Abstract
Fasciola hepatica (F. hepatica) is a well-known zoonotic parasite that is crucial for economic and public health worldwide. Quantitative proteomics studies have been performed on proteins expressed by F. hepatica to investigate the differential expression of proteomes in different growth phases. And the screening of several marker proteins for use as early diagnostic antigens is essential. In this study, high-performance liquid chromatography-tandem mass spectrometry (LC-MS/MS) was conducted to analyze the differences in the expression of F. hepatica somatic proteins in different growth phases. Furthermore, gene ontology (GO) functional annotation, KEGG metabolic pathway, and clustering analyses were also performed. LC-MS/MS identified 629, 2286, 2254, and 2192 proteins in metacercariae, juvenile flukes 28dpi, immature flukes 59dpi, and adult phases, respectively. GO analysis revealed that differentially expressed proteins (DEPs) were mainly involved in transport, localization, metabolism, enzyme regulation, protein folding and binding, and nucleoside and nucleotide binding. The DEPs were enriched in cells, intracellular components, organelles, cytoplasm, vesicles, and membranes. KEGG pathway annotation results showed that the DEPs were involved in metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, and other processes. These findings provide a theoretical basis for vaccine development and establishing early diagnostic methods in the future.
Electronic supplementary material
The online version of this article (10.1007/s00436-020-06833-x) contains supplementary material, which is available to authorized users.
Keywords: Fasciola hepatica, Bioinformatics, Proteomics, High-performance liquid chromatography-tandem mass spectrometry
Introduction
Fasciola hepatica (F. hepatica) is widely distributed worldwide, especially in Latin America, Europe, and Africa. It can cause fasciolosis. It is also an important livestock pathogen and a neglected zoonotic pathogen that exerts a profound influence on the economy and public health (Furst et al. 2012; Keiser and Utzinger 2009).
The definitive hosts of F. hepatica are infected by ingesting the infective metacercariae that are immobilized in aquatic plants or floating in water. The lifecycle of F. hepatica starts by the ingestion of metacercariae that excyst within the host intestine as newly excysted juveniles (NEJ) and then migrate to the liver. After growth and development in the liver, the immature flukes migrate to the bile duct, where they develop, mature, and lay eggs. These conditions result in the destruction of liver tissue and the blockage and hardening of the bile duct, thereby affecting digestion and appetite, and causing hepatitis. The proteins secreted by worms are released into the blood, dissolve blood cells, and cause poisoning phenomena, such as anemia, weight loss, and edema in livestock (Mas-Coma et al. 2019; Mas-Coma et al. 2009).
NEJ secrete large amounts of proteins and other compounds in their intestinal tracts, excretory pores, and cyst surface during their development into adult worms. These compounds protect the parasites from host defense responses and are essential in the interaction between parasites and hosts (Bennuru et al. 2018; Hewitson et al. 2008). Proteomics analysis is used to study the interactions between parasites and hosts, especially when pathogens are difficult to obtain (Robinson and Connolly 2005; Robinson et al. 2007). Di Maggio et al. (2016) identified 575 proteins in the soluble NEJ extract, 90 proteins in NEJ E/S products, and 202 proteins in adult E/S products using proteomics analysis. Robinson et al. (2009) predicted 14,031 expressed sequence tags (ESTs) of F. hepatica secretory proteins. And the proteins secreted by infective larvae, immature flukes, and adult F. hepatica are mainly proteases and an array of antioxidants that are highly regulated in their migration through host tissues. Cwiklinski et al. (2018) found that major proteins of the total NEJ secretome include proteases, protease inhibitors, antioxidants, and an array of immunomodulators that likely to disarm host innate immune cells.
In this study, we used the label-free quantitative proteomics analysis method to determine the differentially expressed proteins (DEPs) in the metacercariae (MC), juvenile flukes 28dpi (JF), immature flukes 59dpi (IF), and adult (AD) phases of F. hepatica to further analyze the differences in somatic protein expression at different developmental stages. The somatic proteome was analyzed and revealed a variety of proteins participated in modulating the growth and development of F. hepatica and the immune response of the hosts. Meanwhile, these proteins represent key targets for future diagnosis, treatment, and vaccine development for fasciolosis.
Materials and methods
Sample preparation
MC (the infected Galba pervia were cultured under suitable conditions for 30–45 days, when cercariae escaped from the Galba pervia, the process of cyst formation was observed and the metacercariae were collected), JF (worms obtained from the livers of sheep that were artificially infected the infective metacercariae for 28 days), IF (worms taken from the livers of sheep that were artificially infected the infective metacercariae for 59 days), and AD (worms obtained from the livers of sheep that were artificially infected the infective metacercariae for 118 days) were collected and used as samples. At least three samples from each group were ground into powder in liquid nitrogen and added with 400 μl of SDT buffer (4% SDS, 1 mM DTT, 150 mM Tris-HCL, pH 8.0). Protein was extracted through 5 min of boiling and 2 min of sonication on ice. The crude extract was boiled again for 5 min and centrifuged at 4 °C for 20,000×g to obtain the supernatant. Protein concentration was determined through the BCA method.
SDS-page
The protein extracted using the method described in Section 2.1 was mixed with SDS-PAGE loading buffer (5×) (Beyotime Biotechnology, China) and boiled for 5 min. A total of 20 μg of the boiled sample was run on 5% concentrated gel for 30 min at 80 V and then on 12% separated gel at 120 V until the strip reached the bottom. The gel was stained with Coomassie Brilliant Blue R-250 (Beyotime Biotechnology, China) for 10 min and washed with a destaining solution. Afterward, images were captured with an image scanner (GS800, Bio-Rad).
Protein digestion
Protein digestion was implemented in accordance with the filter-aided sample preparation method described in previous literature (Cox and Mann 2008; Cox et al. 2014). To prepare somatic extracts, worms were crushed and homogenized in PBS buffer with sterile pestle and mortar on ice at 4 °C. After centrifuging at 4 °C and 10,000g for 30 min, the supernatant of the solutions were collected and stored until required (Afshin et al. 2019). Exactly 300 μg of the somatic protein sample was diluted with 30 μl of solution containing 4% SDS, 100 mM Tris-HCl pH 8.0, and 100 mM dithiothreitol then heated at 95 °C for 5 min. After each sample was cooled to room temperature, the sample was loaded onto an ultrafiltration filter (cutoff 10 kDa; Sartorius, Goettingen, Germany), and 200 μl of UT buffer (8 M Urea and 150 mM Tris-HCl, pH 8.0) was added to the filter and centrifuged at 12,000×g for 15 min. Subsequently, 100 μl of iodoacetamide solution (50 mM iodoacetamide in UT buffer) was added, and the sample was incubated at room temperature in the dark for 30 min then centrifuged at 12,000×g for 10 min. The filter was washed by adding 100 μl of UT buffer and centrifuging at 12,000×g for 10 min. This step was repeated twice. Then, 100 μl of NH4HCO3 buffer was added to the filter and centrifuged at 12,000×g for 10 min (also repeated twice). Subsequently, 40 μl of trypsin buffer (6 μg of trypsin in 40 μl of dissolution buffer) was added, and the samples were incubated at 37 °C for 16–18 h. Each filter unit was transferred to a new tube and centrifuged at 12,000×g for 10 min. The filtrate was collected, and an appropriate amount of 0.1% TFA solution was added to the filtrate. After enzymatic hydrolysis, a C18 Cartridge was used for desalination and vacuum drying for liquid chromatography-mass spectrometry (LC-MS) analysis.
