Peritoneal dialysis (PD) patients have a distinct uremic toxin profile and metabolic abnormalities, which could be related in part to the differences in gut microbiota.1 Prebiotics are microbial feed supplements that beneficially affect the host by improving its intestinal microbial balance and re-establishing symbiosis. Treatment with the prebiotic, oligofructose-enriched p-inulin (p-inulin) is believed to enable proliferation of beneficial gut bacteria such as Bifidobacteria, to attenuate inflammation, and to improve metabolic functions.2,3 The aims of this pilot study are (i) to compare the microbiome profile in PD patients and individuals without kidney disease, and (ii) to evaluate the effect of p-inulin treatment on inflammatory biomarkers and microbiome community composition and to associate it with host metabolomic profile.
Results
Patients
In a nonrandomized crossover study, 8 PD patients were followed during 3 sequential 8-week phases of no intervention, p-inulin (16 g/d) administration, and a post-intervention phase with no intervention (Supplementary Table S1). We also studied 7 individuals without kidney disease. Patient characteristics are summarized in Supplementary Table S2. Patients’ dietary intake and gastrointestinal symptoms are summarized in Supplementary Tables S3 and S4.
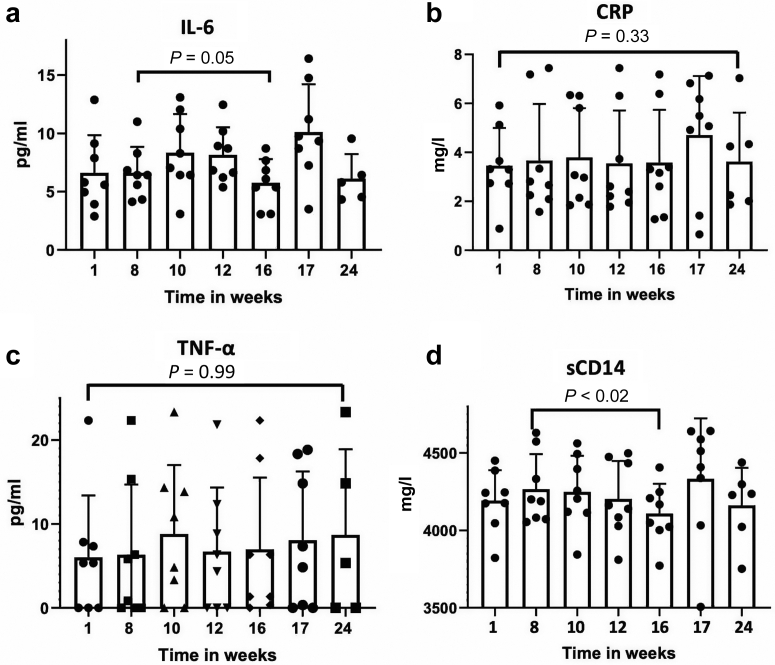
We measured several markers of inflammation, but only plasma-level soluble CD14 (sCD14) was significantly lower at week 16 compared to that in the pretreatment period at week 8 (4267 ± 195.8 vs. 4110 ± 191.6 mg/ml, P < 0.02) (Figure 1a−d).
Figure 1.
Plasma levels of inflammatory biomarkers. (a) Interleukin-6 (IL-6). (b) C-reactive protein (CRP). (c) Tumor necrosis factor alpha (TNF-α). (d) Soluble CD14 (sCD14).
