Abstract
Domestic animal populations are often characterised by high rates of inbreeding and low effective population sizes due to selective breeding practices. These practices can result in otherwise rare recessive deleterious alleles drifting to high frequencies, resulting in reduced fertility rates. This study aimed to identify potential recessive lethal haplotypes in the Thoroughbred horse breed, a closed population that has been selectively bred for racing performance. In this study, we identified a haplotype in the LY49B gene that shows strong evidence of being homozygous lethal, despite having high frequencies of heterozygotes in Thoroughbreds and other domestic horse breeds. Variant analysis of whole-genome sequence data identified two SNPs in the 3′UTR of the LY49B gene that may result in loss of function. Analysis of transcriptomic data from equine embryonic tissue revealed that LY49B is expressed in the trophoblast during placentation stage of development. These findings suggest that LY49B may have an essential, but as yet unknown function in the implantation stage of equine development. Further investigation of this region may allow for the development of a genetic test to improve fertility rates in horse populations. Identification of other lethal variants could assist in improving natural levels of fertility in horse populations.
Subject terms: Computational biology and bioinformatics, Genetics
Introduction
There is estimated to be a high rate of natural embryonic mortality in mammals. A large proportion of these embryonic losses occur soon after fertilisation, such that pregnancies often go undetected, with the only sign being reduced fertility1. Mutation screens in mice reveal that many genes are essential for development, with knockout of 29% of genes tested resulting in embryonic death by day 142,3. Although mutations in these genes are expected to be under strong negative selection due to being completely deleterious, many species are estimated to carry between one and two recessive lethal mutations per genome4. However, single mutations are often uncommon in a population, such that unrelated individuals are unlikely to carry the same recessive lethal mutations5–7. The likelihood of an individual inheriting two copies of the same lethal mutation is dramatically increased by inbreeding events, whereby alleles that are identical by descent are inherited from a common ancestor8–10.
In recent years, a number of studies in livestock have identified embryonic lethal mutations at high frequencies due to intensive selective breeding practices11–18. This is often due to a limited number of sires with desirable characteristics making large genetic contributions to the population11,19. Moreover, population bottlenecks due to domestication and breed formation have also resulted in increased deleterious mutation loads and diminished gene pools in many domestic breeds20–23. These processes lead to a reduction in genetic diversity indices including effective population size, which is defined as the theoretical population size that shows the same rate of loss in genetic diversity as the study population (Ne)24,25. Reduced genetic diversity can increase the risk of drift and inbreeding events in future generations of a population. Lethal mutations that have reached high frequencies are often detected by deviations from the Hardy–Weinberg equilibrium with a lack of homozygotes for one allele12. Characterisation of such mutations can assist in improving breeding decisions to increase fertility rates in these populations and prevent these mutations from drifting to higher frequencies26,27.
The identification of high frequency lethal variants is of particular interest in domestic horse populations. Although a recent study has identified some candidate mutations28, to date there has been no published comprehensive characterisation of common embryonic lethal alleles in horse populations. Despite the large variety of domestic horse breeds found throughout the world, many breeds suffer from low within-breed diversity and small Ne29–31. Some horse breeds with large census population sizes also experience low genetic diversity due to intense artificial selective breeding practices and closed population structures30,31. Maintaining good fertility rates is particularly important for horse populations due to the seasonal nature of breeding and the low individual fertility output, as mares produce only one foal from an eleven month gestation period32. Despite the extensive use of hormonal therapies to increase covering success in many domestic horse populations, per cycle pregnancy rates in some breeds only average around 65%, suggesting the presence of unknown variables that may reduce fertility33.
In this study, we aimed to characterise variants at high frequencies that may cause lethality in the Thoroughbred horse population. The Thoroughbred breed is of particular interest due to the closed population structure since the foundation of the studbook in the eighteenth century34. The population has since been intensely selected for the improvement of athletic abilities35,36, resulting in contemporary Thoroughbred horses being characterised by high levels of inbreeding and a small Ne31, 37–39. Due to selective breeding practices, all Thoroughbred horses can trace their ancestry back to a small number of individuals from the foundation of the breed37,38. Genetic diversity in the Thoroughbred breed has been reduced in recent decades due to the increased commercialisation of popular stallions providing large genetic contributions to the population40. Although such practices are in line with selective breeding principles41, they could also inadvertently increase the frequency of embryonic lethal variants in the population. Reproductive technologies such as artificial insemination are banned in the Thoroughbred population, making the maintenance of high levels of natural fertility imperative. Additionally, Thoroughbred horses have been used as foundation stock for other popular horse breeds including The Quarter Horse, Standardbred, and many Warmblood breeds30. Therefore, identification of lethal variants in Thoroughbreds is also likely to assist in the breeding management of these populations. We also aimed to determine the frequency of any potentially lethal variants identified in the Thoroughbred population in other horse breeds and examine their transcriptomic profile in embryonic tissue.
Results
Identifying candidate lethal SNPs at high frequencies in Thoroughbred horses
Analysis of genotype data from Thoroughbred horses (n = 156) identified only two adjacent, linked SNPs that significantly deviated from the Hardy–Weinberg equilibrium with an absence of homozygotes (Table 1). Under Hardy–Weinberg equilibrium, seven minor allele homozygotes were expected for both of these SNPs in the dataset. Genotype data from Japanese Thoroughbred horses (n = 370) also showed an absence of homozygotes for these SNPs (Table 1). In this dataset, the expected number of minor homozygotes for these SNPs under Hardy–Weinberg equilibrium was nine. Despite a complete absence of homozygotes, almost 35% of Thoroughbreds across both of these datasets were heterozygous for this two-SNP haplotype. These SNPs also showed an absence or reduction of minor homozygotes in genotype data for other domestic horse breeds (n = 2030, Table 1).
