Highlights
-
•
Estimation of the isoelectric point and size of Vero cell-derived Orf virus.
-
•
Limited dynamic binding capacity of tested Orf virus to sulfated cellulose.
-
•
Purification of Orf virus by steric exclusion chromatography lead to 84 % recovery.
-
•
Hydrophobic interaction chromatography suitable for Orf virus purification.
-
•
Promising unit operations for a scalable DSP to produce Orf virus viral vectors.
Abbreviations: DBC, dynamic binding capacity; DLS, dynamic light scattering; HIC, hydrophobic interaction chromatography; HICP, HIC membrane adsorber with phenyl ligand; IEX, ion exchange chromatography; IEX-Q, strong anion exchanger (Quaternary ammonium); IEX-S, strong cation exchanger (Methyl sulfonate); IEX-STPA, salt tolerant polyamide anion exchanger; IU, infective units; MVA, Modified Vaccinia Ankara virus; PEG, polyethylene glycol; pI, isoelectric point; SCMA, sulfated cellulose membrane adsorber; SEC, size exclusion chromatography; SXC, steric exclusion chromatography; TCID50, fifty-percent tissue culture infective dose
Keywords: Parapoxvirus, Viral vector, Steric exclusion chromatography, Isoelectric point
Abstract
In recent years, the Orf virus has become a promising tool for protective recombinant vaccines and oncolytic therapy. However, suitable methods for an Orf virus production, including up- and downstream, are very limited. The presented study focuses on downstream processing, describing the evaluation of different chromatographic unit operations. In this context, ion exchange-, pseudo-affinity- and steric exclusion chromatography were employed for the purification of the cell culture-derived Orf virus, aiming at a maximum in virus recovery and contaminant depletion. The most promising chromatographic methods for capturing the virus particles were the steric exclusion- or salt-tolerant anion exchange membrane chromatography, recovering 84 % and 86 % of the infectious virus. Combining the steric exclusion chromatography with a subsequent Capto™ Core 700 resin or hydrophobic interaction membrane chromatography as a secondary chromatographic step, overall virus recoveries of up to 76 % were achieved. Furthermore, a complete cellular protein removal and a host cell DNA depletion of up to 82 % was possible for the steric exclusion membranes and the Capto™ Core 700 combination.
The study reveals a range of possible unit operations suited for the chromatographic purification of the cell culture-derived Orf virus, depending on the intended application, i.e. a human or veterinary use, and the required purity.
1. Introduction
The first viral vector-based therapy drugs have already received marketing approval, and numerous products are at different stages of clinical and preclinical testing to treat a variety of both inherited and acquired diseases (Shirley et al., 2020). Among the currently used viral vectors for gene therapy and vaccines, recombinant poxviruses have a large packaging capacity and a favorable safety and efficacy (Pastoret and Vanderplasschen, 2003; Verheust et al., 2012). One example of these viral vectors is the Orf virus (ORFV), a member of the genus Parapoxvirus of the family Poxviridae. It contains a linear double-stranded DNA genome of approx. 140 kbp in length, and is an enveloped ovoid-shaped virus measuring approximately 260 nm x 160 nm (Nagington et al., 1964). The envelope is surrounded by a characteristic tubule-like structure in a spiral fashion, resembling a ball of wool (Spehner et al., 2004).
Previously, we reported the successful use of the attenuated ORFV for the generation of different recombinants with diverse modifications able to protect against various infectious viral diseases (Rohde et al., 2011, 2013; Rziha et al., 2016; van Rooij et al., 2010). Also other ORFV strains were successfully used for the generation of recombinant vaccines (Hain et al., 2016; Tan et al., 2012), and inactivated ORFV showed immunomodulatory properties in different preclinical models (Bergqvist et al., 2017; Fleming et al., 2015; Wang et al., 2019). Furthermore, Rintoul et al. demonstrated an oncolytic activity for the wild-type ORFV strain NZ2, causing a significantly reduced tumor growth in immune-competent and xenograft human tumor models (Rintoul et al., 2012). These and other reports show the excellent potential of the ORFV vector and its broad field of possible applications (Amann et al., 2013; Friebe et al., 2018; O’Leary et al., 2018).
Knowledge on ORFV production processes is very limited and mainly focuses on upstream processing (Pohlscheidt et al., 2008; Wang et al., 2019). Downstream processing (DSP) procedures generally employ sucrose gradient ultracentrifugation (Rziha et al., 2016) and do not satisfy regulatory demands for the purity of vaccines and gene therapeutic products (European Pharmacopoeia, 2020; World Health Organization, 2017). In order to identify promising chromatographic methods for the purification of ORFV, a direct comparison of the methods used for the DSP of other poxviruses could indicate a feasible approach. For the Modified Vaccinia Ankara virus (MVA), the utilization of anion exchange chromatography (Wolff et al., 2010b) or hydrophobic interaction chromatography (Wolff et al., 2010a) has been reported and could be carried forward for the ORFV purification. Alternatively, Wolff et al. described the successful purification by pseudo-affinity stationary phases based on sulfated carbohydrates with virus yields exceeding 60 % (Wolff et al., 2010a, b) for MVA. Similar methods have previously been shown for the purification of Influenza A virus, using sulfated-cellulose membranes as a stationary phase (Carvalho et al., 2018; Fortuna et al., 2018, 2019) with recoveries of up to 81 %. As Scagliarini and co-workers described the affinity of ORFV to heparin (Scagliarini et al., 2004), the application of sulfated cellulose ligands appears possible, as sulfated carbohydrates are known to mimic heparin ligands (Gallagher et al., 2020; Paluck et al., 2016). Generally, as for nearly all larger macromolecules and nanoplexes, size-dependent chromatographic methods, e.g. size exclusion chromatography (SEC) or steric exclusion chromatography (SXC), should be applicable for ORFV. Examples for the purification of viruses by SEC are the turkey coronavirus (Loa et al., 2002), vesicular stomatitis virus-pseudotyped retrovial particles (Transfiguracion et al., 2003), and the Influenza virus (Heyward et al., 1977; Kalbfuss et al., 2007b; Nayak et al., 2005). More recently, SXC has also been proven to be a valuable tool for the purification of large proteins and bacteriophages (Gagnon et al., 2014; Lee et al., 2012; Tao et al., 2015; Wang et al., 2014). Furthermore, it was successfully applied for the purification of the Influenza A virus (Marichal-Gallardo et al., 2017) and baculovirus particles (Lothert et al., 2020), using regenerated cellulose membranes. Moreover, the Capto™ Core 700 resin (named CC700 hereafter) has also been described as an approach for the purification of infectious virus particles, allowing an efficient impurity removal (James et al., 2016).
Here, we describe the evaluation of different chromatographic applications that can be used for the purification of the ORFV strain D1701-V. One aim was to identify feasible unit operations that have a high specificity for the virus. Secondly, it is desired to use these methods, either individually or in a combination, for a virus purification with regard to product yields and respective purity requirements. Among the tested methods are the membrane-based ion exchange-, hydrophobic interaction-, pseudo-affinity-, and the steric exclusion chromatography, as well as CC700.
2. Materials and methods
2.1. Virus production and initial clarification
For all chromatographic capture steps, a clarified virus harvest was required. Virus amplification was done in an adherent cell culture. Briefly, Vero cells (ATCC, CCL-81) were seeded at 2 × 104 cells per cm2 in T-225 CytoOne® flasks (STARLAB International). Infection was done at a multiplicity of infection of 0.05. After five days, the supernatant was harvested and frozen/thawed (−80 °C, 22 °C) for a cell disruption and a subsequent virus release. For an initial clarification, the supernatant was centrifuged at 4 °C and 6000 x g for 10 min. The clarified virus suspension was used for following chromatographic steps. For a reliable comparability, one production batch was used for all subsequent experiments.
2.2. Screening of feasible chromatographic process unit operations
The chromatographic experiments were done using an Äkta™ Pure 25 liquid chromatography system (GE Healthcare Life Sciences) with UV (280 nm), and conductivity monitoring was carried out at room temperature. Additionally, the light scattering signal was measured online with a Nano DLS Particle Size Analyzer (Brookhaven Instruments). All buffers were filtered with a 0.2 μm filter (Corning) and degassed by ultra-sonication (USC500 THD, VWR) prior to usage.
2.2.1. SXC
The SXC was performed exclusively as a capture step. Regenerated cellulose membranes with a nominal pore size of 1 μm (Whatman RC60, GE Healthcare Life Sciences) were applied as stationary phases. The membranes were punched to discs of 13 or 24 mm and assembled into stainless steel filter holders with 13 mm (#4042; PALL Life Sciences) or 25 mm diameter (XX4502500, Merck), respectively. For each device, 10 membrane layers were stacked, providing a total surface area of 13.3 cm² (∼0.09 mL) for the 13 mm device, and 45.2 cm² (0.32 mL) for the 24 mm membranes. At first, the membrane stack was equilibrated, using either PBS (Thermo Fisher Scientific) or 20 mM TRIS-HCl (Carl Roth) at a pH of 7.4 containing 8 % polyethylene glycol (PEG) 8000 (Carl Roth). The TRIS buffer was additionally supplemented with 180 mM NaCl (Carl Roth). The clarified virus harvest was conditioned with PBS, or 20 mM TRIS-HCl buffer (pH 7.4, 180 mM NaCl) containing an appropriate PEG concentration to meet the criteria of the equilibration buffer. Samples were applied using either a 10 mL or a 150 mL super-loop, depending on the employed membrane volume. After sample application, the column was washed with equilibration buffer (at least 30 column volumes) and the virus was finally eluted using PBS, or 20 mM TRIS-HCl buffer pH 7.4 without PEG, but supplemented with 0.4 M NaCl (Carl Roth). A new stack of membranes was used for every chromatographic experiment. For the characterization of the SXC performance, the 13 mm filter holder was operated with a PBS buffer at a flow rate of 0.5 mL min−1. To evaluate subsequent purification steps, larger amounts of SXC-purified material were prepared. Therefore, the 25 mm filter holder module was used, and TRIS-buffer was employed during loading, wash, and elution to reduce buffer exchanges between process units. Furthermore, these runs were conducted at a flow rate of 3 mL min−1 to reduce processing time. The chromatographic experiments were carried out three times, and the elutions were pooled, aliquoted, and frozen at −80 °C until further usage. In parallel, freeze/thaw stabilities in the elution buffer were evaluated for four freeze/thaw cycles after SXC purification (see Section 2.5).
