Abstract
Malaria is a devastating infectious disease and the immune response is complex and dynamic during a course of a malarial infection. Rodent malaria models allow detailed time-series studies of the host response in multiple organs. Here, we describe two comprehensive datasets containing host transcriptomic data from both the blood and spleen throughout an acute blood stage infection of virulent or avirulent Plasmodium chabaudi infection in C57BL/6 mice. The mRNA expression profiles were generated using Illumina BeadChip microarray. These datasets provide a groundwork for comprehensive and comparative studies on host gene expression in early, acute and recovering phases of a blood stage infection in both the blood and spleen, to explore the interaction between the two, and importantly to investigate whether these responses differ in virulent and avirulent infections.
Subject terms: Parasite host response, Malaria, Transcriptomics
| Measurement(s) | transcriptome • gene expression • malaria |
| Technology Type(s) | Microarray |
| Factor Type(s) | blood versus spleen • virulent versus avirulent malaria infection |
| Sample Characteristic - Organism | Mus musculus |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12619232
Background & Summary
Malaria is a mosquito-borne disease caused by Plasmodium parasites, inflicting nearly half a million deaths annually, mostly in low and middle income countries (World Malaria Report 2018). The deaths are mainly caused by malaria complications that particularly affect young children and pregnant women. Clinical manifestations of malaria take place during the blood stages of the infection, during which host-parasite interactions occur mainly within the vasculature and most importantly in the spleen1,2. As the infection progresses, the parasite also interacts with, and damages multiple host organs via the process of sequestration3. This adherence of infected red blood cells to the endothelium of capillaries and venues, causes complications such as cerebral malaria and acute lung injury1,4. Leukocytes that are tissue resident, or that are recruited into the inflamed/damaged organs are in contact with the parasite or parasite product such as hemozoin (byproduct of hemoglobin degradation)5 and other pathogen-associated molecular patterns (PAMPs)6, resulting in activation of downstream immune genes. It has long been established that spleen is the most important immune organ that generate anti-malarial immune responses1,2. It has no afferent lymph vessels and collects its leukocytes directly from blood. The circulating immune cells continuously migrate into and out of the spleen, with their changed transcriptional activities during the course of malaria infection. In support of this, a recent study showed that parasite specific CD8+ T cell were primed in the spleen and migrated into the lungs7; another study showed that the ‘lung pathology’ signature can be picked up by analysing whole blood transcriptome8.
Genome-wide expression profiling is being increasingly applied to dissect the complex details of the host response to malaria infection9,10. As blood is the most accessible tissue in field studies, numerous field studies analyse blood transcriptomes as read-outs for anti-malarial immunity11–13. Therefore, it is very important to understand whether the immune responses detected in the blood serve as a reliable proxy for immune responses occurring in the spleen; if so, to what extent and at which stage of infection are they most closely related. To date, few studies have performed transcriptomic analyses of the blood in the mouse model9 and only one study carried out by us attempted to investigate the similarities between blood and spleen transcriptome14.
Here we describe two comprehensive, time-series analyses of the blood and spleen transcriptomic changes throughout the acute phase of blood stage infection (Fig. 1a) using a well-established rodent malaria model, Plasmodium chabaudi. This parasite is widely used to study host responses as it mirrors many pathological manifestations associated with P. falciparum infection, the most deadly species infecting humans, including parasite sequestration, severe malarial anemia, and chronic infection8,15,16. Time-series gene expression analysis is most helpful in identifying genes with transient expression changes and in investigation of gene regulation profiles during an infection. In our previous studies, we showed that pathology signatures can be picked up from blood transcriptome and they are quite distinct in the avirulent P. chabaudi AS or virulent P. chabaudi CB infection8; further analysing the blood and spleen transcriptome from the avirulent P. chabaudi AS infection, we identified only a small set of immune genes shared between them14. Here we report a new dataset of spleen transcriptome from the virulent P. chabaudi CB infection, which were collected from the same mice as the published blood transcriptome8. Our datasets, including the published PcAS/PcCB blood8, PcAS spleen14 and this new PcCB spleen transcriptome, offer a unique possibility to identify the complete set of activated or suppressed genes during an acute blood stage infection, to infer their rates of change and their causal effects. Further, it would be of high interest to investigate whether the interaction between blood and spleen differ in these two infections or whether more subtle relationship can be unearthed using more elaborate time modeling methods.
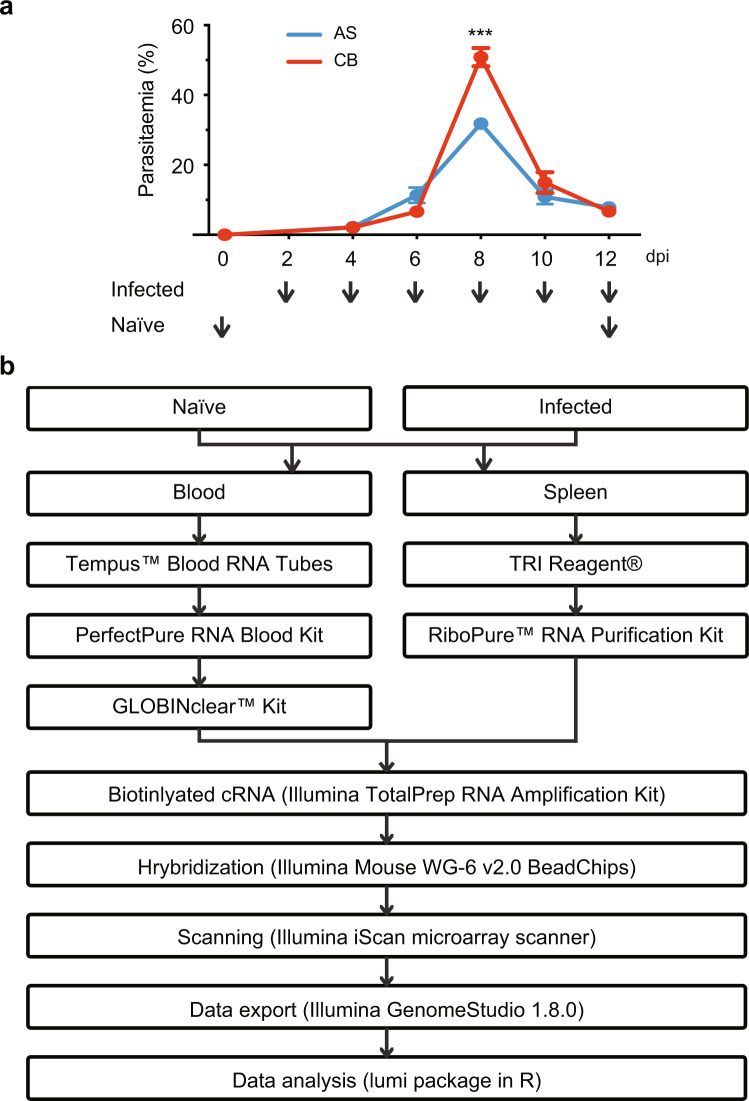
Fig. 1.
Sample collection and workflow. (a) Parasitemia (percentage of infected erythrocytes) of infected mice during the acute phase of blood stage infection and the time points (arrow heads) when the samples were collected. The mice were intraperitoneally infected with 105 erythrocytes that were infected with P. chabaudi parasite. (b) The flow chart illustrating the steps of microarray analysis.
Methods
Mice and parasites
Female C57BL/6 aged 6–8 weeks from the SPF (Specific Pathogen Free) unit at the Francis Crick Institute Mill Hill Laboratory were housed under reverse light conditions (light 19.00–07.00, dark 07.00–19.00 GMT) at 20–22 °C, and were allowed access to diet and water ad libitum. This study was carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 (Home Office license 80/2538 and 70/8326), and was approved by the Francis Crick Institute Ethical Committee.