Table 1.
The allele frequencies of two adjacent SNPs with an absence of minor homozygotes in genotype data from two Thoroughbred horse datasets.
| Population | Sample size | Reference | 6:38278097 | 6:38278874 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Expected GG | GG | AG | AA | Expected CC | CC | AC | AA | |||
| Australian Thoroughbreds | 156 | Own data | 7* | 0 | 66 | 90 | 7* | 0 | 66 | 90 |
| Japanese Thoroughbreds | 370 | Fawcett et al.69 | 9* | 0 | 117 | 253 | 9* | 0 | 117 | 253 |
| Swedish Warmblood | 380 | Privately provided, Ablondi et al.70 | 4* | 0 | 75 | 304 | 4* | 0 | 74 | 306 |
| Coldblooded Trotter | 646 | Privately provided, Velie et al.84 | 26* | 0 | 258 | 388 | 28 | 22 | 226 | 393 |
| Quarter Horse | 137 | Petersen et al.65 | 17* | 0 | 97 | 40 | 17* | 0 | 97 | 40 |
| Exmoor Pony | 285 | Velie et al.71 | 0 | 0 | 1 | 279 | 0 | 0 | 1 | 282 |
| Various breeds | 582 | Petersen et al.30 | 15 | 0 | 85 | 497 | 15 | 0 | 85 | 497 |
The expected number of minor homozygotes in each population was calculated under Hardy–Weinberg equilibrium. Observed genotype frequencies that significant deviate from Hardy–Weinberg equilibrium frequency expectations (p < 0.05) are denoted with an asterisk.
These two candidate SNPs mapped to the coordinates of 6:38278097 (rs68661802) and 6:38278874 (rs68663106), which are found in an intronic region of the LY49B gene on chromosome 6. This gene is part of the LY49 gene family, which plays an important role in innate immunity. There are five functional members of the LY49 gene family in Equus caballus, all of which closely grouped together on chromosome 6. Since both of the SNPs mapped to a non-coding region of the LY49B gene, the likelihood of either being a causal variant for lethality is low.
Phylogenetic origin of the candidate SNPs
According to the phylogenetic tree generated by Petersen et al.29,30, and their associated SNP data, the SNPs of interest were present in heterozygous state across most phylogenetic branches of domestic horse breeds. Of the 32 breeds in this dataset, 23 had at least one heterozygote for both SNPs of interest. Notably, this two-SNP haplotype was not found in genotype data from one branch of the tree which contains the North Swedish Horse (n = 19), Norwegian Fjord Horse (n = 21) and Exmoor Pony (n = 24) (Table S1). A larger sample of Exmoor Pony data (n = 274, Table 1) found only one heterozygote for this haplotype.
Frequency of the candidate SNPs in other breeds
Analysis of SNP data from other domestic breeds showed that heterozygotes for the SNPs of interest were at a particularly high frequency in the Quarter Horse population (71%, n = 137) (Table 1, Table S2). The proportion of heterozygotes was also high in Swedish Warmbloods (n = 380) and Norwegian-Swedish Coldblooded Trotters (n = 641), being 20% and 40% respectively (Table 1). Smaller datasets also revealed that Belgian Draft (n = 19), French Trotter (n = 17), Paint (n = 15), Morgan (n = 19), Mongolian Paulista (n = 19) and Tuva (n = 15) breeds may also have a high proportion of heterozygotes for this haplotype in their populations (Table S1).
Identifying candidate causal variants using whole genome sequence data
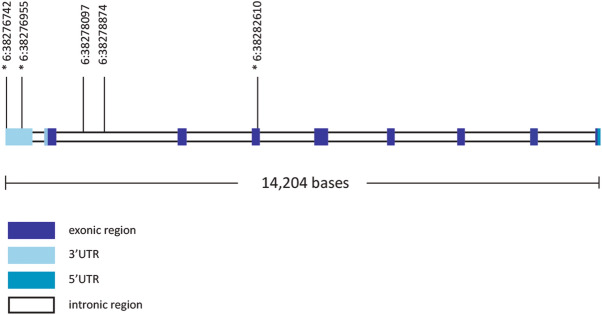
To further investigate SNP frequencies in this region, variants were called from whole-genome sequence data of 90 domestic horses. The two SNPs identified in the preliminary analysis showed a complete absence of homozygotes for their minor alleles in these individuals (Table S3). Additionally, a number of variants closely linked to these SNPs were identified (Table S3, Figure S1). Annotation of these loci using SIFT42 identified three variants that may result in changes to protein structure or expression, so these represent the most likely candidates to cause lethality in homozygous state (Fig. 1).
Figure 1.
The equine LY49B gene structure and SNP positions. The two variants in the intronic region (6:38278097 and 6:38278874) were identified in preliminary analysis as showing a significant absence of homozygotes for one allele. The three variants marked with a * are in linkage disequilibrium to these SNPs and may cause a loss of function in homozygous state. The structure of the gene is based on the EquCab 2.0 reference genome where the LY49B gene is on the reverse strand.
