Abstract
Brazil currently has one of the fastest growing SARS-CoV-2 epidemics in the world. Owing to limited available data, assessments of the impact of non-pharmaceutical interventions (NPIs) on virus spread remain challenging. Using a mobility-driven transmission model, we show that NPIs reduced the reproduction number from >3 to 1–1.6 in São Paulo and Rio de Janeiro. Sequencing of 427 new genomes and analysis of a geographically representative genomic dataset identified >100 international virus introductions in Brazil. We estimate that most (76%) of the Brazilian strains fell in three clades that were introduced from Europe between 22 February11 March 2020. During the early epidemic phase, we found that SARS-CoV-2 spread mostly locally and within-state borders. After this period, despite sharp decreases in air travel, we estimated multiple exportations from large urban centers that coincided with a 25% increase in average travelled distances in national flights. This study sheds new light on the epidemic transmission and evolutionary trajectories of SARS-CoV-2 lineages in Brazil, and provide evidence that current interventions remain insufficient to keep virus transmission under control in the country.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel betacoronavirus with a 30-kb genome that was first reported in December 2019 in Wuhan, China (1, 2). SARS-CoV-2 was declared a public health emergency of international concern on 30 January 2020. As of 12 July 2020, coronavirus disease 2019 (COVID-19) has caused over 12.5 million cases and 561 thousand deaths globally (3). The virus can be classified into two main phylogenetic lineages, namely A and B, that spread from Wuhan before strict travel restrictions were enacted (4, 5) and now co-circulate around the world (6). The case fatality ratio of SARS-CoV-2 infection has been estimated between 1.2 and 1.6% (7–9) with substantially higher ratios in those aged above 60 years (8). Some estimates suggest that 18-56% of SARS-CoV-2 transmission is from asymptomatic or pre-symptomatic individuals (10–13), complicating epidemiological assessments and public health efforts to curb the pandemic.
Challenges of real-time assessment of transmission
While the SARS-CoV-2 epidemics in several countries including China, Italy, and Spain have been brought under control through non-pharmaceutical interventions (NPIs) (3), the number of SARS-CoV-2 cases and deaths in Brazil continues to increase (14) (Fig. 1A). As of 12 July 2020, Brazil has now reported 1,800,827 SARS-CoV-2 cases, the second largest number in the world, and 70,398 deaths. Over a third of the cases (34%) in Brazil are concentrated in the southeast region which includes São Paulo city (Fig. 1B), the world’s fourth largest conurbation, where the first case in Latin America was reported on 25 February 2020 (15). Diagnostic assays for SARS-CoV-2 molecular detection were widely distributed across the regional reference centres of the national public health laboratory network from 21 February 2020 onwards (16, 17). However, several factors, including delays in reporting, changes in notification, and heterogeneous access to testing across populations, obfuscate the real-time assessment of virus transmission using SARS-CoV-2 case counts (15). Consequently, a more accurate measure of SARS-CoV-2 transmission in Brazil is provided by reported deaths due to severe acute respiratory infections (SARI), provided by the Sistema Único de Saúde (SUS) (18). Changes in the opportunity for SARS-CoV-2 transmission are strongly associated with changes in average mobility (18–20), and can typically be measured by calculating the effective reproduction number, R, defined as the average number of secondary infections caused by an infected person. R >1 indicates a growing epidemic while R <1 is needed to achieve a decrease in transmission.
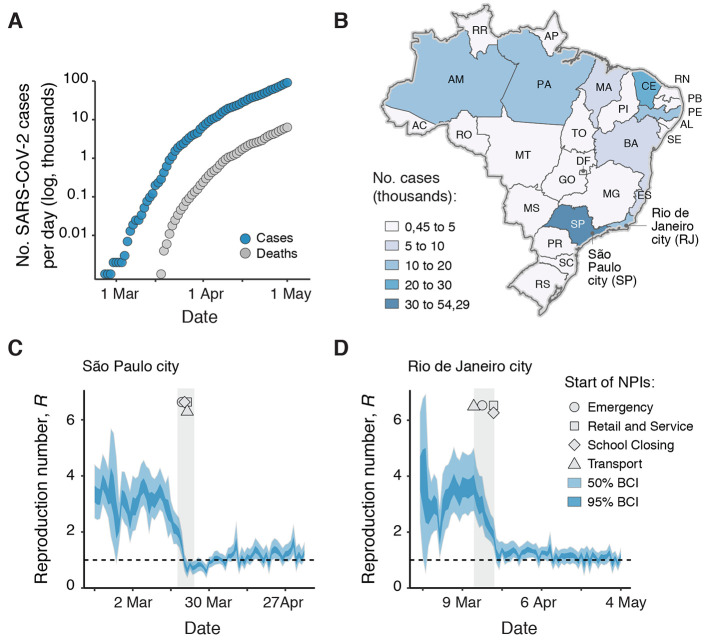
Fig. 1. SARS-CoV-2 epidemiology and epidemic spread in Brazil.
(A) Cumulative number of SARS-CoV-2 reported cases (blue) and deaths (grey) in Brazil. (B) States are colored according to the number of cumulative confirmed cases by 30 April 2020. (C and D) Reproduction number (R) over time for the cities of São Paulo (C) and Rio de Janeiro (D). R were estimated using a Bayesian approach incorporating daily number of deaths and four variables related to mobility data (a social isolation index from Brazilian geolocation company InLoco, and Google mobility indices for time spent in transit stations, parks, and the average between groceries and pharmacies, retail and recreational, and workspaces). Dashed horizontal line indicates R = 1. Grey area and geometric symbols show the times at which NPIs interventions were implemented. Bayesian credible intervals (BCIs, 50 and 95%) are shown as shaded areas. The 2-letter ISO 3166-1 codes for the 27 federal units in Brazil are provided in Supplementary Information.
We used a Bayesian semi-mechanistic model (21, 22) to analyze SARI mortality statistics and human mobility data to estimate daily changes in R in São Paulo city (12,2 million inhabitants) and Rio de Janeiro city (6,7 million inhabitants), the largest urban metropoles in Brazil (Fig. 1, C and D). NPIs in Brazil consisted of school closures implemented between 12 and 23 March 2020 across the country’s 27 federal units/states, and store closures implemented between 13 and 23 March 2020. In São Paulo city, schools started closing on 16 March and stores closed four days later. At the start of the epidemics, we found R >3 in São Paulo and Rio de Janeiro, and that concurrent with the timing of state mandated NPIs, R values fell close to 1.
Mobility driven reproduction number changes
Analysis of the reproduction number after NPI implementation highlights several notable mobility-driven features. There was a period immediately following NPIs, between 21 and 31 March 2020, when R was consistently <1 in São Paulo city (Fig. 1C). However, after this initial decrease, the R value for São Paulo rises >1 and increases through time, a trend associated with increased population mobility. This can be seen in the Google transit stations index, which rises from -60% to -52%, and by a decrease in the social isolation index from 54% to 47%. By 4 May 2020, we estimate R = 1.3 (BCI 95%: 1.0-1.6) in both São Paulo and Rio de Janeiro cities (table S1). However, we note that there were instances in the previous 7 days when the 95% credible intervals for R included values below 1, drawing attention to the fluctuations and uncertainty in the estimated reproduction number for both cities.