Cloned lines of Plasmodium chabaudi chabaudi AS and CB were originally obtained from David Walliker, University of Edinburgh, United Kingdom. Infections were initiated by intraperitoneal injection of 105 parasitised erythrocytes derived from cryopreserved stocks. The course of infection was monitored on Giemsa-stained thin blood films by enumerating the percentage of erythrocytes infected with asexual parasites (parasitemia). The limit of detection for patent parasitemia was 0.01% infected erythrocytes. During the experiments, mouse condition were closely monitored. Core body temperature was measured with an infrared surface thermometer (Fluke); body weight was calculated relative to a baseline measurement taken before infection; and erythrocyte density was determined on a VetScan HM5 haematology system (Abaxis). The animals were euthanized upon reaching humane end points by showing the following signs: emaciation (more than 25% weight loss), persistent labored breathing, severe hypothermia (body temperature below 28 °C), inability to remain upright when conscious or lack of natural functions, or continuous convulsions lasting more than 5 min.
RNA isolation and preparation for microarray analysis
The sample collection and processing workflow is summarised in Fig. 1. These methods are expanded versions of descriptions in our related studies8,14. Female C57BL/6 mice aged between 6–8 weeks were injected intraperitoneally with 105 infected red blood cells of P. chabaudi AS or CB strain. At 2, 4, 6, 8, 10 and 12 days post infection (dpi), 0.5 mL of blood was collected via cardiac puncture into 1 mL Tempus RNA stabilising solution (Applied Biosystems). Spleens were aseptically removed and were homogenised immediately in TRI reagent (Ambion) by pulsing with a Polytron homogenising unit (Kinematic). An extra day 9 group was collected from PcCB infected mice that had reached humane end points. Naïve control samples were also collected on day 0 (the day of infection) and day 12 (the end of the experiment). Samples were snap frozen in dry ice and stored at −80 °C until RNA isolation.
Total blood RNA was extracted using PerfectPure RNA Blood Kit (5 PRIME), and Globin mRNA was removed from 2 µg of total isolated RNA using GLOBINclear 96-well Mouse/Rat Whole Blood Globin Reduction Kit (Ambion) according to the manufacturer’s instructions. Total splenic RNA was extracted using RiboPure RNA Purification Kit (Ambion) following the manufacture’s protocol. All RNA samples derived from the same experiment were isolated altogether at the end of the experiment, in 2–3 batches within a day. Globin mRNA reduction was performed in 2 batches, one for all PcAS blood samples and the other for all PcCB blood. Batch information for RNA isolation and subsequent processing was provided in Online-only Tables 1–4.
Online-only Table 1.
Batch information, RNA quality and concentration and related GEO accession numbers for PcAS blood (GSE9363119).
| Series GSE93631 | PRJNA361313 | Caliper LabChip | Nanodrop | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GEO accession ID | Sample Name in GEO | BeadChip No. | RQS | rRNA 28 s/18 s | [ng/ul] | 260/280 | 260/230 | RNA isolation batch | Globin mRNA reduction | cRNA preparation | BeadChip hybridasation |
| GSM2459164 | AS_naive_D0 rep 1 | 8762536135_E | 9 | 2.85 | 147.7 | 2.1 | 2.25 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459165 | AS_naive_D0 rep 2 | 8762536084_A | 8.2 | 2.27 | 164.28 | 2.12 | 2.24 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459166 | AS_naive_D0 rep 3 | 8762536052_F | 8.3 | 2.33 | 244.74 | 2.1 | 2.24 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459167 | AS_naive_D12 rep 1 | 8762536072_E | 8.2 | 2.18 | 325.34 | 2.1 | 2.22 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459168 | AS_naive_D12 rep 2 | 8784170061_E | 8 | 2.03 | 372.4 | 2.08 | 2.23 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459169 | AS_naive_D12 rep 3 | 8784170059_F | 8.3 | 2.13 | 305.17 | 2.1 | 2.17 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459170 | AS_D2 rep 1 | 8762536072_F | 8.7 | 2.58 | 210.18 | 2.13 | 2.2 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459171 | AS_D2 rep 2 | 8762536084_B | 7.6 | 2.62 | 343.31 | 2.09 | 2.21 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459172 | AS_D2 rep 3 | 8762536135_C | 6.9 | 3.32 | 269.34 | 2.1 | 2.28 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459173 | AS_D2 rep 4 | 8784170059_C | 7.5 | 2.8 | 504.89 | 2.17 | 2.27 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459174 | AS_D4 rep 1 | 8762536052_B | 7.9 | 1.9 | 314.44 | 2.12 | 2.26 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459175 | AS_D4 rep 2 | 8762536084_F | 7.9 | 1.91 | 315.67 | 2.1 | 2.26 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459176 | AS_D4 rep 3 | 8784170061_A | 7.8 | 2.06 | 258.9 | 2.14 | 2.27 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch |
| GSM2459177 | AS_D6 rep 1 | 8762536052_E | 7.1 | 1.97 | 658.79 | 2.25 | 2.38 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459178 | AS_D6 rep 2 | 8762536135_A | 6.5 | 2.24 | 737.49 | 2.23 | 2.38 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459179 | AS_D6 rep 3 | 8784170059_B | 7.2 | 2.54 | 669.17 | 2.28 | 2.41 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459180 | AS_D8 rep 1 | 8762536084_D | 7.2 | 1.15 | 1112.8 | 2.18 | 2.26 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459181 | AS_D8 rep 2 | 8762536135_F | 7 | 1.94 | 415.4 | 2.19 | 2.4 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459182 | AS_D8 rep 3 | 8784170059_E | 7.4 | 3.35 | 625.92 | 2.29 | 2.39 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459183 | AS_D10 rep 1 | 8762536052_C | 7.9 | 0.18 | 2845.83 | 2.07 | 2.16 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459184 | AS_D10 rep 2 | 8762536135_D | 7.8 | 0.21 | 3054.76 | 2.07 | 2.16 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459185 | AS_D10 rep 3 | 8784170061_B | 7.7 | 0.22 | 3857.18 | 1.88 | 1.95 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459186 | AS_D12 rep 1 | 8762536072_C | 7.4 | 0.94 | 4074.58 | 1.75 | 1.82 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459187 | AS_D12 rep 2 | 8762536084_E | 7.2 | 0.97 | 3442.65 | 2.02 | 2.1 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
| GSM2459188 | AS_D12 rep 3 | 8784170061_C | 7.9 | 1.02 | 3104.6 | 2.05 | 2.14 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch |
Online-only Table 4.
Batch information, RNA quality and concentration and related GEO accession numbers for PcCB spleen (GSE14578121).