The first of these variants, 6:38282610G > A (rs68663123), was located in an exonic region of the LY49B gene and resulted in an amino acid change from a phenylalanine to a serine residue. This substitution is located next to a tryptophan residue that appears to be highly conserved across members of the LY49 family and across species. However, there is little conservation of the phenylalanine residue across taxa; some species have a phenylalanine and others a serine at this position. This SNP is annotated as being “tolerated” in SIFT.
Two other variants that were closely linked to the candidate SNPs (6:38276742A > T and 6:38276955G > A) were found within the 3′UTR (3′untranslated region) of the LY49B gene. Alignment of the 3′UTR of the five functional LY49 genes in Equus caballus revealed that the region containing the SNP 6:38276955G > A (rs1139567427) is highly conserved in all members of the LY49 gene family (Table 2). This region may be important for mRNA stability and translation into a functional protein. The other variant 6:38276742A > T (rs1137325172) was found in an AU-rich region at the end of the LY49B mRNA transcript, which is often associated with polyadenylation and post translation stability.
Table 2.
Amino acid residue sequence in a conserved area of the 3′UTR found in all Equus caballus LY49 genes as mapped in the EquCab2.0 assembly.
| Gene | Sequence |
|---|---|
| LY49B | AAAGACTTTCTCAGGGCCATTAAAGAGATGGGAAACTGCTTTCCAAAGAC |
| LY49C | AGAGAATTTCCCAGGGCCATTAAAGAGAAGAGCAACTGATTTCCAAAGAC |
| LY49D | AGAGAATTTCTCAGGGCCATTAAAGAGAAGGGCAACTGATTTCCAAAGAC |
| LY49E | AGAGAATTTCTCAGGGCCATTAAAGAGAAGGGCAACTGATTTCCAAAGAC |
| LY49F | AGAGAATTTTGCAGGGTCATTAAAGAGAGGGGTAACTGCTTTCCAAAGAC |
The SNP position of 6:38276955G > A is highlighted in bold.
Transcriptomic analysis of RNA sequence data
Measurable levels of LY49B mRNA were not detected in equine trophectoderm tissue collected on day 16 of development. However, LY49B mRNA was observed in trophectoderm tissue collected on days 23 and 24 of development (Table 3). Additionally, LY49B mRNA transcripts were detected in microarray data from equine chorion and chorionic girdle tissue between days 27 and 34 of development (Table S4). Inner cell mass tissue collected on days 15, 22 and 25 of development did not show any measurable transcription of LY49B (Table 3). The genotypes of the candidate SNPs in the mRNA samples analysed were unknown.
Table 3.
Gene counts from RNA sequence data of three trophectoderm and three inner cell mass tissue samples from equine embryos.
| Tissue | Gene count (FPKM) | ||
|---|---|---|---|
| Trophectoderm | Day 15 | Day 22 | Day 25 |
| 0.000 | 0.031 | 0.024 | |
| Inner cell mass | Day 16 | Day 23 | Day 24 |
| 0.00 | 0.00 | 0.00 | |
Transcript counts are in fragments per kilobase/million (FPKM).
Discussion
Analysis of genotype data identified a two-SNP haplotype as a strong candidate for harbouring a variant that causes lethality in homozygous state. The SNPs identified in this preliminary analysis mapped to an intronic region in the LY49B gene on ECA6 (Table 1). The LY49B gene belongs to the LY49 (Killer cell lectin-like receptor subfamily A) family of receptors, which consists of five functional members in Equus caballus43. Other species (including humans) have a functionally similar, but structurally different gene family called KIR (Killer cell immunoglobin line receptors)44. The LY49/KIR gene family are expressed across various types of immune cells, and mediate their function through bindings to MHC-145. The LY49B gene is expressed in myeloid cells where it regulates their activity through an inhibitory effect, possibly to prevent their spontaneous activation46. Despite the important role that they play in immunity, the function of LY49 genes in development is currently unknown. In humans, incompatibilities between foetal KIR and maternal MHC (HLA) genotypes are associated with an increased risk of miscarriage and preeclampsia47–49. Additionally, knockdown of LY49 in mice showed a high rate of implantation failure50,51. These findings indicate that LY49B may play an important role in maternal/foetal compatibility and implantation success in horses.
Analysis of transcriptomic data found that LY49B was first expressed in equine trophoblast tissue during the placental development stage. The first evidence of LY49B expression was found on day 23–24 of development (Table 3), during which the glycoprotein capsule surrounding the embryo is broken down and placental tissue starts to develop52. Measurable expression of LY49B was also found in chorion and chorionic girdle tissues between days 27 and 34 of development (Table S4). During this time, trophoblast cells rapidly proliferate to form the chorionic girdle, which then invades the endometrium to form epithelial cups53. It is possible that LY49B is important for successful implantation of the embryo by mediating the action of MHC-1 which is expressed during this time54,55. Further investigations into the role of LY49B in equine development would confirm whether impaired function causes lethality and the stage of development at which this occurs.
Variant calling in whole-genome sequence data from 90 domestic horses further confirmed an absence of minor homozygotes for the two SNPs of interest. Three variants closely linked to these SNPs were also identified in these data as the most likely candidates to cause loss of function in the LY49B gene and result in lethality in homozygous state (Table S3). One SNP was a missense variant in the coding region of the LY49B gene that results in the substitution of a negatively charged serine for an aromatic phenylalanine residue. However, lack of conservation of this SNP in LY49 genes across taxa makes it seem unlikely to be a causative variant for embryonic lethality. Two other variants found in the 3′UTR of the LY49B gene were also closely linked to the SNPs identified in the preliminary analysis, and seemed more likely candidates to cause embryonic lethality in homozygous state.
