Abstract
Understanding why some species accumulate more deleterious substitutions than others is an important question relevant in evolutionary biology and conservation sciences. Previous studies conducted in terrestrial taxa suggest that life history traits correlate with the efficiency of purifying selection and accumulation of deleterious mutations. Using a large genome data set of 76 species of teleostean fishes, we show that species with life history traits associated with vulnerability to fishing have an increased rate of deleterious mutation accumulation (measured via dN/dS, i.e., nonsynonymous over synonymous substitution rate). Our results, focusing on a large clade of aquatic species, generalize previous patterns found so far in few clades of terrestrial vertebrates. These results also show that vulnerable species to fishing inherently accumulate more deleterious substitutions than nonthreatened ones, which illustrates the potential links among population genetics, ecology, and fishing policies to prevent species extinction.
Keywords: mutational meltdown, dN/dS ratio, extinction, effective population size
Determining the factors explaining why some species are more vulnerable than others is urgent given the acceleration of species extinction rates in the last decades (Barnosky et al. 2011). Although it is accepted that a combination of demographic, ecological (Jennings et al. 1998; Reynolds et al. 2001), and genetic factors (Spielman et al. 2004) contribute to species extinction, their relative importance is still unclear.
Based on metrics such as geographical range or population size, the widely used red list IUCN index relies nearly exclusively on demographic and population dynamics criteria (Strona et al. 2014). Such an approach reliably measures the conservation status of a species under ongoing threats but is not designed to assess the intrinsic vulnerability of a species to potential future threats (Miranda 2017). To fill this gap and better manage the increasing problem of overfishing, an alternative vulnerability index has been developed for fish species: the Fishbase vulnerability index (Cheung et al. 2005). Instead of relying on demographic criteria, this index exploits the well-known fact that population size and population growth depend on life history and ecological species features, such as body length, longevity, fecundity, or sexual maturity, as populations of large and long-lived species being smaller and less resilient than small and short-lived ones (Cheung et al. 2005).
Because the effective population size and genome evolution are related, the genomic sequence of a given species keeps track of past variations in population size (Nadachowska-Brzyska et al. 2015). Therefore, long-term population size can be tracked down by measuring species genetic diversity (Romiguier et al. 2014) or the ratio of nonsynonymous substitutions over synonymous substitutions (dN/dS; Nikolaev et al. 2007; Popadin et al. 2007; Romiguier et al. 2013; Figuet et al. 2016; Botero-Castro et al. 2017). In this study, we propose to test whether species more likely to be vulnerable to fishing have also been accumulated more deleterious alleles than nonthreatened ones. We choose to focus our study on teleostean fishes, a large taxa with various life history traits, for which 1) several genomes have been recently sequenced (Malmstrøm et al. 2017) and 2) unmatched resources in terms of vulnerability index are available due to their economic importance for fishing activities. Furthermore, studies trying to link effective population size, life history traits, and genome evolution focused so far exclusively on terrestrial species (mammals: Nikolaev et al. 2007; Popadin et al. 2007; Romiguier et al. 2012, 2013; birds: Botero-Castro et al. 2017; reptiles: Figuet et al. 2016). These studies concluded that species with a “K-strategy” (as described by MacArthur and Wilson 1967 for a species with large body size, high longevity, and low fecundity) are more likely to exhibit small effective population sizes on the long term. By comparing the life history traits and species vulnerability to the genomewide efficiency of purifying selection in 76 teleostean species, we test here whether commonly assumed patterns in terrestrial habitats can be generalized to aquatic environments.