Early sharing of genomic sequences, including the first SARS-CoV-2 genome, Wuhan-Hu-1, released on 10 January (23), has enabled unprecedented global levels of molecular testing for an emerging virus (24, 25). However, despite the thousands of virus genomes deposited on public access databases, there is a lack of consistent sampling structure, and limited data from Brazil (26–28), which hampers accurate reconstructions of virus movement and transmission using phylogenetic analyses. To investigate how SARS-CoV-2 became established in the country, and to quantify the impact of NPIs on virus spatiotemporal spread, we tested a total of 26,732 samples from public and private laboratories using real-time PCR assays and found 7,944 (29%) to be positive for SARS-CoV-2. We then focused our sequencing efforts on generating a large and spatially representative genomic dataset with curated metadata in order to maximise the association between the number of sequences and the number of SARS-CoV-2 confirmed cases per state.
Spatially representative sequencing efforts
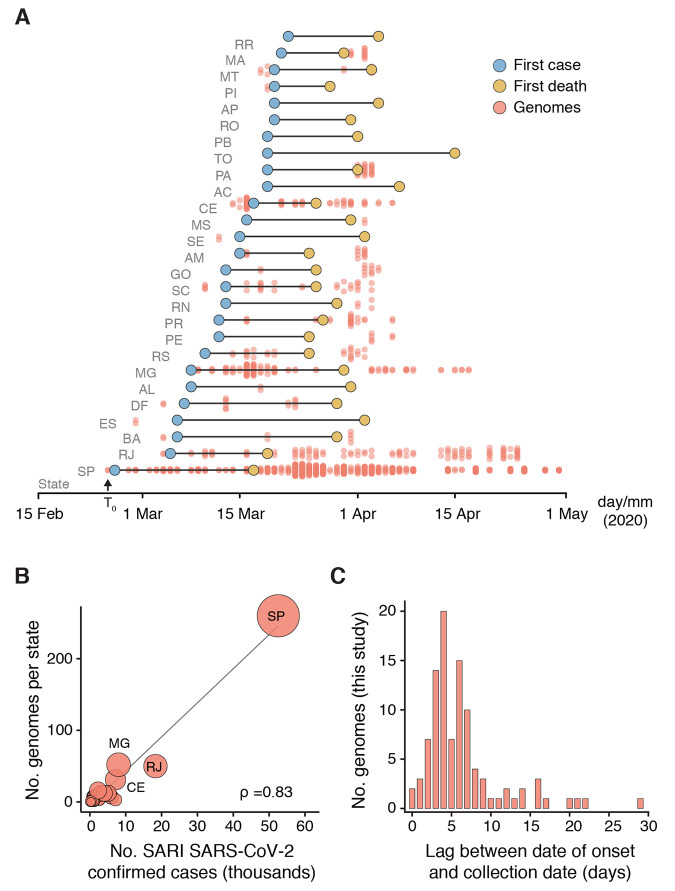
We generated 427 new SARS-CoV-2 genomes with >75% genome coverage from Brazilian samples collected between 5 March and 30 April 2020 (figs. S1 to S3 and data S1). For each state, the time between the date of the first reported case and the collection date of the first sequence analyzed in that state was only 4.5 days on average (Fig. 2A). For eight federal states, genomes were obtained from samples collected up to 6 days before the first case notifications. The genomes generated here were collected in 85 municipalities across 18 of 27 federal units spanning all regions in Brazil (Fig. 2A and fig. S2). Sequenced genomes were obtained from samples collected on average 4 days (median, range: 0 to 29 days) after onset of symptoms and were generated in 3 laboratories using harmonized sequencing and bioinformatic protocols (table S2). When we include 63 additional available sequences from Brazil deposited in GISAID (29) (see data S1 and S2), we find the data set is representative of the spatial heterogeneity of the Brazilian epidemic. Specifically, the number of genomes per state strongly correlates with SARI SARS-CoV-2 confirmed cases and SARI cases with unknown aetiology per state (n = 490 sequences from 21 states, Spearman’s correlation, ρ = 0.83; Fig. 2A). This correlation varied from 0.70 to 0.83 when considering SARI cases and deaths caused by SARS-CoV-2, and SARI cases and deaths from unknown aetiology (fig. S4). Most (n = 485/490) Brazilian sequences belong to SARS-CoV-2 lineage B, with only 5 strains belonging to lineage A (2 from Amazonas, 1 from Rio Grande do Sul, 1 from Minas Gerais and 1 from Rio de Janeiro; data S1 and fig. S5 show detailed lineage information for each sequence). Moreover, we used an in-silico assessment of diagnostic assay specificity for Brazilian strains (n = 490) to identify potential mismatches in some assays targeting Brazilian strains. We find that the forward primers of the Chinese CDC and Hong Kong University nucleoprotein-targeting RT-qPCR may be less appropriate for use in Brazil than other diagnostic assays, for which few or no mismatches were identified (fig. S6 and table S3). The impact of these mismatches on the sensitivity of these assays should be confirmed experimentally. If sensitivity is affected, the use of duplex RT-qPCR assays that concurrently target different genomic regions may help the detection of viruses with variants in primer or probe binding regions.
Fig. 2. Spatially-representative genomic sampling.
(A) Dumbbell plot showing the time intervals between date of collection of sampled genomes, notification of first cases and first deaths in each state. Red lines indicate the lag between the date of collection of first genome sequence and first reported case. The key for the 2-letter ISO 3166-1 codes for Brazilian federal units (or states) are provided in Supplementary Information. (B) Spearman’s rank (ρ) correlation between the number of SARI SARS-CoV-2 confirmed and SARI cases with unknown aetiology against number of sequences for each of the 21 Brazilian states included in this study (see also fig. S4). Circle sizes are proportional to the number of sequences for each federal unit. (C) Interval between the date of symptom onset and date of sample collection for the sequences generated in this study.
Phylogenetic analyses and international introductions
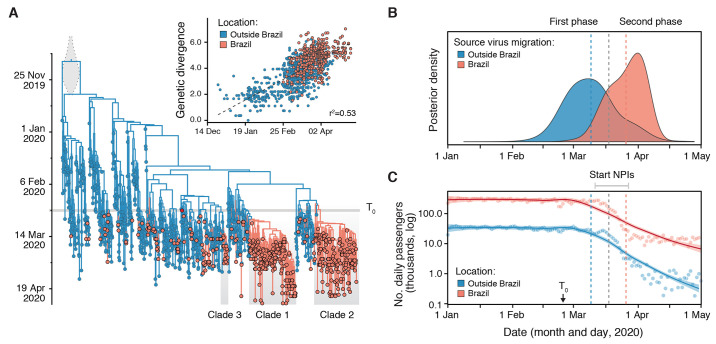
We estimated maximum likelihood and molecular clock phylogenies for a global dataset with a total of 1,182 genomes sampled from 24 Dec 2019 to 30 Apr 2020 (root-to-tip genetic distance correlation with sampling dates, r2 = 0.53; Fig. 3A and fig. S7). We inferred a median evolutionary rate of 1.13 × 10−3 (95% BCI: 1.03–1.23 × 10−3) substitutions per site per year (s/s/y), using an exponential growth coalescent model, equating to 33 changes per year on average across the virus genome. This is within the range of evolutionary rates estimated for other human coronaviruses (30–33). We estimate the date of the common ancestor (TMRCA) of the SARS-CoV-2 pandemic to around mid-Nov 2019 (median = 19 Nov 2019, 95% BCI: 26 Oct 2019 to 6 Dec 2019), in line with recent findings (34, 35).
Fig. 3. Evolution and spread of SARS-CoV-2 in Brazil.