The 3′UTR of a gene is responsible for transcriptional stability through the binding of miRNAs and RNA binding proteins56. The addition of the polyadenylation tail to the 3′UTR is also essential to ensure proper processing and translation of the mRNA strand57. Mutations in the 3′UTR can lead to degradation of the mRNA, resulting in reduced or inhibited translation even when the gene is transcribed58. Variation in the 3′UTR of genes are associated with a number of diseases including Huntington’s and breast cancer in humans59,60. Additionally, SNPs in the 3′UTR are associated with traits in livestock including milk production in cows, muscularity in sheep and obesity in horses58,61,62.
Despite the importance of the 3′UTR for the mRNA stability and normal expression of a gene, little is known about how specific polymorphisms can affect post-transcriptional processing. This makes it difficult to identify how the 3′UTR variants identified in this study could affect the translation of LY49B mRNA into a functional protein. The 3′UTR variant 6:38276955G > A was identified as a possible candidate for embryonic lethality because it is highly conserved between all members of the LY49B family (Table 2) in horses, so may play an important role in mRNA stability. The other 3′UTR variant (6:38276742A > T) is found in an AU-rich region at the end of the transcript, so may be important for the addition of the polyadenylation tail. Further examination of the effects that these variants have on post-transcriptional processing would determine if they impact the normal expression of LY49B in horses.
Despite an absence of homozygotes, the two intronic SNPs identified in this study were found at high heterozygote frequencies in the Thoroughbred population. Mares are often covered multiple times in a season, which may explain why a more discernible reduction in fertility has not been observed as a result of the high frequency of this variant. However, the presence of lethal variants at high frequencies may result in more coverings being required for each mare in a season. Currently, there is no evidence that variation in the LY49B gene is associated with phenotypic advantages in horses. However, it is possible that one of the variants linked to these SNPs results in a phenotypic advantage in heterozygotes, which could explain why they have reached such high frequencies in the breed. It is also possible that selective breeding practices favouring a limited number of stallion bloodlines are responsible for this potentially lethal haplotype drifting to high frequencies in the Thoroughbred population. This would be most likely to occur if a stallion that made a large genetic contribution to the population was a carrier. A similar instance has recently been documented in cattle, where a lethal variant at a high frequency was traced back to a sire with an extensive genetic influence on the population19.
The presence of this potentially lethal haplotype across many diverse breeds of domestic horses indicates that it may not be the result of a recent mutation present only in the Thoroughbred population. Rather, heterozygotes for this haplotype may have been present in pre-domesticated horses as a rare variant, and have become more frequent in some domestic breeds as the result of population bottlenecks due to breed formations and selective breeding practices. Domestication and breed formation events have been well documented to result in increased deleterious mutation loads in horses and other domestic species22,31,63,64. A high proportion of heterozygotes for this haplotype were found in some breeds closely related to the Thoroughbred including the Paint, French Trotter, Morgan and Quarter Horse. Notably, over 70% of Quarter Horse samples included in this study were heterozygous for these SNPs (Table 1). The Quarter Horse has an open stud book, and higher genetic diversity than the Thoroughbred population65, making the high frequency of a potentially lethal haplotype at first surprising. The Quarter Horse dataset reportedly did not contain full or half siblings65, but the collection of samples from one geographical area may not fully reflect the diversity of the worldwide population. An average relatedness analysis of these samples noted the large genetic influence of one particular Thoroughbred stallion65, which may explain the high frequency of heterozygotes observed in this population. However, the extremely high frequency of heterozygotes in this breed may be due to selective breeding favouring these individuals.
The Belgian Draft, Mangalarga Paulista and Tuva breeds also show a high proportion of heterozygotes, but are more distantly related to the Thoroughbred and to each other. Therefore, the high frequency of heterozygotes in these breeds may be due to independent genetic drift events. Heterozygotes for this haplotype were notably absent from one branch of the tree containing small heavy horses from Northern Europe, which are more distantly related to the Thoroughbred. A larger dataset of Exmoor Pony samples from this phylogenetic branch revealed one heterozygote for this haplotype (Table 1). This could be due to a calling error, but it is also possible that these SNPs exist at very low frequencies in these breeds. The small sample size of the genotype data for many individual breeds in this study means that heterozygote frequencies across all subpopulations found throughout the world may deviate from that reported here. However, these data provide an indication of breeds with high proportions of heterozygotes for this region. Analysis of SNP data from Northern-Swedish Coldblooded Trotters identified 22 homozygotes for the SNP at position 6:38278874. It is likely that there has been recombination between this SNP and the causal variant, and may appear more frequent in this population due to differences in breed history and recombination patterns66. However, additional analyses are required to explore this further. Overall, our findings suggest that this region shows evidence harbouring a homozygous lethal variant, yet a high proportion of heterozygotes are found across many domestic horse breeds.
In this study, we identified a haplotype at high heterozygote frequencies in the Thoroughbred horse population that is a strong candidate for harbouring a variant causing lethality in homozygous state. Similar analyses on larger datasets in other livestock populations have identified multiple lethal haplotypes, so it is likely that other such variants are present at high frequencies in the Thoroughbred population but were not captured in this study. Additionally, the use of commercial SNP arrays only allows for the identification of variants with high minor allele frequencies in populations. Analysis of larger sample sizes, and using higher density genotype data could allow for identification of other variants associated with lethality in domestic horses. The identification of this potentially lethal haplotype demonstrates the potential implications of heavily favouring a limited number of bloodlines in selective breeding practices. Further characterisation of lethal haplotypes in other breeds would also assist in breeding management to increase per covering fertility rates in domestic horse populations.
