Abstract
Remdesivir was shown to inhibit RNA-dependent RNA-polymerases (RdRp) from distinct viral families such as from Filoviridae (Ebola) and Coronaviridae (SARS-CoV, SARS-CoV-2, MERS). In this study, we tested the ability of remdesivir to inhibit RdRps from the Flaviviridae family. Instead of remdesivir, we used the active species that is produced in cells from remdesivir, the appropriate triphosphate, which could be directly tested in vitro using recombinant flaviviral polymerases. Our results show that remdesivir can efficiently inhibit RdRps from viruses causing severe illnesses such as Yellow fever, West Nile fever, Japanese and Tick-borne encephalitis, Zika and Dengue. Taken together, this study demonstrates that remdesivir or its derivatives have the potential to become a broad-spectrum antiviral agent effective against many RNA viruses.
Keywords: Remdesivir, Flavivirus, RNA-dependent RNA polymerase, Inhibitor
Graphical abstract
Highlights
-
•
Remdesivir triphosphate inhibits flaviviral polymerases.
-
•
Remdesivir is a low micromolar inhibitor of flaviviral polymerases.
-
•
Remdesivir has the potential to cure diseases caused by flaviviruses such as Yellow fever or Zika.
Flaviviruses are anthropod-borne single-stranded positive sense RNA viruses (+RNA viruses) that cause numerous human diseases. These pathogens are usually transmitted by mosquitoes or ticks. A genome of flaviviruses is about 11 kb long and it has only one open reading frame, which is transcribed into a single polyprotein, that is subsequently cut in three structural and seven nonstructural viral proteins either by viral or host proteases (Kok, 2016; Weaver and Reisen, 2010) Among the nonstructural proteins (NS), NS5 has a unique position since it has two very important functions connected to two distinct protein domains. It has RNA-dependent RNA polymerase (RdRp) functionality, which is connected to the C-terminus of the protein. The N-terminal domain acts as viral RNA methyltransferase. While RdRp is essential for copying + RNA to and from -RNA, the methyltransferase is responsible for RNA cap creation, which is indispensable for normal function of viral RNA and shields it from innate immunity (Dong et al., 2014; Garcia-Blanco et al., 2016; Lescar et al., 2012; Lim et al., 2015; Ray et al., 2006; Sampath and Padmanabhan, 2009). Importantly, NS5 protein possesses highly conserved drug targets shared among flaviviruses (Dubankova and Boura, 2019).
Among the numerous other flaviviruses, special attention has been paid to viruses that caused larger outbreaks in recent years. First, Dengue virus (DENV) causes approximately 390 million infections each year, of which nearly 100 million have clinical manifestations, which makes it the most common flavivirus infection globally (Behnam et al., 2016; Chan et al., 2015; Lescar et al., 2018). West Nile virus (WNV) affects numerous patients in North America, including USA, every year. Although the occurrence of WNV neuroinvasive disease is rather rare its fatality rate is around 10% (Benjelloun et al., 2016; Farrar, 2013; Kramer et al., 2007). Zika virus caused a significant epidemic in 2015 and 2016, which was associated with a significant increase in concerns over flavivirus diseases and a significant revival of research into new anti-flavivirus substances (Paixao et al., 2016; Weaver et al., 2016). Yellow fever virus (YFV) and Japanese encephalitis virus (JEV) have also continued to be a significant health problem in South and Central America and Africa, and in Asia and Oceania, respectively, despite the availability of an effective vaccine. Similarly, tick-borne encephalitis virus (TBEV) causes numerous infection in Czechia, Germany, Austria and other central European countries as well as in Russia during the period of activity of locally occurring ticks of the genus Ixodes (Chrdle et al., 2016).
Contemporary antiviral therapies are often based on nucleoside and nucleotide derivatives, which target the viral polymerase, the central enzyme of the virus. Therefore, compounds targeting RdRp are expected to also be the backbone of anti-flavivirus therapy for numerous diseases caused by members of this genus (Eyer et al., 2016a, 2016b, 2017, 2018; Hercík et al., 2017; Malet et al., 2008; Sebera et al., 2018).