| Series GSE145781 | PRJNA608201 | Caliper LabChip | Nanodrop | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GEO accession ID | Sample Name in GEO | BeadChip No. | RIN | rRNA 28 s/18 s | [ng/ul] | 260/280 | 260/230 | RNA isolation batch | cRNA preparation | BeadChip hybridasation |
| GSM4332890 | Spleen, CB-Naïve-D0.rep1 | 9440690042_E | 8.0 | 10.69 | 481.54 | 2.15 | 2.19 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332891 | Spleen, CB-Naïve-D0.rep2 | 9440690065_D | 7.7 | 13.97 | 503.42 | 2.19 | 2.24 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332892 | Spleen, CB-naïve-D0.rep3 | 9440690046_F | 9.3 | 10.02 | 390.79 | 2.15 | 2.23 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332893 | Spleen, CB-Naïve-D12.rep1 | 9440690056_F | 7.6 | 1.46 | 516.78 | 2.19 | 2.28 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332894 | Spleen, CB-Naïve-D12.rep2 | 9440690059_B | 7.4 | 2.16 | 501.91 | 2.17 | 2.19 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332895 | Spleen, CB-Naïve-D12.rep3 | 9440690061_A | 8.1 | 3.47 | 890.14 | 2.19 | 2.26 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332896 | Spleen, CB-D2.rep1 | 9440690056_B | 8.5 | 3.48 | 504.5 | 2.14 | 2.25 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332897 | Spleen, CB-D2.rep2 | 9440690046_A | 8.2 | 2.29 | 575.9 | 2.16 | 2.24 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332898 | Spleen, CB-D2.rep3 | 9440690061_F | 8.0 | 3.04 | 527.61 | 2.14 | 2.23 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332899 | Spleen, CB-D2.rep4 | 9440690059_C | 8.1 | 2.24 | 633.96 | 2.17 | 2.25 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332900 | Spleen, CB-D4.rep1 | 9440690046_B | 8.0 | 2.29 | 1481.51 | 2.15 | 2.28 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332901 | Spleen, CB-D4.rep2 | 9440690061_C | 7.1 | 3.93 | 1171.25 | 2.14 | 1.84 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332902 | Spleen, CB-D4.rep3 | 9440690056_E | 7.9 | 3.29 | 840.35 | 2.17 | 2.24 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332903 | Spleen, CB-D4.rep4 | 9440690065_F | 7.7 | 2.14 | 1617.19 | 2.15 | 2.27 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332904 | Spleen, CB-D6.rep1 | 9440690046_C | 8.9 | 8.89 | 1627.51 | 2.14 | 2.22 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332905 | Spleen, CB-D6.rep2 | 9440690042_F | 7.4 | 9.01 | 2846.56 | 2.13 | 2.23 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332906 | Spleen, CB-D6.rep3 | 9440690056_A | 8.0 | 4.39 | 1475.99 | 2.16 | 2.25 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332907 | Spleen, CB-D6.rep4 | 9440690061_B | 7.7 | 2.73 | 2511.74 | 2.14 | 2.23 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332908 | Spleen, CB-D8.rep1 | 9440690056_C | 7.6 | 3.14 | 2476.91 | 2.15 | 2.22 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332909 | Spleen, CB-D8.rep2 | 9440690065_A | 8.4 | 3.42 | 3756.04 | 2.03 | 2.13 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332910 | Spleen, CB-D8.rep3 | 9440690061_E | 8.2 | 3.38 | 1248.69 | 2.15 | 2.22 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332911 | Spleen, CB-D8.rep4 | 9440690059_D | 8.2 | 3.22 | 2456.41 | 2.16 | 2.24 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332912 | Spleen, CB-D9.rep1 | 9440690065_C | 8.1 | 2.72 | 1057.83 | 2.16 | 2.22 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332913 | Spleen, CB-D9.rep2 | 9440690059_E | 7.9 | 2.26 | 3901.31 | 2 | 2.11 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332914 | Spleen, CB-D9.rep3 | 9440690061_D | 8.0 | 2.19 | 2278.08 | 2.15 | 2.24 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332915 | Spleen, CB-D10.rep1 | 9440690065_E | 8.6 | 2.86 | 1628.2 | 2.17 | 2.25 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332916 | Spleen, CB-D10.rep2 | 9440690056_D | 8.4 | 2.51 | 1162.55 | 2.17 | 2.24 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332917 | Spleen, CB-D10.rep3 | 9440690046_E | 8.5 | 2.27 | 2509.68 | 2.15 | 2.23 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332918 | Spleen, CB-D10.rep4 | 9440690059_A | 8.7 | 2.54 | 1224.36 | 2.18 | 2.24 | 06/12/2013_Batch1 | In one batch | In one batch |
| GSM4332919 | Spleen, CB-D12.rep1 | 9440690059_F | 8.6 | 2.62 | 1172.01 | 2.17 | 2.26 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332920 | Spleen, CB-D12.rep2 | 9440690046_D | 8.3 | 2.4 | 909.93 | 2.19 | 2.25 | 06/12/2013_Batch2 | In one batch | In one batch |
| GSM4332921 | Spleen, CB-D12.rep3 | 9440690065_B | 8.6 | 2.62 | 1047.02 | 2.18 | 2.26 | 06/12/2013_Batch2 | In one batch | In one batch |
To test whether parasite RNA gives signals in microarray analysis of mouse gene expression, an independent experiment was performed using naïve and infected mouse blood RNA, and purified parasite RNA. RNA from naïve or infected blood was processed as described above. Parasite purification and RNA extraction methods were performed as described previously17. Briefly, infected blood collected at day 8 post infection were depleted of leukocytes by filtration through Plasmodipur filters (EuroProxima) followed by erythrocyte lysis using 0.15% saponin (Sigma) in ice-cold PBS and extensive washes with PBS. Purified parasite pellets were then resuspended in 1 ml TRI reagent (Ambion), snap-frozen on dry ice and kept at −80 °C. Parasite RNA was extracted using RiboPure RNA Purification Kit (Ambion) according to the manufacturer’s protocols. Parasite RNA was also subjected to Globin mRNA removal. Batch information for RNA isolation and processing was provided in Online-only Table 5.
Online-only Table 5.
Batch information, RNA quality and concentration and related GEO accession numbers for parasite RNA (GSE14563422).
| Series GSE145634 | PRJNA607775 | Bioanalyser | Nanodrop | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEO accession ID | Sample Name in GEO | BeadChip No. | RIN | rRNA 28 s/18 s | [ng/ul] | 260/280 | 260/230 | RNA isolation batch | Comments | Globin mRNA reduction | cRNA preparation | BeadChip hybridasation |
| GSM4322547 | naïve blood rep1 | 8697771087_A | 9 | 2.85 | 147.7 | 2.1 | 2.25 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM4322548 | Infected D8 rep1 | 8697771087_B | 7 | 1.94 | 415.4 | 2.19 | 2.4 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM4322549 | Infected D8 rep2 | 8697771087_C | 7.2 | 1.15 | 1112.8 | 2.18 | 2.26 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM4322550 | naïve blood rep2 | 8697771087_D | 8.3 | 2.33 | 244.74 | 2.1 | 2.24 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM4322551 | Infected D8 rep3 | 8697771087_F | 9.1 | 2.2 | 1050.82 | 2.15 | 2.42 | 31/07/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM4322552 | Infected D8 rep4 | 8697771096_A | N/A | 2.5 | 667.16 | 2.22 | 2.55 | 31/07/2013_Batch1 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM4322553 | naïve blood rep3 | 8697771096_B | 8.2 | 2.27 | 164.28 | 2.12 | 2.24 | 03/06/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM4322554 | Parasite RNA rep1 | 8697771096_C | N/A | 1.1 | 351.09 | 2.14 | 2.31 | 31/07/2013_Batch1 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM4322555 | Infected D8 rep5 | 8697771096_D | 7.4 | 3.35 | 625.92 | 2.29 | 2.39 | 03/06/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM4322556 | Parasite RNA rep2 | 8697771096_E | N/A | 0 | 248.67 | 2.14 | 2.54 | 31/07/2013_Batch1 | Parasite rRNA detected | In one batch | In one batch | In one batch |
Biotinylated, amplified antisense complement RNA (cRNA) samples were prepared from 300 ng of either globin reduced blood/parasite RNA, or splenic total RNA using Illumina TotalPrep RNA Amplification Kit (Ambion). cRNA was prepared in 4 batches: PcAS blood, PcCB blood, all spleen samples and blood/parasite RNA.
At each step, the quantity of the RNA samples was measured using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific) and the quality of RNA was verified using Agilent 2100 bioanalyzer (Agilent Technologies) or Caliper LabChip GX (Caliper Life Sciences), provided as RNA Integrity Number (RIN) or RNA Quality Score (RQS), respectively. Only RNA samples with RIN/RQS above 7 were used for subsequent treatment and analysis; however, for blood samples with high parasite load, parasite rRNA affected RIN/RQS determination, and the quality of these samples were determined by examining electropherograms. RNA concentration and RIN/RQS of each sample were also provided in Online-only Tables 1–5.