2.2.2. Ion exchange chromatography (IEX)
The IEX was tested for both the capture and the second chromatographic purification step. Therefore, Sartobind® S devices were tested for the cation exchange chromatography (named IEX-S hereafter), and Sartobind® Q (IEX-Q) and Sartobind® STIC®-PA (IEX-STPA) were applied as an anion exchanger and salt-tolerant anion exchanger, respectively (all membrane devices were “pico”-scale modules obtained from Sartorius Stedim Biotech). For all IEX experiments, the columns were equilibrated with 20 mM TRIS-HCl pH 7.4, supplemented with 180 mM NaCl. The samples were mixed with the appropriate buffer to meet these conditions. Afterwards, the samples were applied and the columns subsequently washed with equilibration buffer, until the UV- and light scattering signal reached the baseline. Using 2 M NaCl in 20 mM TRIS-HCl pH 7.4, the bound fraction was finally eluted. In the case of the STPA membrane, an additional 150 mM sodium phosphate (Sigma-Aldrich) was added to the elution buffer. For all steps, a flow rate of 1 mL min−1 was applied. The Q and the STPA membranes were tested as a capture step. Furthermore, the SXC-purified material was processed with all described anion exchange membranes for a secondary purification step. All anion exchange experiments were performed in triplicates. Cation exchange experiments were performed only once for both processing steps, as the isoelectric point (pI) of the ORFV (see Section 3.1) suggests a low binding potential for the virus with a high level of impurities in the flow-through fraction at the given buffer conditions.
2.2.3. Hydrophobic interaction chromatography (HIC)
Sartobind® Phenyl Pico membrane modules (Sartorius Stedim Biotech GmbH) were used for the secondary chromatography of SXC-purified material (named HICP hereafter). Using similar conditions as previously described for Modified Vaccinia Ankara virus (MVA, (Wolff et al., 2010a), the column was equilibrated with 20 mM TRIS-HCl pH 7.4, supplemented with 180 mM NaCl and 1.7 M ammonium sulfate (VWR International GmbH). SXC eluates were adjusted to these conditions by adding the required amounts of salts. After sample loading, the membrane stack was washed, maintaining the high salt concentration. The elution was achieved by removing the ammonium sulfate from the system, using 20 mM TRIS-HCl pH 7.4 and 180 mM NaCl. HICP-secondary chromatographic steps were performed in triplicates, using flow rates of 1 mL min−1.
2.2.4. Pseudo-affinity chromatography (sulfated cellulose)
Sulfated cellulose membrane adsorbers (SCMA) were used in a bind-and-elute mode, and tested for capture and secondary purification. SCMA membranes were obtained as a DIN A4 format sheet (#94SC--04-001, Sartorius Stedim Biotech) and punched to discs of 13 mm diameter. The discs were assembled into the 13 mm stainless steel filter holder mentioned above, with stacks comprising 10 membrane layers. Both the clarified virus broth (capture) and SXC-purified material (secondary purification) were subjected to SCMA purification with triplicate runs for each case. Prior to chromatography, the respective sample was mixed with 20 mM TRIS-HCl pH 7.4 in order to reduce its conductivity below 5 mS cm−1. Conductivities were in the range between 3–4.2 mS cm−1 for all runs. After the equilibration of the membrane stack with 20 mM TRIS-HCl pH 7.4, the sample was applied and subsequently washed with equilibration buffer. The elution was achieved by using an equilibration buffer supplemented with 2 M NaCl. Each experiment was conducted in triplicates at a flow rate of 1 mL min−1 during all runs and steps of the experiments.
2.2.5. CC700 purification
As a secondary purification, the CC700 resin (readily packed 1 mL columns, GE Healthcare Life Sciences) was tested in the flow-through mode. The general method equals the conditions for the ion exchange membranes (see Section 2.2.2). The SXC-purified material was adjusted to meet the conditions of the equilibration buffer, which was 20 mM TRIS-HCl pH 7.4 and 180 mM NaCl, respectively. After sample loading and washing with the equilibration buffer, the bound material was eluted, using additional 2 M NaCl in the same buffer. All CC700 runs were done in triplicates, using a flow rate of 1 mL min−1.
2.3. Dynamic binding capacity (DBC) determination
DBC experiments were performed for all membranes operated in the bind-and-elute mode. The virus feed of a known concentration was loaded onto the column until a complete breakthrough was observed, based on the light scattering detection. The amount of virus at 10 % breakthrough was subsequently calculated in relation to the maximum signal. Where necessary, sample loading was stopped prior 100 % breakthrough, if the pressure exceeded the maximum operational limit for the tested membrane modules or the system. For a better comparability, capacities were provided in relation to the applied bed volume.
2.4. Characterization of the virus
SXC-purified virus particles were analyzed for their size and isoelectric point. Therefore, SXC virus elutions were subjected to size (diameter) and to surface potential measurements, using a Zetasizer Nano ZS90 (Malvern Panalytical). For the diameter measurements, disposable semi-micro cuvettes (#67.742, Sarstedt) were used at a 90° light scattering angle. The dispersant refractive index and viscosity of the dispersant were set to 1.45 and 0.954 cP, respectively. For the zeta potential measurement, the sample was mixed with 20 mM Tris at pH values between 3 and 13. The pH was checked and adjusted to the respective value prior to the measurement, and reassessed after analysis. Folded capillary zeta cells (#DTS1070, Malvern) were applied to determine the viral zeta potentials at different pH values. The data analysis was conducted with the Malvern Panalytical Zetasizer Software (version 7.12).
2.5. Virus stability evaluation
Additionally, the virus stability was evaluated by using SXC elution fractions (see Section 2.2.1). The samples were supplemented with 0 %, 5 % or 10 % sucrose (Carl Roth) and subjected to up to three freeze/thaw cycles (alternating between -80 and +20 degrees). After each cycle, the infective virus titer was determined (see Section 2.6.1) for each sample composition in triplicates.
2.6. Analytics of chromatographic fractions
Chromatographic fractions were analyzed for their content of infective virus particles as well as with regard to the levels of protein and DNA. The chromatographic fractions considered during analytics included feed, flow-through, wash, and elution for the evaluation of bind-and-elute methods (SXC, IEX-Q, IEX-STPA, HICP and SCMA). In contrast, the flow-through and wash fractions were pooled and analyzed as a single fraction when using IEX-S and CC700.
2.6.1. Flow cytometric titration
Virus quantification was done by flow cytometry. Initially, Vero cells were seeded in a 24-well format with 100,000 cells per well in a culture volume of 1 mL DMEM (Gibco™, Thermo Fisher Scientific), supplemented with 5 % FCS (Capricorn Scientific). Directly after seeding the cells, the infection of each well was performed using 100 μL of the respective chromatographic sample, the medium blank, or standard virus stock (within the range of 1.5 × 105 to 1 × 107 infective units (IU) mL−1). The cells were incubated for 16 h, washed with PBS, and harvested by using Trypsin/EDTA (Gibco™, Thermo Fisher Scientific). Detached cells were supplemented with FCS to stop a trypsin activity, and transferred to a 96-well U-bottom plate (Nunc, Thermo Fisher Scientific). The cells were washed two times by centrifugation at 400 g for 2 min, removing the supernatant, and re-suspending the pellet in 100 μL PBS. Finally, the cells were analyzed using a flow cytometer (Guava® easyCyte HT, Merck). The percentage of GFP positive (equals infected) cells, compared to the total cell number, was determined. The assays’ standard deviation was less than 10 %.
2.6.2. Total protein assay
The content of total protein was evaluated using the Pierce™ BCA Protein Assay Kit (#23225; Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, 25 μL of each sample were mixed with 200 μL of working reagent and incubated at 37 °C for 30 min in a 96-well plate (Nunc, Thermo Fisher Scientific). Afterwards, the absorbance was measured at 562 nm, using a plate reader (BioTek™ Cytation 3™, Fisher Scientific). Bovine gamma globulin (contained in kit) was applied to prepare a standard calibration curve in the range of 20−2,000 μg mL−1 with a relative standard deviation of about 10 %.
2.6.3. DNA assay
The total double-stranded DNA (referred to as “DNA” in this work) content was quantified, using the Quant-iT™ PicoGreen® dsDNA Kit (#P11496, Thermo Fisher Scientific) according to the manufacturer’s instructions. Samples were analyzed in duplicates in a 96-well format, using black plates (Nunc, Thermo Fisher Scientific) and the same plate reader as for the total protein assay (see Section 2.6.2). Each plate contained two standard calibration curves prepared from λ-DNA contained in the kit in the range of 0.025 ng mL−1 to 25 ng mL−1 (low-range) and 1 ng mL−1 to 1000 ng mL−1 (high-range). Fluorescence excitation was at 480 nm, and the emission intensity was measured at 520 nm, with an assay standard deviation of less than 15 %.