Remdesivir, a monophosphate prodrug of a modified adenosine, has recently received a lot of attention due to its applicability for treatment of coronavirus infection caused by severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) (Brown et al., 2019; de Wit et al., 2020; Sheahan et al., 2017). The compound was originally developed as a potential treatment for the Ebola virus and it also proved to be active against numerous other RNA viruses including Filo-, Pneumo-, and Paramyxoviruses (Lo et al., 2017; Siegel et al., 2017; Warren et al., 2016).
Since remdesivir was reported to inhibit distinct viral polymerases we aimed to test its ability to inhibit various flaviviral polymerases. Expression plasmids encoding NS5s from TBEV, DENV3, JEV, and WNV were ordered from the European Virus Archive (EVAg), expression plasmids encoding the ZIKV and YFV NS5s were described previously (Dubankova and Boura, 2019; Hercik et al., 2017). We have expressed all the NS5s proteins in E. coli and purified them using our previous protocol developed for ZIKV NS5 protein (Hercik et al., 2017) as detailed in Supplementary Information (SI Fig. 1 ).
Fig. 1.
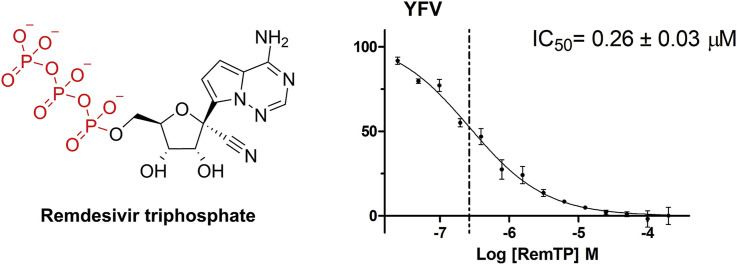
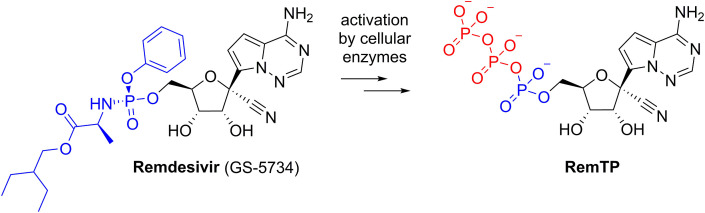
Remdesivir and remdesivir triphosphate. Remdesivir (left) is enzymatically transformed to remdesivir triphosphate (RemTP) upon human cell entry.
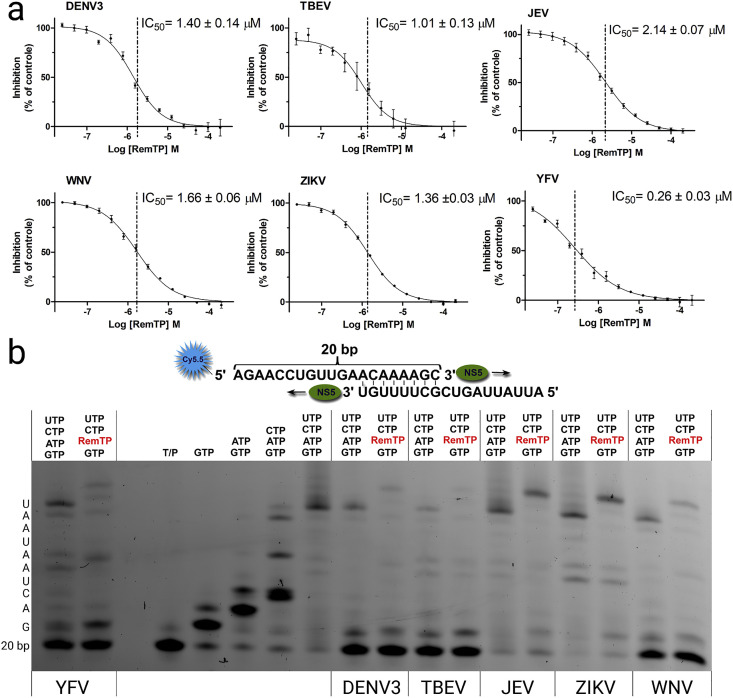
We aimed to test remdesivir directly in a polymerase assay in vitro. The prodrug form of remdesivir is not suitable because the active form is remdesivir triphosphate (RemTP) unveiled enzymatically from remdesivir once remdesivir is inside mammalian cells (Warren et al., 2016). We have used a previously published protocol to synthetize RemTP chemically (Cho et al., 2012) and obtained sufficient amounts for our experiments. Next, we performed the polymerase assay using the alkaline phosphatase-coupled polymerase assay (FAPA) (Niyomrattanakit et al., 2011) as detailed in SI. The assay revealed that remdesivir inhibits all the tested flaviviral polymerases with an IC50 in the range 0.2–2.2 μM. The best IC50 value (0.26 ± 0.03 μM) was observed for the YFV polymerase. All the other polymerases tested (DENV3, TBEV, JEV, WNV, ZIKV) were inhibited similarly with IC50 ranging from 1.3 to 2.2 μM (Fig. 2 A). Next we used a gel-based RNA polymerization assay to gain deeper insight into the remdesivir mechanism of action. We expected to observe the delayed termination of the transcription mechanism of action that was reported for Ebola and coronavirus polymerases (Gordon et al., 2020a, Gordon et al., 2020b; Tchesnokov et al., 2019). However, all