Results and Discussion
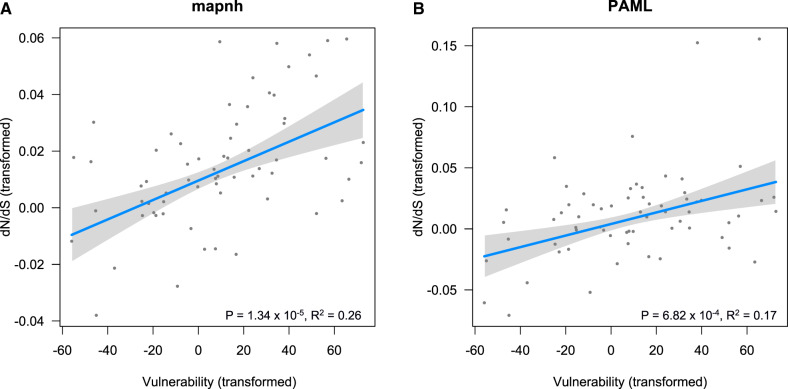
For several decades, studies in conservation sciences have aimed to identify the causes of species vulnerability to extinction. Large body size, low fecundity, and low rate of niche evolution have already been identified in previous studies as correlates of high extinction risk in mammals or birds (e.g., Fritz et al. 2009; Lavergne et al. 2013). Based on a data set of 76 species of fish, our genomic analyses showed that the rate of nonsynonymous over synonymous substitutions (dN/dS) was strongly associated to vulnerability to fishing (P = 1.34 × 10−5, R2 = 0.26, fig. 1 and table 1). This relationship was found when dN/dS was estimated with mapnh and PAML and when we controlled for phylogenetic correlation (fig. 1 and table 1, supplementary material S2, Supplementary Material online). We also found a strong relationship between dN/dS and life history traits (dN/dS and body length: P = 1.67 × 10−5, R2 = 0.26; dN/dS and longevity: P = 4.03 × 10−2, R2 = 0.14; table 1), which is consistent with previous studies on terrestrial clades in mammals, birds, and metazoa (Nikolaev et al. 2007; Popadin et al. 2007; Romiguier et al. 2012, 2013, 2014; Botero-Castro et al. 2017; Figuet et al. 2016). Our analyses indicate that these results are not due to substitution saturation that could bias dN/dS estimations (supplementary material S3, S8, and S9, Supplementary Material online). Additional tests showed that the relationship between dN/dS and vulnerability remained significant even when we removed the 15% of the longest branches that are potentially more affected by saturation (P < 0.05, supplementary material S4, Supplementary Material online).
Fig. 1.
dN/dS is associated with vulnerability. dN/dS was computed using either (A) mapnh or (B) PAML. Regression lines, P values, and R2 of the phylogenetically generalized least squares regressions are shown on each panel, respectively, for all terminal branches of more than 10,000 synonymous substitutions. The shaded gray represents the 95% CIs. x and y axes have been transformed in order to account for the phylogenetic relatedness between variables (script available in supplementary material S6, Supplementary Material online).
Table 1.
Phylogenetic Generalized Least Squares Regressions between dN/dS, Vulnerability, Latitude, Body Length, and Longevity.
| Variable | Intercept (SE) | Estimate (SE) | t Value | P Value | R 2 |
|---|---|---|---|---|---|
| Vulnerability | 5.16 × 10−2 (8.43 × 10−3) | 3.43 × 10-4 (7.26 × 10−5) | 4.72 | 1.34 × 10−5 | 0.26 |
| Latitude | 5.18 × 10−2 (9.50 × 10−3) | 3.82 × 10-4 (1.21 × 10−4) | 3.16 | 2.39 × 10−3 | 0.14 |
| Log (body length) | 4.05 × 10−2 (9.50 × 10−3) | 6.90 × 10−3 (1.48 × 10−3) | 4.66 | 1.67 × 10−5 | 0.26 |
| Log (longevity) | 5.46 × 10−2 (1.10 × 10−2) | 5.45 × 10−3 (2.54 × 10−3) | 2.15 | 4.03 × 10−2 | 0.14 |
Note.—Our analyses were run with phylogenetic generalized least squares regressions (cf., supplementary material S6, Supplementary Material online) which allow to model the relationship between variables accounting for phylogenetical nonindependence. dN/dS Was computed using mapnh.
A strong relationship between dN/dS and vulnerability can be explained by the fact that life history traits (e.g., body size), vulnerability, and dN/dS are all correlated (supplementary material S1, Supplementary Material online and table 1) because they are all different proxies of effective population size (Jennings and Blanchard 2004). It is, however, difficult to know whether more direct links exist between the accumulation of deleterious mutations and vulnerability to extinction. Indeed, studies have already proposed that the decrease of the efficiency of purifying selection in populations with small effective sizes (Ohta 1992) could lead to a snow-ball mechanism for species extinction known as mutational meltdown (Lynch et al. 1995). Such negative demographic/genetic feedbacks loops toward extinction have been hypothesized as a potential explanation for the local extirpation of large vertebrates (e.g., woolly mammoth, Rogers and Slatkin 2017). If such a genetic component to species extinction actually exists in teleosts, our results would suggest that vulnerable species to fishing are also more vulnerable to genetic risks of extinction. However, no clear evidence demonstrate that deleterious mutations increase species extinction risks of fishes for now. Future studies directly testing the correlation between average extinction rates (estimated using paleontological data or diversification models) and the average dN/dS of a clade could potentially help to better disentangle causal links among life history traits, accumulation of deleterious mutation, effective population sizes, and species extinction risks.
