j.chom.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pieper K., Tan J., Piccoli L., Foglierini M., Barbieri S., Chen Y., Silacci-Fregni C., Wolf T., Jarrossay D., Anderle M., Abdi A., Ndungu F. M., Doumbo O. K., Traore B., Tran T. M., Jongo S., Zenklusen I., Crompton P. D., Daubenberger C., Bull P. C., Sallusto F., Lanzavecchia A., Public antibodies to malaria antigens generated by two LAIR1 insertion modalities. Nature 548, 597–601 (2017). 10.1038/nature23670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setliff I., McDonnell W. J., Raju N., Bombardi R. G., Murji A. A., Scheepers C., Ziki R., Mynhardt C., Shepherd B. E., Mamchak A. A., Garrett N., Karim S. A., Mallal S. A., Crowe Jr. J. E., Morris L., Georgiev I. S., Multi-donor longitudinal antibody repertoire sequencing reveals the existence of public antibody clonotypes in HIV-1 infection. Cell Host Microbe 23, 845–854.e6 (2018). 10.1016/j.chom.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu N. C., Wilson I. A., Structural insights into the design of novel anti-influenza therapies. Nat. Struct. Mol. Biol. 25, 115–121 (2018). 10.1038/s41594-018-0025-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanzavecchia A., Frühwirth A., Perez L., Corti D., Antibody-guided vaccine design: Identification of protective epitopes. Curr. Opin. Immunol. 41, 62–67 (2016). 10.1016/j.coi.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 9.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letko M., Marzi A., Munster V., Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569 (2020). 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020). 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- 13.W. Li, A. Drelich, D. R. Martinez, L. Gralinski, C. Chen, Z. Sun, X. Liu, D. Zhelev, L. Zhang, E. C. Peterson, A. Conard, J. W. Mellors, C. T. Tseng, R. S. Baric, D. S. Dimitrov, Potent neutralization of SARS-CoV-2 in vitro and in an animal model by a human monoclonal antibody. bioRxiv 2020.05.13.093088 [Preprint] (2 June 2020); 10.1101/2020.05.13.093088. 10.1101/2020.05.13.093088 [DOI]
- 14.E. Andreano, E. Andreano, E. Nicastri, I. Paciello, P. Pileri, N. Manganaro, G. Piccini, A. Manenti, E. Pantano, A. Kabanova, M. Troisi, F. Vacca, D. Cardamone, C. De Santi, C. Agrati, M. R. Capobianchi, C. Castilletti, A. Emiliozzi, M. Fabbiani, F. Montagnani, E. Montomoli, C. Sala, G. Ippolito, R. Rappuoli, Identification of neutralizing human monoclonal antibodies from Italian Covid-19 convalescent patients. bioRxiv 2020.05.05.078154 [Preprint] (9 May 2020); 10.1101/2020.05.05.078154. 10.1101/2020.05.05.078154 [DOI]
- 15.S. J. Zost et al., Nat. Med. 10.1038/s41591-020-0998-x (2020). 10.1101/2020.05.12.091462 [DOI]
- 16.Wec A. Z., Wrapp D., Herbert A. S., Maurer D. P., Haslwanter D., Sakharkar M., Jangra R. K., Dieterle M. E., Lilov A., Huang D., Tse L. V., Johnson N. V., Hsieh C.-L., Wang N., Nett J. H., Champney E., Burnina I., Brown M., Lin S., Sinclair M., Johnson C., Pudi S., Bortz 3rd R., Wirchnianski A. S., Laudermilch E., Florez C., Fels J. M., O’Brien C. M., Graham B. S., Nemazee D., Burton D. R., Baric R. S., Voss J. E., Chandran K., Dye J. M., McLellan J. S., Walker L. M., Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science eabc7424 (2020). 10.1126/science.abc7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan M., Wu N. C., Zhu X., Lee C. D., So R. T. Y., Lv H., Mok C. K. P., Wilson I. A., A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020). 10.1126/science.abb7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto D., Park Y.-J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., Chen R. E., Havenar-Daughton C., Snell G., Telenti A., Virgin H. W., Lanzavecchia A., Diamond M. S., Fink K., Veesler D., Corti D., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 19.B. Ju, Q. Zhang, X. Ge, R. Wang, J. Yu, S. Shan, B. Zhou, S. Song, X. Tang, J. Yu, J. Ge, J. Lan, J. Yuan, H. Wang, J. Zhao, S. Zhang, Y. Wang, X. Shi, L. Liu, X. Wang, Z. Zhang, L. Zhang, Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. bioRxiv 2020.03.21.990770 [Preprint] (26 March 2020); 10.1101/2020.03.21.990770. 10.1101/2020.03.21.990770 [DOI] [PubMed]