LC–tandem MS (LC-MS/MS) analysis
Lyophilized powder was re-dissolved with 0.1% FA, and the resulting peptide concentration was estimated at OD280. Exactly 5 μg of the peptide mixture was loaded onto Thermo Easy-nLC 1200 (Thermo Finnigan, San Jose, CA, USA) for further separation and identification. A column (75 μm × 150 mm, 3 μm) was balanced with 95% solution A (0.1% formic acid). The samples were added to the balanced column and separated with a linear gradient of solution B (80% acetonitrile with 0.1% v/v formic acid) at a flow rate of 300 nL/min over 120 min as follows: 0–2 min with 4–7% solution B, 2–92 min with 7%–20% solution B, and 92–120 min with 20–35% solution B. The peptides were separated and subjected to data-dependent acquisition mass spectrometry by using a Q-Exactive Plus mass spectrometer (Thermo Scientific). Data acquisition was conducted in positive ion mode with a selected mass range of 350–1800 m/z. Q-Exactive survey scans were set as 70,000 at 200 m/z for MS scans and 17,500 at 200 m/z for HCD spectra. MS/MS data were acquired using the top 15 most abundant precursor ions. The dynamic exclusion of the selected precursor ions was set at 30 s.
Protein identification and quantification
Protein identification and quantification were conducted as previously described (Cox and Mann 2008; Cox et al. 2014). Protein identification was performed using the Max Quant 1.6.0.16 mass spectral database and by searching against the Uniport protein database of F. hepatica (liver fluke) (downloaded on 2019/05/10, containing 15,315 protein items). The Max Quant search software analysis parameter settings are shown in Table 1. The quantified proteins in each group were analyzed through one-way ANOVA, and Tukey’s test was implemented to evaluate the differences between groups. P < 0.05 and fold-charge >1.5 were defined as significant.
Table 1.
MaxQuant software analysis parameter list
| Item 2 | Value |
|---|---|
| Enzyme | Trypsin |
| Max missed cleavages | 2 |
| Main search peptide tolerance | 4.5 ppm |
| First search peptide tolerance | 20 ppm |
| MS/MS tolerance | 20 ppm |
| Fixed modifications | Carbamidomethyl (C) |
| Variable modifications | Oxidation (M), Acetyl (Protein N-term) |
| Database | uniport-Fasciola hepatica (Liver fluke)-15315-20190510 |
| Database pattern | Target-Reverse |
| PSM (peptide-spectral matching) FDR | 0.01 |
| Protein FDR | 0.01 |
| Protein quantification | Razor and unique peptides were used for protein |
| Quantification | LFQ True |
| LFQ min. ratio count | 1 |
| Match between runs | True |
Bioinformatic and multivariate analysis
The annotated functions of the somatic proteome of F. hepatica were analyzed by applying gene ontology (GO) annotation software (http://david.abcc.ncifcrf.gov/home.jsp). The identified proteins were subjected to principal component analysis (PCA) by utilizing Unscrambler software (Camo, version 9.8, Norway). The identified proteins were hierarchically clustered with Cluster 3.0 software, and pathway analysis of the identified proteins was conducted using the KEGG pathway database (http://www.genome.jp/kegg). Protein–protein interactions were evaluated using the STRING database (http://string-db.org/).
Data availability
The mass spectrometry proteomics data have been deposited to the iProX with the dataset identifier IPX0002165000 (https://www.iprox.org//page/project.html?id=IPX0002165000).
Results
Statistical analysis of identified proteins
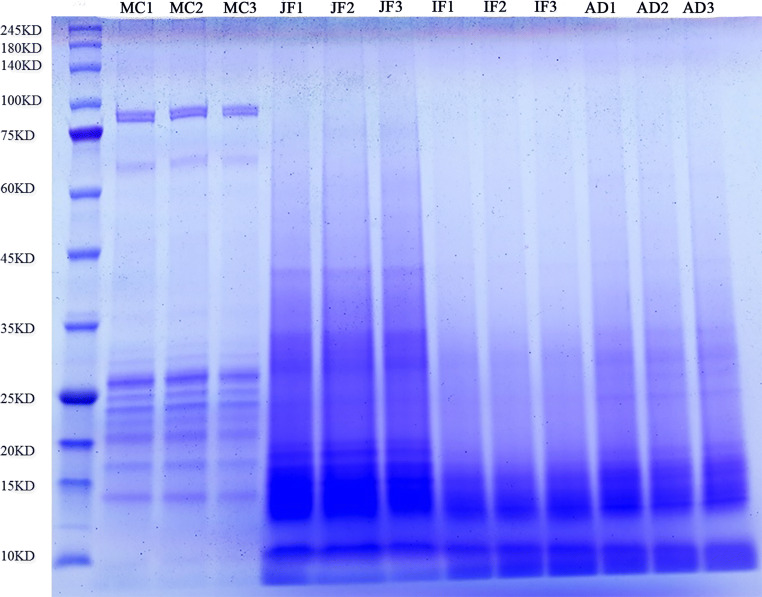
SDS-PAGE was performed with 20 μg total protein samples. Electrophoresis revealed that the four growth phases of F. hepatica showed distinct protein bands, indicating that the total protein was successfully extracted. The protein bands from MC were clearer and more specific than those from JF, IF, and AD. The results are presented in Fig. 1.
Fig. 1.
SDS-PAGE results of total protein in four phases of F. hepatica. Metacercariae are represented by MC, juvenile flukes 28dpi are represented by JF, immature flukes 59dpi are represented by IF, and adults are represented by AD
The statistical results of protein identification are provided in Table 2. A total of 3248 peptide segments and 629 proteomics were identified in MC. A total of 33,591 peptides and 2286 proteomics were identified in JF. A total of 34,955 peptides and 2254 proteomics were identified in IF, and 35,707 peptides and 2192 proteomics were identified in AD. Overall, 107,501 peptides and 2406 proteomics were identified in MC, JF, IF, and AD. The total number of peptides and proteomics identified in MC was significantly lower than that identified in JF, IF, and AD.
Table 2.
Protein identification results statistics
| Sample | Unique peptides | PSM | Protein groups |
|---|---|---|---|
| MC 1 | 748 | 908 | 397 |
| MC 2 | 999 | 1183 | 487 |
| MC 3 | 979 | 1157 | 468 |
| MC total | 1414 | 3248 | 629 |
| SS 1 | 8647 | 11,149 | 2143 |
| SS 2 | 8101 | 10,597 | 2117 |
| SS 3 | 8948 | 11,845 | 2198 |
| SS total | 9827 | 33,591 | 2286 |
| LV 1 | 8719 | 11,694 | 2153 |
| LV 2 | 8517 | 11,482 | 2078 |
| LV 3 | 8669 | 11,779 | 2125 |
| LV total | 9840 | 34,955 | 2254 |
| AD 1 | 8677 | 11,866 | 2063 |
| AD 2 | 8547 | 11,951 | 2023 |
| AD 3 | 8615 | 11,890 | 2036 |
| AD total | 9740 | 35,707 | 2192 |
| Sum | 11,172 | 107,501 | 2406 |
The proteomics data were subjected to non-labeled quantitative calculation using the label-free algorithm in Max Quant software, and a significant difference analysis was conducted on the quantitative results. Proteins that met the screening criteria with a difference of more than 1.5 times and P < 0.05 were considered significant DEPs. The number of DEPs is reported in Table 3.