six flaviviral polymerases tested were able to incorporate several (five) remdesivir molecules into the newly synthesized RNA chain and finish the primer elongation reaction (Fig. 2B). We also noted that the 20 bp band corresponding to the primer disappeared in the case of JEV and ZIKV polymerase, we attribute it to endonuclease activity of these polymerases under these reactions conditions (3 mM MnCl2), which was not observed under the same conditions but in 1 mM MnCl2 (SI Fig. 2), however, we cannot exclude the possibility that JEV and ZIKV enzymes were contaminated by an exonuclease that is active only in 3 mM MnCl2. Our results confirm and explain the observed effect of remdesivir on the TBEV in cell culture-based experiments (Lo et al., 2017) albeit they suggest that the mechanism of action, in case of flaviviruses, cannot be ascribed to delayed chain termination.
Fig. 2.
Inhibitions of selected flaviviral polymerases by remdesivir triphosphate. A) IC50 values were established for each flaviviral RdRp tested using the fluorescence based alkaline phosphatase-coupled polymerase assay. B) Gel-based polymerase assay - The incorporation of RemTP incorporation was monitored for each flaviviral RdRp tested (substrate: GTP, RemTP, CTP, UTP) and compared with the natural condition (substrate: GTP, ATP, CTP, UTP). Left - polymerization reaction performed with YFV polymerase with four NTP s followed by a reaction performed with RemTP instead of ATP, next is just the template/primer (T/P) followed by control reactions performed with JEV polymerase in the presence of indicated NTPs, followed by comparison of reactions with ATP/RemTP using indicated flaviviral polymerases. 20 bp band corresponds to unreacted primer, the sequence of nucleotides added by the polymerase is shown on the left side of the gel.
Remdesivir is known to be a potent inhibitor of Ebola RdRp (Tchesnokov et al., 2019) and several coronaviral RdRps (Brown et al., 2019; Gordon et al., 2020a, Gordon et al., 2020b). In this study, we show that remdesivir also efficiently inhibits flaviviral RdRps using pure recombinant RdRps and RemTP. The IC50s range from hundreds of nanomolar to single digit micromolar suggesting that flaviviral polymerases are a weaker target for remdesivir than the coronaviral polymerases where the reported IC50 values are in the tens of nanomoles (Gordon et al., 2020a). Taken together, our data show that remdesivir is a single-digit-micromolar inhibitor of flaviviral RdRps and suggest that its structure, which was originally designed to combat Ebola, could be optimized to combat multiple, if not all, diseases caused by flaviviruses.