(A) Time-resolved maximum clade credibility phylogeny of 1182 SARS-CoV-2 sequences, 490 from Brazil (red) and 692 from outside Brazil (blue). The largest Brazilian clades are highlighted by grey boxes (Clade 1, Clade 2 and Clade 3). The panel A inset shows a root-to-tip regression of genetic divergence against dates of sample collection. (B) Dynamics of SARS-CoV-2 import events in Brazil. Dates of international and national (between federal states) migration events were estimated from virus genomes using a phylogeographic approach. The first phase was dominated by virus migrations from outside Brazil while the second phase is marked by virus spread within Brazil. Dashed vertical lines correspond to the mean posterior estimate for migration events from outside Brazil (blue) and within Brazil (red). (C) Locally estimated scatterplot smoothing of the daily number of international (blue) and national (red) air passengers in Brazil in 2020. T0 = date of first reported case in Brazil (25 February 2020).
Phylogenetic analysis revealed that the majority of the Brazilian genomes (76%, n = 370/490) fell into three clades hereafter named as Clade 1 (n = 186/490, 38% of Brazilian strains), Clade 2 (n = 166, 34%) and Clade 3 (n = 18/490, 4%; Fig. 3A and figs. S8 and S9), which were largely in agreement with those identified in a phylogenetic analysis using 13,833 global genomes. The most recent common ancestors of the three main Brazilian clades (Clades 1 to 3) were dated from 28 February (21 Feb to 4 Mar 2020) (Clade 1), 22 February (17 to 24 Feb 2020) (Clade 2) to 11 March (9 to 12 Mar 2020) (Clade 3) (Fig. 3A and fig. S10). This indicates that community-driven transmission was already established in Brazil by early March, suggesting that international travel restrictions initiated after this period would have had limited impact. Brazilian Clade 1 is characterized by a nucleotide substitution in the spike protein (G25088T, numbering relative to GenBank reference NC_045512.2) and circulates predominantly in São Paulo state (n = 159, 85.4%; figs. S9 and S11). Clade 2 is defined by two nucleotide substitutions in ORF6 (T27299C) and nucleoprotein (T29148C); this is the most spatially widespread lineage, with sequences from a total of 16 states in Brazil. Clade 3 is concentrated in Ceará state (n = 16, 89%) and falls in a global cluster with sequences mainly from Europe. In the Amazon region, where the epidemic is expanding rapidly (14, 22), we find evidence for multiple national and international introductions, with 37% (n = 7/19) of sequences from Pará and Amazonas states clustering in Clade 1 and 32% (n = 6/19) in Clade 2.
Time-measured phylogeographic analyses revealed at least 102 (95% BCI: 95–109) international introductions of SARS-CoV-2 in Brazil (Fig. 3A and figs. S8 and S12). This represents an underestimate of the real number of introductions, as we have sequenced, on average, only 1 out of 200 confirmed cases. Most of these estimated introductions were directed to internationally well-connected states (36) such as São Paulo (36% of all imports), Minas Gerais (24%), Ceará (10%) and Rio de Janeiro (8%) (fig. S12). We further assessed the contribution of international vs. national virus lineage movement events through time (Fig. 3B). In the first phase of the epidemic, we find an increasing number of international introductions until 10 Mar 2020 (Fig. 2B). Limited available travel history data (15) suggests that these early cases were predominantly acquired from Italy (26%, n = 70 of 266 unambiguously identified country of infection) and the USA (28%, n = 76 of 266). After this initial phase, we find that the estimated number of international imports decreased concomitantly with the decline in the number of international passengers travelling to Brazil (Fig. 3, B and C, and S13). In contrast, despite the declines in the number of passengers travelling on national flights (Fig. 3C), we detected an increase in virus lineage movement events between Brazilian regions at least until early April 2020.
Modelling spatiotemporal spread within Brazil
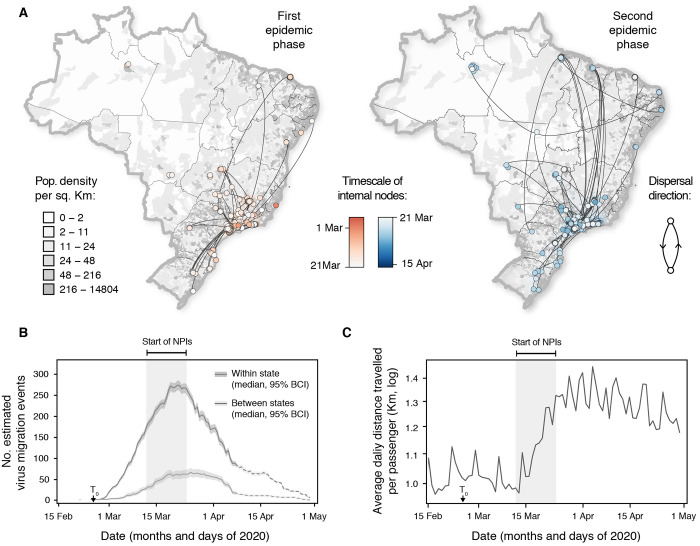
To better understand virus spread across spatiotemporal scales within Brazil, we use a continuous phylogeographic model that maps phylogenetic nodes to their inferred origin locations (37) (Fig. 4). We distinguish branches that remain within a state versus those that cross a state to infer the proportion of within versus between state observed virus movement.
Fig. 4. Spread of SARS-CoV-2 in Brazil.
(A) Spatiotemporal reconstruction of the spread of Brazilian SARS-CoV-2 clusters containing >2 sequences during the first (left) and the second epidemic phase (right) epidemic phase (Fig. 3B). Circles represent nodes of the MCC phylogeny and are colored according to their inferred time of occurrence. Shaded areas represent the 80% highest posterior density (HPD) interval and depict the uncertainty of the phylogeographic estimates for each node. Solid curved lines denote the links between nodes and the directionality of movement. Sequences belonging to clusters with <3 sequences were also plotted on the map with no lines connecting them. Background population density for each municipality was obtained from the Brazilian Institute of Geography (https://www.ibge.gov.br/). See fig. S14 for details of virus spread in the Southeast region. (B) Estimated number of within state (or within a given federal unit) and between-state (or between federal units) virus migrations over time. Dashed lines indicate estimates obtained during period of limited sampling (fig. S2). (C) Average distance in kilometres travelled by an air passenger per day in Brazil. Number of daily air passengers is shown in Fig. 3B. Light grey boxes indicate starting dates of NPIs across Brazil.
We estimate that during the first epidemic phase, SARS-CoV-2 spread mostly locally and within-state borders. In contrast, the second phase was characterized by long-distance movement events and the ignition of the epidemic outside the southeast region of Brazil (Fig. 4A). Throughout the epidemic, we find that within-state virus lineage movement was, on average, 5.1-fold more frequent than between-state movement. Moreover, our data suggests that within-state virus spread, and to a lesser extent, between-state virus spread, decreased after the implementation of NPIs (Fig. 4B). However, it is useful to note that the more limited sampling after April 6 2020 (see fig. S2) decreases inferred virus lineage movement toward present (Figs. 3B and 4B).
Interestingly, we find that the average route length travelled by passenger increased by 25% during the second phase of the epidemic (Fig. 4C), despite a concomitant reduction in the number of passengers flying within Brazil (Fig. 3C). The increase in the average route length post-NPI implementation results from a larger reduction in the number of air passengers flying on shorter distance journeys compared to those flying longer distance journeys. For example, we find an 8.8-fold reduction in the number of passengers flying in flight legs < 1000 km, compared to a 4.4-fold reduction in those flying >2000 km (fig. S15). These findings emphasize the roles of within and between-state mobility as a key driver of both local and inter-regional virus spread, with highly populated and well-connected urban conurbations in the southeast region acting as main sources of virus exports within the country (fig. S12).