Table 3.
Differential expression protein statistics result
| Total DEP number | Upregulated DEP number | Downregulated DEP number | |
|---|---|---|---|
| MC vs JF | 250 | 93 | 157 |
| MC vs IF | 237 | 87 | 150 |
| MC vs AD | 240 | 92 | 147 |
| JF vs IF | 502 | 248 | 254 |
| JF vs AD | 717 | 446 | 271 |
| IF vs AD | 407 | 263 | 144 |
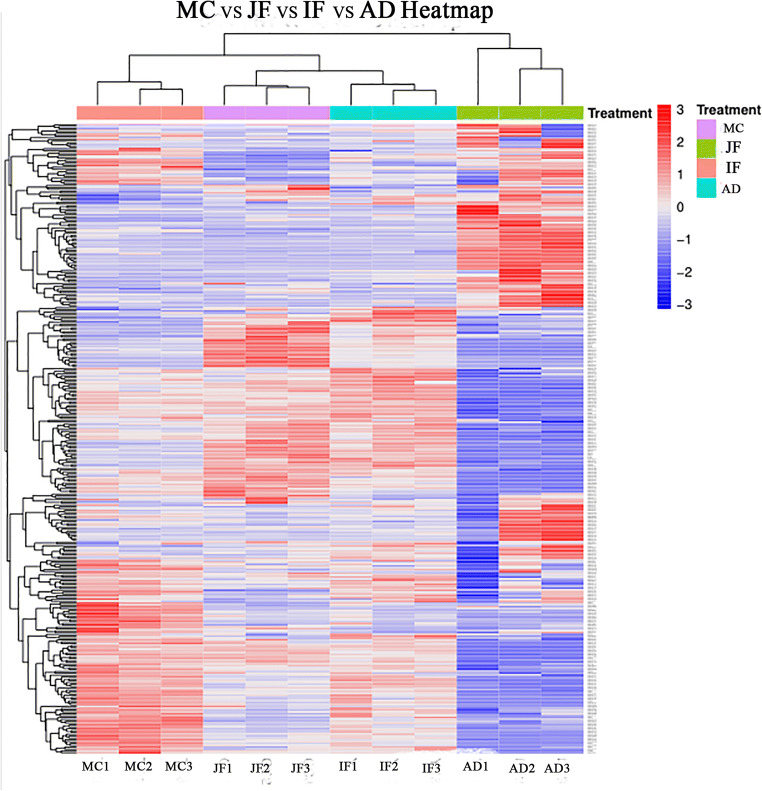
Cluster analysis
The clustering results for DEPs in MC, JF, IF, and AD are shown in Fig. 2. Red represents upregulated proteins, and blue represents downregulated proteins. The cluster analysis chart indicates that most of the upregulated proteins in MC were downregulated in JF, IF, and AD, whereas most of the proteins that were downregulated in MC worms were upregulated in JF, IF, and AD. Therefore, we speculate that altering the level of protein expression in different phases can influence the interaction between F. hepatica and the host.
Fig. 2.
Differential protein clustering results for metacercariae, juvenile flukes 28dpi, immature flukes 59dpi, and adults. Red is the upregulated protein, and blue is the downregulated protein
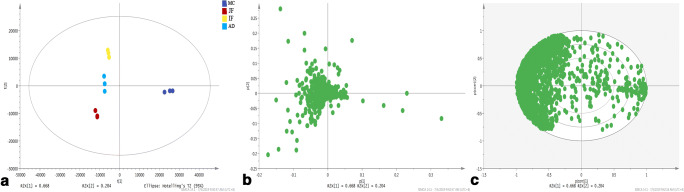
PCA
In this study, the protein expression of F. hepatica in different growth phases was analyzed, and all proteins were defined as variables in PCA. The PCA score diagram is depicted in Fig. 3a. The PCA results revealed that somatic proteome from JF, IF, and AD formed distinct clusters, and those from MC clustered separately. The loading plots and the corresponding correlation loading plots of PCA are presented in Fig. 3b, c, respectively. These plots show a significant difference in protein expression between MC and the three other phases, but the difference among the three other phases was not significant.
Fig. 3.
a PCA score plots, b loading plots, and c corresponding correlation loading plots of the principal components of proteins in metacercariae, juvenile flukes 28dpi, immature flukes 59dpi, and adults
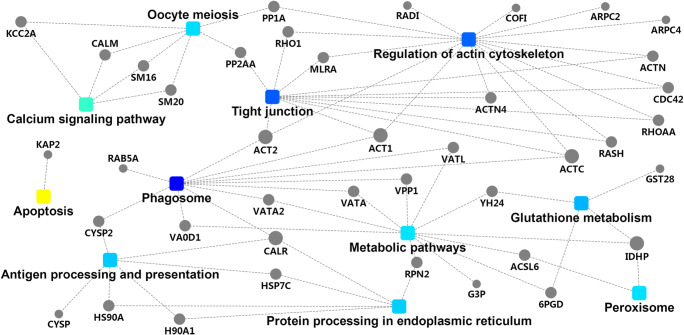
Protein–protein interaction analysis
The DEPs were submitted to the STRING database for protein–protein interaction analysis. A color-coded network was generated to represent different types of evidence for the associations between DEPs (Fig. 4). The network identified the unique features of the functional relationship between proteins. In the network analysis diagram, the dots denote proteins, and the squares represent pathways. Large dots indicate that a protein is connected to numerous pathways and has numerous functions. Large squares indicate that a large number of proteins are connected to pathways. We discovered that 40 proteins were involved in 11 pathways that formed a complex regulatory network.
Fig. 4.
Protein–protein interactions of proteins in metacercariae, juvenile flukes 28dpi, immature flukes 59dpi, and adults. The dots are proteins, and the squares are pathways
Bioinformatics analysis
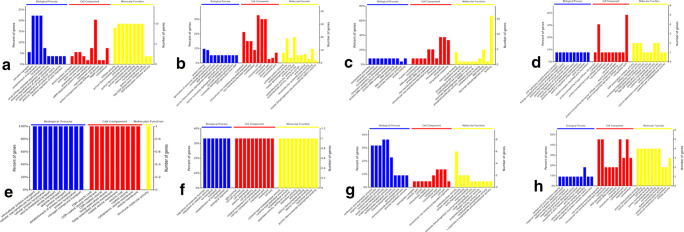
The DEPs in the four phases were subjected to GO component enrichment analysis, and the results are shown in Fig. 5. The upregulated DEPs in MC were primarily involved in the biological processes of transport, establishment of localization, and localization. Macromolecular complexes in the cell components were the most abundant proteins. Proteins were primarily involved in the following molecular functions: purine ribonucleoside binding, GTP binding, guanyl ribonucleotide binding, purine nucleoside binding, ribonucleside binding, and guanyl nucleotide binding (Fig. 5a). The downregulated DEPs in MC were primarily involved in the biological processes of protein folding and the generation of precursor metabolites. Most were enriched in cells, intracellular parts, and intracellular organelles. The molecular functions of most proteins were protein binding and oxidoreductase activity (Fig. 5b). The upregulated DEPs in JF generally participated in a series of processes from nucleosome assembly to protein folding. Most proteins were enriched in intracellular parts and intracellular organelles. And most proteins were involved in the molecular binding function (Fig. 5c). The downregulated DEPs in JF were primarily involved in energy transport, positive regulation, and actin nucleation. Most proteins were enriched in intracellular parts, followed by the cytoplasm. The proteins were generally involved in the biological process of enzyme regulation (Fig. 5d). The upregulated DEPs in IF were mainly involved in transport and localization, and most proteins were enriched in vesicles and membranes and involved in the molecular function of structural molecule activity (Fig. 5e). The downregulated DEPs in IF participated primarily in transport, localization, and biological regulation. Most proteins were enriched in the junction, membrane, and cell parts. The molecular functions of most proteins included peptidase activity, hydrolase activity, and binding (Fig. 5f). The upregulated DEPs in AD were primarily involved in the metabolic process, and most proteins were enriched in the intracellular and cell parts. The proteins were mostly involved in the molecular function of oxidoreductase activity (Fig. 5g). The downregulated DEPs in AD generally participated in cellular macromolecular complex assembly, and most proteins were enriched in non-membrane-bound organelles and the organelle part. These proteins were mostly involved in the molecular functions of nucleotide and nucleoside binding (Fig. 5h). Given that the upregulated DEPs in MC were involved in transport and localization, we speculated that these DEPs are conducive for the parasitism of MC in the host. Meanwhile, the upregulated DEPs in JF, IF, and AD were involved in protein biosynthesis. Thus, we speculated that these DEPs help worms escape the host immune response, thereby achieving long-term survival in the host.