Microarray hybridisation and raw data export
The following procedures for microarray hybridisation and data acquisition was done for each sample. Briefly, 1.5 µg of labelled cRNA was hybridised to Illumina Mouse WG-6 v2.0 Expression BeadChip (consisting of 45,281 probe sets representing 30,854 genes) according to the manufacturer’s protocols. The arrays were then washed, blocked, stained and scanned on an Illumina iScan, following the manufacturer’s instructions. Illumina BeadStudio/GenomeStudio 1.8.0 software was used to generate signal intensity values, quality control values, and to subtract background. Hybridisation was performed in 4 batches: PcAS blood, PcCB blood, all spleen samples and blood/parasite RNA.
Microarray data preparation and analysis
Data input, quality control, variance stabilisation, log transformation and quantile normalisation were performed using the lumi package18. The full feature set (a total of 45,281 probes) of each sample was used for the following analyses including hierarchical clustering, principle component analysis (PCA) and Euclidean distance, all conducted using R 3.6.0 (www.r-project.org). For hierarchical clustering, agglomerative clustering with average linkage was used.
Data Records
Gene expression data were deposited at the Gene Expression Omnibus database (GEO) under the following accession numbers: GSE9363119 (AS and CB blood) and GSE12339120 (AS spleen) which were published previously8,14; GSE14578121 (CB spleen) and GSE14563422 (the raw data of parasite RNA experiment) which were new datasets.
GEO accession numbers of blood or spleen samples that were derived from the same mouse were provided in Tables 1 and 2. Batch information, RNA quality and concentration and related GEO accession numbers were provided in Online-only Tables 1–5.
Table 1.
GEO accession numbers of blood or spleen samples that were derived the same PcAS infected mouse (Data Source: GSE9363119 and GSE12339120).
| mouse No. | Blood (GSE9363119) | Spleen (GSE12339120) | ||
|---|---|---|---|---|
| GEO accession | BeadChip No. | GEO accession | BeadChip No. | |
| naïve_D0_m1 | GSM2459164 | 8762536135_E | GSM3502544 | 9440690022_B |
| naïve_D0_m2 | GSM2459165 | 8762536084_A | GSM3502545 | 9440690030_C |
| naïve_D0_m3 | GSM2459166 | 8762536052_F | GSM3502546 | 9440690035_B |
| naïve_D12_m1 | GSM2459167 | 8762536072_E | GSM3502568 | 9440690030_F |
| naïve_D12_m2 | GSM2459168 | 8784170061_E | GSM3502569 | 9440690037_C |
| naïve_D12_m3 | GSM2459169 | 8784170059_F | GSM3502570 | 9440690042_A |
| AS_D2_m1 | GSM2459172 | 8762536135_C | GSM3502547 | 9440690022_C |
| AS_D2_m2 | GSM2459171 | 8762536084_B | GSM3502548 | 9440690035_A |
| AS_D2_m3 | GSM2459173 | 8784170059_C | GSM3502549 | 9440690037_D |
| AS_D2_m4 | GSM2459170 | 8762536072_F | GSM3502550 | 9440690042_C |
| AS_D4_m1 | N/A | N/A | GSM3502551 | 9440690022_D |
| AS_D4_m2 | GSM2459175 | 8762536084_F | GSM3502552 | 9440690030_A |
| AS_D4_m3 | GSM2459174 | 8762536052_B | GSM3502553 | 9440690037_E |
| AS_D4_m4 | GSM2459176 | 8784170061_A | GSM3502554 | 9440690042_B |
| AS_D6_m1 | N/A | N/A | GSM3502555 | 9440690022_E |
| AS_D6_m2 | GSM2459177 | 8762536052_E | GSM3502556 | 9440690035_C |
| AS_D6_m3 | GSM2459179 | 8784170059_B | GSM3502557 | 9440690042_D |
| AS_D6_m4 | GSM2459178 | 8762536135_A | GSM3502558 | 9440690037_A |
| AS_D8_m1 | GSM2459182 | 8784170059_E | GSM3502559 | 9440690022_F |
| AS_D8_m2 | GSM2459181 | 8762536135_F | GSM3502560 | 9440690030_B |
| AS_D8_m3 | GSM2459180 | 8762536084_D | GSM3502561 | 9440690035_D |
| AS_D10_m1 | GSM2459185 | 8784170061_B | GSM3502562 | 9440690035_E |
| AS_D10_m2 | GSM2459184 | 8762536135_D | GSM3502563 | 9440690030_D |
| AS_D10_m3 | GSM2459183 | 8762536052_C | GSM3502564 | 9440690037_F |
| AS_D12_m1 | GSM2459186 | 8762536072_C | GSM3502565 | 9440690030_E |
| AS_D12_m2 | GSM2459188 | 8784170061_C | GSM3502566 | 9440690035_F |
| AS_D12_m3 | GSM2459187 | 8762536084_E | GSM3502567 | 9440690037_B |
Table 2.
GEO accession numbers of blood or spleen samples that were derived the same PcCB infected mouse (Data Source: GSE9363119 and GSE14578121).
| mouse No. | Blood (GSE9363119) | Spleen (GSE14578121) | ||
|---|---|---|---|---|
| GEO accession | BeadChip No. | GEO accession | BeadChip No. | |
| naïve_D0_m1 | GSM2459189 | 8762536055_D | GSM4332890 | 9440690042_E |
| naïve_D0_m2 | GSM2459190 | 8762536056_E | GSM4332891 | 9440690065_D |