Our estimate of dN/dS was also significantly higher in species distributed at higher latitude (P = 2.39 × 10−3, R2 = 0.14, table 1). This result suggests that species distributed at high latitude may have generally lower or less stable population sizes. The same analysis ran with PAML did lead to similar result (although the results were significant only when removing two outlier points, supplementary material S7, Supplementary Material online). We also found no significant relationship between latitude and vulnerability (P > 0.05, R2 = 3.00 × 10−4, supplementary material S1, Supplementary Material online), body size (P > 0.05, R2 = 4.85 × 10−3), or longevity (P > 0.05, R2 = 0.01). We thus hypothesize that if latitude is associated to population size, it may be by other mechanisms than an increase of body size and longevity. For example, population dynamics at high latitude might be related to other ecological factors, such as strong seasonal and long-term climatic oscillations which reduce periodically the carrying capacity of high latitude ecosystems (Dynesius and Jansson 2000). This result also potentially implies that particular attention should be paid to vulnerable species at high latitude, given that they might have lower long-term effective population sizes, which would lead to accumulate more deleterious mutations, compared with tropical species.
The strong correlations found between vulnerability and dN/dS and between body size and dN/dS suggest that vulnerability and body size might be generally better proxies of effective population size than latitude or longevity. In theory, longevity should be also related to population size and dN/dS (Romiguier et al. 2013), but in practice, the difficulty to measure longevity in the wild may lead to spurious life span measures that may blur the relationship with dN/dS. Similarly, latitude might be related to population size but species distribution is also depending on other factors such as dispersal contingencies and species interactions (Sexton et al. 2009), which are not expected to be directly related to the effective population size.
Conclusion
Based on genomes of 76 species of fishes, our study shows that large, long-lived, and vulnerable to fishing species accumulate more deleterious mutations than small, short-lived, and not vulnerable species. Our results extend to aquatic environments the relationship between the efficiency of natural selection and species life history strategies previously found in few clades of terrestrial vertebrates. Finally, this work also highlights for the first time a positive relationship between dN/dS and latitude, suggesting that species distributed at higher latitude might have small long-term effective population sizes and consequently accumulates more deleterious mutations than tropical species.
Materials and Methods
Molecular Data and Phylogeny
We used the alignments and the phylogenetic tree provided by the authors of Malmstrøm et al. (2017). The authors aligned 1938 exons, from 120 to 1764 bp (supplementary material S10, Supplementary Material online), in 76 fish species using a multi-step blast procedure with 33,737 annotated zebrafish genes (from the Ensembl release 78; Cunningham et al. 2015). All alignments provided by Malmstrøm et al. are available in the zenodo data set repository (https://zenodo.org/record/3624895) and correspond to the less strictly filtered data set before removal of third codon position (Malmstrøm et al. 2017). The phylogenetic tree was constructed with RAxML v. 8.1.17 (Stamatakis 2014) and BEAST v.2.2 (Bouckaert et al. 2014) using 567 exons from 111 genes, with a total alignment length of 71,418 bp and 17 fossil calibrations (see Malmstrøm et al. 2017 for more details).
Data
Body length, latitude, and vulnerability data were obtained for all 76 species in Fishbase (http://www.fishbase.org/, last accessed April 2018, Froese and Pauly 2018). Longevity was obtained for 33 species. The Fishbase vulnerability index is calculated from a method averaging several traits related to the rate of reproduction, such as body size and generation time (Cheung et al. 2005).
Molecular Evolution and dN/dS
We estimated the number of synonymous and nonsynonymous substitutions in the terminal branches of the 76 teleost species from the alignment of 1938 concatenated genes (378,663 bp for the concatenated alignment) using the probabilistic substitution mapping implemented in the software mapnh (Romiguier et al. 2012, https://github.com/BioPP/testnh, YN98 codon model) after a first run of the software bppml for estimating branch lengths and parameter values under an homogeneous YN98 codon model (Nielsen and Yang 1998). Examples of the bppml and mapnh control files are available on the zenodo data set depository (https://zenodo.org/record/3624895). The mapnh analysis estimates a probabilistic count of synonymous and nonsynonymous substitutions. We used the ratio of the sum of nonsynonymous substitutions and the sum of synonymous substitutions then divided it by the ratio of total nonsynonymous sites and total synonymous sites (computed via the script pNpSdNdS.py, available here: https://github.com/popgenomics/pNpSdNdS) as our dN/dS measure. For the PAML analysis, we used the free-ratio branch model to estimate one omega parameter estimated per branch for each individual genes (see supplementary material S10, Supplementary Material online, for the details) and then computed the average dN and average dS by weighting with gene length.