Fig. 5.
Annotation of the GO functions of the four growth phases of F. hepatica. a The function of the upregulated DEPs in metacercariae compared with the three other phases. b The function of the downregulated DEPs in metacercariae compared with the three other phases. c The function of the upregulated DEPs in juvenile flukes 28dpi compared with the three other phases. d The function of the downregulated DEPs in juvenile flukes 28dpi compared with the three other phases. e The function of the upregulated DEPs in immature flukes 59dpi compared with the three other phases. f The function of the downregulated DEPs in immature flukes 59dpi compared with the three other phases. g The function of the upregulated DEPs in adults compared with the three other phases. h The function of the downregulated DEPs in adults compared with the three other phases
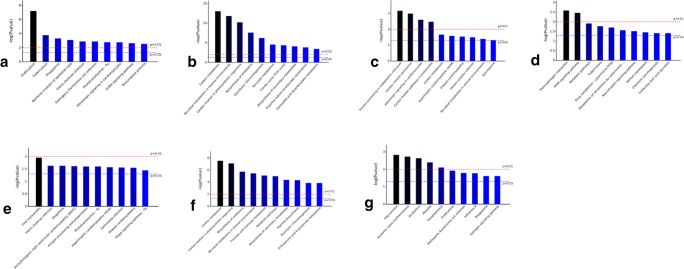
Figure 6 presents the KEGG pathway enrichment analysis of the DEPs. The upregulated DEPs in MC were mainly involved in endocytosis (Fig. 6a), and the downregulated DEPs in MC were mainly involved in carbon metabolism, microbial metabolism in diverse environments, and carbon fixation in photosynthetic organisms (Fig. 6b). The upregulated DEPs in JF were primarily involved in protein processing in the endoplasmic reticulum and cardiac muscle contraction (Fig. 6c), whereas the downregulated DEPs in JF were mainly involved in plant-pathogen interaction and the PPAR signaling pathway (Fig. 6d). The upregulated DEPs in IF lacked KEGG annotations, and the downregulated DEPs in IF primarily participated in viral myocarditis (Fig. 6e). The upregulated DEPs in AD were mostly involved in carbon metabolism and carbon fixation in photosynthetic organisms (Fig. 6f), and the downregulated DEPs in AD were mainly involved in the gap junction, systemic lupus erythematosus, and alcoholism (Fig. 6g).
Fig. 6.
KEGG pathway enrichment analysis of DEPs. a The pathway in which the upregulated DEPs in metacercariae are involved in compared with the three other phases. b The pathway in which the downregulated DEPs in metacercariae are involved in compared with the three other phases. c The pathway in which the upregulated DEPs in juvenile flukes 28dpi are involved in compared with the three other phases. d The pathway in which the downregulated DEPs in juvenile flukes 28dpi are involved in compared with the three other phases. e The pathway in which the downregulated DEPs in immature flukes 59dpi are involved in compared with the three other phases. f The pathway in which the upregulated DEPs in adults are involved in compared with the three other phases. g The pathway in which the downregulated DEPs in adults are involved in compared with the three other phases
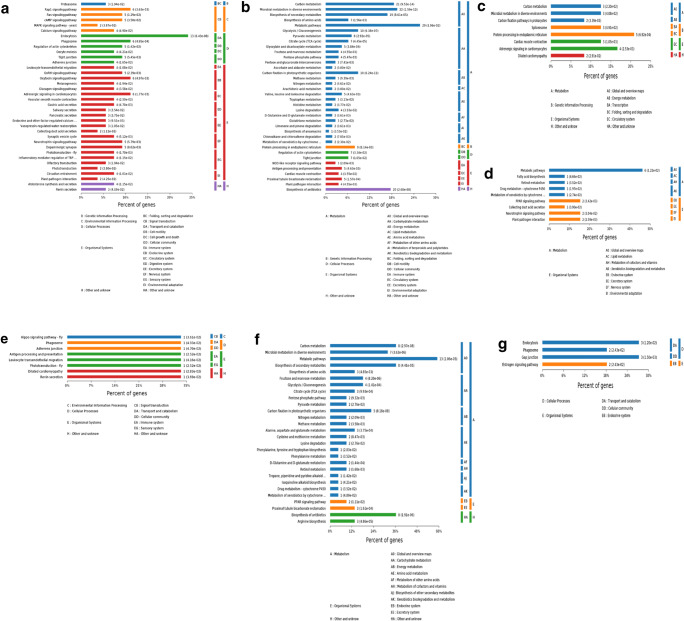
A detailed analysis of the upregulated and downregulated DEPs in the four phases is provided in Fig. 7. The upregulated DEPs in MC were mainly involved in genetic information processing, environmental information processing, cellular processes, organismal systems, and others (Fig. 7a), and the downregulated DEPs participated primarily involved in metabolism, genetic information processing, cellular processes, organismal systems, and other processes (Fig. 7b). The upregulated DEPs in JF were mainly involved in metabolism, genetic information processing, organismal systems, and others (Fig. 7c), and the downregulated DEPs were generally involved in metabolism and organismal systems (Fig. 7d). The upregulated DEPs in IF lacked KEGG annotations, whereas the downregulated DEPs mainly participated in environmental information processing, cellular processes, organismal systems, and other processes (Fig. 7e). The upregulated DEPs in AD were mainly involved in metabolism, organismal systems, and other processes (Fig. 7f), whereas the downregulated DEPs primarily participated involved in cellular processes and organismal systems (Fig. 7g).
Fig. 7.