| naïve_D0_m3 | GSM2459191 | 8762536079_E | GSM4332892 | 9440690046_F |
| naïve_D12_m1 | GSM2459192 | 8762536054_F | GSM4332893 | 9440690056_F |
| naïve_D12_m2 | GSM2459193 | 8762536055_E | GSM4332894 | 9440690059_B |
| naïve_D12_m3 | GSM2459194 | 8762536056_A | GSM4332895 | 9440690061_A |
| CB_D2_m1 | GSM2459195 | 8762536049_B | GSM4332896 | 9440690056_B |
| CB_D2_m2 | GSM2459196 | 8762536054_C | GSM4332897 | 9440690046_A |
| CB_D2_m3 | GSM2459197 | 8762536055_F | GSM4332898 | 9440690061_F |
| CB_D2_m4 | GSM2459198 | 8762536079_C | GSM4332899 | 9440690059_C |
| CB_D4_m1 | GSM2459199 | 8762536049_F | GSM4332900 | 9440690046_B |
| CB_D4_m2 | GSM2459200 | 8762536054_D | GSM4332901 | 9440690061_C |
| CB_D4_m3 | GSM2459201 | 8762536056_B | GSM4332902 | 9440690056_E |
| CB_D4_m4 | GSM2459202 | 8762536078_A | GSM4332903 | 9440690065_F |
| CB_D6_m1 | GSM2459206 | 8762536097_B | GSM4332904 | 9440690046_C |
| CB_D6_m2 | GSM2459203 | 8762536055_A | GSM4332905 | 9440690042_F |
| CB_D6_m3 | GSM2459204 | 8762536056_F | GSM4332906 | 9440690056_A |
| CB_D6_m4 | GSM2459205 | 8762536079_A | GSM4332907 | 9440690061_B |
| CB_D8_m1 | GSM2459207 | 8762536049_D | GSM4332908 | 9440690056_C |
| CB_D8_m2 | GSM2459208 | 8762536054_E | GSM4332909 | 9440690065_A |
| CB_D8_m3 | GSM2459209 | 8762536078_D | GSM4332910 | 9440690061_E |
| CB_D8_m4 | GSM2459210 | 8762536097_F | GSM4332911 | 9440690059_D |
| CB_D9_m1 | GSM2459221 | 8762536054_A | GSM4332912 | 9440690065_C |
| CB_D9_m2 | GSM2459218 | 8762536055_B | GSM4332913 | 9440690059_E |
| CB_D9_m3 | GSM2459219 | 8762536097_D | N/A | N/A |
| CB_D9_m4 | GSM2459220 | 8762536078_F | GSM4332914 | 9440690061_D |
| CB_D10_m1 | GSM2459211 | 8762536049_C | GSM4332915 | 9440690065_E |
| CB_D10_m2 | GSM2459212 | 8762536056_C | GSM4332916 | 9440690056_D |
| CB_D10_m3 | GSM2459213 | 8762536078_B | GSM4332917 | 9440690046_E |
| CB_D10_m4 | GSM2459214 | 8762536079_D | GSM4332918 | 9440690059_A |
| CB_D12_m1 | GSM2459215 | 8762536049_E | GSM4332919 | 9440690059_F |
| CB_D12_m2 | GSM2459216 | 8762536078_C | GSM4332920 | 9440690046_D |
| CB_D12_m3 | GSM2459217 | 8762536079_B | GSM4332921 | 9440690065_B |
Technical Validation
Sample preparations and quality control
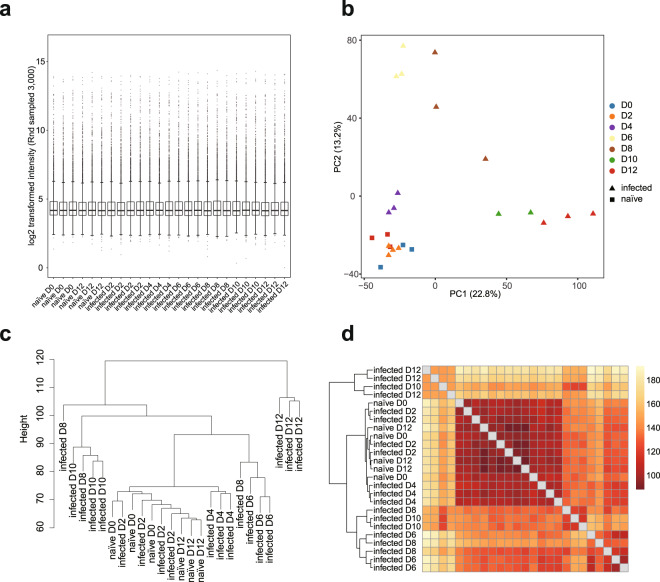
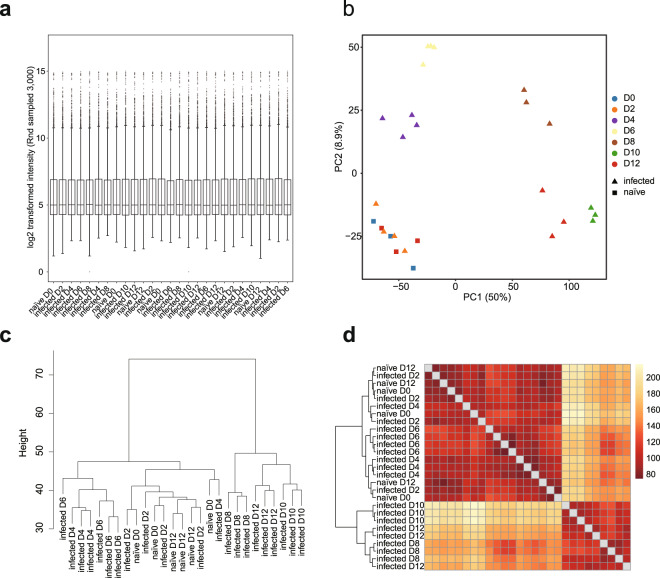
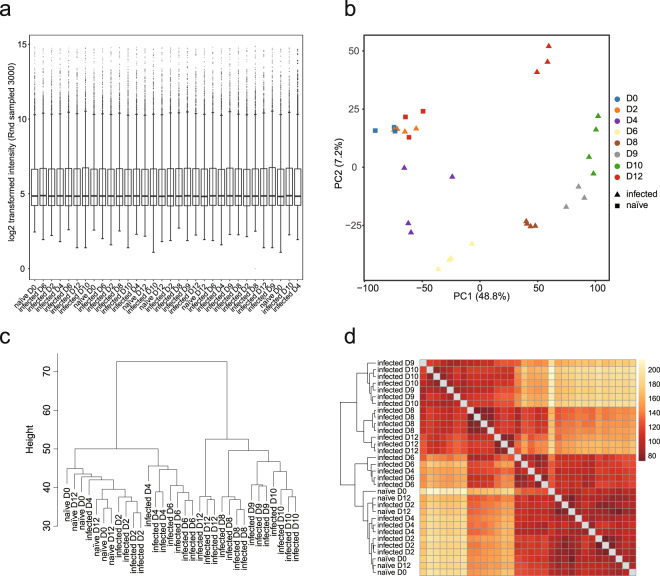
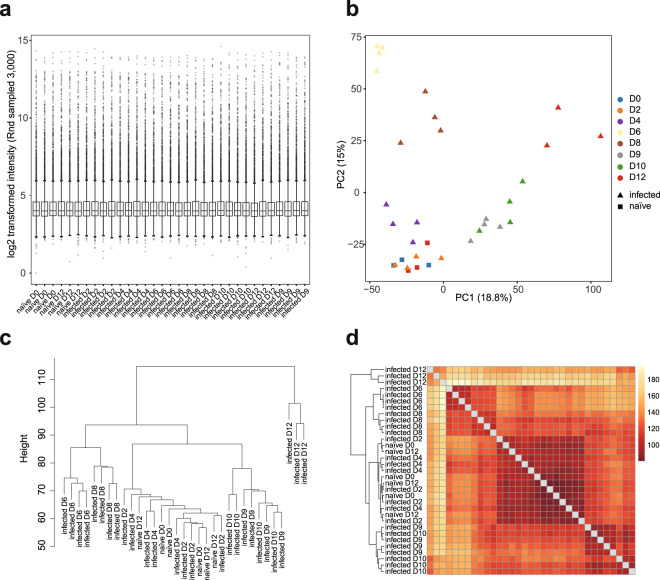
Several aspects of the experiment were designed to ensure the quality of the data. For example, the control naïve mice were randomly selected from the same batch of age-matched mice, 3 of which were sacrificed at the same day of infection, and 3 of which were housed under the same conditions as the infected mice and were sacrificed along with mice after 12 days of infection. All mice in the infected group were infected at the same time and were randomly selected for sample collection at each time point. Overall, both blood and spleen samples collected from either PcAS or PcCB infections showed uniformed normalised intensities (Figs. 2a and 3a). Importantly, high similarities were observed between biological replicates (Figs. 2 and 3).
Fig. 2.
Quality check of BeadChip gene expression data of PcAS blood samples. (a) Box plot showing distribution of 3,000 randomly sampled probe signals for normalised PcAS infected blood expression data. The median, two hinges, two whiskers and outlying points were shown. (b) Principal component analysis of normalised expression data of naïve and infected blood samples. (c) Hierarchical clustering plot of normalised intensity data among the samples was generated using agglomerative clustering with average linkage. (d) Heatmap of Euclidean distance. A full feature set was used for (b–d). This dataset was submitted to GEO (GSE93631).
Fig. 3.
Quality check of BeadChip gene expression data of PcAS spleen samples.(a) Box plot showing distribution of 3,000 randomly sampled probe signals for normalised PcAS spleen expression data. The median, two hinges, two whiskers and outlying points were shown. (b) Principal component analysis of normalised expression data of naïve and infected spleen samples. (c) Hierarchical clustering plot of normalised intensity data among the samples was generated using agglomerative clustering with average linkage. (d) Heatmap of Euclidean distance. A full feature set was used for (b-d). This dataset was submitted to GEO (GSE123391).