p-Inulin Alters the Microbial Composition
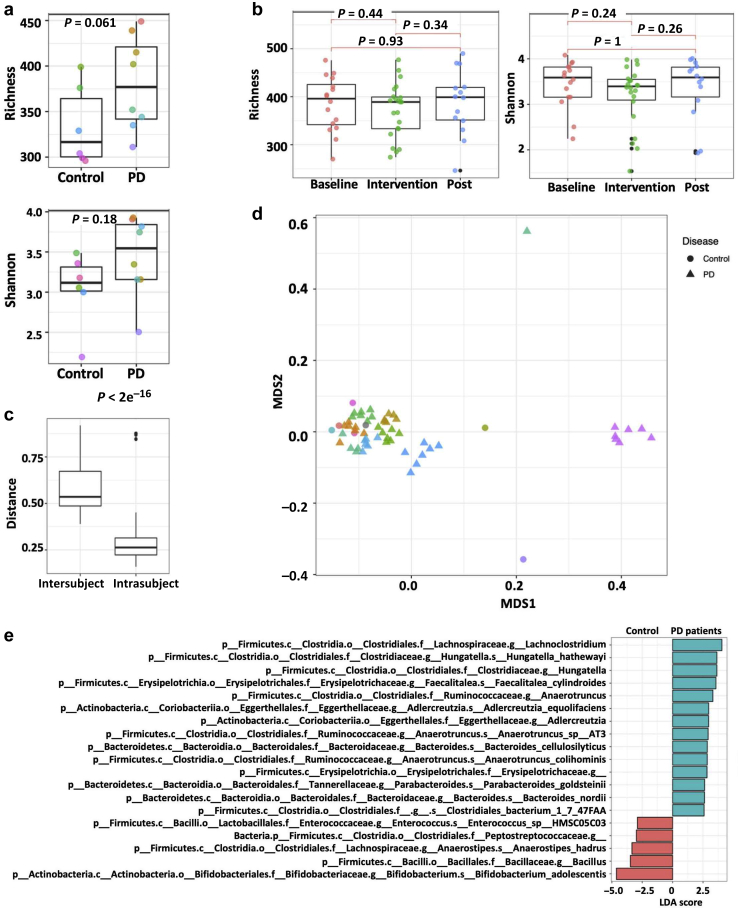
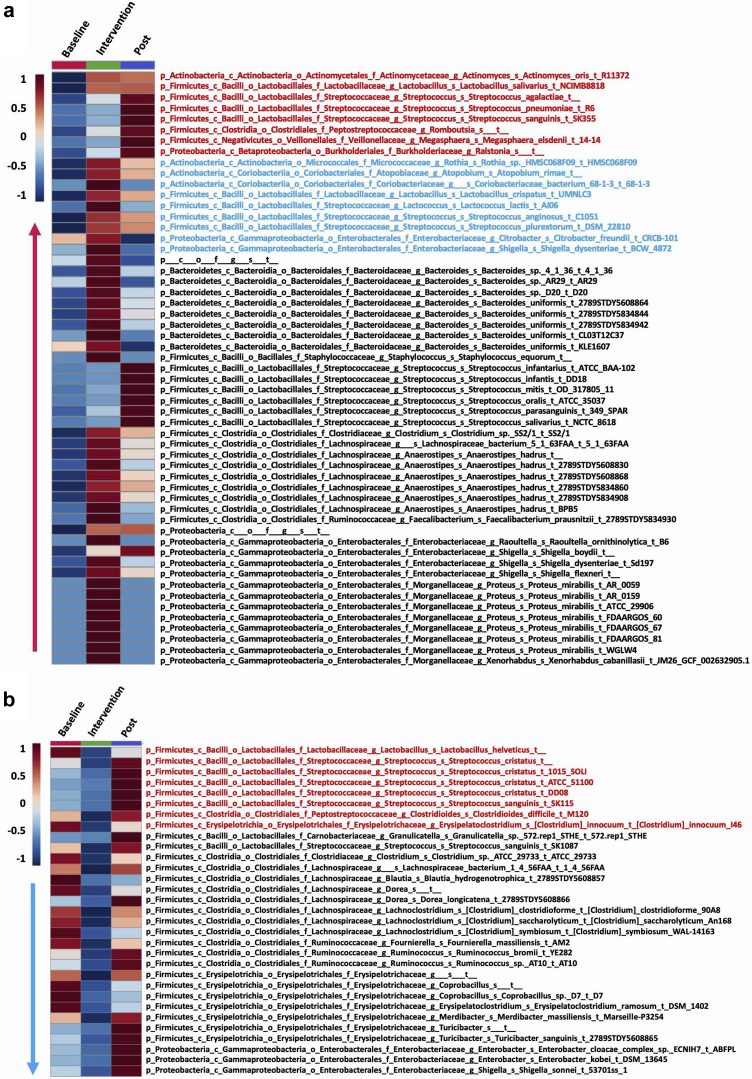
The 5 most abundant phyla identified in PD patients, in decreasing order, were Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Verrucomicrobia (Supplementary Figure S1A). Alpha diversity was not significantly different between controls and PD patients (Supplementary Figure 2A). We also did not observe a significant change in alpha diversity with p-inulin treatment (Supplementary Figure 2B). Intersubject variability in community composition was significantly higher than intrasubject variability (Wilcoxon rank-sum, P < 2e−16) (Figure 2c). With some exceptions, the overall gut microbiome composition of PD patients did not differ from that of controls (adonis P = 1) (Figure 2d). Linear discriminant analysis effect size (LEfSe) analysis revealed that 14 taxa were more abundant in PD patients at baseline compared to healthy controls, whereas 5 taxa were more abundant in controls (Figure 2e). Eight weeks of p-inulin treatment did not cause a reduction in cluster density, indicating that p-inulin intervention did not overcome intersubject variation (Figure 2d). To further evaluate the effect of p-inulin treatment, we performed random-effects mixed models controlling for within-subject autocorrelation. A total of 86 bacterial strains (P < 0.05) were significantly altered by p-inulin intervention, with 55 increased and 31 reduced during the intervention phase compared with baseline (Figure 3). Among 55 bacterial strains that increased during the intervention phase, 17 were significantly altered during the post-intervention phase compared with intervention phase, with 8 still increased and 9 decreased (Figure 3a). Among 31 bacterial strains that were reduced during p-inulin treatment, 8 increased significantly during the post-intervention phase (Figure 3b). To assess the effect of diet on the gut microbiota, we performed multivariate analysis with linear models (MaAsLin2) using default parameters, and found a significant positive correlation among Bacteroides, dietary fiber, and carbohydrate consumption (Supplementary Figure S1B and Supplementary Table S5).