Enrichment analysis of significant DEPs. a The function of the upregulated DEPs in metacercariae compared with the three other phases. b The function of the downregulated DEPs in metacercariae compared with the three other phases. c The function of the upregulated DEPs in juvenile flukes 28dpi compared with the three other phases. d The function of the downregulated DEPs in juvenile flukes 28dpi compared with the three other phases. e The function of the downregulated DEPs in immature flukes 59dpi compared with the three other phases. f The function of the upregulated DEPs in adults compared with the three other phases. g The function of the downregulated DEPs in adults compared with the three other phases
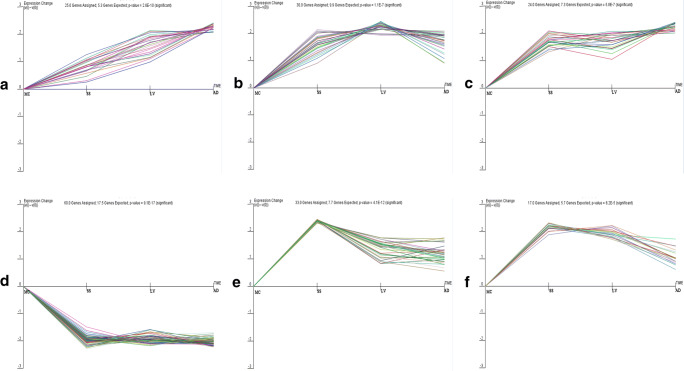
After screening and analyzing the DEP data, we discovered that all DEPs could be divided into 50 profiles with distinct trends. Profiles 42, 48, 49, 12, 45, and 47 were selected for further analysis because of their good trends (Fig. 8). Profiles 42, 49, and 48 exhibited an upward trend. Profile 12 showed a downward trend, and profiles 45 and 47 increased then decreased. Thioredoxin glutathione reductase, thioredoxin peroxidase, glutathione transferase, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were the major components of profile 42. Profile 49 included heat shock protein (HSP) 90, HSP 70, and actin. Profile 48 involved Ca2+-insensitive EF hand and aldehyde dehydrogenase family proteins. Profile 12 primarily comprised Ras family proteins, calmodulin-like protein 1, and calmodulin-like protein 3. Profile 45 contained secreted cathepsin L1, fatty acid-binding protein (FABP), protein disulfide-isomerase, calreticulin family protein, and myosin tail. Myosin heavy chain, myosin head, and myosin regulatory light chain were found in profile 47. Through analysis, we discovered that the main proteins in profile 12 were DEPs that were upregulated in MC, and the primary proteins in profiles 42, 45, 47, 48, and 49 were DEPs that were downregulated in MC. The specific upregulated and downregulated DEPs in MC are shown in Supplementary File 1.
Fig. 8.
Six profiles of DEPs with distinct trends. a Profile 42, b Profile 49. c Profile 48. d Profile 12. e Profile 45. f Profile 47
Discussion and conclusion
The growth and maturation of parasites are related to the changes in the expression of somatic, secretory, and/or surface proteins. All the growth stages of F. hepatica contribute to the pathology observed during fasciolosis, particularly the immature stages are responsible for extensive damage as they migrate through the liver (Taibi et al. 2019). Therefore, the identification of proteins with different characteristics and expression patterns in different phases is also essential for understanding the pathophysiological mechanisms of fasciolosis. In recent years, vaccine candidates with diverse protection rates against fasciolosis have been identified. Since many candidate proteins are part of protein superfamilies, proteomics can be utilized to explore individual proteins as vaccine candidate antigens.
Protein information enrichment is considerably useful in describing biological functions (Wang and Tang 2017). MS can be utilized for the multivariate analysis of novel biomarkers to help fill gaps between genome and protein phenotypes; this approach is conducive to the development of novel diagnostic and therapeutic techniques (Bantscheff et al. 2007). Cluster analysis is a commonly used exploratory data analysis method (Santoni et al. 2014). Clustering results of target proteins can help in distinguishing protein subsets with altered expression patterns (Melchiotti et al. 2017). Proteins with similar expression patterns may have similar functions, participate in the same biological pathway, or be in a nearly regulatory position in the pathway (Zhang et al. 2017).
In this study, we examined the function of DEPs in the four growth phases of F. hepatica through somatic proteomics analysis. These DEPs were involved in numerous biological processes and exerted profound effects on the host-parasite interaction. Through the analysis by GO and KEGG, we found that the upregulated DEPs in MC were playing an important role in transportation and localization, while the upregulated DEPs in other three phases were mainly involved in protein folding and metabolic process. We also discovered the DEPs in MC exhibited clear trends. Thus, analyzing the DEPs in MC is reasonable and several proteins with typical characteristics and functions in MC were selected for further analysis and discussion.
A battery of antioxidant molecules and proteases secreted by F. hepatica was examined in this study. As the parasite invades and migrates through the mammalian host, it encounters a variety of different aerobic/anaerobic micro-environments that generated by immune cells. And the parasite has developed an antioxidant system to protect them against lethal radicals and ensure its continued survival in the host. Peroxiredoxin (PRX) and the related protein, thioredoxin (TRX), are the key components of redox reaction-based antioxidant system. A growing body of evidence indicates that TRX can act as a redox regulator to maintain the redox state of cells and participate in the biological functions associated with the immune evasion mechanism (Alger et al. 2002). TRX has been observed in numerous parasites, including F. hepatica (Salazar-Calderon et al. 2001). Shoda et al. (1999) confirmed that recombinant TRX showed specifically but weakly stimulate in bovine T cells; therefore, TRX is not a promising candidate for inducing resistance to F. hepatica. And they also identified TRX of F. hepatica in both the juvenile and the adult fluke. Cwiklinski et al. (2018) proved that TRX was the most abundant protein within the NEJ secretomes. However, the expression of TRX (B6DT35) was all found in MC, JF, IF, and AD in our study, and it was downregulated in MC. Intriguingly, the FhTRX-reducing enzyme, thioredoxin glutathione reductase (TGR), was also downregulated in the somatic proteome of MC. These dates indicate that once parasite invades and migrates in the host, FhTRX will increase to carry out antioxidant reaction to escape the host’s immune attack. In general, FhTRX may play housekeeping role in F. hepatica throughout its development. Therefore, we may be able to regulate the growth and development of F. hepatica by regulating the expression of FhTRX and FhTGR. FhTRX may be a potential target for drugs to damage F. hepatica.
The enzyme glyceraldehyde-3-P-dehydrogenase (GAPDH) plays a key role in glycolysis, and this reaction precedes the two ATP generating steps. Therefore, inhibition of GAPDH will result in a significant reduction in energy production. Consequently, scholars are interested in developing GAPDH inhibitors as anti-parasitic agents (Zinsser et al. 2014). Worms are constantly excysted and encysted to escape the immune defense mechanisms of their hosts (Morphew et al. 2013). And the presence of GAPDH on the surface of worms may help protect them from oxygen-mediated attack by phagocytes in the host (Waine et al. 1993). These dates have led many researchers to speculate that GAPDH may also be a useful vaccine against worms. However, the primary sequence of FhGAPDH is similar to that from other trematodes, and the predicted structure is also similar to those from other animals including mammalian hosts, so GAPDH as a vaccine candidate is theoretically challenging (Zinsser et al. 2014). Meanwhile, we found that the expression of GAPDH (A0A2H1BWY6) which was involved in glycerol lipid metabolism pathway and biosynthesis pathway of secondary metabolites was downregulated in MC. This data indicates that MC may be in a low activity state, so the energy consumption and the GAPDH expression are significantly lower than the developmental stage in the host.