Quality check of time dependent responses
In the naïve control group, mice collected at day 0 or day 12 clustered together in all 4 datasets. Interestingly, samples collected at 2 dpi at which time point the infection rate was below microscopic detection level, also cluster with naïve groups; and this was observed in both the blood and spleen in either infection (Figs. 2–5). In the avirulent PcAS infection, from day 4 onwards, the expression profiles changed significantly, showing clear time-dependent responses in both the blood and the spleen. At 4 dpi, spleen showing longer distance from the naïve groups than the blood, 46.4 vs 21.6 distance on PC2 (Figs. 2b and 3b), which indicates higher host responses in the spleen than in the blood. Similar responses took place in the virulent PcCB infection, showing 4 dpi-naïve distance on PC2 of 18.5 in the blood vs 32.0 in the spleen (Figs. 4b and 5b). This is in line with the current view that parasite-host interaction mainly take place in the spleen.
Fig. 5.
Quality check of BeadChip gene expression data of PcCB spleen samples. (a) Box plot showing distribution of 3,000 randomly sampled probe signals for normalised PcCB spleen expression data. The median, two hinges, two whiskers and outlying points were shown. (b) Principal component analysis of normalised expression data of naïve and infected spleen samples. (c) Hierarchical clustering plot of normalised intensity data among the samples was generated using agglomerative clustering with average linkage. (d) Heatmap of Euclidean distance. A full feature set was used for (b-d). This dataset was submitted to GEO (GSE145781).
Fig. 4.
Quality check of BeadChip gene expression data of PcCB blood samples. (a) Box plot showing distribution of 3,000 randomly sampled probe signals for normalised PcCB blood expression data. The median, two hinges, two whiskers and outlying points were shown. (b) Principal component analysis of normalised expression data of naïve and infected blood samples. (c) Hierarchical clustering plot of normalised intensity data among the samples was generated using agglomerative clustering with average linkage. (d) Heatmap of Euclidean distance. A full feature set was used for (b-d). This dataset was submitted to GEO (GSE93631).
An interesting difference between the blood and spleen is the divergence between day 6 and 8 post infection. In the avirulent PcAS infection the distances between 6 and 8 dpi on PC1 were 36.4 in the blood and 91.8 in the spleen (Figs. 2b and 3b). This is slightly less striking in the virulent PcCB infection, with 21.1 in the blood and 66.8 in the spleen. These differences are also apparent in hierarchical clustering and heatmaps of euclidean distance (Figs. 2 and 3).
The striking differences between PcAS and PcCB infections were the responses took place between day 10 and 12 post infection. In the avirulent PcAS infection, while day 10 and 12 were clearly different from previous infected samples, they clustered tightly together in both blood and spleen samples (Figs. 2b and 3b). By contrast, in the virulent PcCB infection the two days differed in both PC1 and PC2 (Figs. 4b and 5b), and the heatmaps of Euclidean distance showed that 12 dpi clearly separate from other samples (Figs. 4d and 5d).
Parasite RNA does not affect BeadChip gene expression results
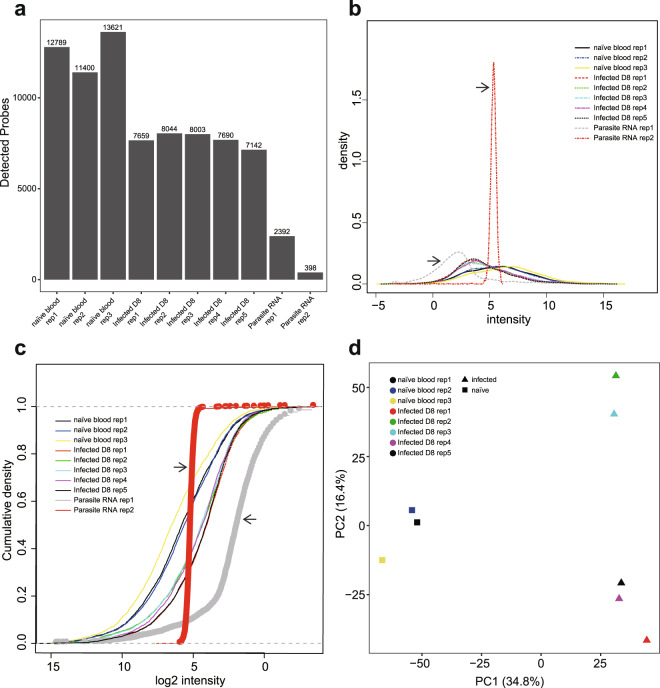
Because the malaria parasite infects erythrocytes, RNA isolated from the infected blood contains both mouse and Plasmodium RNA. We therefore performed an independent experiment to rule out the interference of parasite RNA in downstream analysis using Mouse WG-6 v2.0 Expression BeadChip. We prepared purified P. chabaudi AS parasite RNA by passing infected blood through a leukocyte filter, usually removing more than 99% leukocytes, followed by erythrocyte lysis and extensive washes. Globin mRNA removal was also performed as for infected blood samples. As shown in Fig. 6, the numbers of detectable probes in parasite samples were significantly lower (Fig. 6a), and this hindered the normalisation step. Moreover, the non-normalised expression data of parasite samples showed very different density or cumulative density profiles (Fig. 6b,c). After removing parasite data from the dataset, the subsequent analyses can be easily performed and it was clear that the infected blood collected at 8 dpi significantly differed from naïve blood (Fig. 6d), validating our previous finding.
Fig. 6.
Validation of parasite RNA does not affect BeadChip gene expression results. (a) Bar chart showing the number of probes detected in each sample. (b) Density plot of non-normalised expression data showing the signal density distribution. (c) Cumulative distribution function plot of non-normalised expression data of each sample. Arrowheads indicate parasite samples. (d) PCA plot of normalised expression data from infected and naïve blood samples after excluding parasite samples. This dataset was submitted to GEO (GSE145634).
Usage Notes
One major advantage of this study is that we collected both the blood and spleen simultaneously from the same mouse (GEO accession numbers of blood or spleen samples that were derived from the same mouse were provided in Tables 1 and 2) throughout the acute phase of blood stage infection, from as early as day 2 post infection when the infection rate was below microscopic detection, till day 12 post infection when the parasite load was controlled. Moreover, the samples were collected at 2-day intervals to allow a more detailed analysis of the time-dependent transcriptional changes. It is hoped that this will facilitate the users to investigate in detail the interaction between the blood and the spleen. It would also provide some answers to the question of whether some of the responses in the blood happen before or after the spleen responses, for example using time series modelling. And importantly, we collected samples from both the virulent PcCB and the avirulent PcAS infections. It would be of high interest to investigate whether the interaction between blood and spleen differ in these two infections.
Acknowledgements
The authors would like to thank Dr. Lu Chen for critical assistance in data analysis. This work was supported by the Wellcome Trust (WT102907) and Francis Crick Institute (FC001101), and a Wellcome Trust Senior Investigator award to Jean Langhorne (104777/Z/14/Z).
Online-only Tables
Online-only Table 2.