Figure 2.
Microbiota analysis in peritoneal dialysis (PD) patients and control subjects. (a) Alpha diversity of taxonomy in PD patients at baseline and control subjects. Different colors represent different subjects. (b) The impact of p-inulin intervention on alpha diversity of taxonomy in PD patients. Post, post-intervention. (c) Inter- and intrasubject variability. (d) Clustering of microbial composition data by individual in the nonmetric multidimensional scaling (NMDS) ordination. Different colors represent different subjects. (e) Linear discriminant analysis (LDA) effect size analysis of bacteria in PD patients at baseline and control subjects. LDA > 2.5 are shown.
Figure 3.
Impact of p-inulin intervention on bacterial strains. (a) Increased bacterial strains at intervention phase compared with baseline (overall P < 0.05; intervention vs. baseline P < 0.05). Red indicates increased strains at post-intervention (Post) phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05). Blue indicates decreased strains at Post phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05). (b) Decreased bacterial strains at intervention phase compared with baseline (overall P < 0.05; intervention vs. baseline P < 0.05). Red text indicates increased strains at Post phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05). Blue text indicates decreased strains at Post phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05).
KEGG Modules Influenced by p-Inulin
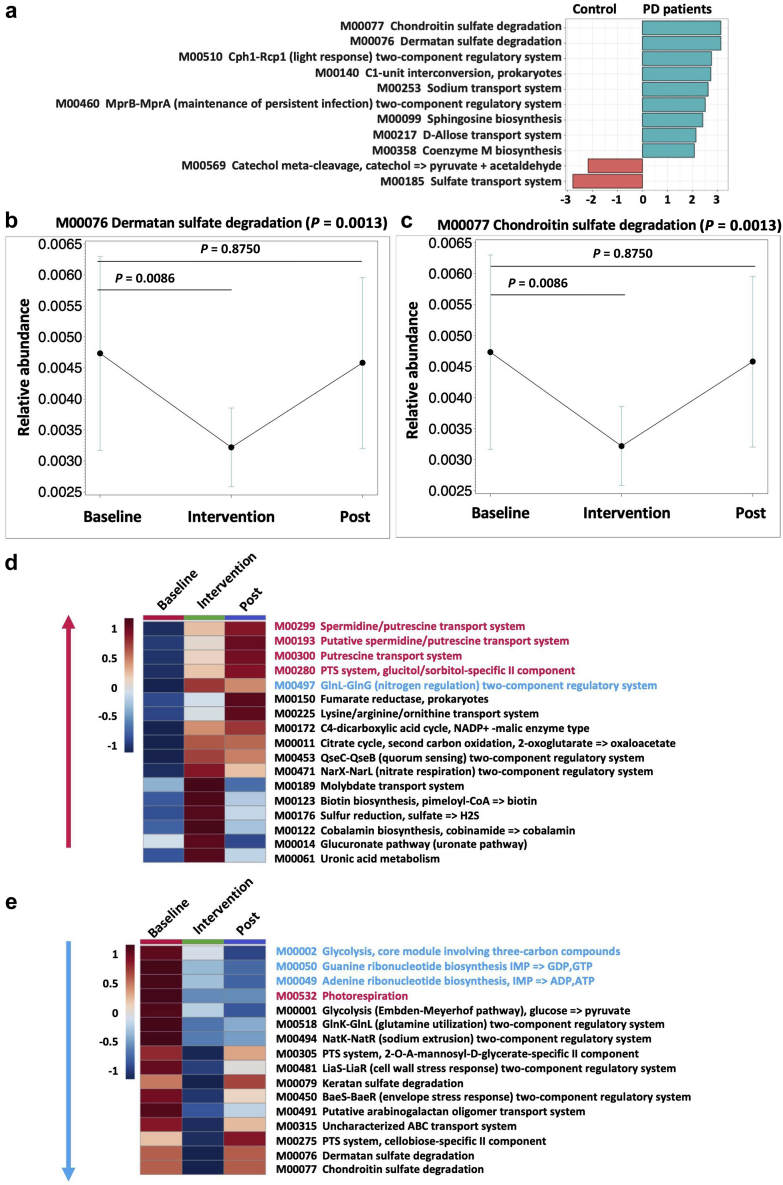