3. Results
3.1. Virus characterization
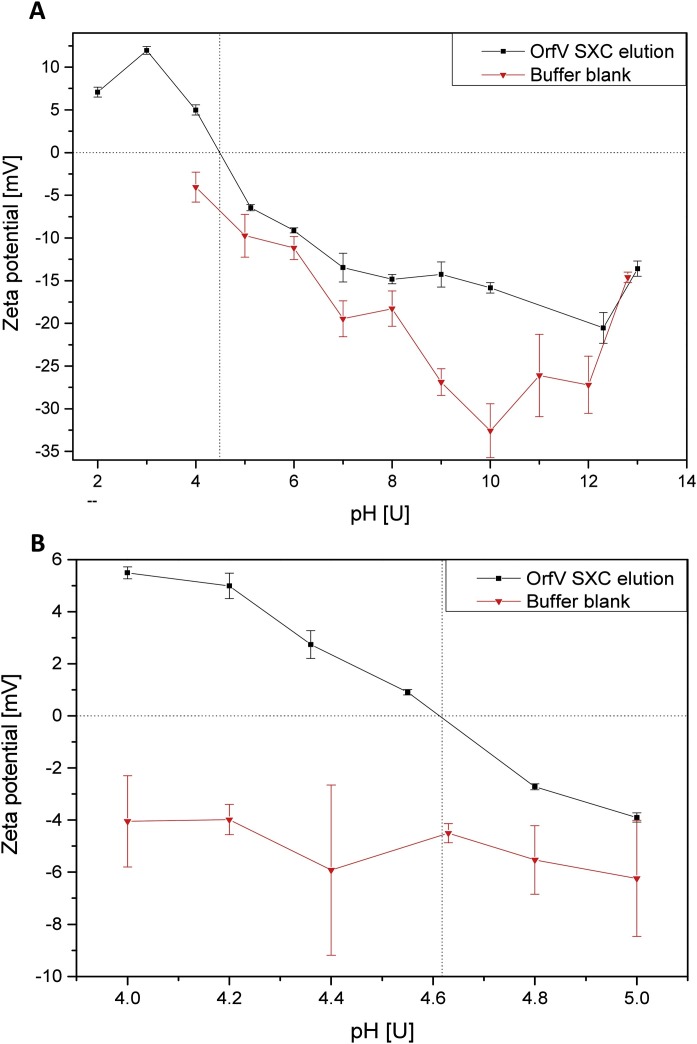
DLS measurements of SXC-purified virus particles showed a size distribution between 130 nm and 370 nm with two main size populations at about 150 ± 14 nm and 250 ± 32 nm. Zeta potential measurements detected a negative viral surface charge above pH values of 5, and a positive surface charge at a pH below or equal to 4 (Fig. 1 A). A more detailed analysis of the pH range between 4 and 5 revealed the transition from a positive to a negative surface charge at pH 4.6, which was not detected for the negative control (Fig. 1B). Hence, the viruses’ isoelectric point was determined with pH 4.6 under the given environmental conditions.
Fig. 1.
Determination of the isoelectric point of the Orf virus (ORFV). Zeta potential measurements of virus purified by steric exclusion chromatography (SXC) and the corresponding buffer over a pH range from 2 to 13 (A), and more detailed resolution for the apparent transition from positive to negative zeta potential between pH 4 and 5 (B). At a pH of 4.61 the net charge is zero. Error bars represent standard deviations of triplicate measurements.
3.2. Determination of the dynamic binding capacities
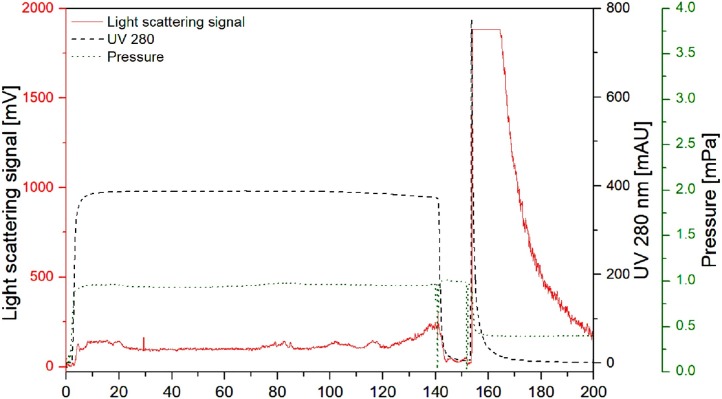
The DBC was determined for all bind-and-elute methods at similar conditions, using a clarified harvest. The highest DBC10 % was measured for the SXC, using the 24 mm filter holder with 1.1E + 09 IU mL−1 (7.78E + 06 IU cm-2), and for IEX-Q membranes with 5.93E + 08 IU mL−1 (1.63E + 07 IU cm-2, Table 1 ). Compared to the 24 mm device used for SXC, the 13 mm filter holder allowed a DBC10 % of 1.81E + 08 IU mL−1 (1.27E + 06 IU cm-2) during SXC, depicting a non-linear correlation between the filter area (volume resp.) and the binding capacity. Comparable results could be determined for the IEX-STPA, HICP and SCMA membranes, with 1.42E + 08, 1.31E + 08, and greater than 1.26E + 08 IU mL−1 (3.91E + 06, 3.57E + 06 and greater than 8.40E + 05 IU cm-2), respectively. A representative chromatogram is shown in Fig. 2 for the SXC (24 mm membranes), indicating a contaminant breakthrough during sample loading (UV signal), and a virus desorption in the elution fraction (light scattering signal). During loading of the HICP, at 10 % breakthrough, the pressure increased rapidly above 0.5 MPa, which is the operating limit of the HICP according to the manufacturers´ instructions. For the SCMA, a constant breakthrough of 20 % of the maximum signal was observed (data not shown). As a result, a 10 % breakthrough was permanently exceeded during the loading process, and sample loading was stopped once the pressure increased above one MPa.
Table 1.
Overview of the DSP-screening for infective ORFV particles. Different stationary phases were tested for virus and impurity recovery. Capture steps were conducted with clarified virus harvests, whereas SXC-purified material was used for secondary steps. The numbers in brackets show an overall recovery of the respective analyte after secondary chromatography, employing SXC as a capture step. For bind-and-elute methods (SXC, IEX-Q, IEX-STPA, HIC-P, SCMA), the DBC is given in infective units per cm². The DBC was not evaluated with regard to methods operated in a flow-through mode, as no binding of the virus was supposed to occur. Recovery values are means of technical triplicates.
| SXC | IEX-Q | IEX-STPA |
IEX-S |
HICP | CC700 | SCMA |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Capture | SP | Capture | SP | Capture | SP | SP | SP | Capture | SP | ||
| Recovery in product fraction [%] | Virus | 84 | 68 (57) | 86 | 57 (48) | 39 | 46 (39) | 76 (64) | 90 (76) | 34 | 54 (45) |
| Total protein | <1 | 0 | 29 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | |
| DNA | 37 | 100 (37) | 64 | 100 (37) | 89 | 84 (31) | 23 (8) | 36 (13) | 5 | 80 (30) | |
| DBC10% [Infective Units mL−1] |
1.81E + 08 (13 mm) 1.1E + 09 (24 mm) |
5.93E + 08 | 1.42E + 08 | Not determined (flow-through method) |
1.31E + 08 | Not determined (flow-through method) |
>1.26E + 08 | ||||
SXC – Steric exclusion chromatography; IEX – Ion exchanger; STPA – Salt tolerant polyamide; HICP – Hydrophobic interaction chromatography with phenyl ligand; CC700 – Capto™ Core 700; SCMA Sulfated cellulose membrane adsorber; DBC10 % – dynamic binding capacity (at 10 % breakthrough), SP – Secondary purification.
Fig. 2.
Determination of the dynamic binding capacity (DBC) during SXC. Representative chromatogram for the SXC using the 25 mm filter holder device, monitoring of UV at 280 nm, light scattering, and pressure. Loading was stopped at 10 % DBC based on light scattering detection.
3.3. Capture step – virus recovery and impurity removal
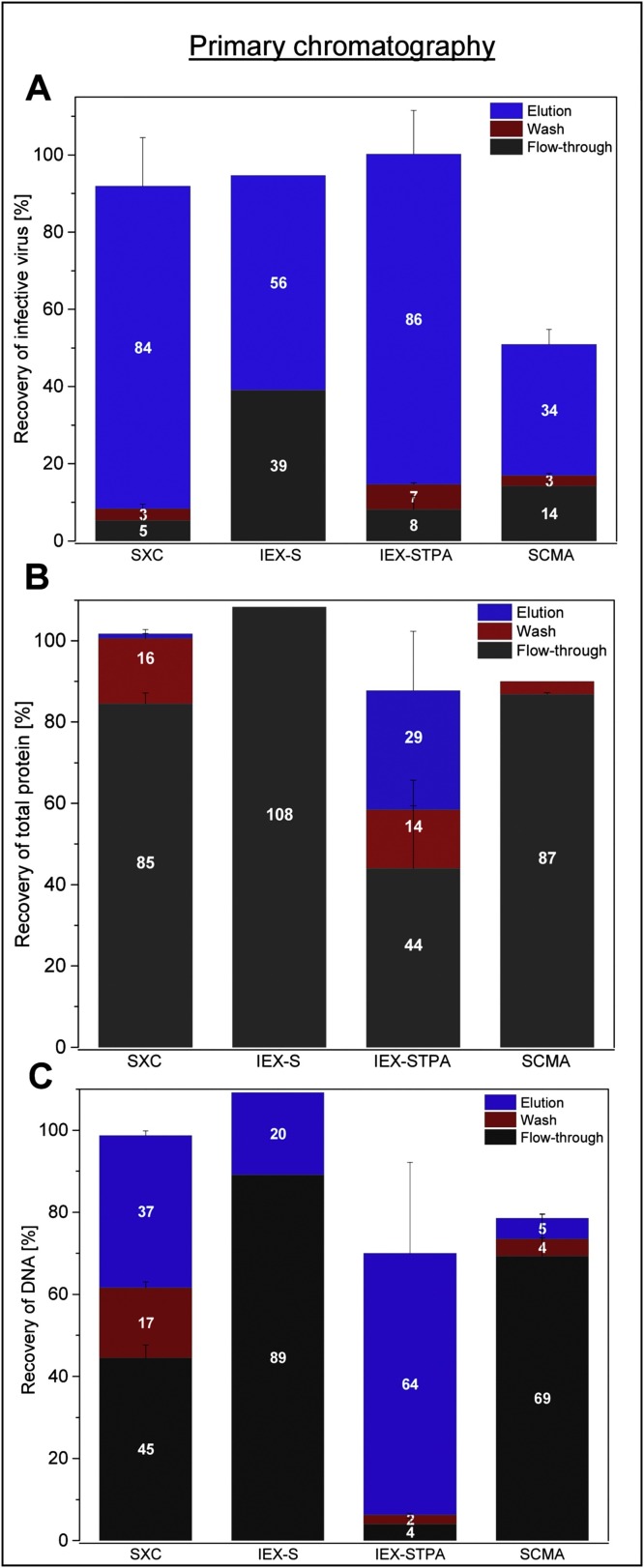
Of the four tested membranes for the capture step (SXC, IEX-S, IEX-STPA and SCMA), the best virus recovery was achieved by SXC and STPA with viral yields of 84 % and 86 %, respectively (Fig. 3 , A). In contrast, using the SCMA or the IEX-S membrane, only 34 % and 39 % of the infective virus particles were collected in the product fraction. Additionally, for the SCMA, only approximately 51 % of the virus could be recovered collectively in all chromatographic fractions, whereas the remaining 49 % could not be accounted for. For the cation exchanger (IEX-S), it has to be noted that the majority of the viruses was found in the elution fraction (56 %) after an application of 2 M NaCl, and not in the flow-through.