- 20.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X. D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X. S., Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 10.1016/j.cell.2020.05.025 (2020). 10.1016/j.cell.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brouwer P. J. M., Caniels T. G., van der Straten K., Snitselaar J. L., Aldon Y., Bangaru S., Torres J. L., Okba N. M. A., Claireaux M., Kerster G., Bentlage A. E. H., van Haaren M. M., Guerra D., Burger J. A., Schermer E. E., Verheul K. D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M. J., Bijl T. P. L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N. A., Wiersinga W. J., Vidarsson G., Haagmans B. L., Ward A. B., de Bree G. J., Sanders R. W., van Gils M. J., Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science eabc5902 (2020). 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H.-H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West Jr. A. P., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 10.1038/s41586-020-2456-9 (2020). 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G. F., Gao F., Liu L., A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368, 1274–1278 (2020). 10.1126/science.abc2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.X. Chi et al., Science 369, 650–655 (2020). 10.1101/2020.05.08.083964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seydoux E., Homad L. J., MacCamy A. J., Parks K. R., Hurlburt N. K., Jennewein M. F., Akins N. R., Stuart A. B., Wan Y.-H., Feng J., Whaley R. E., Singh S., Boeckh M., Cohen K. W., McElrath M. J., Englund J. A., Chu H. Y., Pancera M., McGuire A. T., Stamatatos L., Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 10.1016/j.immuni.2020.06.001 (2020). 10.1016/j.immuni.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W. J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F. S., Wang Q., Gao G. F., Yuan Z., Yan J., A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature 10.1038/s41586-020-2381-y (2020). 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- 27.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W. T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M. J., Rawlings S. A., Wu N. C., Yuan M., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Wilson I. A., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science eabc7520 (2020). 10.1126/science.abc7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes C. O., West Jr. A. P., Huey-Tubman K. E., Hoffmann M. A. G., Sharaf N. G., Hoffman P. R., Koranda N., Gristick H. B., Gaebler C., Muecksch F., Cetrulo Lorenzi J. C., Finkin S., Hägglöf T., Hurley A., Millard K. G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P. D., Caskey M., Robbiani D. F., Nussenzweig M. C., Bjorkman P. J., Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 10.1101/2020.05.28.121533 (2020). 10.1016/j.cell.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyd S. D., Gaëta B. A., Jackson K. J., Fire A. Z., Marshall E. L., Merker J. D., Maniar J. M., Zhang L. N., Sahaf B., Jones C. D., Simen B. B., Hanczaruk B., Nguyen K. D., Nadeau K. C., Egholm M., Miklos D. B., Zehnder J. L., Collins A. M., Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J. Immunol. 184, 6986–6992 (2010). 10.4049/jimmunol.1000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briney B., Inderbitzin A., Joyce C., Burton D. R., Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 566, 393–397 (2019). 10.1038/s41586-019-0879-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31 The highly related IGHV3-66–encoded RBD-targeting antibodies are also frequently found and reported in five out of 12 studies and in 11 of 32 COVID-19 patients (17–28).