Kyoto Encyclopedia of Genes and Genomes (KEGG) modules were used to characterize the gene sets linked to microbial metabolic capabilities. Functional alpha diversity was not significantly different (Supplementary Figure S1C). While assessing beta diversity, we noted that that the donor effect persisted irrespective of the study phase (Supplementary Figure S1D). LEfSe analysis showed that 9 KEGG modules were significantly enriched in PD patients compared to control subjects, whereas 2 KEGG modules were significantly decreased (Figure 4a). Furthermore, we examined the time effects on KEGG modules during the 3 phases of the study. Among the 11 KEGG modules, which were different between PD patients at baseline and controls, p-inulin treatment effects were significant (overall P < 0.05; intervention vs. baseline P < 0.05) for 2 KEGG modules (Figure 4b and c). In addition, we noted significant treatment effects on 33 KEGG modules, with 17 KEGG modules increased (Figure 4d) and 16 reduced after p-inulin treatment compared to baseline (Figure 4e). During the post-intervention phase, some of these changes were reversed, whereas others persisted (Figure 4d and e).
Figure 4.
p-Inulin treatment effects on Kyoto Encyclopedia of Genes and Genomes (KEGG) modules. (a) Linear discriminant analysis (LDA) effect size analysis of KEGG modules in peritoneal dialysis (PD) patients at baseline and control subjects. LDA > 2.0 are shown. (b) M00076 dermatan sulfate degradation. (c) M00077 chondroitin sulfate degradation. (d) Increased KEGG modules at intervention phase compared with baseline (overall P < 0.05; intervention vs. baseline P < 0.05). Red indicates increased KEGG modules at post-intervention (Post) phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05). Blue indicates decreased KEGG modules at Post phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05). (e) Decreased KEGG modules at intervention phase compared with baseline (overall P < 0.05; intervention vs. baseline P < 0.05). Red text indicates increased KEGG modules at Post phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05). Blue text indicates decreased KEGG modules at Post phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05).