Fig. 3.
Virus and impurity recovery for different unit operations during capture. Shown are the relative recoveries for ORFV (A), protein (B) and DNA (C) using SXC, cation exchange membrane adsorbers (IEX-S), anion exchangers (IEX-STPA) and sulfated cellulose membrane adsorbers (SCMA). Depicted are the quantities contained in the flow-through, wash and elution fractions, whereas for the IEX-S (operated in flow-through-mode), flow-through and wash fractions are combined. Error bars reflect the standard deviation of technical triplicates, except for the IEX-S membrane, which was only tested once to show the proof of concept.
Concerning the protein removal, SXC and SCMA performed equally with an almost complete protein depletion (Fig. 3, B). In contrast, almost no protein was removed in the IEX-S flow-through fraction, and about 29 % were found in the STPA elution.
The best unit operation to remove DNA during the capture step is the SCMA, with only 5 % of the initial DNA content to be detected in the product fraction, and 95 % of the DNA being removed during flow-through and wash or remaining bound to the membrane (Fig. 3, C). For the SXC, about 37 % of the DNA were recovered in the elution. Furthermore, most of the contaminating DNA remained in the product fractions of the IEX-S (89 %) and the IEX-STPA membrane (64 %). About 30 % of the DNA was bound to the IEX-STPA membrane and could not be eluted under any of the tested conditions.
3.4. Secondary purification step – virus recovery and impurity removal after SXC
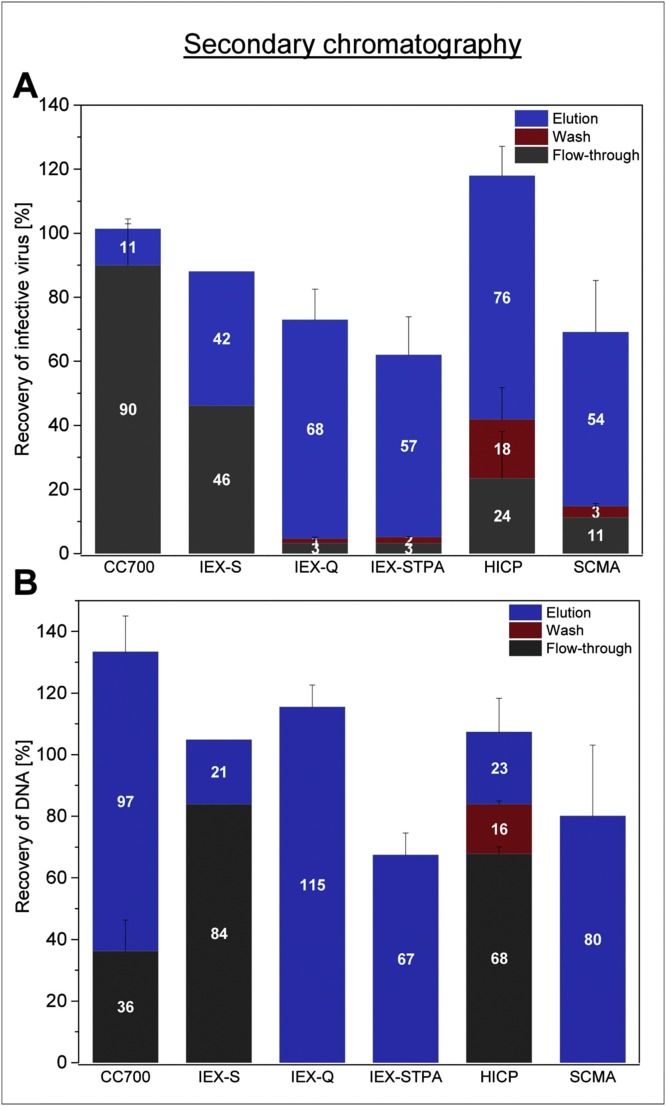
After SXC, a subsequent purification step was evaluated. In the course of these experiments, the highest virus recovery was achieved using CC700 with about 90 % (Fig. 4 , A). The second-best performing unit operation was the HICP membrane adsorber, resulting in virus yields of about 76 % (elution fraction) and in about 42 % of losses in the flow-through and wash fractions. Similar to the capture step using the IEX-S purification, approximately half of the virus amount was found in the flow-through product fraction, whereas the other half bound to the membrane and eluted at 2 M NaCl. The tested anion exchange membranes all performed similarly. Approximately 68 % of the virus was found in the product fraction for the Q membrane adsorber, and 57 % for the STPA membrane adsorber. Notably, a distinct amount of virus could not be eluted at 2 M NaCl, with roughly 30 % and 40 % not being recovered from the IEX-Q and IEX-STPA membranes adsorbers, respectively. The SCMA showed less retained viruses on the device than in the previous capture step. However, the overall recovery of all fractions was at about 70 %, with 54 % of the viruses being detected in the product fraction.
Fig. 4.
Virus and impurity recovery for different unit operation during secondary chromatography. Shown are the relative recoveries for ORFV (A) and DNA (B) during secondary purification, evaluating the Capto™ Core 700 resin (CC700), IEX-S, strong and salt-tolerant anion exchange membrane adsorbers (IEX-Q and IEX-STPA), as well as hydrophobic interaction membrane adsorbers (HICP) and the SCMA. Depicted are the quantities contained in the flow-through, wash and elution fractions, whereas for methods in the flow-through-mode (CC700 and IEX-S), flow-through and wash fractions are combined. Error bars reflect the standard deviation of technical triplicates, (IEX-S membrane was only tested once). Protein recovery is not shown for the secondary purification as already after SXC capture no protein quantities were remaining.
For none of the applications used for the secondary purification, proteins could be determined in any fraction.
The most efficient DNA depletion was achieved using the CC700 and the HICP membrane adsorber, with 36 % and 23 % of the initial DNA amount remaining in the product fraction, respectively, (Fig. 4, B). For all other applications, most of the contained DNA was co-eluted with the product virus, with DNA amounts of 84 % (S-membrane), more than 100 % (IEX-Q), 67 % (STPA), and 80 % (SCMA) being left in the product fraction. Following Q, STPA, and SCMA purification, no DNA was found during flow-through and wash. Last it should be noted that for STPA and SCMA the material balance could not be closed with 33 % (STPA), and up to 20 % (SCMA) of the DNA remaining unaccounted for.
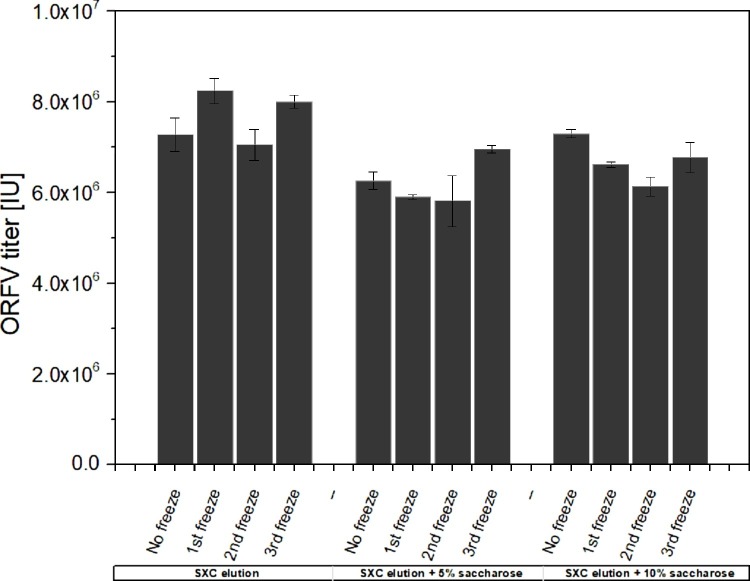
3.5. Virus stability
Virus particles after SXC purification were stable for at least four freeze/thaw cycles, without any loss of infectivity (Fig. 5 ). An additional application of sucrose did not affect the viruses’ infectivity during freezing at −80 °C.