A number of proteomics analyses have revealed that proteins belonging to the glutathione S-transferase (GST) family from liver fluke have been demonstrated to contain protective vaccine candidates (Chemale et al. 2010). Abath and Werkhauser (1996) confirmed that glutathione S-transferase (GST) can be expressed instantaneously on the surface of worms and plays an essential role in the detoxification of xenobiotic and endogenously derived toxins within the host bile environment. In particular, the liver is an organ of intense metabolic activity and is precisely the place where juvenile and adult stages of F. hepatica localize. Therefore, the parasite may be damaged by the attack of xenobiotics during the normal metabolism of the host, so GST is essential for the survival of F. hepatica in the liver of the host (Vasti et al. 2018). Our results showed that GST was not only expressed in somatic proteins of JF, IF, and AD of F. hepatica which colonized in the liver of the host, but also expressed in MC which survived in the external water environment. Numerous studies have shown that F. hepatica infection suppressed Th1 responses to demonstrate its anti-inflammatory effect. Aguayo et al. (2019) proposed that FhGST could play relevant roles in parasite immunomodulation. Thus, FhGST could be one of the antigens that F. hepatica employs to suppress the Th1 immune response of the host, which may also benefit for the parasite survival. We found that the expression level of GST (A0A2H1BT36) in MC in vitro was significantly downregulated. In other words, the expression level of GST in JF, IF, and AD in the host was significantly increased, so as to maintain its long-term survival in the host. Conversely, if the expression of GST is inhibited, the toxins and a series of immune molecules secreted by the liver of the host will damage the parasite. Thus, GST could serve as a potential target for drugs and vaccines against F. hepatica.
The soluble superfamily of fatty acid-binding proteins (FABPs) is small (14–15 kDa) proteins that bind or sequestrate hydrophobic ligands such as anthelmintics (Esteves and Ehrlich 2006). Previous studies have shown that cytosolic FABPs are potential vaccine candidates (Dalton et al. 2003; McManus and Dalton 2006). Hillyer (2005) confirmed that in different animal models, native and recombinant F. hepatica FABPs induce significant levels of protection against infection with F. hepatica. In general, their secretion is highly regulated during the migration of F. hepatica. And growing evidence support the importance of FABPs for the establishment of F. hepatica in the host. Intracellular FABPs act as fatty acid (FA) transporters (Esteves and Ehrlich 2006; Glatz and van der Vusse 1996). Because adult F. hepatica does not synthesize FA, high levels of FABP may absorb FA from the host environment through uncharacteristic tegumental mechanisms (Dalton et al. 2004). Importantly, FABP is an abundant component of the soluble tegumental proteome of adult F. hepatica (Morphew et al. 2013; Wilson et al. 2011). However, there is a high level of preformed/stored lipid in NEJs to support initial survival in the host (Thorpe 1968), and the requirement for FA uptake and FABP transporters is likely reduced. The observations of Morphew et al. (2016) revealed that FABPs were relatively lower in juvenile liver fluke compared to adults. Meanwhile, we also found that the expression of fatty acid-binding protein type 3 (FABP 3) (Q9U1G6) was downregulated in MC and the greatest abundance of FABP 3 was found in adults, suggesting FABP 3 is important for development. In addition to be involved in intracellular transport and detoxification, FhFABP 3 also function as FA transport proteins. Bile contains high levels of FAs; therefore, high levels of FhFABP 3 in adults may take up FAs from the bile. However, there may be a high level of lipid in MC to support survival and MC is likely in a low activity state. Thereby, FhFABP 3 is downregulated in MC. At the same time, the higher expression of FhFABP 3 in adults may be a potential vaccine candidate.
In general, metacercariae obtain oxygen from the environment, but once the parasite enters the host tissue, rapid growth and development ensues, and the diffusion of oxygen throughout the parasite is limited by the size of the parasite. This results in the gradual transformation of aerobic energy metabolism to anaerobic energy metabolism (Tielens et al. 1982). Yoo et al. (2011) showed that antioxidant molecules and detoxification proteins offered sufficient protection for the adult flukes residing within the bile ducts. Our dates are consistent with the previous conclusion. As the parasite migrates and colonizes in the host, we found that the expression of TRX, GAPDH, GST, and FABP 3 gradually increases in juvenile flukes 28dpi, immature flukes 59dpi, and adults. Therefore, FhTRX, FhGAPDH, FhGST, and FhFABP 3 can be used as potential targets for drugs and vaccines of fasciolosis. Meanwhile, the treatment may be more effective in the later stages of infection.
Cathepsin L is the most studied trematode peptidase owing to its importance in host-parasite interactions. It is a potential target for the development of novel chemical drugs and vaccines and a selective diagnostic marker of diseases (Perez-Sanchez et al. 2006). Numerous cathepsin L proteases have been isolated from adult flukes which form three distinct clades, that is FhCL1, FhCL2, and FhCL5 (Irving et al. 2003; Robinson et al. 2008), while several diverse sequences have also been isolated from metacercariae and NEJ flukes (Cancela et al. 2008; Harmsen et al. 2004). The enzymes expressed in metacercariae could be involved in excystment. Robinson et al. (2009) showed that excystment of the F. hepatica infective metacercariae was dependent on the cathepsin L proteases, and switching on the genes encoding these enzymes was one of the first steps to activate the dormant metacercariae. And those cathepsins expressed in NEJ would be involved in invasion, whereas immature and adults may participate in migration and feeding in the liver of the host (Morphew et al. 2013). Three cathepsins were identified as DEPs in this study. The three sequences showed 95.92% similarity after comparison. Once ingested, the metacercariae need to trigger a mechanism to ensure they can quickly escape from the cyst. The outer cyst wall is removed by the contents of the host stomach. And the parasites quickly emerge from the inner cyst wall by releasing their cecal contents that are loaded with cathepsin cysteine proteases (Cwiklinski et al. 2018). Since the metacercariae we detected are still encysted and have not been stimulated by the host digestive tract, they will not be excysted by releasing cathepsin L. Therefore, cathepsin L (Q7KYH5) which exists in metacercariae was upregulated. While cathepsin L is encoded by stage-specific genes and has distinct enzymatic properties, the gene encoding cathepsin L (Q7KYH5) which we found upregulated in MC has yet to be determined. Meanwhile, Cancela et al. (2008) showed the fact that CL1 is not produced by NEJ; NEJ produce a novel CL-like protein named CL3. However, the expression levels of secreted cathepsin L1 (Q7JNQ9) and cathepsin L1 (Fragment) (Q9GRW5) were significantly higher in JF, IF, and adults than that in MC in our study. This indicated that cathepsin L1 started to be synthesized in F. hepatica after metacercariae completed the process of excystment and continued to the adult stage, so as to facilitate the migration of F. hepatica in the host. A recombinant version of the cysteine protease cathepsin L1 (rmFhCL1) from F. hepatica has been a vaccine candidate for many years. Our experiments confirmed that secreted cathepsin L1 and fragment of cathepsin L1 could also be expected to be vaccine candidate antigens.