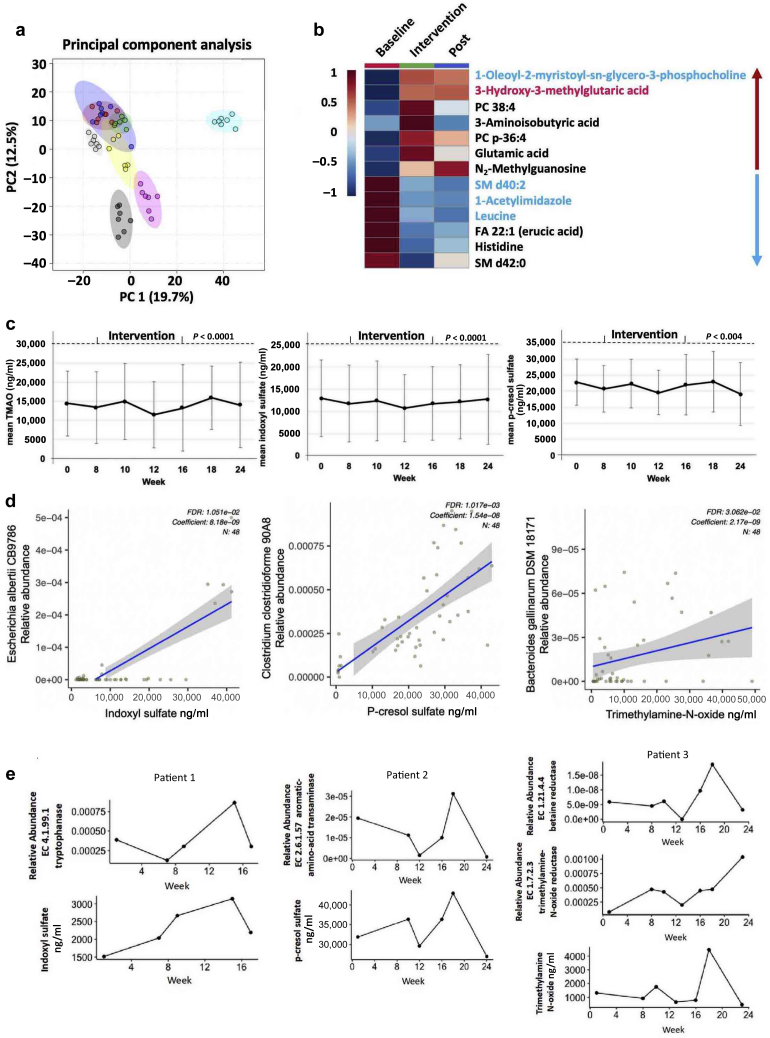
Plasma Metabolites, TMAO, IS, and PCS
The interpatient variability was greater than the intrapatient variability (Figure 5a). p-Inulin treatment significantly altered 13 metabolites (overall P < 0.05; intervention vs. baseline P < 0.05), with 7 increased and 6 decreased during the treatment phase compared to baseline (Figure 5b). Using weekly data, significant overall time effects were found for TMAO, IS (both P < 0.0001), and PCS (P = 0.004) (Figure 5c). Top bacterial strain correlated with each metabolite is shown in Figure 5d. Next, we examined the bacterial enzymes involved in IS, PCS, and TMAO biosynthesis pathways in each patient. The trends between bacterial enzymes and respective metabolite changes during 3 phases of the study varied among patients. Interestingly, the changing trajectory of tryptophanase and IS were similar in selected patient (Figure 5e). In addition, a similar changing trajectory was found among amino acid transaminase and PCS, betaine reductase, TMAO reductase, and TMAO (Figure 5e). These results suggest that the functional changes in microbial gene abundance affects the levels of IS, PCS, and TMAO to some extent in some patients.
Figure 5.
Effects of p-inulin treatment on plasma metabolites. (a) Principal component analysis of plasma metabolome. Different colors represent different subjects. (b) Significantly altered plasma metabolites influenced by p-inulin intervention (overall P < 0.05; intervention vs. baseline P < 0.05). Red text indicates increased metabolites at post-intervention (Post) phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05). Blue text indicates decreased metabolites at Post phase compared with intervention phase (overall P < 0.05; Post vs. baseline P < 0.05). (c) Plasma levels of 3 uremic toxins by week. Left, trimethylamine N-oxide; middle, indoxyl sulfate; right, p-cresol sulfate. A significant overall time effect on indoxyl sulfate, trimethylamine N-oxide (both P < 0.001), and p-cresol sulfate (P = 0.004) was noted. (d) Correlation between bacterial strains with 3 microbial metabolites in 8 patients. Left, indoxyl sulfate; middle, p-cresol sulfate; right, trimethylamine N-oxide. (e) Trend of bacterial enzymes and microbial metabolites change over time. Left, tryptophanase and indoxyl sulfate in patient 1; middle, amino-acid transaminase and p-cresol sulfate in patient 2; right, betaine reductase, trimethylamine-N-oxide reductase, and trimethylamine N-oxide in patient 3. FDR, false discovery rate.