Fig. 5.
ORFV stability after freeze/thawing with and without sugar supplementation. Infective ORFV titers in SXC elution buffer (20 mM TRIS-HCL pH 7.4 with 0.4 M NaCl), supplemented with 0 %, 5 % and 10 % sucrose. The quantification was done by flow cytometry, directly after the purification and after each of up to three freeze/thaw cycles. Error bars indicate the standard deviation of technical triplicates.
3.6. Overview and summary
In summary, the combinations of SXC and CC700 or HICP showed the highest overall virus recovery of approximately 76 % and 64 % after both chromatography steps, respectively. Furthermore, these combinations offered a complete protein removal, and final DNA concentrations of 24 ng mL−1 (CC700) and 47 ng mL-1 (HICP) at virus titers of 2.0E + 06 IU mL−1 and 1.1E + 06 IU mL−1, respectively.
4. Discussion
Medical applications of viral vectors for gene therapy and vaccines are continuously increasing (Shirley et al., 2020). One promising viral vector that is currently undergoing preclinical evaluations, the ORFV, lacks an efficient and economic production process. In this study, we focused on its downstream processing and, in particular, on the development of chromatographic methods for the purification of the cell culture-derived ORFV.
4.1. Virus characterization
So far, the pI has not yet been determined for ORFV strain D1701-V particles. Previously reported values for vaccinia viruses range between 2.3 and 5.9, depending on the analytical method and on the virus strain analyzed (Douglas et al., 1966, 1969; Resch et al., 2007; Taylor and Bosmann, 1981; Wolff and Reichl, 2011). Hence, the determined pI of 4.6 for ORFV is within the expected range. The same applies for the determined main size populations after SXC purification of 150 and 250 nm, representing width and length of the ORFV in accordance with literature (Nanington et al. 1964).
4.2. Determination of the dynamic binding capacities
In general, membrane adsorbers have been widely used for the purification of viruses and virus-like particles due to their superior performance, especially in view of their flow properties and binding capacities for nanoplexes (Hoffmann et al., 2019). In this study, we evaluated ion exchange-, pseudo-affinity-, and hydrophobic interaction chromatography membrane adsorbers, as well as size-dependent techniques such as the CC700 and SXC.
At present, the membrane-based SXC was only described for Influenza A (Marichal-Gallardo et al., 2017) and baculovirus (Lothert et al., 2020) purifications, yielding dynamic binding capacities of up to 1.2E + 07 plaque-forming units cm−2, which exceed our observations for the ORFV (1.63E + 06 IU cm−2 to 7.78E + 06 IU cm−2). In the presented study, two membrane holders with a diameter of 13 mm and 25 mm (24 mm membranes) were tested with a corresponding volumetric increase between the two membrane holders by a factor of 3.7. The amount of retained viruses between the two membrane volumes was increased by a factor of approximately 6 (Table 1). The slight deviation between the volumetric increase and the retained amount of virus suggests a potential non-linear behavior when scaling up the SXC. However, this could be explained by the error of the analytical method for virus quantification, and the differences in the membrane housing devices in terms of the assembly and housing materials. For the assembly, both require a sealing ring in order to prevent a leakage. This O-ring is relatively large for the 13 mm device, probably resulting in an inefficient perfusion of the membrane stack. This could reduce the accessible membrane volume, thus reducing the overall capacity compared to the larger filter holder. Additionally, most of the interactions for the SXC occur in the upper layers of the membrane stack (unpublished data), suggesting, that a comparison of the membrane areas accessible for the virus particles might be more appropriate. However, this cannot be reliably determined.
The 4.2-fold higher DBC observed for the Q membrane adsorber, in comparison to the STPA membrane adsorber (Table 1), was not expected. Based on the manufacturer´s product sheet, the ligand density is nearly six times higher for the STPA membrane than for the Q membrane, resulting in BSA binding capacities of 0.8 mg cm−2 (Q membrane) and 1.4 mg cm-2 (STPA). However, for virus molecules as large as the ORFV particles, an increased ligand density might not affect the binding capacity, but rather the binding strength, as multiple ligands bind to a single virus particle.
Anion exchange membranes were frequently applied for the purification of different viruses with varying results on binding capacities. For example, Grein et al. reported total capacities on similar Q membranes of 1.7E + 08 plaque-forming units per cm² using a recombinant baculovirus (Grein et al., 2012) whereas McNally et al. captured retroviral vectors on Mustang Q membranes with 1.2E + 08 IP per mL membrane (McNally et al., 2014). Hence, data on binding capacities is frequently difficult to compare. Reasons for this are differences in virus morphology and surface properties, varying analytical quantification methods, and the deviations between membrane adsorbers from different manufactures. However, the data we obtained can be considered within the same range as described by Grein et al. and McNally et al. For MVA, Wolff et al. achieved binding capacities of greater than 1.2E + 07 fifty-percent tissue culture infective dose (TCID50) per cm² using Q and D anion exchangers (Wolff et al., 2010b), which were also exceeded for the Q device.
For the SCMA, a constant breakthrough of a small virus fraction was observed by the light scattering detector from the moment that sample loading started (data not shown). This was confirmed by offline analytics, as approximately 10 %–15 % of the viruses were found in the flow-through fraction. Previous publications described the application of SCMA for the purification of the Influenza A virus, Influenza virus-like particles, and MVA (Carvalho et al., 2018; Fortuna et al., 2018, 2019; Opitz et al., 2009; Wolff et al., 2010a), with the virus content varying between 2.4 % and 30 % in the flow-through fraction during the loading procedure. The precise interaction of different viruses and sulfated cellulose or sulfated dextran is currently not yet fully understood. However, several studies reported antiviral activities (Chattopadhyay et al., 2008; Mitsuya et al., 1988; Nelson and Rosowsky, 2002; Piret et al., 2000) and the binding of viruses to sulfated cellulose (O’Neil and Balkovic, 1993), suggesting that sulfated cellulose can mimic a heparin or heparin sulfate ligand (Gallagher et al., 2020; Paluck et al., 2016). The purification of ORFV via a heparin affinity chromatography has already been described in literature (Scagliarini et al., 2002, 2004), thus, ORFV could be suitable for SCMA purification. Scagliarini and co-workers reported the ORF virus’ F1L protein being mainly responsible for heparin binding (Scagliarini et al., 2004). It is located on the viral envelope, presenting a glycosaminoglycan binding motive (Lin et al., 2000; Scagliarini et al., 2002). During the life cycle of the ORFV, different infectious progeny of ORFV exist with altered surface structures and properties (Spehner et al., 2004; Tan et al., 2009), and F1L could not be detected on all progenies (Tan et al., 2009). For a complete virus recovery, the host cells were disrupted after harvesting, releasing all progeny regardless of their maturity level, which might explain the diminished performance during SCMA for these virus particles. Besides, for Influenza virus particles, Fortuna et al. showed a dependency of the SCMA performance on the virus titer of the feed stream and on its ionic strength (Fortuna et al., 2018, 2019). Different virus titers were not examined, therefore no statement can be made on this. However, the conductivity of the tested feed stream (3.5–4 mS cm−2) matched their recommendation, and therefore should not have influenced the adsorption behavior of the virus during the sample loading process.
4.3. Evaluation of the capture step
Due to the estimated isoelectric point of the virus and the applied buffer pH of 7.4, the IEX-S membrane was intended to be used in flow-through mode (contaminant adsorption). In contrast to the expectation, about 50 % were retained in the applied neutral pH process condition and only eluted at higher (2 M NaCI) salt concentrations (Fig. 3). Virus purification, using cation- and anion exchangers under comparable process conditions, has been described repeatedly in the literature for various cell culture-derived viruses (Wolff and Reichl, 2011), such as the Influenza A virus (Opitz et al., 2009), the adeno-associated virus (Okada et al., 2009), and for MVA (Wolff et al., 2010b), where virus particles can be adsorbed up to a certain degree to both types of ion exchangers. Based on the varying protein compositions, it can be assumed that individual patches on the virus surface have different physicochemical properties, leading to a complex adsorption behavior.
The tested anion exchange membrane adsorber resulted in a virus recovery for the IEX-STPA of 86 % during primary chromatography (Fig. 3). Anion exchange chromatography is widely used for virus purifications, such as adenoviruses (Nestola et al., 2015) and the Influenza A virus (Weigel et al., 2016). Generally, the main disadvantage of the method is the co-elution of process-related nucleic acids with the product virus (Wolff and Reichl, 2011). As a result, the obtained virus yield depends on the level of DNA depletion and must be optimized with regard to the economics of the entire production process.
During the capture of the ORFV from the clarified cell culture material by SCMA, the losses, which were observed in the DBC studies throughout column loading, were confirmed (Fig. 3). In the course of the capture experiments, virus losses throughout loading and wash amounted to 17 %. This was also the case for the secondary chromatographic purification via SCMA, where 14 % of the virus was detected in the flow-through and wash fraction (see Sections 3.4 and 4.4). However, during the SCMA capture only 51 % (Fig. 3) of the virus could be recovered in total, excluding this method as a potential capture step. For the SCMA purification of MVA, Wolff et al. described virus yields ranging from 65 % to 75 %, while at the same time achieving a DNA and protein depletion greater than 90 % and 95 %, respectively (Wolff et al., 2010b, a). MVA also belongs to the poxvirus family and, thus, allows a limited basis for comparison despite certain differences in morphology. For the capturing step in the presented study, the protein and DNA depletions are comparable to the studies of Wolff et al. According to the literature (Fortuna et al., 2019; Wolff et al., 2010b) however, the virus recovery is below expectations. This might be attributed to aspects already discussed in 3.2. Besides, it could also be due to the fact, that custom-made membranes with pore sizes of 3−5 μm were employed in the cited MVA study. Here, we used commercially available membranes with 0.8 μm pores, leading to an increased filtration effect, and thus, losses in the total virus recovery.