Discussion
We provide a comprehensive analysis of SARS-CoV-2 spread in Brazil that shows the importance of community and nation-wide measures to control the COVID-19 epidemic Brazil. Although NPIs initially reduced virus transmission and spread, the continued increase in the number of cases and deaths in Brazil highlights the urgent need to prevent future virus transmission by implementing rapid and accessible diagnostic screening, contact tracing, quarantining of new cases and coordinated social and physical distancing measures across the country (38). With the recent relaxation of NPIs in Brazil and elsewhere, continued molecular, immunological and genomic surveillance are required for real-time data-driven decisions. Our analysis shows how changes in mobility may impact global and local transmission of SARS-CoV-2, and demonstrates how combining genomic and mobility data can complement traditional surveillance approaches.
Acknowledgments
A full list acknowledging those involved in the diagnostic and generation of new sequences as part of the CADDE-Genomic-Network can be found in the Supplementary Material. We thank GISAID database for supporting rapid and transparent sharing of genomic data during the COVID-19 pandemic. A full list acknowledging the authors submitting data used in this study can be found in data S2. We thank Paola Resende (FIOCRUZ), Talita Adelino (FUNED), Claudio Sacchi (IAL), Valdinete Nascimento (FIOCRUZ Amazonia), and their colleagues for submitting Brazilian data to GISAID. We thank Adriano Pinter (SUCEN), Nelson Gouveia (USP) and Izabel Marcílio de Souza (HCFM-USP) for fruitful discussions. We thank Lucy Matkin and Josh Quick for logistic support. We thank the UNICAMP Task Force against Covid-19 for support in generating genomes from Campinas. The analysis of openly available epidemiological data from https://covid.saude.gov.br/ has benefited from the COVID-19 surveillance efforts by the Secretaria de Vigilância em Saúde, Ministry of Health, Brazil. Funding: This project was supported by the Medical Research Council-São Paulo Research Foundation (FAPESP) CADDE partnership award (MR/S0195/1 and FAPESP 18/14389-0) (http://caddecentre.org/). FAPESP further supports IMC (2018/17176-8 and 2019/12000-1), JGJ (2018/17176-8 and 2019/12000-1, 18/14389-0), FCSS (2018/25468-9), WMS (2017/13981-0, 2019/24251-9), MF (2018/09383-3), TMC (2019/07544-2), CAMS (2019/21301-5), HIN (2018/14933-2), PSP (16/18445-7), MLN (20/04836-0), JLM (2020/04558-0 and 2016/00194-8). NRF is supported by a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (204311/Z/16/Z). DSC is supported by the Clarendon Fund and by the Department of Zoology, University of Oxford. SD is supported by the Fonds National de la Recherche Scientifique (FNRS, Belgium). JT and PL supported by European Union's Horizon 2020 project MOOD (874850). This project was supported by CNPq (MTM and MLN; ATRV: 303170/2017-4; RSA: 312688/2017-2 and 439119/2018-9; RPS: 310627/2018-4; and WMS: 408338/2018-0), FAPERJ (ATRV: E-26/202.826/2018 and RSA: 202.922/2018). MSR is supported by FMUSP. CAP, GMF, JH, MRA are supported by CAPES. OJB is supported by Sir Henry Wellcome Fellowship funded by the Wellcome Trust (206471/Z/17/Z). RPS is supported by FAPEMIG (APQ-00475-20). MMT is supported by Instituto Nacional de Ciância e Tecnologia em Dengue (INCT Dengue 465425/2014-3). ATRV is supported by FINEP (01.16.0078.00). PL and NJL are supported by Wellcome Trust ARTIC network (Collaborators Award 206298/Z/17/Z). PL and AR are supported by the European Research Council (grant agreement no. 725422 -ReservoirDOCS). OGP, NRF and LdP are supported by the Oxford Martin School. This work received funding from the UK Medical Research Council, under a concordat with the UK Department for International Development. We additionally acknowledge support from Community Jameel and the NIHR Health Protection Research Unit in Modelling Methodology. Author Contributions: Conception: DSC, IMC, JGJ, ECS, NRF; Acquisition: DSC, IMC, JGJ, WMS, FRRM, RHMP, FCSS, ERM, MTM, CMV, MJF, TMC, CAMS, MSR, MRA, JA, HN, PSP, AT, ADR, CKVB, ALG, APG, NG, CSA, ACSF, CXL, JEL, CG, GMF, RSF, FG, MTG, MLM, MWP, TMPPC, CSL, AASS, CLS, JF, ACS, AZS, MNNS, CZS, RPS, LCRM, MMT, JH, PAFL, RGM, MLN, SFC, JLPM, ATRV, RSA, ECS, NRF; Analysis: DSC, IMC, JGJ, WMS, FRRM, SD, TAM, LP, RHMP, JT, LA, CMV, HH, SM, MSG, LMC, LFB, CAP, OJB, SMN, SCH, JLPM, ATRV, SB, OGP, PL, CHW, RSA, NRF; Interpretation: DSC, IMC, JGJ, WMS, FRRM, SD, TAM, LP, RHMP, SCH, AASS, NMF, ATRV, SB, PL, CHW, AR, RSA, OGP, ECS, NRF; Drafting: DSC, IMC, JGJ, WMS, FRRM, SD, TAM, RSA, OGP, ECS, NRF; Revising: All authors have read and approved the final version of the manuscript. Funding: WMS, MLN, NMF, JLPM, ATRV, NJL, RSA, OGP, ECS and NRF. Competing interests: Authors declare no competing interests. Data and materials availability: The 427 SARS-CoV newly generated genomes from this study can be found on GISAID under the accession IDs: EPI_ISL_470568-470655 and EPI_ISL_476152-476490. An interactive visualization of the temporal, geographic and mutational patterns in our data can be found at https://microreact.org/project/rKjKLMrjdPVHKR1erUzKyi (39). Reads have been deposited to accession numbers PRJEB39487 (IMT-USP and UNICAMP) and PRJNA640656 (UFRJ-LNCC). All data, code, and materials used in the analysis are available on DRYAD (40). The IRB protocol number is CAAE 30127020.0.0000.0068 as currently described in Material and Methods. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Supplementary Materials
science.sciencemag.org/cgi/content/full/science.abd2161/DC1
Materials and Methods
Figs. S1 to S15
Tables S1 to S3
List of Members of the CADDE Genomic Network
Data S1 and S2
MDAR Reproducibility Checklist
References and Notes
- 1.Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020). 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization, Coronavirus Disease (COVID-2019) Situation Reports (2020); www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