Methods
DNA extractions
DNA was extracted from the hair samples of Australian Thoroughbred horses using the Qiagen Gentra Puregene Tissue Kit (Qiagen, Redwood City, CA, USA). DNA was extracted from the hair samples of Norwegian-Swedish Coldblooded Trotters and Swedish Warmbloods by incubating the samples for 2 h at 56 °C with Chelex 100 Resin (Bio-Rad Laboratories, Hercules, CA) and Proteinase K (20 mg/mL; Merck KgaA, Darmstadt, Germany). The Proteinase K was then inactivated by incubating for 10 min at 95 °C and DNA resuspended in low TE (1 mM Tris, 0.1 mM EDTA). DNA was extracted from blood samples using the Qiasymphony DSP DNA mini kit (Qiagen, Hilden, Germany).
Initial genotyping
Genotype data from a representative sample of Thoroughbreds were used to identify SNPs with a high proportion of heterozygotes, but an absence of homozygotes for one allele. Genome-wide SNP data were generated for 156 Australian Thoroughbred horses by genotyping samples on either the Illumina 70 K Chip (65,102 SNPs) (n = 102) or the Affymetrix 670 K Chip (670,796 SNPs) (n = 54). Common genotyped SNPs between the two arrays were scanned for deviations from the Hardy–Weinberg equilibrium with an absence of homozygotes for one allele using PLINK (version 1.9)67. The p values were adjusted using a false discovery rate correction with the R package “qvalue”68. Since SNPs with an absence of homozygotes could indicate a calling error, the search was narrowed to only include adjacent SNPs that fit such criteria.
The frequencies of the candidate SNPs were then examined in publicly available genotype data from Japanese Thoroughbreds (n = 370) typed on the Affymetrix 670 K Chip69 and these were added the Thoroughbred sample. The SNP frequencies were then characterised from genotype data from Swedish Warmbloods (n = 380)70 and Norwegian-Swedish Coldblooded Trotters (n = 646)71 typed on the Affymetrix 670 K Chip. Publicly available data from Exmoor Ponies (n = 285, typed on the Affymetrix 670 K Chip)71, Quarter Horses (n = 137, typed on the Illumina 70 K Chip)72 and horses of 32 different domestic breeds (n = 582, typed on the Illumina 50 K Chip)29,30 were also included in this preliminary scan for SNP frequencies. In these data, raw intensities were plotted to check for calling errors. If potential calling errors were detected, SNPs were recalled using a mixture model fitted with an expectation–maximization algorithm in R.
Variant discovery and mapping
Publicly available whole-genome sequence data were used to further examine the frequencies of the candidate SNPs identified in the initial genotype analysis, and to identify linked variants. Paired end whole-genome sequence data from 90 horses of different domestic breeds were used in this analysis (Table S5). The whole genome datasets were downloaded from the European Nucleotide Archives (ENA, https://www.ebi.ac.uk/ena) which included horses of different domestic breeds (PRJEB14779, n = 70) and additional Thoroughbred samples (PRJNA168142, n = 16 and PRJNA184688, n = 4) (Table S5).
The SNP array used in the initial genotyping analysis was developed based on coordinates of the EquCab2.0 reference genome. For consistency, we used the EquCab2.0 assembly as a reference for the whole-genome sequence analysis. The EquCab2.0 assembly was also used because of an issue with resolution in the area of interest on the newer EquCab3.0 assembly. The raw reads were mapped to the EquCab2.0 reference genome using BWA-MEM algorithm from Burrows-Wheeler Alignment Tool (version 0.7.17)73. Duplicate reads were flagged using Samblaster (version 0.1.22)74, and base recalibration was performed using Genome Analysis Toolkit (GATK) (version 4.0.8.1)75. Variants (SNPs and INDELs (insertions and deletions)) were called using Haplotype Caller and then filtered using the standard hard filtering recommendations in GATK76. The individual SNPs were then filtered to only include high quality allele calls with an average filtered depth over 10 and a Phred score over 20.
Variants that were linked to the SNPs identified from the genotype data were produced using the LD function in PLINK (version 1.9) with a window size of 5 Mb67. Only SNPs with an r2 value of over 0.8 and a D′ value > 0.9 were shortlisted. The effects of each SNP on gene structure and function was characterised using SIFT (version 4G)42. The conservation of variants across taxa was analysed using the NCBI Conserved Domain Database Search77.
Transcriptomic analysis
Publicly available RNA sequence data were used to examine expression levels of the genes of interest in embryonic tissue. The data included equine inner cell mass tissue (collected at day 15, 22 and 25, n = 3) and trophectoderm tissue (collected at day 16, 23 and 24, n = 3) from the Functional Annotation of ANimal Genomes (FAANG) equine biobank (available from ENA under the project name PRJNA223157)78. Adaptors were trimmed using bbduk from BBtools (version 37.98)79. Reads were aligned to the EquCab 2.0 genome using STAR (version 2.7.2b)80. Counts were generated using featurecounts from Subread package(version 1.5.1)81, then quantified in fragments per kilobase/million (FPKM) using the R package “edgeR”82 with the Equus_caballus_Ensembl_94 file used for annotation. Microarray data for chorion (n = 19) and chorionic girdle (n = 19) tissue collected from horse embryos between days 27–34 of development83 were also examined for gene expression levels.