4.4. Evaluation of the secondary chromatographic step after SXC
After the purification of the virus particles by SXC, several membrane adsorbers (IEX-S, IEX-Q, IEX-STPA, HICP and SCMA) as well as the CC700 resin were tested for a subsequent chromatographic step to remove residual contaminants.
For the cation exchange membrane adsorber, a comparable performance of the secondary chromatography and the capture step (see Sections 3.3 and 3.4) was achieved. Nearly half of the total virus amount was not retained, and the product fractions did not show a sufficient contaminant depletion (Fig. 4). Thus, the cation exchange membrane adsorber was not suitable for a purification process of Vero cell culture-derived ORFV under the tested conditions. The use of anion exchangers as a secondary chromatographic step resulted in lower virus yields compared to the capture step. More accurately, the virus yield was reduced by 29 % (Fig. 3, Fig. 4) for the IEX-STPA membrane adsorber. Compared to the IEX-Q membrane adsorber, though, the IEX-STPA membrane adsorber had an increased ligand density and virus losses were insignificantly higher. The yields for the Q membrane adsorber were comparable to values previously described for other virus applications, such as for Influenza A (77–86 %, (Kalbfuss et al., 2007a), Parvovirus-like particles (59 %, (Ladd Effio et al., 2016)) and MVA (77 %, (Wolff et al., 2010b)). Thus, virus losses in the course of anion exchange chromatography have already been described previously. The above-mentioned higher virus losses, using the IEX-STPA in the secondary step compared to the capture step (complete virus recovery, Fig. 3), could not have been caused by an infectivity loss as the process conditions were the same in both approaches. Instead, the effect might be explained by a shielding effect of contaminants in the solution, preventing interactions of viruses with the surface ligands (Weigel et al., 2016), with higher contaminant levels during the primary chromatography. Under the applied elution buffer conditions, the DNA could not be separated from the virus in any of the anion exchange methods, resulting in DNA levels above 120 ng mL−1, thus rendering these methods to be less suitable for the ORFV purification. If possible at all, an extensive method optimization for an appropriate functionality would be required.
While the absence of contaminating proteins reduces the performance of the anion exchangers, the opposite effect is observable for the SCMA membrane. When applied as a secondary step, the total virus recovery is approximately 17 % higher than during capture (Table 1, Fig. 3, Fig. 4). This increase is mainly attributed to a higher amount of virus being eluted from the membrane adsorber, as the amounts in flow-through and wash were unchanged. Possibly, the membrane fouling effect is reduced when performed as a secondary purification in the absence of proteins. As described in 3.3. the membranes’ pores are at 0.8 μm, generally leading to higher filtration effects than in previous reports on MVA with larger pore membranes (Wolff et al., 2010b, a). Thus, the filtration effects could be increased at higher protein concentrations during the capture step. Total virus recoveries in all fractions accounted for 68 %, ruling out SCMA for an ORFV purification.
Concerning the HICP membrane, high losses (about 40 %) were observed during sample loading and wash (Fig. 4). HIC processes for virus purification are less well characterized and applied than the use of anion exchange methods. However, Weigel et al. described a resin-based HIC purification for Influenza A with > 90 % virus yield and about 99 % DNA removal (Weigel et al., 2019). Furthermore, also MVA was previously purified with the HICP membranes as a secondary purification, yielding up to 76 % of the virus (Wolff et al., 2010a), matching the data of 76 % virus in the elution pool that we obtained (Fig. 4). However, the DNA removal was higher in the referred study, with less than 1 % of the initial DNA content being found in the product (23 % in this work). Relative DNA amounts are difficult to compare, and no values on absolute DNA concentrations are given in the published data. In the presented study, the final DNA concentration in the product fraction was about 47 ng mL−1 (containing total virus amounts of 1.1E + 06 IU). Thus, when considering this unit operation for process implementation, a further optimization of the buffer composition must be evaluated. The virus recovery in the elution could potentially be increased for higher ammonium sulfate concentrations in the binding buffer, presuming that the virus activity is not affected. The total amount of recovered virus in all fractions can be attributed to the analytical error, accounting up to 15 % for each fraction.
All IEX and HIC methods described here, were performed using elution buffer conditions that support a high virus recovery using high salt concentrations (2 M NaCl). As under these circumstances most of the DNA is co-eluted with the virus, the product purity was not optimized. However, the intention here was the identification of possible chromatographic methods for the purification of ORFV, and optimizations would have exceeded the scope of this study.
Among the techniques tested as secondary chromatographic methods, CC700 allowed the highest DNA removal with only 24 ng mL−1 of the DNA remaining in the product fraction at virus titers of 2.0E + 06 IU mL−1. Accordingly, this combination offers the highest potential for a further process optimization in order to comply with a production process in agreement with regulatory guidelines (European Pharmacopoeia, 2020; World Health Organization, 2017). The use of CC700 and HICP both showed a high virus yield (90 % and 76 %) and a high DNA depletion in the product fraction (64 % and 77 %, Fig. 4). The latter is based on a total DNA assay, which needs to be further characterized for a production process in order to discriminate between viral DNA and host cell DNA. Furthermore, the combination SXC and CC700 or HICP allowed a protein depletion below the quantification limit (25 μg/mL) of the applied assay and, thus, meets the requirements of the regulatory guidelines. Nonetheless, it will be necessary for a production process to further characterize the remaining proteins in order to determine their source.
As carried out in the experiments shown here, the final arrangement of the two chromatographic purification methods should start with the SXC. This method is mainly independent of the loading- and the elution buffer, reducing the need for additional buffer exchanges, which would be necessary for HICP (Weigel et al., 2019; Wolff et al., 2010a). Furthermore, SXC allows a concentration of the virus (Marichal-Gallardo et al., 2017), whereas the CC700 rather leads to a sample dilution. Also the polishing effect of the CC700 resin might be reduced, if overburdened with higher contaminant amounts of the clarified feed (James et al., 2016).
The experiments described here were all conducted on a laboratory scale. However, most of the evaluated techniques are membrane-based and available, either commercially in different scales, or can be custom-made from commercial flat sheet membranes (SXC, SCMA) to accommodate the required scale. The only applied resin-based technique, the CC700, is used to bind and remove remaining impurities in a flow-through mode for the virus. It is therefore uncritical for upscaling.
4.5. Evaluation of the virus stability during freeze/thaw cycles
The data suggests that the virus can be stored at −80 °C after the first chromatographic purification, without affecting the virus stability for up to three freeze/thaw cycles (Fig. 5). Additionally, a supplementation of 5% or 10 % sucrose is not required and can be omitted. Previously, the addition of stabilizers, such as sugars, was described to be beneficial for storage and formulation of various viruses (Adebayo et al., 1998; Kumru et al., 2018). In our case, an intermediate storage and freeze/thawing in a SXC elution buffer (20 mM Tris pH 7.4, 180 mM NaCl) was uncritical; for a long term storage, though, additional studies are required.
5. Conclusion
This study evaluates the general performance and applicability of different chromatographic unit operations for the purification of cell culture-derived ORFV. In summary, anion exchangers, HIC, and SXC are suitable possibilities with satisfying product recoveries and contaminant depletions. Cation exchange membrane chromatography and SCMA appeared less eligible for such a process. SXC and IEX-STPA membranes present the most promising capture steps with 84 % and 86 % virus recovery, respectively, whereas the impurity removal was better for SXC (greater than 99 % protein and 67 % DNA reduction). The combination of SXC chromatography with a subsequent CC700 or HIC membrane adsorption chromatography resulted in an overall virus yield in the two combined chromatographic steps in 90 % and 76 %, respectively. Hence, these unit operations or the combination of the SXC capture step with a CC700 or HIC chromatography are promising candidates for a DSP of cell culture-derived ORFV. In addition, the utilized membranes or chromatographic modules are well suited for upscaling and single use applications, providing the possibility for the development of an economic production process, which will be the focus of upcoming experiments.
Funding
The work was supported by the EXIST-Forschungstransfer grand # 03EFKBW171 of the German Federal Ministry for Economic Affairs and Energy.
CRediT authorship contribution statement
Keven Lothert: Conceptualization, Methodology, Data curation, Formal analysis, Writing - original draft. Felix Pagallies: Methodology, Investigation, Writing - review & editing. Thomas Feger: Methodology, Data curation, Writing - review & editing. Ralf Amann: Supervision, Funding acquisition, Writing - review & editing. Michael W. Wolff: Supervision, Project administration, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
None.
Acknowledgements
The authors would like to thank Hanns-Joachim Rziha for scientific advice on the manuscript. Additionally, we want to thank Catherine Meckel-Oschmann and Friederike Eilts for proofreading. This work is part of a dissertation under the aegis of the Justus Liebig University of Giessen, Germany in cooperation with the Technische Hochschule Mittelhessen (University of Applied Sciences), Giessen, Germany.