Estimates of dN/dS can be poorly estimated when the total number of substitutions is small (short branches). We controlled for this bias by running all analyses after the removal of the terminal branches with <10,000 synonymous substitutions (∼15% shortest branches). We chose this threshold because it allowed to eliminate atypical short branches, which were difficult to compare with the others branches for accurate dN/dS estimations. These short branches belonged mostly to closely related species inside taxa that were oversampled compared with the other clades, such as the Gadinae subfamily. We replicated the analysis with the dN/dS estimations produced by codeml from the PAML package (Yang 2007).
Relationship between dN/dS, Life History Traits, and Vulnerability
We tested whether dN/dS was associated with life history traits and vulnerability with phylogenetic generalized least squares regressions (code provided in the supplementary material S6, Supplementary Material online). This allows to model a linear relationship while taking into account the phylogenetic structure of correlation between variables. For plotting the phylogenetic generalized least squares regressions, we transformed the x- and y-axis in order to take into account the phylogenetic correlation in variables (code provided in supplementary material S6, Supplementary Material online, fig. 1). We then tested the robustness of the association between dN/dS and life history traits/vulnerability with two other sets of analyses: one set of analyses with all species (including branches with <10,000 synonymous substitutions, supplementary material S5, Supplementary Material online) and one set of analyses removing the ∼15% longest branches (branches with more than 35,000 synonymous substitution) to test whether substitution saturation was affecting our results (supplementary material S4, Supplementary Material online).
Testing for Potential Bias Related to Substitution Saturation
The synonymous substitution saturation effect (i.e., several substitutions occurring sequentially at the same site on the same branch of the phylogenetic tree) may potentially bias our analyses. This bias may particularly affect dN/dS estimation on very long branches, for which dN/dS may be increased in the case of saturation (Cannarozzi and Schneider 2012). We tested if our analyses were affected by saturation using the entropy index of substitution saturation (Iss) of Xia et al. 2003 in the software DAMBE 7 (supplementary material S3, Supplementary Material online, procedure described in Xia and Lemey 2009). This index indicates if there is substitution saturation when the Iss is significantly smaller (P < 0.05) than the critical Iss value at which the sequences will begin to fail to recover the true tree.
Saturation should particularly impact our results by spuriously increasing dN/dS values for longer branches of the phylogenetic tree. We thus also tested whether our results were also found when removing the longest branches (supplementary material S4, Supplementary Material online), and we plotted the relationship between dS, dN, and dN/dS with branch lengths (supplementary material S8, Supplementary Material online).
Supplementary Material
Acknowledgments
We thank Konstantin Popadin and two anonymous reviewers for insightful comments. We also thank Camille Roux and Etienne Loire for providing the code pNpSdNdS.py and for useful discussions and advices, and finally Tom Booker, Xuhua Xia, Matthew Hahn for advices on the analyses. J.Rol. received funding from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie (Grant No. 785910) and the Banting Postdoctoral Fellowship (Grant No. 151042).
Author Contributions
J.Rol. and J.Rom. designed the study, ran the analyses, and wrote the manuscript. D.S. provided the code for the phylogenetic generalized least squares, helped for the analyses, and provided comments on the manuscript.
References
- Barnosky AD, Matzke N, Tomiya S, Wogan GOU, Swartz B, Quental TB, Marshall C, McGuire JL, Lindsey EL, Maguire KC, et al. 2011. Has the Earth’s sixth mass extinction already arrived? Nature 471(7336):51–57. [DOI] [PubMed] [Google Scholar]
- Botero-Castro F, Figuet E, Tilak MK, Nabholz B, Galtier N.. 2017. Avian genomes revisited: hidden genes uncovered and the rates versus traits paradox in birds. Mol Biol Evol. 34(12):3123–3131. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarozzi GM, Schneider A.. 2012. Codon evolution: mechanisms and models. Oxford: Oxford University Press. [Google Scholar]
- Cheung WW, Pitcher TJ, Pauly D.. 2005. A fuzzy logic expert system to estimate intrinsic extinction vulnerabilities of marine fishes to fishing. Biol Cons. 124(1):97–111. [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2015. Ensembl 2015. Nucleic Acids Res. 43:D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynesius M, Jansson R.. 2000. Evolutionary consequences of changes in species’ geographical distributions driven by Milankovitch climate oscillations. Proc Natl Acad Sci U S A. 97(16):9115–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figuet E, Nabholz B, Bonneau M, Mas Carrio E, Nadachowska-Brzyska K, Ellegren H, Galtier N.. 2016. Life history traits, protein evolution, and the nearly neutral theory in amniotes. Mol Biol Evol. 33(6):1517–1527. [DOI] [PubMed] [Google Scholar]
- Fritz SA, Bininda‐Emonds OR, Purvis A.. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol Lett. 12(6):538–549. [DOI] [PubMed] [Google Scholar]
- Froese R, Pauly D.. 2018. FishBase Available from: https://www.fishbase.org/. World Wide Web Electron. Publ. version. (04/2018).