A well-developed muscular system is essential for the survival of both endo- and ecto-parasitic flatworms. The previous study has demonstrated the presence of two contractile proteins, actin and myosin, in the musculature of F. hepatica (Kumar et al. 2003). Actin and myosin are located in the muscles surrounding the teguments and abdominal suckers (Kumar et al. 2003; Tansatit et al. 2006). They are primarily involved in the regulation of the actin cytoskeleton, tight junctions, adhesion, platelet activation, and other related pathways and contribute to adhesion to the host intestinal tract, liver, and bile duct (Boukli et al. 2011). The somatic soluble NEJ sample possesses a series of actin-binding proteins. Their role may be related to vesicular trafficking in the tegumental syncytium of NEJ, as well as to other intracellular events which require formation and/or depolimerization of actin in muscle tissue or nervous system development. At the same time, they might be used in morphogenesis, tissue remodeling, and/or in exocytosis/endocytosis events during its development to the immature stages (Di Maggio et al. 2016). Similarly, the current study revealed that the expression levels of the myosin tail (A0A2H1CQG2), myosin head (A0A2H1CNZ5), and myosin regulatory light chain (D0VAX0) were downregulated in MC, while the highest expression levels were found in juvenile flukes 28dpi. This indicated that F. hepatica has colonized in host tissue in about 28 days after the host infected with infective metacercariae. However, given that actin expression was both upregulated and downregulated in MC, we further determine which subtype is upregulated (A0A2H1CC81) or downregulated (A0A2H1C3Q6). Nevertheless, whether the two different subtypes play the same function and, if they play the opposite function, which function dominates remains to be explored. These results can provide a theoretical basis for screening landmark proteins and establishing early diagnostic methods. Apart from their vaccine potential, proteomics analysis of the major DEPs in F. hepatica might contribute to a better understanding of drug resistance mechanisms and the host-parasite relationship. Our analysis of the F. hepatica proteomes provides a comprehensive and dynamic view of contemporary anti-F. hepatica treatments which target the molecules described in the four developmental stages of F. hepatica.
Electronic supplementary material
(PDF 152 kb)
Acknowledgements
We would like to thank Prof. Wang from Heilongjiang Bayi Agricultural University for the different developmental stages of flukes. And we also want to thank all of the staff members who provided laboratory assistance.
Funding information
This work was supported by the State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (SKLVEB2019KFKT011) and the National Key Research and Development Program of China (2017YFD0501200).
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Ethical approval statement
The study protocol was approved by Northeast Agriculture University Veterinary Research Ethics Committee. And all procedures were strictly in accordance with the guidelines of the Chinese National Institute of Health Guide for the Care and Use of Laboratory Animals.
Footnotes
Jingyun Xu and Lijia Wu are co-first authors of the article.
The National Key Research and Development Program of China (2017YFD0501200)
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- ABATH FG, WERKHAUSER RC. The tegument of Schistosoma mansoni: functional and immunological features. Parasite Immunol. 1996;18:15–20. doi: 10.1046/j.1365-3024.1996.d01-6.x. [DOI] [PubMed] [Google Scholar]
- Afshin R, Ali F, Hankjmeh Z, Mostafa R, Abolfazl G, Mohammad MR. Protein detection of excretory-secretory products and somatic extracts from Fasciola hepatica and F. gigantica using two-dimensional electrophoresis. Iran J Parasitol. 2019;14:379–386. [PMC free article] [PubMed] [Google Scholar]
- Aguayo V, Valdés FBN, Rodríguez-Valentín M, Ruiz-Jiménez C, Ramos-Benítez MJ, Méndez LB, Espino AM. Fasciola hepatica GST downregulates NF-κB pathway effectors and inflammatory cytokines while promoting survival in a mouse septic shock model. Sci Rep. 2019;9:2275. doi: 10.1038/s41598-018-37652-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALGER HM, AYED AAS, TADECKER MJS, ILLIAMS DLW. Molecular and enzymatic characterisation of Schistosoma mansoni thioredoxin. Int J Parasitol. 2002;32:1285–1292. doi: 10.1016/s0020-7519(02)00108-x. [DOI] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Bennuru S, Semnani R, Meng Z, Ribeiro JM, Veenstra TD, Nutman TB. Brugia malayi excreted/secreted proteins at the host/parasite interface: stage- and gender-specific proteomic profiling. PLoS Negl Trop Dis. 2018;3:410. doi: 10.1371/journal.pntd.0000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukli NM, Delgado B, Ricaurte M, Espino AM. Fasciola hepatica and Schistosoma mansoni: identification of common proteins by comparative proteomic analysis. Parasitol. 2011;97:852–861. doi: 10.1645/GE-2495.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancela M, Acosta D, Rinaldi G, Silva E, Duran R, Roche L, Zaha A, Carmona C, Tort JF. A distinctive repertoire of cathepsins is expressed by juvenile invasive Fasciola hepatica. Biochimie. 2008;90:1461–1475. doi: 10.1016/j.biochi.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Chemale G, Morphew R, Moxon JV, Morassuti AL, Lacourse EJ, Barrett J. Proteomic analysis of glutathione transferases from the liver fluke parasite, fasciola hepatica. Proteomics. 2010;6:6263–6273. doi: 10.1002/pmic.200600499. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, Termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cwiklinski K, Jewhurst H, McVeigh P, Barbour T, Maule AG, Tort J, O’Neill SM, Robinson MW, Donnelly S, Dalton JP. Infection by the helminth parasite fasciola hepatica requires rapid regulation of metabolic, virulence, and invasive factors to adjust to its mammalian host. Mol Cell Proteomics. 2018;17(4):792–809. doi: 10.1074/mcp.RA117.000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton JP, Brindley PJ, Knox DP, Brady CP, Hotez PJ, Donnelly S, O’Neill SM, Mulcahy G, Loukas A. Helminth vaccines: from mining genomic information for vaccine targets to systems used for protein expression. Int J Parasitol. 2003;33:621–640. doi: 10.1016/s0020-7519(03)00057-2. [DOI] [PubMed] [Google Scholar]
- Dalton JP, Skelly P, Halton DW. Role of the tegument and gut in nutrient uptake by parasitic platyhelminths. Can J Zool. 2004;82:211–232. [Google Scholar]
- Esteves A, Ehrlich R. Invertebrate intracellular fatty acid binding proteins. Comp Biochem Phys C. 2006;142:262–274. doi: 10.1016/j.cbpc.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Furst T, Duthaler U, Sripa B, Utzinger J, Keiser J. Trematode infections: liver and lung flukes. Infect Dis Clin N Am. 2012;26:399–419. doi: 10.1016/j.idc.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Glatz JFC, van der Vusse GJ. Cellular fatty acid-binding proteins: their function and physiological significance. Prog Lipid Res. 1996;35:243–282. doi: 10.1016/s0163-7827(96)00006-9. [DOI] [PubMed] [Google Scholar]
- Harmsen MM, Cornelissen JB, Buijs HE, Boersma WJ, Jeurissen SH, van Milligen FJ. Identification of a novel Fasciola hepatica cathepsin L protease containing protective epitopes within the propeptide. Int J Parasitol. 2004;34:675–682. doi: 10.1016/j.ijpara.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Hewitson JP, Harcus YM, Curwen RS, Dowle AA, Atmadja AK, Ashton PD, Wilson A, Maizels RM. The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-se-cretory products. Mol Biochem Parasitol. 2008;160:8–21. doi: 10.1016/j.molbiopara.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Hillyer GV. Fasciola antigens as vaccines against fascioliasis and schistosomiasis. Helminthol. 2005;79:241–247. doi: 10.1079/joh2005304. [DOI] [PubMed] [Google Scholar]