References
- Adebayo A.A., Sim-Brandenburg J.W., Emmel H., Olaleye D.O., Niedrig M. Stability of 17D yellow fever virus vaccine using different stabilizers. Biologicals. 1998;26(4):309–316. doi: 10.1006/biol.1998.0157. [DOI] [PubMed] [Google Scholar]
- Amann R., Rohde J., Wulle U., Conlee D., Raue R., Martinon O., Rziha H.-J. A new rabies vaccine based on a recombinant ORF virus (parapoxvirus) expressing the rabies virus glycoprotein. J. Virol. 2013;87(3):1618–1630. doi: 10.1128/JVI.02470-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergqvist C., Kurban M., Abbas O. Orf virus infection. Rev. Med. Virol. 2017;27(4) doi: 10.1002/rmv.1932. [DOI] [PubMed] [Google Scholar]
- Carvalho S.B., Fortuna A.R., Wolff M.W., Peixoto C.M., Alves P., Reichl U., Jt Carrondo M. Purification of influenza virus-like particles using sulfated cellulose membrane adsorbers. J. Chem. Technol. Biotechnol. 2018;93(7):1988–1996. doi: 10.1002/jctb.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay K., Ghosh T., Pujol C.A., Carlucci M.J., Damonte E.B., Ray B. Polysaccharides from Gracilaria corticata: sulfation, chemical characterization and anti-HSV activities. Int. J. Biol. Macromol. 2008;43(4):346–351. doi: 10.1016/j.ijbiomac.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Douglas H.W., Rondle C.J., Williams B.L. Micro-electrophoresis of cowpox and vaccinia viruses in molar sucrose. J. Gen. Microbiol. 1966;42(1):107–113. doi: 10.1099/00221287-42-1-107. [DOI] [PubMed] [Google Scholar]
- Douglas H.W., Williams B.L., Rondle C.J.M. Micro-electrophoresis of pox viruses in molar sucrose. J. Gen. Virol. 1969;5(3):391–396. doi: 10.1099/0022-1317-5-3-391. [DOI] [PubMed] [Google Scholar]
- European Pharmacopoeia, 2020. Ed. 10.2.
- Fleming S.B., Wise L.M., Mercer A.A. Molecular genetic analysis of orf virus: a poxvirus that has adapted to skin. Viruses. 2015;7(3):1505–1539. doi: 10.3390/v7031505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortuna A.R., Taft F., Villain L., Wolff M.W., Reichl U. Optimization of cell culture-derived influenza A virus particles purification using sulfated cellulose membrane adsorbers. Eng. Life Sci. 2018;18(1):29–39. doi: 10.1002/elsc.201700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortuna A.R., van Teeffelen S., Ley A., Fischer L.M., Taft F., Genzel Y., Villain L., Wolff M.M., Reichl U. Use of sulfated cellulose membrane adsorbers for chromatographic purification of cell cultured-derived influenza A and B viruses. Sep. Purif. Technol. 2019;(226):350–358. doi: 10.1016/j.seppur.2019.05.101. [DOI] [Google Scholar]
- Friebe A., Siegling A., Weber O. Inactivated Orf-virus shows disease modifying antiviral activity in a guinea pig model of genital herpesvirus infection. J. Microbiol. Immunol. Infect. 2018;51(5):587–592. doi: 10.1016/j.jmii.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Gagnon P., Toh P., Lee J. High productivity purification of immunoglobulin G monoclonal antibodies on starch-coated magnetic nanoparticles by steric exclusion of polyethylene glycol. J. Chromatogr. A. 2014;1324:171–180. doi: 10.1016/j.chroma.2013.11.039. [DOI] [PubMed] [Google Scholar]
- Gallagher Z.J., Fleetwood S., Kirley T.L., Shaw M.A., Mullins E.S., Ayres N., Foster E.J. Heparin mimic material derived from cellulose nanocrystals. Biomacromolecules. 2020;21(3):1103–1111. doi: 10.1021/acs.biomac.9b01460. [DOI] [PubMed] [Google Scholar]
- Grein T.A., Michalsky R., Vega López M., Czermak P. Purification of a recombinant baculovirus of Autographa californica M nucleopolyhedrovirus by ion exchange membrane chromatography. J. Virol. Methods. 2012;183(2):117–124. doi: 10.1016/j.jviromet.2012.03.031. [DOI] [PubMed] [Google Scholar]
- Hain K.S., Joshi L.R., Okda F., Nelson J., Singrey A., Lawson S., Martins M., Pillatzki A., Kutish G.F., Nelson E.A., Flores E.F., Diel D.G. Immunogenicity of a recombinant parapoxvirus expressing the spike protein of Porcine epidemic diarrhea virus. J. Gen. Virol. 2016;97(10):2719–2731. doi: 10.1099/jgv.0.000586. [DOI] [PubMed] [Google Scholar]
- Heyward J.T., Klimas R.A., Stapp M.D., Obijeski J.F. The rapid concentration and purification of influenza virus from allantoic fluid. Arch. Virol. 1977;55(1-2):107–119. doi: 10.1007/BF01314484. [DOI] [PubMed] [Google Scholar]
- Hoffmann D., Leber J., Loewe D., Lothert K., Oppermann T., Zitzmann J., Weidner T., Salzig D., Wolff M., Czermak P. Purification of new biologicals using membrane-based processes. In: Basile A., Charcosset C., editors. Current Trends and Future Developments on (Bio-) Membranes. Elsevier; 2019. pp. 123–150. [DOI] [Google Scholar]
- James K.T., Cooney B., Agopsowicz K., Trevors M.A., Mohamed A., Stoltz D., Hitt M., Shmulevitz M. Novel high-throughput approach for purification of infectious virions. Sci. Rep. 2016;6:36826. doi: 10.1038/srep36826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfuss B., Wolff M., Geisler L., Tappe A., Wickramasinghe R., Thom V., Reichl U. Direct capture of influenza A virus from cell culture supernatant with Sartobind anion-exchange membrane adsorbers. J. Membr. Sci. 2007;299(1-2):251–260. doi: 10.1016/j.memsci.2007.04.048. [DOI] [Google Scholar]
- Kalbfuss B., Wolff M., Morenweiser R., Reichl U. Purification of cell culture-derived human influenza A virus by size-exclusion and anion-exchange chromatography. Biotechnol. Bioeng. 2007;96(5):932–944. doi: 10.1002/bit.21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumru O.S., Wang Y., Gombotz C.W.R., Kelley-Clarke B., Cieplak W., Kim T., Joshi S.B., Volkin D.B. Physical characterization and stabilization of a lentiviral vector against adsorption and freeze-thaw. J. Pharm. Sci. 2018;107(11):2764–2774. doi: 10.1016/j.xphs.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Ladd Effio C., Hahn T., Seiler J., Oelmeier S.A., Asen I., Silberer C., Villain L., Hubbuch J. Modeling and simulation of anion-exchange membrane chromatography for purification of Sf9 insect cell-derived virus-like particles. J. Chromatogr. A. 2016;1429:142–154. doi: 10.1016/j.chroma.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Lee J., Gan H.T., Latiff S.M.A., Chuah C., Lee W.Y., Yang Y.-S., Loo B., Ng S.K., Gagnon P. Principles and applications of steric exclusion chromatography. J. Chromatogr. A. 2012;1270:162–170. doi: 10.1016/j.chroma.2012.10.062. [DOI] [PubMed] [Google Scholar]
- Lin C.L., Chung C.S., Heine H.G., Chang W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 2000;74(7):3353–3365. doi: 10.1128/jvi.74.7.3353-3365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loa C.C., Lin T.L., Wu C.C., Bryan T.A., Thacker H.L., Hooper T., Schrader D. Purification of turkey coronavirus by Sephacryl size-exclusion chromatography. J. Virol. Methods. 2002;104(2):187–194. doi: 10.1016/S0166-0934(02)00069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lothert K., Sprick G., Beyer F., Lauria G., Czermak P., Wolff M.W. Membrane-based steric exclusion chromatography for the purification of a recombinant baculovirus and its application for cell therapy. J. Virol. Methods. 2020;275 doi: 10.1016/j.jviromet.2019.113756. [DOI] [PubMed] [Google Scholar]
- Marichal-Gallardo P., Pieler M.M., Wolff M.W., Reichl U. Steric exclusion chromatography for purification of cell culture-derived influenza A virus using regenerated cellulose membranes and polyethylene glycol. J. Chromatogr. A. 2017;1483:110–119. doi: 10.1016/j.chroma.2016.12.076. [DOI] [PubMed] [Google Scholar]
- McNally D.J., Darling D., Farzaneh F., Levison P.R., Slater N.K.H. Optimised concentration and purification of retroviruses using membrane chromatography. J. Chromatogr. A. 2014;1340:24–32. doi: 10.1016/j.chroma.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuya H., Looney D.J., Kuno S., Ueno R., Wong-Staal F., Broder S. Dextran sulfate suppression of viruses in the HIV family: inhibition of virion binding to CD4+ cells. Science. 1988;240(4852):646–649. doi: 10.1126/science.2452480. [DOI] [PubMed] [Google Scholar]
- Nagington J., Newton A.A., Horne R.W. The structure of orf virus. Virology. 1964;23(4):461–472. doi: 10.1016/0042-6822(64)90230-2. [DOI] [PubMed] [Google Scholar]
- Nayak D.P., Lehmann S., Reichl U. Downstream processing of MDCK cell-derived equine influenza virus. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005;823(2):75–81. doi: 10.1016/j.jchromb.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Nelson R.G., Rosowsky A. Dicyclic and tricyclic diaminopyrimidine derivatives as potent inhibitors of cryptosporidium parvum dihydrofolate reductase: structure-activity and structure-selectivity correlations. Antimicrob. Agents Chemother. 2002;46(3):940. doi: 10.1128/aac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestola P., Peixoto C., Villain L., Alves P.M., Carrondo M.J.T., Mota J.P.B. Rational development of two flowthrough purification strategies for adenovirus type 5 and retro virus-like particles. J. Chromatogr. A. 2015;1426:91–101. doi: 10.1016/j.chroma.2015.11.037. [DOI] [PubMed] [Google Scholar]