Batch information, RNA quality and concentration and related GEO accession numbers for PcCB blood (GSE9363119).
| Series GSE93631 | PRJNA361313 | Bioanalyser | Nanodrop | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEO accession ID | Sample Name in GEO | BeadChip No. | RIN | rRNA 28 s/18 s | [ng/ul] | 260/280 | 260/230 | RNA isolation batch | Comments | Globin mRNA reduction | cRNA preparation | BeadChip hybridasation |
| GSM2459189 | CB_naive_D0 rep 1 | 8762536055_D | 9.5 | 1.9 | 317.8 | 2.11 | 2.2 | 16/10/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM2459190 | CB_naive_D0 rep 2 | 8762536056_E | 9.3 | 1.7 | 248.7 | 2.05 | 2.2 | 16/10/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM2459191 | CB_naive_D0 rep 3 | 8762536079_E | 9.4 | 1.6 | 251.47 | 2.04 | 2.22 | 16/10/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM2459192 | CB_naive_D12 rep 1 | 8762536054_F | 9.9 | 1.9 | 331.23 | 2.1 | 2.2 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459193 | CB_naive_D12 rep 2 | 8762536055_E | 9.1 | 1.6 | 331.11 | 2.11 | 2.27 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459194 | CB_naive_D12 rep 3 | 8762536056_A | 9.6 | 1.9 | 537.73 | 2.11 | 2.21 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459195 | CB_D2 rep 1 | 8762536049_B | 9.4 | 1.8 | 324.98 | 2.11 | 2.29 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459196 | CB_D2 rep 2 | 8762536054_C | 9.7 | 2 | 334.51 | 2.13 | 2.24 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459197 | CB_D2 rep 3 | 8762536055_F | 9.4 | 1.7 | 285.15 | 2.13 | 2.28 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459198 | CB_D2 rep 4 | 8762536079_C | 9.2 | 1.7 | 210.81 | 2.12 | 2.22 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459199 | CB_D4 rep 1 | 8762536049_F | 9.6 | 2.4 | 437.07 | 2.11 | 2.25 | 16/10/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM2459200 | CB_D4 rep 2 | 8762536054_D | 9.5 | 2.5 | 237.53 | 2.1 | 2.23 | 16/10/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM2459201 | CB_D4 rep 3 | 8762536056_B | 10 | 2.1 | 295.06 | 2.08 | 2.15 | 16/10/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM2459202 | CB_D4 rep 4 | 8762536078_A | 10 | 2.1 | 245.02 | 2.08 | 2.25 | 16/10/2013_Batch1 | In one batch | In one batch | In one batch | |
| GSM2459203 | CB_D6 rep 1 | 8762536055_A | N/A | 0.9 | 819.77 | 2.26 | 2.55 | 16/10/2013_Batch1 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM2459204 | CB_D6 rep 2 | 8762536056_F | N/A | 1.3 | 821.73 | 2.31 | 2.59 | 16/10/2013_Batch1 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM2459205 | CB_D6 rep 3 | 8762536079_A | N/A | 1.7 | 427.12 | 2.17 | 2.42 | 16/10/2013_Batch1 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM2459206 | CB_D6 rep 4 | 8762536097_B | N/A | 0.6 | 713.05 | 2.29 | 2.58 | 16/10/2013_Batch1 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM2459207 | CB_D8 rep 1 | 8762536049_D | N/A | 1.6 | 1512.02 | 2.2 | 2.56 | 16/10/2013_Batch3 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM2459208 | CB_D8 rep 2 | 8762536054_E | N/A | 1.2 | 635.79 | 2.34 | 2.6 | 16/10/2013_Batch3 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM2459209 | CB_D8 rep 3 | 8762536078_D | N/A | 1.1 | 927.2 | 2.22 | 2.59 | 16/10/2013_Batch3 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM2459210 | CB_D8 rep 4 | 8762536097_F | N/A | 1.4 | 1145.57 | 2.25 | 2.56 | 16/10/2013_Batch3 | Parasite rRNA detected | In one batch | In one batch | In one batch |
| GSM2459211 | CB_D10 rep 1 | 8762536049_C | 10 | 2.1 | 1004.02 | 2.13 | 2.18 | 16/10/2013_Batch3 | In one batch | In one batch | In one batch | |
| GSM2459212 | CB_D10 rep 2 | 8762536056_C | 10 | 2 | 1969.42 | 2.1 | 2.22 | 16/10/2013_Batch3 | In one batch | In one batch | In one batch | |
| GSM2459213 | CB_D10 rep 3 | 8762536078_B | 10 | 2.1 | 3733.01 | 1.93 | 1.98 | 16/10/2013_Batch3 | In one batch | In one batch | In one batch | |
| GSM2459214 | CB_D10 rep 4 | 8762536079_D | 10 | 2.1 | 2197.77 | 2.09 | 2.18 | 16/10/2013_Batch3 | In one batch | In one batch | In one batch | |
| GSM2459215 | CB_D12 rep 1 | 8762536049_E | 9.9 | 1.8 | 4052.15 | 1.77 | 1.84 | 16/10/2013_Batch3 | In one batch | In one batch | In one batch | |
| GSM2459216 | CB_D12 rep 2 | 8762536078_C | 9.8 | 1.4 | 4284.33 | 1.52 | 1.56 | 16/10/2013_Batch3 | In one batch | In one batch | In one batch | |
| GSM2459217 | CB_D12 rep 3 | 8762536079_B | 9.9 | 1.9 | 2436.88 | 2.09 | 2.21 | 16/10/2013_Batch3 | In one batch | In one batch | In one batch | |
| GSM2459218 | CB_D9 rep 1 | 8762536055_B | 10 | 2.5 | 825.61 | 2.11 | 2.26 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459219 | CB_D9 rep 2 | 8762536097_D | 10 | 2.1 | 1264.37 | 2.1 | 2.22 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459220 | CB_D9 rep 3 | 8762536078_F | 10 | 2.3 | 675.76 | 2.12 | 2.25 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
| GSM2459221 | CB_D9 rep 4 | 8762536054_A | 10 | 2.4 | 416.6 | 2.1 | 2.24 | 16/10/2013_Batch2 | In one batch | In one batch | In one batch | |
Online-only Table 3.
Batch information, RNA quality and concentration and related GEO accession numbers for PcAS spleen (GSE12339120).
| Series GSE123391 | PRJNA508619 | Caliper LabChip | Nanodrop | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GEO accession ID | Sample Name in GEO | BeadChip No. | RQS | rRNA 28 s/18 s | [ng/ul] | 260/280 | 260/230 | RNA isolation batch | cRNA preparation | BeadChip hybridasation |
| GSM3502544 | AS-naive-day0-rep1 | 9440690022_B | 7.6 | 2.66 | 678.78 | 2.07 | 2.31 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502545 | AS-naive-day0-rep2 | 9440690030_C | 8.2 | 2.79 | 530.39 | 2.08 | 2.32 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502546 | AS-naive-day0-rep3 | 9440690035_B | 8.6 | 2.13 | 605.41 | 2.09 | 2.32 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502568 | AS-naive-day12-rep1 | 9440690030_F | 8.7 | 2.35 | 647.28 | 2.11 | 2.31 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502569 | AS-naive-day12-rep2 | 9440690037_C | 8.4 | 2.34 | 703.11 | 2.13 | 2.31 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502570 | AS-naive-day12-rep3 | 9440690042_A | 8.4 | 2.4 | 677.79 | 2.13 | 2.33 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502547 | AS-infected-day2-rep1 | 9440690022_C | 8.4 | 3.93 | 593.21 | 2.15 | 2.32 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502548 | AS-infected-day2-rep2 | 9440690035_A | 8.1 | 3.4 | 523.12 | 2.15 | 2.31 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502549 | AS-infected-day2-rep3 | 9440690037_D | 7.1 | 4.38 | 794.01 | 2.11 | 2.31 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502550 | AS-infected-day2-rep4 | 9440690042_C | 8.4 | 2.33 | 579.89 | 2.11 | 2.34 | 31/05/2013_batch1 | In one batch | In one batch |
| GSM3502551 | AS-infected-day4-rep1 | 9440690022_D | 7.2 | 3.89 | 854.39 | 2.11 | 2.31 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502552 | AS-infected-day4-rep2 | 9440690030_A | 8.5 | 3.13 | 711.79 | 2.09 | 2.31 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502553 | AS-infected-day4-rep3 | 9440690037_E | 8.2 | 3.36 | 1042.2 | 2.1 | 2.31 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502554 | AS-infected-day4-rep4 | 9440690042_B | 8.8 | 2.66 | 970.62 | 2.1 | 2.31 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502555 | AS-infected-day6-rep1 | 9440690022_E | 6.9 | 4.47 | 1283.88 | 2.11 | 2.29 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502556 | AS-infected-day6-rep2 | 9440690035_C | 7.4 | 2.46 | 1766.58 | 2.11 | 2.28 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502557 | AS-infected-day6-rep3 | 9440690042_D | 7.9 | 2.39 | 2096.84 | 2.1 | 2.27 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502558 | AS-infected-day6-rep4 | 9440690037_A | 8.2 | 2.57 | 2006.02 | 2.1 | 2.25 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502559 | AS-infected-day8-rep1 | 9440690022_F | 7.7 | 0.98 | 2670.24 | 2.07 | 2.2 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502560 | AS-infected-day8-rep3 | 9440690030_B | 7.8 | 0.94 | 2911.04 | 2.05 | 2.18 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502561 | AS-infected-day8-rep4 | 9440690035_D | 7.8 | 1.32 | 2911.33 | 2.04 | 2.17 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502562 | AS-infected-day10-rep1 | 9440690035_E | 7.8 | 1.29 | 3279.66 | 2.01 | 2.11 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502563 | AS-infected-day10-rep2 | 9440690030_D | 7.7 | 1.3 | 3418.2 | 1.98 | 2.08 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502564 | AS-infected-day10-rep3 | 9440690037_F | 7.8 | 1.27 | 3034.67 | 2.03 | 2.13 | 31/05/2013_batch2 | In one batch | In one batch |
| GSM3502565 | AS-infected-day12-rep2 | 9440690030_E | 8.5 | 2.33 | 2993.75 | 2.04 | 2.17 | 31/05/2013_batch3 | In one batch | In one batch |
| GSM3502566 | AS-infected-day12-rep3 | 9440690035_F | 8.4 | 2.21 | 1799.39 | 2.09 | 2.25 | 31/05/2013_batch3 | In one batch | In one batch |
| GSM3502567 | AS-infected-day12-rep4 | 9440690037_B | 8.6 | 2.17 | 2257.28 | 2.07 | 2.23 | 31/05/2013_batch3 | In one batch | In one batch |
Author contributions
Y.-c. Z. analysed the data, created the figures and wrote the manuscript; J.-w. Lin designed the project, performed the experiment and wrote the manuscript; C.H. and D.C. assisted in the animal experiments and sample maintenance; J.L. directed the project and edited the manuscript.