- 32.Ye J., Ma N., Madden T. L., Ostell J. M., IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34-40 (2013). 10.1093/nar/gkt382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33 The amino-acid sequences of IGHV3-53 and IGHV3-66 germline genes only differ by one amino acid at residue 12. Specifically, IGHV3-53 contains Ile12, whereas IGHV3-66 contains Val12.
- 34 CDRH3 lengths with Kabat numbering are two amino acids shorter than those with IMGT numbering.
- 35.Zemlin M., Klinger M., Link J., Zemlin C., Bauer K., Engler J. A., Schroeder Jr. H. W., Kirkham P. M., Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 334, 733–749 (2003). 10.1016/j.jmb.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 36.Yu L., Guan Y., Immunologic basis for long HCDR3s in broadly neutralizing antibodies against HIV-1. Front. Immunol. 5, 250 (2014). 10.3389/fimmu.2014.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37 IGHV3-53 and IGHV3-66 antibodies in general have slightly shorter than average CDR H3s (by around one residue) and also appear to have a few much shorter CDR H3s (<10 amino acids) than average in the baseline antibody repertoire (30).
- 38.Jardine J. G., Ota T., Sok D., Pauthner M., Kulp D. W., Kalyuzhniy O., Skog P. D., Thinnes T. C., Bhullar D., Briney B., Menis S., Jones M., Kubitz M., Spencer S., Adachi Y., Burton D. R., Schief W. R., Nemazee D., HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161 (2015). 10.1126/science.aac5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burton D. R., Walker L. M., Rational vaccine design in the time of COVID-19. Cell Host Microbe 27, 695–698 (2020). 10.1016/j.chom.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbott R. K., Crotty S., Factors in B cell competition and immunodominance. Immunol. Rev. 10.1111/imr.12861 (2020). 10.1111/imr.12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 42.Ekiert D. C., Friesen R. H. E., Bhabha G., Kwaks T., Jongeneelen M., Yu W., Ophorst C., Cox F., Korse H. J. W. M., Brandenburg B., Vogels R., Brakenhoff J. P. J., Kompier R., Koldijk M. H., Cornelissen L. A. H. M., Poon L. L. M., Peiris M., Koudstaal W., Wilson I. A., Goudsmit J., A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850 (2011). 10.1126/science.1204839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 44.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.J. Huo, Y. Zhao, J. Ren, D. Zhou, H. M. E. Duyvesteyn, H. M. Ginn, L. Carrique, T. Malinauskas, R. R. Ruza, P. N. M. Shah, T. K. Tan, P. Rijal, N. Coombes, K. R. Bewley, J. A. Tree, J. Radecke, N. G. Paterson, P. Supasa, J. Mongkolsapaya, G. R. Screaton, M. Carroll, A. Townsend, E. E. Fry, R. J. Owens, D. I. Stuart, Neutralization of SARS-CoV-2 by destruction of the prefusion Spike. bioRxiv 2020.05.05.079202 [Preprint] (6 May 2020); . 10.1101/2020.05.05.079202 [DOI]
- 46.Chen X., Hnida K., Graewert M. A., Andersen J. T., Iversen R., Tuukkanen A., Svergun D., Sollid L. M., Structural basis for antigen recognition by transglutaminase 2-specific autoantibodies in celiac disease. J. Biol. Chem. 290, 21365–21375 (2015). 10.1074/jbc.M115.669895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu N. C., Grande G., Turner H. L., Ward A. B., Xie J., Lerner R. A., Wilson I. A., In vitro evolution of an influenza broadly neutralizing antibody is modulated by hemagglutinin receptor specificity. Nat. Commun. 8, 15371 (2017). 10.1038/ncomms15371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
science.sciencemag.org/content/369/6507/1119/suppl/DC1
Materials and Methods
Figs. S1 to S12
Tables S1 to S3
MDAR Reproducibility Checklist