Ethics statement
Hair samples from Australian Thoroughbred horses were collected under approval from University of Sydney Ethics Committee (Number: N00-2009-3-5109.) Written informed consent to use the animals in this study was obtained from the owners of the animals. The hair samples from Swedish Warmblood horses were originally collected for parentage testing and stored in the biobank at the Animal Genetics Laboratory, SLU so ethics approval was not applicable. Hair and blood samples of Norwegian-Swedish Coldblooded Trotters were collected under approval from the Ethics Committee for Animal Experiments in Uppsala, Sweden (Number: C 121/14). All the methods were performed in accordance with the guidelines set out by the respective Animal Ethics Committees and the guidelines contained in the Guide for the Care and Use of Laboratory Animals. No experimental procedure was performed on live animals. All other data was downloaded from publicly available repositories.
Data availability
The whole-genome sequence data used in this study is publicly available for The European Nucleotide Archive. Genotype data for Japanese Thoroughbreds, Exmoor Ponies, Quarter Horses and other domestic horse breeds can be found in the supplementary information of their respective papers (https://doi.org/10.1371/journal.pone.0218407, 10.1371/journal.pone.0152966, 10.1093/jhered/est079 and 10.1371/journal.pone.0054997). The exceptions are genotype data from Australian Thoroughbred, Swedish Warmbloods and North Swedish Coldblooded Trotters, which are available on request but restrictions apply to the availability of these data which were used under license for the current study and so are not publicly available.
Supplementary information
Acknowledgements
The authors acknowledge the technical assistance provided by the Sydney Informatics Hub, a Core Research Facility of the University of Sydney.
Author contributions
N.A.H., B.D.V., R.A.A., G.L., A.V., S.E., S.M. and E.S. assisted in the collection and genotyping of the DNA samples. E.T.T., B.D.V., P.C.T. and N.A.H. designed the project. E.T.T. analysed the data and wrote the manuscript. All authors edited, read and approved the final manuscript.
Competing interests
N.A.H. is supported by Racing Australia in the form of salary. All other authors declare that they have no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-68946-8.
References
- 1.Spencer TE. Early pregnancy: concepts, challenges, and potential solutions. Anim. Front. 2013;3:48–55. doi: 10.2527/af.2013-0033. [DOI] [Google Scholar]
- 2.White JK, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mary ED, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016 doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Z, Waggoner D, Stephens M, Ober C, Przeworski M. An estimate of the average number of recessive lethal mutations carried by humans. Genetics. 2015;199:1243–1254. doi: 10.1534/genetics.114.173351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballinger MA, Noor MAF. Are lethal alleles too abundant in humans? Trends Genet. 2018;34:87–89. doi: 10.1016/j.tig.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Mukai T, Chigusa SI, Mettler LE, Crow JF. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics. 1972;72:335. doi: 10.1093/genetics/72.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sulem P, et al. Identification of a large set of rare complete human knockouts. Nat. Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 8.Curik I, Ferencakovic M, Solkner J. Inbreeding and runs of homozygosity: a possible solution to an old problem. Livest. Sci. 2014;166:26–34. doi: 10.1016/j.livsci.2014.05.034. [DOI] [Google Scholar]
- 9.Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. doi: 10.1016/S0169-5347(02)02489-8. [DOI] [Google Scholar]
- 10.Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat. Rev. Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 11.Hoff JL, Decker JE, Schnabel RD, Taylor JF. Candidate lethal haplotypes and causal mutations in Angus cattle. BMC Genom. 2017 doi: 10.1186/s12864-017-4196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.VanRaden PM, Olson KM, Null DJ, Hutchison JL. Harmful recessive effects on fertility detected by absence of homozygous haplotypes. J. Dairy Sci. 2011;94:6153–6161. doi: 10.3168/jds.2011-4624. [DOI] [PubMed] [Google Scholar]
- 13.Fasquelle C, et al. Balancing selection of a frame-shift mutation in the MRC2 gene accounts for the outbreak of the crooked tail syndrome in Belgian Blue cattle. PLoS Genet. 2009;5:e1000666. doi: 10.1371/journal.pgen.1000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, MacNeil MD, Kemp RA, Dyck MK, Plastow GS. Putative loci causing early embryonic mortality in Duroc swine. Front. Genet. 2018;17:655. doi: 10.3389/fgene.2018.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charlier C, et al. NGS-based reverse genetic screen for common embryonic lethal mutations compromising fertility in livestock. Genome Res. 2016;26:1333–1341. doi: 10.1101/gr.207076.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonstegard TS, et al. Identification of a nonsense mutation in CWC15 associated with decreased reproductive efficiency in Jersey cattle. PLoS ONE. 2013;8:e54872. doi: 10.1371/journal.pone.0054872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derks MFL, et al. A systematic survey to identify lethal recessive variation in highly managed pig populations. BMC Genom. 2017 doi: 10.1186/s12864-017-4278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derks MFL, et al. Loss of function mutations in essential genes cause embryonic lethality in pigs. PLoS Genet. 2019;15:e1008055. doi: 10.1371/journal.pgen.1008055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourneuf E, et al. Rapid discovery of de novo deleterious mutations in cattle enhances the value of livestock as model species. Sci. Rep. 2017;7:11466. doi: 10.1038/s41598-017-11523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert M, et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc. Natl. Acad. Sci. USA. 2014;111:E5661–E5669. doi: 10.1073/pnas.1416991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruz F, et al. The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol. Biol. Evol. 2008;25:2331–2336. doi: 10.1093/molbev/msn177. [DOI] [PubMed] [Google Scholar]