- 4.Tian H., Liu Y., Li Y., Wu C.-H., Chen B., Kraemer M. U. G., Li B., Cai J., Xu B., Yang Q., Wang B., Yang P., Cui Y., Song Y., Zheng P., Wang Q., Bjornstad O. N., Yang R., Grenfell B. T., Pybus O. G., Dye C., An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in China. Science 368, 638–642 (2020). 10.1126/science.abb6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraemer M. U. G., Yang C.-H., Gutierrez B., Wu C.-H., Klein B., Pigott D. M., du Plessis L., Faria N. R., Li R., Hanage W. P., Brownstein J. S., Layan M., Vespignani A., Tian H., Dye C., Pybus O. G., Scarpino S. V.; Open COVID-19 Data Working Group , The effect of human mobility and control measures on the COVID-19 epidemic in China. Science 368, 493–497 (2020). 10.1126/science.abb4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rambaut A., Holmes E. C., O’Toole Á., Hill V., McCrone J. T., Ruis C., du Plessis L., Pybus O. G., A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. (2020). 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell T. W., Hellewell J., Jarvis C. I., van Zandvoort K., Abbott S., Ratnayake R., Flasche S., Eggo R. M., Edmunds W. J., Kucharski A. J.; Cmmid Covid-Working Group , Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship, February 2020. Euro Surveill. 25, 2000256 (2020). 10.2807/1560-7917.ES.2020.25.12.2000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verity R., Okell L. C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P. G. T., Fu H., Dighe A., Griffin J. T., Baguelin M., Bhatia S., Boonyasiri A., Cori A., Cucunubá Z., FitzJohn R., Gaythorpe K., Green W., Hamlet A., Hinsley W., Laydon D., Nedjati-Gilani G., Riley S., van Elsland S., Volz E., Wang H., Wang Y., Xi X., Donnelly C. A., Ghani A. C., Ferguson N. M., Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 20, 669–677 (2020). 10.1016/S1473-3099(20)30243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J. T., Leung K., Bushman M., Kishore N., Niehus R., de Salazar P. M., Cowling B. J., Lipsitch M., Leung G. M., Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 26, 506–510 (2020). 10.1038/s41591-020-0822-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arons M. M., Hatfield K. M., Reddy S. C., Kimball A., James A., Jacobs J. R., Taylor J., Spicer K., Bardossy A. C., Oakley L. P., Tanwar S., Dyal J. W., Harney J., Chisty Z., Bell J. M., Methner M., Paul P., Carlson C. M., McLaughlin H. P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L. C., Kay M., Lewis J., Montgomery P., Stone N. D., Clark T. A., Honein M. A., Duchin J. S., Jernigan J. A.; Public Health–Seattle and King County and CDC COVID-19 Investigation Team , Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 382, 2081–2090 (2020). 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., Parker M., Bonsall D., Fraser C., Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 368, eabb6936 (2020). 10.1126/science.abb6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavezzo E., Franchin E., Ciavarella C., Cuomo-Dannenburg G., Barzon L., Del Vecchio C., Rossi L., Manganelli R., Loregian A., Navarin N., Abate D., Sciro M., Merigliano S., De Canale E., Vanuzzo M. C., Besutti V., Saluzzo F., Onelia F., Pacenti M., Parisi S., Carretta G., Donato D., Flor L., Cocchio S., Masi G., Sperduti A., Cattarino L., Salvador R., Nicoletti M., Caldart F., Castelli G., Nieddu E., Labella B., Fava L., Drigo M., Gaythorpe K. A. M., Brazzale A. R., Toppo S., Trevisan M., Baldo V., Donnelly C. A., Ferguson N. M., Dorigatti I., Crisanti A., Suppression of COVID-19 outbreak in the municipality of Vo, Italy. Nature (2020). 10.1038/s41586-020-2488-1 [DOI] [PubMed] [Google Scholar]
- 13.Mizumoto K., Kagaya K., Zarebski A., Chowell G., Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 25, 2000180 (2020). 10.2807/1560-7917.ES.2020.25.10.2000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brazilian Ministry of Health, Painel de Casos de Doença Pelo Coronavírus 2019 (COVID-19) No Brasil Pelo Ministério da Saúde (2020); http://covid.saude.gov.br.
- 15.W. M. de Souza, L. Fletcher Buss, D. da Silva Candido, J. P. Carrera, S. Li, A. Zarebski, M. Vincenti-Gonzalez, J. Messina, F. C. da Silva Sales, P. dos Santos Andrade, C. A. Prete Jr., V. H. Nascimento, F. Ghilardi, R. H. Moraes Pereira, A. A. de Souza Santos, L. Abade, B. Gutierrez, M. U. G. Kraemer, R. Santana Aguiar, N. Alexander, P. Mayaud, O. J. Brady, I. O. M. de Souza, N. Gouveia, G. Li, A. Tami, S. Barbosa Oliveira, V. B. Gomes Porto, F. Ganem, W. Ferreira Almeida, F. Fontana Sutile Tardetti Fantinato, E. Marques Macario, W. Kleber Oliveira, O. Pybus, C.-H. Wu, J. Croda, E. Cerdeira Sabino, N. R. Faria, Epidemiological and clinical characteristics of the early phase of the COVID-19 epidemic in Brazil. medRxiv 10.1101/2020.04.25.20077396 [Preprint]. 29 April 2020; . 10.1101/2020.04.25.20077396 [DOI]
- 16.Croda J., Oliveira W. K., Frutuoso R. L., Mandetta L. H., Baia-da-Silva D. C., Brito-Sousa J. D., Monteiro W. M., Lacerda M. V. G., COVID-19 in Brazil: Advantages of a socialized unified health system and preparation to contain cases. Rev. Soc. Bras. Med. Trop. 53, e20200167 (2020). 10.1590/0037-8682-0167-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croda J., Garcia L., Immediate health surveillance response to COVID-19 epidemic [in Portuguese]. Epidemiol. Ser. Saúde 29, e2020002 (2020). 10.5123/S1679-49742020000100021 [DOI] [PubMed] [Google Scholar]
- 18.S. B. Oliveira, V. Bertollo Gomes Porto, F. Ganem, F. Macedo Mendes, M. Almiron, W. Kleber de Oliveira, F. Fontana Sutile Tardetti Fantinato, W. Aparecida Ferreira de Almeida, A. Pereira de Macedo Borges, H. Natan Batista Pinheiro, R. dos Santos Oliveira, J. R. Andrews, N. R. Faria, M. Barreto Lopes, W. Araujo, F. A. Diaz-Quijano, H. I. Nakaya, J. Croda, Monitoring social distancing and SARS-CoV-2 transmission in Brazil using cell phone mobility data. medRxiv 2020.04.30.20082172 [Preprint] (5 May 2020); . 10.1101/2020.04.30.20082172 [DOI]
- 19.S. M. Kissler, Reductions in commuting mobility predict geographic differences in SARS-CoV-2 prevalence in New York City (Harvard DASH Repository, 2020); https://dash.harvard.edu/bitstream/handle/1/42665370/Kissler_etal_NYC_mobility.pdf?sequence = 1&isAllowed = y. [DOI] [PMC free article] [PubMed]
- 20.H. J. T. Unwin, S. Mishra, V. C. Bradley, A. Gandy, M. Vollmer, T. Mellan, H. Coupland, K. Ainslie, C. Whittaker, J. Ish-Horowicz, S. Filippi, X. Xi, M. Monod, O. Ratmann, M. Hutchinson, F. Valka, H. Zhu, I. Hawryluk, P. Milton, M. Baguelin, A. Boonyasiri, N. Brazeau, L. Cattarino, G. Charles, L. V. Cooper, Z. Cucunuba, G. Cuomo-Dannenburg, B. Djaafara, I. Dorigatti, O. J. Eales, J. Eaton, S. van Elsland, R. FitzJohn, K. Gaythorpe, W. Green, T. Hallett, W. Hinsley, N. Imai, B. Jeffrey, E. Knock, D. Laydon, J. Lees, G. Nedjati-Gilani, P. Nouvellet, L. Okell, A. Ower, K. V. Parag, I. Siveroni, H. A. Thompson, R. Verity, P. Walker, C. Walters, Y. Wang, O. J. Watson, L. Whittles, A. Ghani, N. M. Ferguson, S. Riley, C. A. Donnelly, S. Bhatt, S. Flaxman, Report 23: State-Level Tracking of COVID-19 in the United States (21-05-2020) (Imperial College London, 2020); . 10.25561/79231 [DOI]
- 21.Flaxman S., Mishra S., Gandy A., Unwin H. J. T., Mellan T. A., Coupland H., Whittaker C., Zhu H., Berah T., Eaton J. W., Monod M., Ghani A. C., Donnelly C. A., Riley S., Vollmer M. A. C., Ferguson N. M., Okell L. C., Bhatt S.; Imperial College COVID-19 Response Team , Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature (2020). 10.1038/s41586-020-2405-7 [DOI] [PubMed] [Google Scholar]