Acknowledgment
The project was supported by the Czech Health Reseach Council (Registration No NU20-05-00472). This publication was supported by the European Virus Archive goes Global (EVAg) project that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 653316. The work was also supported from European Regional Development Fund; OP RDE; Project: “Chemical biology for drugging undruggable targets (ChemBioDrug)" (No. CZ.02.1.01/0.0/0.0/16_019/0000729) and by the Academy of Sciences of the Czech Republic (RVO: 61388963). We are grateful to Dr. A. Michael Downey (Max Planck Institute of Colloids and Interfaces) for critical reading of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104899.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Behnam M.A.M., Nitsche C., Boldescu V., Klein C.D. The medicinal chemistry of dengue virus. J. Med. Chem. 2016;59:5622–5649. doi: 10.1021/acs.jmedchem.5b01653. [DOI] [PubMed] [Google Scholar]
- Benjelloun A., El Harrak M., Belkadi B. West Nile disease epidemiology in North-west Africa: bibliographical review. Transboundary and Emerging Diseases. 2016;63:e153–e159. doi: 10.1111/tbed.12341. [DOI] [PubMed] [Google Scholar]
- Brown A.J., Won J.J., Graham R.L., Dinnon K.H., 3rd, Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019;169:104541. doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.W.K., Watanabe S., Kavishna R., Alonso S., Vasudevan S.G. Animal models for studying dengue pathogenesis and therapy. Antivir. Res. 2015;123:5–14. doi: 10.1016/j.antiviral.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Cho A., Saunders O.L., Butler T., Zhang L., Xu J., Vela J.E., Feng J.Y., Ray A.S., Kim C.U. Synthesis and antiviral activity of a series of 1'-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg. Med. Chem. Lett. 2012;22:2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrdle A., Chmelík V., Růžek D. Tick-borne encephalitis: what travelers should know when visiting an endemic country. Hum. Vaccines Immunother. 2016;12:2694–2699. doi: 10.1080/21645515.2016.1218098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H.P., Fink K., Zust R., Lim S.P., Qin C.F., Shi P.Y. Flavivirus RNA methylation. J. Gen. Virol. 2014;95:763–778. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- Dubankova A., Boura E. Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antivir. Res. 2019;169:104536. doi: 10.1016/j.antiviral.2019.104536. [DOI] [PubMed] [Google Scholar]
- Eyer L., Nencka R., de Clercq E., Seley-Radtke K., Růžek D. Nucleoside analogs as a rich source of antiviral agents active against arthropod-borne flaviviruses. Antiviral Chem. Chemother. 2018;26 doi: 10.1177/2040206618761299. 2040206618761299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyer L., Nencka R., Huvarova I., Palus M., Alves M.J., Gould E.A., De Clercq E., Ruzek D. Nucleoside inhibitors of Zika virus. JID (J. Infect. Dis.) 2016;214:707–711. doi: 10.1093/infdis/jiw226. [DOI] [PubMed] [Google Scholar]
- Eyer L., Šmídková M., Nencka R., Neča J., Kastl T., Palus M., De Clercq E., Růžek D. Structure-activity relationships of nucleoside analogues for inhibition of tick-borne encephalitis virus. Antivir. Res. 2016;133:119–129. doi: 10.1016/j.antiviral.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Eyer L., Zouharová D., Širmarová J., Fojtíková M., Štefánik M., Haviernik J., Nencka R., de Clercq E., Růžek D. Antiviral activity of the adenosine analogue BCX4430 against West Nile virus and tick-borne flaviviruses. Antivir. Res. 2017;142:63–67. doi: 10.1016/j.antiviral.2017.03.012. [DOI] [PubMed] [Google Scholar]