- 22.Marsden CD, et al. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. USA. 2016;113:152–157. doi: 10.1073/pnas.1512501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosse M, Derks M, Groenen M. Deleterious alleles in the context of domestication, inbreeding, and selection. Evol. Appl. 2019;12:6–17. doi: 10.1111/eva.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husemann M, Zachos FE, Paxton RJ, Habel JC. Effective population size in ecology and evolution. Heredity. 2016 doi: 10.1038/hdy.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.García-Dorado A. Understanding and predicting the fitness decline of shrunk populations: inbreeding, purging, mutation, and standard selection. Genetics. 2012;190:1461–1476. doi: 10.1534/genetics.111.135541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casas E, Kehrli ME. A review of selected genes with known effects on performance and health of cattle. Front. Vet. Sci. 2016 doi: 10.3389/fvets.2016.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagannathan V, et al. Comprehensive characterization of horse genome variation by whole-genome sequencing of 88 horses. Anim. Genet. 2019;50:74–77. doi: 10.1111/age.12753. [DOI] [PubMed] [Google Scholar]
- 29.Petersen JL, et al. Genome-wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genet. 2013;9:e1003211. doi: 10.1371/journal.pgen.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen JL, et al. Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLOS ONE. 2013;8:e54997. doi: 10.1371/journal.pone.0054997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlando L, Librado P. Origin and evolution of deleterious mutations in horses. Genes. 2019;10:649. doi: 10.3390/genes10090649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aurich C. Reproductive cycles of horses. Anim. Reprod. Sci. 2011;124:220–228. doi: 10.1016/j.anireprosci.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Allen WR, Wilsher S. Half a century of equine reproduction research and application: a veterinary tour de force. Equine Vet. J. 2018;50:10–21. doi: 10.1111/evj.12762. [DOI] [PubMed] [Google Scholar]
- 34.Weatherby and Sons. An Introduction to the General Stud Book. (Weatherby and Sons, 1791).
- 35.Hill EW, Gu J, McGivney BA, MacHugh DE. Targets of selection in the Thoroughbred genome contain exercise-relevant gene SNPs associated with elite racecourse performance. Anim. Genet. 2010;41:56–63. doi: 10.1111/j.1365-2052.2010.02104.x. [DOI] [PubMed] [Google Scholar]
- 36.Gu J, et al. A genome scan for positive selection in Thoroughbred horses. PLoS ONE. 2009;4:e5767. doi: 10.1371/journal.pone.0005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunningham EP, Dooley JJ, Splan RK, Bradley DG. Microsatellite diversity, pedigree relatedness and the contributions of founder lineages to Throughbred horses. Anim. Genet. 2001;32:360. doi: 10.1046/j.1365-2052.2001.00785.x. [DOI] [PubMed] [Google Scholar]
- 38.Todd ET, et al. Founder-specific inbreeding depression affects racing performance in Thoroughbred horses. Sci. Rep. 2018;8:6167. doi: 10.1038/s41598-018-24663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corbin LJ, et al. Linkage disequilibrium and historical effective population size in the Thoroughbred horse. Anim. Genet. 2010;41:8–15. doi: 10.1111/j.1365-2052.2010.02092.x. [DOI] [PubMed] [Google Scholar]
- 40.McGivney BA, et al. Genomic inbreeding trends, influential sire lines and selection in the global Thoroughbred horse population. Sci. Rep. 2020;10:466. doi: 10.1038/s41598-019-57389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woolliams JA, Berg P, Dagnachew BS, Meuwissen THE. Genetic contributions and their optimization. J. Anim. Breed. Genet. 2015;132:89–99. doi: 10.1111/jbg.12148. [DOI] [PubMed] [Google Scholar]
- 42.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Futas J, Horin P. Natural killer cell receptor genes in the family Equidae: not only Ly49. PLoS ONE. 2013;8:e64736. doi: 10.1371/journal.pone.0064736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:129–139. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahim MMA, et al. Ly49 receptors: innate and adaptive immune paradigms. Front. Immunol. 2014;5:145. doi: 10.3389/fimmu.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gays F, et al. Ly49B is expressed on multiple subpopulations of myeloid cells. J. Immunol. Res. 2006;177:5840–5851. doi: 10.4049/jimmunol.177.9.5840. [DOI] [PubMed] [Google Scholar]
- 47.Hiby S, et al. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum. Reprod. 2008;23:972–976. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- 48.Hiby SE, et al. Materal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J. Clin. Investig. 2010;120:4102. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long W, et al. Association of maternal KIR and fetal HLA-C genes with the risk of preeclampsia in the Chinese Han population. Placenta. 2015;36:433–437. doi: 10.1016/j.placenta.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Leon L, et al. Ly49 knockdown in mice results in aberrant uterine crypt formation and impaired blastocyst implantation. Placenta. 2016;39:147–150. doi: 10.1016/j.placenta.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 51.Lima PD, et al. Ly49 receptors activate angiogenic mouse DBA+ uterine natural killer cells. Cell. Mol. Immunol. 2014;11:467–476. doi: 10.1038/cmi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen WR, Wilsher S. A review of implantation and early placentation in the mare. Placenta. 2009;30:1005–1015. doi: 10.1016/j.placenta.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 53.Noronha LE, Antczak DF. Maternal immune responses to trophoblast: the contribution of the horse to pregnancy immunology. Am. J. Reprod. Immunol. 2010;64:231–244. doi: 10.1111/j.1600-0897.2010.00895.x. [DOI] [PubMed] [Google Scholar]