- 22.T. A. Mellan, H. H. Hoeltgebaum, S. Mishra, C. Whittaker, R. P. Schnekenberg, A. Gandy, H. J. T. Unwin, M. A. C. Vollmer, H. Coupland, I. Hawryluk, N. Rodrigues Faria, J. Vesga, H. Zhu, M. Hutchinson, O. Ratmann, M. Monod, K. Ainslie, M. Baguelin, S. Bhatia, A. Boonyasiri, N. Brazeau, G. Charles, L. V. Cooper, Z. Cucunuba, G. Cuomo-Dannenburg, A. Dighe, B. Djaafara, J. Eaton, S. L. van Elsland, R. FitzJohn, K. Fraser, K. Gaythorpe, W. Green, S. Hayes, N. Imai, B. Jeffrey, E. Knock, D. Laydon, J. Lees, T. Mangal, A. Mousa, G. Nedjati-Gilani, P. Nouvellet, D. Olivera, K. V. Parag, M. Pickles, H. A. Thompson, R. Verity, C. Walters, H. Wang, Y. Wang, O. J. Watson, L. Whittles, X. Xi, L. Okell, I. Dorigatti, P. Walker, A. Ghani, S. Riley, N. M. Ferguson, C. A. Donnelly, S. Flaxman, S. Bhatt, Report 21: Estimating COVID-19 Cases and Reproduction Number in Brazil (2020); . 10.25561/78872 [DOI]
- 23.Y.-Z. Zhang, E. C. Holmes, Novel 2019 coronavirus genome, Virological (2020); https://virological.org/t/novel-2019-coronavirus-genome/319.
- 24.Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K. W., Bleicker T., Brünink S., Schneider J., Schmidt M. L., Mulders D. G. J. C., Haagmans B. L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M. P. G., Drosten C., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25,2000045 (2020). 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thi Nhu Thao T., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., Gultom M., Schmied K., Laloli L., Hüsser L., Wider M., Pfaender S., Hirt D., Cippà V., Crespo-Pomar S., Schröder S., Muth D., Niemeyer D., Corman V. M., Müller M. A., Drosten C., Dijkman R., Jores J., Thiel V., Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 582, 561–565 (2020). 10.1038/s41586-020-2294-9 [DOI] [PubMed] [Google Scholar]
- 26.P. C. Resende, E. Delatorre, T. Gräf, D. Mir, F. do Couto Motta, L. Reis Appolinario, A. C. Dias da Paixão, M. Ogrzewalska, B. Caetano, M. Cordeiro dos Santos, J. de Almeida Ferreira, E. Costa Santos Junior, S. Patroca da Silva, S. Bianchini Fernandes, L. A. Vianna, L. da Costa Souza, J. F. G. Ferro, V. B. Nardy, J. Croda, W. K. Oliveira, A. Abreu, G. Bello, M. M. Siqueira, Genomic surveillance of SARS-CoV-2 reveals community transmission of a major lineage during the early pandemic phase in Brazil. bioRxiv 020.06.17.158006 [Preprint] (2020); . 10.1101/2020.06.17.158006 [DOI]
- 27.J. Xavier, M. Giovanetti, T. Adelino, V. Fonseca, A. V. Barbosa da Costa, A. Aparecida Ribeiro, K. Nascimento Felicio, C. Guerra Duarte, M. V. Ferreira Silva, A. Salgado, M. Teixeira Lima, R. de Jesus, A. Fabri, C. Franco Soares Zoboli, T. Gutemberg Souza Santos, F. Iani, A. M. Bispo de Filippis, M. Agudo Mendonca Teixeira de Siqueira, A. L. de Abreu, V. de Azevedo, D. Brock Ramalho, C. F. Campelo de Albuquerque, T. de Oliveira, E. C. Holmes, J. Lourenco, L. C. Junior Alcantara, M. Aparecida Assuncao Oliveira, The ongoing COVID-19 epidemic in Minas Gerais, Brazil: insights from epidemiological data and SARS-CoV-2 whole genome sequencing. medRxiv 2020.2005.2005.20091611 [Preprint] (2020); . 10.1101/2020.05.05.20091611 [DOI]
- 28.Nascimento V. A., Corado A. L. G., Nascimento F. O., Costa A. K. A., Duarte D. C. G., Jesus M. S., Luz S. L. B., Goncalves L. M. F., Costa C. F., Delatorre E., Naveca F. G., Genomic and phylogenetic characterization of an imported case of SARS-CoV-2 in Amazonas State, Brazil. Memoirs of the Oswaldo Cruz Institute 10.1590/0074-02760200310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu Y., McCauley J., GISAID: Global initiative on sharing all influenza data – from vision to reality. Euro. Surveill. 22, 30494 (2017) 10.1016/S0140-6736(13)61887-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cotten M., Watson S. J., Kellam P., Al-Rabeeah A. A., Makhdoom H. Q., Assiri A., Al-Tawfiq J. A., Alhakeem R. F., Madani H., AlRabiah F. A., Al Hajjar S., Al-nassir W. N., Albarrak A., Flemban H., Balkhy H. H., Alsubaie S., Palser A. L., Gall A., Bashford-Rogers R., Rambaut A., Zumla A. I., Memish Z. A., Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: A descriptive genomic study. Lancet 382, 1993–2002 (2013). 10.1016/S0140-6736(13)61887-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cotten M., Watson S. J., Zumla A. I., Makhdoom H. Q., Palser A. L., Ong S. H., Al Rabeeah A. A., Alhakeem R. F., Assiri A., Al-Tawfiq J. A., Albarrak A., Barry M., Shibl A., Alrabiah F. A., Hajjar S., Balkhy H. H., Flemban H., Rambaut A., Kellam P., Memish Z. A., Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 5, e01062-13 (2014). 10.1128/mBio.01062-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudas G., Carvalho L. M., Rambaut A., Bedford T., MERS-CoV spillover at the camel-human interface. eLife 7, e31257 (2018). 10.7554/eLife.31257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Z., Li H., Wu X., Zhong Y., Zhang K., Zhang Y.-P., Boerwinkle E., Fu Y.-X., Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 4, 21 (2004). 10.1186/1471-2148-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. Duchene, L. Featherstone, M. Haritopoulou-Sinanidou, A. Rambaut, P. Lemey, G. Baele, Temporal signal and the phylodynamic threshold of SARS-CoV-2. bioRxiv 2020.05.04.077735 [Preprint] (2020); . 10.1101/2020.05.04.077735 [DOI] [PMC free article] [PubMed]
- 35.Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., Sun J., François S., Kraemer M. U. G., Faria N. R., McCrone J. T., Peng J., Xiong Q., Yuan R., Zeng L., Zhou P., Liang C., Yi L., Liu J., Xiao J., Hu J., Liu T., Ma W., Li W., Su J., Zheng H., Peng B., Fang S., Su W., Li K., Sun R., Bai R., Tang X., Liang M., Quick J., Song T., Rambaut A., Loman N., Raghwani J., Pybus O. G., Ke C., Genomic Epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell 181, 997–1003.e9 (2020). 10.1016/j.cell.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Candido D. D. S., Watts A., Abade L., Kraemer M. U. G., Pybus O. G., Croda J., de Oliveira W., Khan K., Sabino E. C., Faria N. R., Routes for COVID-19 importation in Brazil. J. Travel Med. 27, taaa042 (2020). 10.1093/jtm/taaa042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.S. Dellicour, K. Durkin, S. L. Hong, B. Vanmechelen, J. Martí-Carreras, M. S. Gill, C. Meex, S. Bontems, E. André, M. Gilbert, C. Walker, N. De Maio, N. R. Faria, J. Hadfield, M.-P. Hayette, V. Bours, T. Wawina-Bokalanga, M. Artesi, G. Baele, P. Maes, A phylodynamic workflow to rapidly gain insights into the dispersal history and dynamics of SARS-CoV-2 lineages. bioRxiv 2020.05.05.078758 [Preprint] (2020); . 10.1101/2020.05.05.078758 [DOI] [PMC free article] [PubMed]
- 38.World Health Organization, Coronavirus disease 2019 (COVID-19): Situation report –72 (WHO, 2020); https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200401-sitrep-72-covid-19.pdf?sfvrsn = 3dd8971b_2 .