- Jennings S, Blanchard JL.. 2004. Fish abundance with no fishing: predictions based on macroecological theory. J Anim Ecol. 73(4):632–642. [Google Scholar]
- Jennings S, Reynolds JD, Mills SC.. 1998. Life history correlates of responses to fisheries exploitation. Proc R Soc Lond B. 265(1393):333–339. [Google Scholar]
- Lavergne S, Evans ME, Burfield IJ, Jiguet F, Thuiller W.. 2013. Are species’ responses to global change predicted by past niche evolution? Philos Trans R Soc B. 368(1610):20120091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery J, Burger R.. 1995. Mutation accumulation and the extinction of small populations. Am Nat. 146(4):489–518. [Google Scholar]
- MacArthur R, Wilson EO.. 1967. The theory of island biogeography. Princeton (NJ): Princeton University Press. [Google Scholar]
- Malmstrøm M, Matschiner M, Tørresen OK, Jakobsen KS, Jentoft S.. 2017. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci Data. 4(1):160132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda R. 2017. The misguided comparison of vulnerability and conservation status. Aquatic Conserv Mar Freshw Ecosyst. 27(4):898–899. [Google Scholar]
- Nadachowska-Brzyska K, Li C, Smeds L, Zhang G, Ellegren H.. 2015. Temporal dynamics of avian populations during Pleistocene revealed by whole-genome sequences. Curr Biol. 25(10):1375–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z.. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148:929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Montoya-Burgos JI, Popadin K, Parand L, Margulies EHNational Institutes of Health Intramural Sequencing Center Comparative Sequencing ProgramAntonarakis SE.. 2007. Life-history traits drive the evolutionary rates of mammalian coding and noncoding genomic elements. Proc Natl Acad Sci U S A. 104(51):20443–20448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 1992. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 23(1):263–286. [Google Scholar]
- Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K.. 2007. Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc Natl Acad Sci U S A. 104(33):13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JD, Jennings S, Dulvy NK.. 2001. Life histories of fishes and population responses to exploitation. In: Reynolds JD, Mace GM, Redford KH, Robinson JG, editors. Conservation of exploited species. Cambridge: Cambridge University Press. [Google Scholar]
- Rogers RL, Slatkin M.. 2017. Excess of genomic defects in a woolly mammoth on Wrangel Island. PLoS Genet. 13(3):e1006601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Figuet E, Galtier N, Douzery EJP, Boussau B, Dutheil JY, Ranwez V.. 2012. Fast and robust characterization of time-heterogeneous sequence evolutionary processes using substitution mapping. PLoS One. 7(3):e33852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Gayral P, Ballenghien M, Bernard A, Cahais V, Chenuil A, Chiari Y, Dernat R, Duret L, Faivre N, et al. 2014. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515(7526):261–263. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, Galtier N.. 2013. Genomic evidence for large, long-lived ancestors to placental mammals. Mol Biol Evol. 30(1):5–13. [DOI] [PubMed] [Google Scholar]
- Sexton JP, McIntyre PJ, Angert AL, Rice KJ.. 2009. Evolution and ecology of species range limits. Annu Rev Ecol Evol Syst. 40(1):415–436. [Google Scholar]
- Spielman D, Brook BW, Frankham R.. 2004. Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci U S A. 101(42):15261–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strona G. 2014. Assessing fish vulnerability: IUCN vs FishBase. Aquatic Conserv Mar Freshw Ecosyst. 24(2):153–154. [Google Scholar]
- Xia X, Lemey P.. 2009. Assessing substitution saturation with DAMBE. In: Lemey P, Salemi M, Vandamme A-M, editors. The phylogenetic handbook: a practical approach to DNA and protein phylogeny. Vo1. 2. 2nd ed Cambridge: Cambridge University Press; p. 615–630. [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y.. 2003. An index of substitution saturation and its application. Mol Phylogenet Evol. 26(1):1–7. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.