- O’Leary M.P., Choi A.H., Kim S.-I., Chaurasiya S., Lu J., Park A.K., Woo Y., Warner S.G., Fong Y., Chen N.G. Novel oncolytic chimeric orthopoxvirus causes regression of pancreatic cancer xenografts and exhibits abscopal effect at a single low dose. J. Transl. Med. 2018;16(1):110. doi: 10.1186/s12967-018-1483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil P.F., Balkovic E.S. Virus harvesting and affinity-based liquid chromatography. A method for virus concentration and purification. Biotechnology. 1993;11(2):173–178. doi: 10.1038/nbt0293-173. [DOI] [PubMed] [Google Scholar]
- Okada T., Nonaka-Sarukawa M., Uchibori R., Kinoshita K., Hayashita-Kinoh H., Nitahara-Kasahara Y., Takeda S.’i., Ozawa K. Scalable purification of adeno-associated virus serotype 1 (AAV1) and AAV8 vectors, using dual ion-exchange adsorptive membranes. Hum. Gene Ther. 2009;20(9):1013–1021. doi: 10.1089/hum.2009.006. [DOI] [PubMed] [Google Scholar]
- Opitz L., Lehmann S., Reichl U., Wolff M.W. Sulfated membrane adsorbers for economic pseudo-affinity capture of influenza virus particles. Biotechnol. Bioeng. 2009;103(6):1144–1154. doi: 10.1002/bit.22345. [DOI] [PubMed] [Google Scholar]
- Paluck S.J., Nguyen T.H., Maynard H.D. Heparin-mimicking polymers: synthesis and biological applications. Biomacromolecules. 2016;17(11):3417–3440. doi: 10.1021/acs.biomac.6b01147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastoret P.-P., Vanderplasschen A. Poxviruses as vaccine vectors. Comp. Immunol. Microbiol. Infect. Dis. 2003;26(5-6):343–355. doi: 10.1016/S0147-9571(03)00019-5. [DOI] [PubMed] [Google Scholar]
- Piret J., Lamontagne J., Bestman-Smith J., Roy S., Gourde P., Désormeaux A., Omar R.F., Juhász J., Bergeron M.G. In vitro and in vivo evaluations of sodium lauryl sulfate and dextran sulfate as microbicides against herpes simplex and human immunodeficiency viruses. J. Clin. Microbiol. 2000;38(1):110–119. doi: 10.1128/jcm.38.1.110-119.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlscheidt M., Langer U., Minuth T., Bödeker B., Apeler H., Hörlein H.-D., Paulsen D., Rübsamen-Waigmann H., Henzler H.-J., Reichl U. Development and optimisation of a procedure for the production of Parapoxvirus ovis by large-scale microcarrier cell culture in a non-animal, non-human and non-plant-derived medium. Vaccine. 2008;26(12):1552–1565. doi: 10.1016/j.vaccine.2008.01.032. [DOI] [PubMed] [Google Scholar]
- Resch W., Hixson K.K., Moore R.J., Lipton M.S., Moss B. Protein composition of the vaccinia virus mature virion. Virology. 2007;358(1):233–247. doi: 10.1016/j.virol.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Rintoul J.L., Lemay C.G., Tai L.-H., Stanford M.M., Falls T.J., Souza C.Tde, Bridle B.W., Daneshmand M., Ohashi P.S., Wan Y., Lichty B.D., Mercer A.A., Auer R.C., Atkins H.L., Bell J.C. ORFV: a novel oncolytic and immune stimulating parapoxvirus therapeutic. Mol. Ther. 2012;20(6):1148–1157. doi: 10.1038/mt.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J., Schirrmeier H., Granzow H., Rziha H.-J. A new recombinant Orf virus (ORFV, Parapoxvirus) protects rabbits against lethal infection with rabbit hemorrhagic disease virus (RHDV) Vaccine. 2011;29(49):9256–9264. doi: 10.1016/j.vaccine.2011.09.121. [DOI] [PubMed] [Google Scholar]
- Rohde J., Amann R., Rziha H.-J. New Orf virus (Parapoxvirus) recombinant expressing H5 hemagglutinin protects mice against H5N1 and H1N1 influenza A virus. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rziha H.-J., Rohde J., Amann R. Generation and selection of orf virus (ORFV) recombinants. Methods Mol. Biol. 2016;1349:177–200. doi: 10.1007/978-1-4939-3008-1_12. [DOI] [PubMed] [Google Scholar]
- Scagliarini A., Ciulli S., Battilani M., Jacoboni I., Montesi F., Casadio R., Prosperi S. Characterisation of immunodominant protein encoded by the F1L gene of orf virus strains isolated in Italy. Arch. Virol. 2002;147(10):1989–1995. doi: 10.1007/s00705-002-0850-2. [DOI] [PubMed] [Google Scholar]
- Scagliarini A., Gallina L., Dal Pozzo F., Battilani M., Ciulli S., Prosperi S. Heparin binding activity of orf virus F1L protein. Virus Res. 2004;105(2):107–112. doi: 10.1016/j.virusres.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Shirley J.L., de Jong Y.P., Terhorst C., Herzog R.W. Immune responses to viral gene therapy vectors. Mol. Ther. 2020;28(3):709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehner D., de Carlo S., Drillien R., Weiland F., Mildner K., Hanau D., Rziha H.-J. Appearance of the bona fide spiral tubule of ORF virus is dependent on an intact 10-kilodalton viral protein. J. Virol. 2004;78(15):8085–8093. doi: 10.1128/JVI.78.15.8085-8093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.L., Ueda N., Mercer A.A., Fleming S.B. Investigation of orf virus structure and morphogenesis using recombinants expressing FLAG-tagged envelope structural proteins: evidence for wrapped virus particles and egress from infected cells. J. Gen. Virol. 2009;90(Pt 3):614–625. doi: 10.1099/vir.0.005488-0. [DOI] [PubMed] [Google Scholar]
- Tan J.L., Ueda N., Heath D., Mercer A.A., Fleming S.B. Development of orf virus as a bifunctional recombinant vaccine: surface display of Echinococcus granulosus antigen EG95 by fusion to membrane structural proteins. Vaccine. 2012;30(2):398–406. doi: 10.1016/j.vaccine.2011.10.079. [DOI] [PubMed] [Google Scholar]
- Tao S.-P., Zheng J., Sun Y. Grafting zwitterionic polymer onto cryogel surface enhances protein retention in steric exclusion chromatography on cryogel monolith. J. Chromatogr. A. 2015;1389:104–111. doi: 10.1016/j.chroma.2015.02.051. [DOI] [PubMed] [Google Scholar]
- Taylor D.H., Bosmann H.B. Measurement of the electrokinetic properties of vaccinia and reovirus by laser-illuminated whole-particle microelectrophoresis. J. Virol. Methods. 1981;2(5):251–260. doi: 10.1016/0166-0934(81)90023-9. [DOI] [PubMed] [Google Scholar]
- Transfiguracion J., Jaalouk D.E., Ghani K., Galipeau J., Kamen A. Size-exclusion chromatography purification of high-titer vesicular stomatitis virus G glycoprotein-pseudotyped retrovectors for cell and gene therapy applications. Hum. Gene Ther. 2003;14(12):1139–1153. doi: 10.1089/104303403322167984. [DOI] [PubMed] [Google Scholar]
- van Rooij E.M.A., Rijsewijk F.A.M., Moonen-Leusen H.W., Bianchi A.T.J., Rziha H.-J. Comparison of different prime-boost regimes with DNA and recombinant Orf virus based vaccines expressing glycoprotein D of pseudorabies virus in pigs. Vaccine. 2010;28(7):1808–1813. doi: 10.1016/j.vaccine.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Verheust C., Goossens M., Pauwels K., Breyer D. Biosafety aspects of modified vaccinia virus Ankara (MVA)-based vectors used for gene therapy or vaccination. Vaccine. 2012;30(16):2623–2632. doi: 10.1016/j.vaccine.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Wang C., Bai S., Tao S.-P., Sun Y. Evaluation of steric exclusion chromatography on cryogel column for the separation of serum proteins. J. Chromatogr. A. 2014;1333:54–59. doi: 10.1016/j.chroma.2014.01.059. [DOI] [PubMed] [Google Scholar]
- Wang R., Wang Y., Liu F., Luo S. Orf virus: a promising new therapeutic agent. Rev. Med. Virol. 2019;29(1) doi: 10.1002/rmv.2013. [DOI] [PubMed] [Google Scholar]
- Weigel T., Solomaier T., Wehmeyer S., Peuker A., Wolff M.W., Reichl U. A membrane-based purification process for cell culture-derived influenza A virus. J. Biotechnol. 2016;220:12–20. doi: 10.1016/j.jbiotec.2015.12.022. [DOI] [PubMed] [Google Scholar]
- Weigel T., Soliman R., Wolff M.W., Reichl U. Hydrophobic-interaction chromatography for purification of influenza A and B virus. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019;1117:103–117. doi: 10.1016/j.jchromb.2019.03.037. [DOI] [PubMed] [Google Scholar]
- Wolff M.W., Reichl U. Downstream processing of cell culture-derived virus particles. Expert Rev. Vaccines. 2011;10(10):1451–1475. doi: 10.1586/ERV.11.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff M.W., Siewert C., Hansen S.P., Faber R., Reichl U. Purification of cell culture-derived modified vaccinia ankara virus by pseudo-affinity membrane adsorbers and hydrophobic interaction chromatography. Biotechnol. Bioeng. 2010;107(2):312–320. doi: 10.1002/bit.22797. [DOI] [PubMed] [Google Scholar]
- Wolff M.W., Siewert C., Lehmann S., Hansen S.P., Djurup R., Faber R., Reichl U. Capturing of cell culture-derived modified Vaccinia Ankara virus by ion exchange and pseudo-affinity membrane adsorbers. Biotechnol. Bioeng. 2010;105(4):761–769. doi: 10.1002/bit.22595. [DOI] [PubMed] [Google Scholar]
- World Health Organization . Repo; Geneva: 2017. WHO Expert Committee on Biological Standardization. [Google Scholar]