- 54.Donaldson WL, Oriol JG, Pelkaus CL, Antczak DF. Paternal and maternal major histocompatibility complex class I antigens are expressed co-dominantly by equine trophoblast. Placenta. 1994;15:123–135. doi: 10.1016/S0143-4004(05)80449-7. [DOI] [PubMed] [Google Scholar]
- 55.Bacon SJ, Ellis SA, Antczak DF. Control of expression of major histocompatibility complex genes in horse trophoblast. Biol. Reprod. 2002;66:1612–1620. doi: 10.1095/biolreprod66.6.1612. [DOI] [PubMed] [Google Scholar]
- 56.Steri M, Idda ML, Whalen MB, Orrù V. Genetic variants in mRNA untranslated regions. Wiley Interdiscip. Rev. RNA. 2018 doi: 10.1002/wrna.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis SL, et al. Genomewide association study reveals a risk locus for equine metabolic syndrome in the Arabian horse 1. J. Anim. Sci. 2017;95:1071–1079. doi: 10.2527/jas.2016.1221. [DOI] [PubMed] [Google Scholar]
- 59.Dorairaj J, et al. A germline mutation in the BRCA1 3'UTR predicts Stage IV breast cancer. BMC Cancer. 2014;14:421. doi: 10.1186/1471-2407-14-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim KH, et al. Full sequence of mutant huntingtin 3′-untranslated region and modulation of its gene regulatory activity by endogenous microRNA. J. Hum. Genet. 2019;64:995–1004. doi: 10.1038/s10038-019-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou J, et al. Two mutations in the caprine MTHFR 3'UTR regulated by microRNAs are associated with milk production traits. PLoS ONE. 2015;10:e0133015–e0133015. doi: 10.1371/journal.pone.0133015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alex C, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006;38:813. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 63.Librado P, et al. Ancient genomic changes associated with domestication of the horse. Science. 2017;356:442–445. doi: 10.1126/science.aam5298. [DOI] [PubMed] [Google Scholar]
- 64.Fages A, et al. Tracking five millennia of horse management with extensive ancient genome time series. Cell. 2019;177:1419–1435.e1431. doi: 10.1016/j.cell.2019.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petersen JL, Mickelson JR, Cleary KD, McCue ME. The American Quarter Horse: population structure and relationship to the Thoroughbred. J. Hered. 2014;105:148–162. doi: 10.1093/jhered/est079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beeson SK, Mickelson JR, McCue ME. Exploration of fine-scale recombination rate variation in the domestic horse. Genome Res. 2019;29:1744. doi: 10.1101/gr.243311.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Storey, J., Bass, A., Dabney, A. & Robinson, D. Qvalue: q-value estimation for false discovery rate control v. R package version 2.18.0. (Accessed 6 September 2018); https://github.com/jdstorey/qvalue.
- 69.Fawcett JA, et al. Genome-wide SNP analysis of Japanese Thoroughbred racehorses. PLoS ONE. 2019 doi: 10.1371/journal.pone.0218407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ablondi M, Viklund A, Lindgren G, Eriksson S, Mikko S. Signatures of selection in the genome of Swedish Warmblood horses selected for sport performance. BMC Genom. 2019 doi: 10.1186/s12864-019-6079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Velie BD, et al. Using an inbred horse breed in a high density genome-wide scan for genetic risk factors of insect bite hypersensitivity (IBH) PLoS ONE. 2016 doi: 10.1371/journal.pone.0152966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petersen JL, Valberg SJ, Mickelson JR, McCue ME. Haplotype diversity in the equine myostatin gene with focus on variants associated with race distance propensity and muscle fibre type proportions. Anim. Genet. 2014;45:827–835. doi: 10.1111/age.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faust G, Hall I. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data (Report) Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Depristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu S, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iqbal K, Chitwood JL, Meyers-Brown GA, Roser JF, Ross PJ. RNA-Seq transcriptome profiling of equine inner cell mass and trophectoderm. Biol. Reprod. 2014 doi: 10.1095/biolreprod.113.113928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bushnell, B. BBMap short read aligner. (Accessed 29 August 2019); https://sourceforge.net/projects/bbmap.
- 80.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 82.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Read JE, et al. Dynamic changes in gene expression and signalling during trophoblast development in the horse. Reproduction. 2018 doi: 10.1530/REP-18-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Velie BD, et al. A genome-wide association study for harness racing success in the Norwegian-Swedish coldblooded trotter reveals genes for learning and energy metabolism. BMC Genet. 2018;19:1–13. doi: 10.1186/s12863-018-0670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome sequence data used in this study is publicly available for The European Nucleotide Archive. Genotype data for Japanese Thoroughbreds, Exmoor Ponies, Quarter Horses and other domestic horse breeds can be found in the supplementary information of their respective papers (https://doi.org/10.1371/journal.pone.0218407, 10.1371/journal.pone.0152966, 10.1093/jhered/est079 and 10.1371/journal.pone.0054997). The exceptions are genotype data from Australian Thoroughbred, Swedish Warmbloods and North Swedish Coldblooded Trotters, which are available on request but restrictions apply to the availability of these data which were used under license for the current study and so are not publicly available.