Code availability
R scripts for raw data reading, normalisation, QC, and plotting were available at https://github.com/LuChenLab/Rscript_for_BeadChip.git.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jean Langhorne, Email: jean.langhorne@crick.ac.uk.
Jing-wen Lin, Email: lin.jingwen@scu.edu.cn.
References
- 1.Buffet PA, et al. The pathogenesis of Plasmodium falciparum malaria in humans: insights from splenic physiology. Blood. 2011;117:381–392. doi: 10.1182/blood-2010-04-202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Portillo HA, et al. The role of the spleen in malaria. Cellular microbiology. 2012;14:343–355. doi: 10.1111/j.1462-5822.2011.01741.x. [DOI] [PubMed] [Google Scholar]
- 3.Cunnington AJ, Riley EM, Walther M. Stuck in a rut? Reconsidering the role of parasite sequestration in severe malaria syndromes. Trends Parasitol. 2013;29:585–592. doi: 10.1016/j.pt.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van den Steen PE, et al. Pathogenesis of malaria-associated acute respiratory distress syndrome. Trends Parasitol. 2013;29:346–358. doi: 10.1016/j.pt.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Deroost K, et al. Hemozoin induces lung inflammation and correlates with malaria-associated acute respiratory distress syndrome. Am J Respir Cell Mol Biol. 2013;48:589–600. doi: 10.1165/rcmb.2012-0450OC. [DOI] [PubMed] [Google Scholar]
- 6.Gazzinelli RT, Kalantari P, Fitzgerald KA, Golenbock DT. Innate sensing of malaria parasites. Nat Rev Immunol. 2014;14:744–757. doi: 10.1038/nri3742. [DOI] [PubMed] [Google Scholar]
- 7.Claser C, et al. Lung endothelial cell antigen cross-presentation to CD8(+)T cells drives malaria-associated lung injury. Nat Commun. 2019;10:4241. doi: 10.1038/s41467-019-12017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin JW, et al. Signatures of malaria-associated pathology revealed by high-resolution whole-blood transcriptomics in a rodent model of malaria. Sci Rep. 2017;7:41722. doi: 10.1038/srep41722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, H. J. et al. Transcriptomic Studies of Malaria: a Paradigm for Investigation of Systemic Host-Pathogen Interactions. Microbiol Mol Biol Rev82 (2018). [DOI] [PMC free article] [PubMed]
- 10.Smith ML, Styczynski MP. Systems Biology-Based Investigation of Host-Plasmodium Interactions. Trends Parasitol. 2018;34:617–632. doi: 10.1016/j.pt.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothen J, et al. Whole blood transcriptome changes following controlled human malaria infection in malaria pre-exposed volunteers correlate with parasite prepatent period. PLoS One. 2018;13:e0199392. doi: 10.1371/journal.pone.0199392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bediako Y, et al. Repeated clinical malaria episodes are associated with modification of the immune system in children. BMC Med. 2019;17:60. doi: 10.1186/s12916-019-1292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boldt ABW, et al. The blood transcriptome of childhood malaria. EBioMedicine. 2019;40:614–625. doi: 10.1016/j.ebiom.2018.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talavera-Lopez C, et al. Comparison of whole blood and spleen transcriptional signatures over the course of an experimental malaria infection. Sci Rep. 2019;9:15853. doi: 10.1038/s41598-019-52388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brugat T, et al. Sequestration and histopathology in Plasmodium chabaudi malaria are influenced by the immune response in an organ-specific manner. Cellular microbiology. 2014;16:687–700. doi: 10.1111/cmi.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brugat T, et al. Antibody-independent mechanisms regulate the establishment of chronic Plasmodium infection. Nat Microbiol. 2017;2:16276. doi: 10.1038/nmicrobiol.2016.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JW, et al. Genomic and transcriptomic comparisons of closely related malaria parasites differing in virulence and sequestration pattern. Wellcome Open Res. 2018;3:142. doi: 10.12688/wellcomeopenres.14797.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du P, Kibbe WA, Lin S. M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, 2017. Whole blood transcriptome during acute phase of infecion in avirulent and virulent Plasmodium chabaudi chabaudi infection. Gene Expression Omnibus. GSE93631
- 20.Talavera-Lopez C, Lin J, Bediako Y, Langhorne J. 2019. Whole spleen transcriptome during acute phase of infection in an avirulent Plasmodium chabaudi chabaudi AS infection. Gene Expression Omnibus. GSE123391
- 21.Lin J, Langhorne J. 2020. Whole spleen transcriptome during acute phase of infection in a virulent Plasmodium chabaudi chabaudi CB infection. Gene Expression Omnibus. GSE145781
- 22.Lin J, Langhorne J. 2020. Rodent malaria parasite RNA does not affect mouse BeadChip results. Gene Expression Omnibus. GSE145634
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Lin J, 2017. Whole blood transcriptome during acute phase of infecion in avirulent and virulent Plasmodium chabaudi chabaudi infection. Gene Expression Omnibus. GSE93631
- Talavera-Lopez C, Lin J, Bediako Y, Langhorne J. 2019. Whole spleen transcriptome during acute phase of infection in an avirulent Plasmodium chabaudi chabaudi AS infection. Gene Expression Omnibus. GSE123391
- Lin J, Langhorne J. 2020. Whole spleen transcriptome during acute phase of infection in a virulent Plasmodium chabaudi chabaudi CB infection. Gene Expression Omnibus. GSE145781
- Lin J, Langhorne J. 2020. Rodent malaria parasite RNA does not affect mouse BeadChip results. Gene Expression Omnibus. GSE145634
Data Availability Statement
R scripts for raw data reading, normalisation, QC, and plotting were available at https://github.com/LuChenLab/Rscript_for_BeadChip.git.