Discussion
The symbiotic relationship between humans and microbiota allows for co-metabolism of diverse substrates. As the final readout, the plasma metabolome revealed the host and microbiota co-metabolism. We integrated the untargeted metabolomics, lipidomics, and targeted metabolomics (IS, PCS, TMAO) with metagenomics to reveal the microbiota-host co-metabolism. Previous studies have shown that oligofructose inulin significantly reduced PCS generation rates and serum concentrations in hemodialysis patients, but had no effect on IS.4 A randomized controlled trial showed that resistant starch decreased IS level in patients treated with hemodialysis.5 In patients with CKD, synbiotics (a combination of prebiotic and probiotic) did not significantly reduce serum IS but did decrease serum PCS.6 However, in another small study in hemodialysis patients, synbiotic treatment reduced PCS but not IS.7 In this small but well-designed study with stringent patient selection criteria, we demonstrate that the host co-metabolic pathways are modulated by p-inulin treatment in PD patients.
To our knowledge this is one of the first studies to investigate microbial and host metabolic responses to p-inulin supplementation in PD patients. Other strengths include rigorous patient selection criteria, study design, shotgun metagenomic sequencing, and use of targeted and untargeted metabolomic approaches. We show that p-inulin treatment was associated with an array of changes in microbiome, their metabolic pathways, and also plasma metabolome in PD patients. However, there are some study limitations as well. First, the sample size is small. Second, a large number of tests were done, and it is likely that some results will prove to be false positive; however, as we were in a hypothesis-generation mode, we did not try to adjust for multiple testing. The findings reported here are preliminary and need to be validated in a study involving larger numbers of patients with longer duration of follow-up.
Disclosure
All the authors declared no competing interests.
Acknowledgments
The study is supported in part by Young Investigator Award from the National Kidney Foundation, Kaleido Biosciences, Jackson GI Medical. DSR is supported by National Institutes of Health grants 1U01DK099924- 01 and 1U01DK099914-01.
Footnotes
Supplementary Methods.
Table S1. Study design.
Table S2. Baseline participant characteristics by treatment group.
Table S3. Dietary intake assessed by food frequency questionnaire.
Table S4. Gastrointestinal symptom rating scale during the study.
Table S5. MaAsLin2 analysis of dietary intake and gut microbes.
Figure S1. Gut microbiota in PD patients.
Supplementary Material
References
- 1.Choi J.-Y., Yoon Y.J., Choi H.J. Dialysis modality-dependent changes in serum metabolites: accumulation of inosine and hypoxanthine in patients on haemodialysis. Nephrol Dial Transplant. 2011;26:1304–1313. doi: 10.1093/ndt/gfq554. [DOI] [PubMed] [Google Scholar]
- 2.Gibson G.R., Beatty E.R., Wang X., Cummings J.H. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. 1995;108:975–982. doi: 10.1016/0016-5085(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 3.Cani P.D., Neyrinck A.M., Fava F. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 4.Meijers B.K.I. p-Cresyl Sulfate and Indoxyl Sulfate in Hemodialysis Patients. Clin J Am Soc Nephrol. 2009;4:1932–1938. doi: 10.2215/CJN.02940509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sirich T.L., Plummer N.S., Gardner C.D. Effect of increasing dietary fiber on plasma levels of colon-derived solutes in hemodialysis patients. Clin J Am Soc Nephrol. 2014;9:1603–1610. doi: 10.2215/CJN.00490114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi M. Synbiotics Easing Renal Failure by Improving Gut Microbiology (SYNERGY): a randomized trial. Clin J Am Soc Nephrol. 2016;11:223–231. doi: 10.2215/CJN.05240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakabayashi I., Nakamura M., Kawakami K. Effects of synbiotic treatment on serum level of p-cresol in haemodialysis patients: a preliminary study. Nephrol Dial Transplant. 2011;26:1094–1098. doi: 10.1093/ndt/gfq624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.