- 39.Centre for Genomic Pathogen Surveillance, Imperial College London, Report of 427 novel genomes from Brazil and the associated metadata, Microreact (2020); https://microreact.org/project/rKjKLMrjdPVHKR1erUzKyi.
- 40.Data and code for: D. S. Candido, I. M. Claro, J. G. de Jesus, W. M. Souza, F. R. R. Moreira, S. Dellicour, T. A. Mellan, L. du Plessis, R. H. M. Pereira, F. C. S. Sales, E. R. Manuli, J. Thézé, L. Almeida, M. T. Menezes, C. M. Voloch, M. J. Fumagalli, T. M. Coletti, C. A. M. da Silva, M. S. Ramundo, M. R. Amorim, H. Hoeltgebaum, S. Mishra, M. S. Gill, L. M. Carvalho, L. F. Buss, C. A. Prete Jr., J. Ashworth, H. I. Nakaya, P. S. Peixoto, O. J. Brady, S. M. Nicholls, A. Tanuri, Á. D. Rossi, C. K. V. Braga, A. L. Gerber, A. P. de C. Guimarães, N. Gaburo Jr., C. Salete Alencar, A. C. S. Ferreira, C. X. Lima, J. E. Levi, C. Granato, G. M. Ferreira, R. S. Francisco Jr., F. Granja, M. T. Garcia, M. L. Moretti, M. W. Perroud Jr., T. M. P. P. Castiñeiras, C. S. Lazari, S. C. Hill, A. A. de Souza Santos, C. L. Simeoni, J. Forato, A. C. Sposito, A. Z. Schreiber, M. N. N. Santos, C. Zolini de Sá, R. P. Souza, L. C. Resende-Moreira, M. M. Teixeira, J. Hubner, P. A. F. Leme, R. G. Moreira, M. L. Nogueira, CADDE-Genomic-Network, N. M. Ferguson, S. F. Costa, J. L. Proenca-Modena, A. T. R. Vasconcelos, S. Bhatt, P. Lemey, C.-H. Wu, A. Rambaut, N. J. Loman, R. S. Aguiar, O. G. Pybus, E. C. Sabino, N. Rodrigues Faria, Evolution and epidemic spread of SARS-CoV-2 in Brazil, Dryad (2020); . 10.5061/dryad.rxwdbrv5z [DOI]
- 41.A. Aktay, S. Bavadekar, G. Cossoul, J. Davis, D. Desfontaines, A. Fabrikant, E. Gabrilovich, K. Gadepalli, B. Gipson, M. Guevara, C. Kamath, M. Kansal, A. Lange, C. Mandayam, A. Oplinger, C. Pluntke, T. Roessler, A. Schlosberg, T. Shekel, S. Vispute, M. Vu, G. Wellenius, B. Williams, R. J. Wilson, Google COVID-19 community mobility reports: Anonymization process description (version 1.0). arXiv:2004.04145 [cs.CR] (8 April 2020).
- 42.inloco, Mapa Brasileiro da COVID-19 (2020); https://mapabrasileirodacovid.inloco.com.br/pt/.
- 43.Peixoto P. S., Marcondes D., Peixoto C., Oliva S. M., Modeling future spread of infections via mobile geolocation data and population dynamics. An application to COVID-19 in Brazil. PLoS One 15, e0235732 (2020). 10.1093/jtm/taaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y., Gayle A. A., Wilder-Smith A., Rocklöv J., The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 27, taaa021 (2020). 10.1093/jtm/taaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waggoner J. J., Stittleburg V., Pond R., Saklawi Y., Sahoo M. K., Babiker A., Hussaini L., Kraft C. S., Pinsky B. A., Anderson E. J., Rouphael N., Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 26, 1633–1635 (2020). 10.3201/eid2607.201285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quick J., Grubaugh N. D., Pullan S. T., Claro I. M., Smith A. D., Gangavarapu K., Oliveira G., Robles-Sikisaka R., Rogers T. F., Beutler N. A., Burton D. R., Lewis-Ximenez L. L., de Jesus J. G., Giovanetti M., Hill S. C., Black A., Bedford T., Carroll M. W., Nunes M., Alcantara Jr L. C., Sabino E. C., Baylis S. A., Faria N. R., Loose M., Simpson J. T., Pybus O. G., Andersen K. G., Loman N. J., Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 12, 1261–1276 (2017). 10.1038/nprot.2017.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milne I., Bayer M., Cardle L., Shaw P., Stephen G., Wright F., Marshall D., Tablet: Next generation sequence assembly visualization. Bioinformatics 26, 401–402 (2010). 10.1093/bioinformatics/btp666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K., Standley D. M., MAFFT: Iterative refinement and additional methods. Methods Mol. Biol. 1079, 131–146 (2014). 10.1007/978-1-62703-646-7_8 [DOI] [PubMed] [Google Scholar]
- 50.Hasegawa M., Kishino H., Yano T., Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174 (1985). 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- 51.Yang Z., Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 39, 306–314 (1994). 10.1007/BF00160154 [DOI] [PubMed] [Google Scholar]
- 52.Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., von Haeseler A., Lanfear R., IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 37, 1530–1534 (2020). 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rambaut A., Lam T. T., Max Carvalho L., Pybus O. G., Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2, vew007 (2016). 10.1093/ve/vew007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singer J., Gifford R., Cotten M., Robertson D., CoV-GLUE: A web application for tracking SARS-CoV-2 genomic variation (2020); . 10.20944/preprints202006.0225.v1 [DOI]
- 55.Jesus J. G., Sacchi C., Candido D. D. S., Claro I. M., Sales F. C. S., Manuli E. R., Silva D. B. B. D., Paiva T. M., Pinho M. A. B., Santos K. C. O., Hill S. C., Aguiar R. S., Romero F., Santos F. C. P. D., Gonçalves C. R., Timenetsky M. D. C., Quick J., Croda J. H. R., Oliveira W., Rambaut A., Pybus O. G., Loman N. J., Sabino E. C., Faria N. R., Importation and early local transmission of COVID-19 in Brazil, 2020. Rev. Inst. Med. Trop. São Paulo 62, e30 (2020). 10.1590/s1678-9946202062030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Centers for Disease Control and Prevention, Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Primers and Probes (2020); https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html.