- Irving JA, Spithill TW, Pike RN, Whisstock JC, Smooker PM. The evolution of enzyme specificity in Fasciola spp. J Mol Evol. 2003;57:1–15. doi: 10.1007/s00239-002-2434-x. [DOI] [PubMed] [Google Scholar]
- Keiser J, Utzinger J. Food-borne trematodiases. Clin Microbiol Rev. 2009;22:466–483. doi: 10.1128/CMR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, MCGEOWN JG, REYNOSO-DUCOING O, AMBROSIO JR, AIRWEATHER IF. Observations on the musculature and isolated muscle fibres of the liver fluke, Fasciola hepatica. Parasitology. 2003;127:457–473. doi: 10.1017/s0031182003003925. [DOI] [PubMed] [Google Scholar]
- Maggio D, Sánchez L, Tirloni L, Pinto AFM, Diedrich JK, Yates JR, Benavides U, Carmona C, Berasain P. Across intra-mammalian stages of the liver fluke Fasciola hepatica: a proteomic study. Sci Rep. 2016;6:32796. doi: 10.1038/srep32796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas-Coma S, Valero MA, Bargues MD. Fascioliasis. Adv Exp Med Biol. 2019;766:77–114. doi: 10.1007/978-1-4939-0915-5_4. [DOI] [PubMed] [Google Scholar]
- McManus DP, Dalton JP. Vaccines against the zoonotic trematodes Schistosoma japonicum, Fasciola hepatica and Fasciola gigantica. Parasitology. 2006;133:S43–S61. doi: 10.1017/S0031182006001806. [DOI] [PubMed] [Google Scholar]
- Melchiotti R, Gracio F, Kordasti S, Todd AK, de Rinaldis E. Cluster stability in the analysis of mass cytometry data. Cytometry A. 2017;91:73–84. doi: 10.1002/cyto.a.23001. [DOI] [PubMed] [Google Scholar]
- Morphew RM, Hamilton CM, Wright HA, Dowling DJ, O’Neill SM, Brophy PM. Identification of the major proteins of an immune modulating fraction from adult Fasciola hepatica released by Nonidet P40. Vet Parasitol. 2013;191:379–385. doi: 10.1016/j.vetpar.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morphew RM, Wilkinson TJ, Mackintosh N, Jahndel V, Paterson S, Mcveigh P, Abidi SMA, Saifullah K, Raman M, Ravikumar G, LaCourse J, Maule AG, Brophy PM. Exploring and expanding the fatty-acid-binding protein superfamily in fasciola species. J Proteome Res. 2016;15:3308–3321. doi: 10.1021/acs.jproteome.6b00331. [DOI] [PubMed] [Google Scholar]
- Perez-Sanchez R, Ramajo-Hernandez A, Ramajo-Martin V, Oleaga A. Proteomic analysis of the tegument and excretory-secretory products of adult Schistosoma bovis worms. Proteomics. 2006;6:S226–S236. doi: 10.1002/pmic.200500420. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Connolly B. Proteomic analysis of the excre-tory-secretory proteins of the Trichinella spiralis L1 larva, a nematode parasite of skeletal muscle. Proteomics. 2005;5:4525–4532. doi: 10.1002/pmic.200402057. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Greig R, Beattie KA, Lamon DJ, Connolly B. Comparative analysis of the excretory-secretory proteome of the muscle larva of Trichinella pseudospiralis and Trichinella spiralis. Int J Parasitol. 2007;37:139–148. doi: 10.1016/j.ijpara.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Tort JF, Lowther J, Donnelly SM, Wong E, Xu W, Stack CM, Padula M, Herbert B, Dalton JP. Proteomics and phylogenetic analysis of the cathepsin L protease family of the helminth pathogen Fasciola hepatica: expansion of a repertoire of virulence-associated factors. Mol Cell Proteomics. 2008;7:1111–1123. doi: 10.1074/mcp.M700560-MCP200. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol Cell Proteomics. 2009;8:1891–1907. doi: 10.1074/mcp.M900045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Calderon M, Martin-Alonso JM, Ruiz De Eguino AD, Parra F. Heterologous expression and functional characterization of thioredoxin from Fasciola hepatica. Parasitol Res. 2001;87:390–395. doi: 10.1007/s004360000353. [DOI] [PubMed] [Google Scholar]
- Santoni D, Swiercz A, Zmieńko A, Kasprzak M, Blazewicz M, Bertolazzi P, Felici G. An integrated approach (CLuster Analysis Integration Method) to combine expression data and protein-protein interaction networks in agrigenomics: application on Arabidopsis thaliana. OMICS. 2014;18:155–165. doi: 10.1089/omi.2013.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoda LK, Rice-Ficht AC, Zhu D, McKown RD, Brown WC. Bovine T cell responses to recombinant thioredoxin of Fasciola hepatica. Vet Parasitol. 1999;82:35–47. doi: 10.1016/s0304-4017(98)00268-4. [DOI] [PubMed] [Google Scholar]
- Taibi A, Aissi M, Harhoura K, Zenia S, Zait H, Hamrioui B. Evaluation of Fasciola hepatica infections in cattle in northeastern Algeria and the effects on both enzyme and hepatic damage, confirmed by scanning electron microscopy. Acta Parasitol. 2019;64:112–128. doi: 10.2478/s11686-018-00013-9. [DOI] [PubMed] [Google Scholar]
- Tansatit T, SAHAPHONG S, RIENGROJPITAK S, VIYANANT V, SOBHON P. Immunolocalization of cytoskeletal components in the tegument of the 3-week-old juvenile and adult Fasciola gigantica. Vet Parasitol. 2006;135:269–278. doi: 10.1016/j.vetpar.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Thorpe E. Comparative enzyme histochemistry of immature and mature stages of Fasciola hepatica. Exp Parasitol. 1968;22:150–159. doi: 10.1016/0014-4894(68)90088-x. [DOI] [PubMed] [Google Scholar]
- Tielens AGM, Heuvel JMVD, Bergh SGVD. Changes in energy metabolism of the juvenile Fasciola hepatica during its development in the liver parenchyma. Mol Biochem Parasitol. 1982;6:277–286. doi: 10.1016/0166-6851(82)90060-3. [DOI] [PubMed] [Google Scholar]
- Vasti A, Bianca V, Espino AM. Assessment of Fasciola hepatica glutathione S-transferase as an antigen for serodiagnosis of human chronic fascioliasis. Acta Trop. 2018;186:41–49. doi: 10.1016/j.actatropica.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waine GJ, BECKER M, YANG W, KALINNA B, MCMANUS DP. Cloning, molecular characterization, and functional activity of Schistosoma japonicum glyceraldehyde-3-phosphate dehydrogenase, a putative vaccine candidate against Schistosomiasis japonica. Infect Immun. 1993;61:4716–4723. doi: 10.1128/iai.61.11.4716-4723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tang H. The physical characteristics of human proteins in different biological functions. PLoS One. 2017;12:176–234. doi: 10.1371/journal.pone.0176234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RA, Wright JM, de Castro-Borges W, Parker-Manuel SJ, Dowle AA, Ashton PD, Young ND, Gasser RB, Spithill TW. Exploring the Fasciola hepatica tegument proteome. Int J Parasitol. 2011;41:1347–1359. doi: 10.1016/j.ijpara.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Yoo WG, Kim DW, Ju JW, Cho PY, Kim TI, Cho SH, Choi SH, Park HS, Kim TS, Hong SJ. Developmental transcriptomic features of the carcinogenic liver fluke, Clonorchis sinensis. PLoS Negl Trop Dis. 2011;5:e1208. doi: 10.1371/journal.pntd.0001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Peng L, Zhang Y, Liu Z, Li W, Chen S, Li G. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med Oncol. 2017;34:101. doi: 10.1007/s12032-017-0963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsser VL, Hoey EM, Trudgett A, Timson DJ. Biochemical characterisation of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) from the liver fluke, Fasciola hepatica. BBA-Proteins Proteom. 2014;1844:744–749. doi: 10.1016/j.bbapap.2014.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 152 kb)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the iProX with the dataset identifier IPX0002165000 (https://www.iprox.org//page/project.html?id=IPX0002165000).