- Farrar F. West Nile virus: an infectious viral agent to the central nervous system. Crit. Care Nurs. Clin. 2013;25:191–203. doi: 10.1016/j.ccell.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco M.A., Vasudevan S.G., Bradrick S.S., Nicchitta C. Flavivirus RNA transactions from viral entry to genome replication. Antivir. Res. 2016;134:244–249. doi: 10.1016/j.antiviral.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercik K., Kozak J., Sala M., Dejmek M., Hrebabecky H., Zbornikova E., Smola M., Ruzek D., Nencka R., Boura E. Adenosine triphosphate analogs can efficiently inhibit the Zika virus RNA-dependent RNA polymerase. Antivir. Res. 2017;137:131–133. doi: 10.1016/j.antiviral.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Hercík K., Kozak J., Šála M., Dejmek M., Hřebabecký H., Zborníková E., Smola M., Ruzek D., Nencka R., Boura E. Adenosine triphosphate analogs can efficiently inhibit the Zika virus RNA-dependent RNA polymerase. Antivir. Res. 2017;137:131–133. doi: 10.1016/j.antiviral.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Kok W.M. New developments in flavivirus drug discovery. Expet Opin. Drug Discov. 2016;11:433–445. doi: 10.1517/17460441.2016.1160887. [DOI] [PubMed] [Google Scholar]
- Kramer L.D., Li J., Shi P.Y. West Nile virus. Lancet Neurol. 2007;6:171–181. doi: 10.1016/S1474-4422(07)70030-3. [DOI] [PubMed] [Google Scholar]
- Lescar J., Lim S.P., Shi P.-Y. Structure and function of the flavivirus NS5 protein. Molecular Virology and Control of Flaviviruses. 2012:101–117. [Google Scholar]
- Lescar J., Soh S., Lee L.T., Vasudevan S.G., Kang C.B., Lim S.P. The dengue virus replication complex: from RNA replication to protein-protein interactions to evasion of innate immunity. Dengue and Zika: Control and Antiviral Treatment Strategies. 2018;1062:115–129. doi: 10.1007/978-981-10-8727-1_9. [DOI] [PubMed] [Google Scholar]
- Lim S.P., Noble C.G., Shi P.Y. The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malet H., Masse N., Selisko B., Romette J.L., Alvarez K., Guillemot J.C., Tolou H., Yap T.L., Vasudevan S., Lescar J., Canard B. The flavivirus polymerase as a target for drug discovery. Antivir. Res. 2008;80:23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Niyomrattanakit P., Abas S.N., Lim C.C., Beer D., Shi P.Y., Chen Y.L. A fluorescence-based alkaline phosphatase-coupled polymerase assay for identification of inhibitors of dengue virus RNA-dependent RNA polymerase. J. Biomol. Screen. 2011;16:201–210. doi: 10.1177/1087057110389323. [DOI] [PubMed] [Google Scholar]
- Paixao E.S., Barreto F., Teixeira M.D., Costa M.D.N., Rodrigues L.C. History, epidemiology, and clinical manifestations of Zika: a systematic review. Am. J. Publ. Health. 2016;106:606–612. doi: 10.2105/AJPH.2016.303112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Shah A., Tilgner M., Guo Y., Zhao Y.W., Dong H.P., Deas T.S., Zhou Y.S., Li H.M., Shi P.Y. West nile virus 5 '-cap structure is formed by sequential guanine N-7 and ribose 2 '-O methylations by nonstructural protein 5. J. Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath A., Padmanabhan R. Molecular targets for flavivirus drug discovery. Antivir. Res. 2009;81:6–15. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebera J., Dubankova A., Sychrovsky V., Ruzek D., Boura E., Nencka R. The structural model of Zika virus RNA-dependent RNA polymerase in complex with RNA for rational design of novel nucleotide inhibitors. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-29459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11 doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S.C., Costa F., Garcia-Blanco M.A., Ko A.I., Ribeiro G.S., Saade G., Shi P.Y., Vasilakis N. Zika virus: history, emergence, biology, and prospects for control. Antivir. Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S.C., Reisen W.K. Present and future arboviral threats. Antivir. Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.