- 57.J. Northill, I. Mackay, Wuhan coronavirus (2019-nCoV) real-time RT-PCR N gene 2020 (Wuhan-N) V.1 (2020); https://www.protocols.io/view/wuhan-coronavirus-2019-ncov-real-time-rt-pcr-n-gen-ba86ihze?version_warning = no.
- 58.N. Nao, Shirato, K., Katano, H., Matsuyama, S., Takeda, M., Detection of second case of 2019-nCoV infection in Japan (corrected version) (2020); https://www.niid.go.jp/niid/images/vir3/nCoV/method-niid-20200123-2_erratum.pdf.
- 59.Thailand Ministry of Public Health, Diagnostic detection of novel coronavirus 2019 by real time RT-PCR (2020); https://www.who.int/docs/default-source/coronaviruse/conventional-rt-pcr-followed-by-sequencing-for-detection-of-ncov-rirl-nat-inst-health-t.pdf?sfvrsn = 42271c6d_4.
- 60.Chinese National Institute for Viral Disease Control and Prevention, Specific primers and probes for detection 2019 novel coronavirus (2020); http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html.
- 61.HKU Med, LKS Faculty of Medicine, School of Public Health, Detection of, 2019 novel coronavirus (2019-nCoV) in suspected human cases by RT-PCR (2020); https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf?sfvrsn = af1aac73_4.
- 62.Bruen T. C., Philippe H., Bryant D., A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681 (2006). 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huson D. H., Bryant D., Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 64.Martin D. P., Murrell B., Golden M., Khoosal A., Muhire B., RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 1, vev003 (2015). 10.1093/ve/vev003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A., Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). 10.1093/ve/vey016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ayres D. L., Darling A., Zwickl D. J., Beerli P., Holder M. T., Lewis P. O., Huelsenbeck J. P., Ronquist F., Swofford D. L., Cummings M. P., Rambaut A., Suchard M. A., BEAGLE: An application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 61, 170–173 (2012). 10.1093/sysbio/syr100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gill M. S., Lemey P., Faria N. R., Rambaut A., Shapiro B., Suchard M. A., Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 30, 713–724 (2013). 10.1093/molbev/mss265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferreira M. A. R., Suchard M. A., Bayesian analysis of elapsed times in continuous-time Markov chains. Can. J. Stat. 36, 355–368 (2008). 10.1002/cjs.5550360302 [DOI] [Google Scholar]
- 69.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lemey P., Rambaut A., Drummond A. J., Suchard M. A., Bayesian phylogeography finds its roots. PLOS Comput. Biol. 5, e1000520 (2009). 10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Faria N. R., Rambaut A., Suchard M. A., Baele G., Bedford T., Ward M. J., Tatem A. J., Sousa J. D., Arinaminpathy N., Pépin J., Posada D., Peeters M., Pybus O. G., Lemey P., HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346, 56–61 (2014). 10.1126/science.1256739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O’Brien J. D., Minin V. N., Suchard M. A., Learning to count: Robust estimates for labeled distances between molecular sequences. Mol. Biol. Evol. 26, 801–814 (2009). 10.1093/molbev/msp003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minin V. N., Suchard M. A., Fast, accurate and simulation-free stochastic mapping. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 3985–3995 (2008). 10.1098/rstb.2008.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minin V. N., Suchard M. A., Counting labeled transitions in continuous-time Markov models of evolution. J. Math. Biol. 56, 391–412 (2008). 10.1007/s00285-007-0120-8 [DOI] [PubMed] [Google Scholar]
- 75.Lemey P., Rambaut A., Welch J. J., Suchard M. A., Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 27, 1877–1885 (2010). 10.1093/molbev/msq067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dellicour S., Baele G., Dudas G., Faria N. R., Pybus O. G., Suchard M. A., Rambaut A., Lemey P., Phylodynamic assessment of intervention strategies for the West African Ebola virus outbreak. Nat. Commun. 9, 2222 (2018). 10.1038/s41467-018-03763-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dellicour S., Rose R., Faria N. R., Lemey P., Pybus O. G., SERAPHIM: Studying environmental rasters and phylogenetically informed movements. Bioinformatics 32, 3204–3206 (2016). 10.1093/bioinformatics/btw384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Y.-Z. Zhang, E. C. Holmes, Novel 2019 coronavirus genome, Virological (2020); https://virological.org/t/novel-2019-coronavirus-genome/319.
- Centre for Genomic Pathogen Surveillance, Imperial College London, Report of 427 novel genomes from Brazil and the associated metadata, Microreact (2020); https://microreact.org/project/rKjKLMrjdPVHKR1erUzKyi.
- Data and code for: D. S. Candido, I. M. Claro, J. G. de Jesus, W. M. Souza, F. R. R. Moreira, S. Dellicour, T. A. Mellan, L. du Plessis, R. H. M. Pereira, F. C. S. Sales, E. R. Manuli, J. Thézé, L. Almeida, M. T. Menezes, C. M. Voloch, M. J. Fumagalli, T. M. Coletti, C. A. M. da Silva, M. S. Ramundo, M. R. Amorim, H. Hoeltgebaum, S. Mishra, M. S. Gill, L. M. Carvalho, L. F. Buss, C. A. Prete Jr., J. Ashworth, H. I. Nakaya, P. S. Peixoto, O. J. Brady, S. M. Nicholls, A. Tanuri, Á. D. Rossi, C. K. V. Braga, A. L. Gerber, A. P. de C. Guimarães, N. Gaburo Jr., C. Salete Alencar, A. C. S. Ferreira, C. X. Lima, J. E. Levi, C. Granato, G. M. Ferreira, R. S. Francisco Jr., F. Granja, M. T. Garcia, M. L. Moretti, M. W. Perroud Jr., T. M. P. P. Castiñeiras, C. S. Lazari, S. C. Hill, A. A. de Souza Santos, C. L. Simeoni, J. Forato, A. C. Sposito, A. Z. Schreiber, M. N. N. Santos, C. Zolini de Sá, R. P. Souza, L. C. Resende-Moreira, M. M. Teixeira, J. Hubner, P. A. F. Leme, R. G. Moreira, M. L. Nogueira, CADDE-Genomic-Network, N. M. Ferguson, S. F. Costa, J. L. Proenca-Modena, A. T. R. Vasconcelos, S. Bhatt, P. Lemey, C.-H. Wu, A. Rambaut, N. J. Loman, R. S. Aguiar, O. G. Pybus, E. C. Sabino, N. Rodrigues Faria, Evolution and epidemic spread of SARS-CoV-2 in Brazil, Dryad (2020); . 10.5061/dryad.rxwdbrv5z [DOI]
Supplementary Materials
science.sciencemag.org/cgi/content/full/science.abd2161/DC1
Materials and Methods
Figs. S1 to S15
Tables S1 to S3
List of Members of the CADDE Genomic Network
Data S1 and S2
MDAR Reproducibility Checklist