Abstract
Satellite DNAs (satDNAs) are among the most dynamically evolving components of eukaryotic genomes and play important roles in genome regulation, genome evolution, and speciation. Despite their abundance and functional impact, we know little about the evolutionary dynamics and molecular mechanisms that shape satDNA distributions in genomes. Here, we use high-quality genome assemblies to study the evolutionary dynamics of two complex satDNAs, Rsp-like and 1.688 g/cm3, in Drosophila melanogaster and its three nearest relatives in the simulans clade. We show that large blocks of these repeats are highly dynamic in the heterochromatin, where their genomic location varies across species. We discovered that small blocks of satDNA that are abundant in X chromosome euchromatin are similarly dynamic, with repeats changing in abundance, location, and composition among species. We detail the proliferation of a rare satellite (Rsp-like) across the X chromosome in D. simulans and D. mauritiana. Rsp-like spread by inserting into existing clusters of the older, more abundant 1.688 satellite, in events likely facilitated by microhomology-mediated repair pathways. We show that Rsp-like is abundant on extrachromosomal circular DNA in D. simulans, which may have contributed to its dynamic evolution. Intralocus satDNA expansions via unequal exchange and the movement of higher order repeats also contribute to the fluidity of the repeat landscape. We find evidence that euchromatic satDNA repeats experience cycles of proliferation and diversification somewhat analogous to bursts of transposable element proliferation. Our study lays a foundation for mechanistic studies of satDNA proliferation and the functional and evolutionary consequences of satDNA movement.
Keywords: satellite DNA, Drosophila, genome evolution, repeats, eccDNA
Introduction
Eukaryotic genomes are replete with large blocks of tandemly repeated DNA sequences. Named for their distinct “satellite” bands on cesium chloride density gradients (Kit 1961; Sueoka 1961; Szybalski 1968), these so-called satellite DNAs (satDNAs) can comprise large fractions of eukaryotic genomes (Britten and Kohne 1968; Yunis and Yasmineh 1971). SatDNAs are a major component of heterochromatin; for example, they accumulate in megabase-length blocks in areas of reduced recombination such as centromeres, telomeres, and Y chromosomes (Charlesworth et al. 1986, 1994). The location, abundance, and sequence of these heterochromatic satDNAs can turnover rapidly (Yunis and Yasmineh 1971; Ugarkovic and Plohl 2002) creating divergent repeat profiles between species (Strachan et al. 1982). SatDNAs can be involved in intragenomic conflicts over transmission through the germline as: the driving centromeres that cheat female meiosis (e.g., centromere drive, Henikoff et al. 2001), or the targets of the sperm killers that cheat male meiosis (Larracuente 2014; Courret et al. 2019). These conflicts may fuel satDNA evolution. Changes in satDNA are expected to have broad evolutionary consequences due to their roles in diverse processes, including chromatin packaging (Blattes et al. 2006) and chromosome segregation (Dernburg et al. 1996). For example, variation in satDNA can impact centromere location and stability (Aldrup-MacDonald et al. 2016), meiotic drive systems (Fishman and Willis 2005; Fishman and Saunders 2008; Lindholm et al. 2016), hybrid incompatibilities (Ferree and Barbash 2009), and genome evolution (Britten and Kohne 1968; Hartl 2000; Bosco et al. 2007).
Small blocks of tandem repeats also occur in euchromatic regions of genomes (Feliciello, Akrap, Brajkovic, et al. 2015; Ruiz-Ruano et al. 2016) and are particularly enriched on Drosophila X chromosomes (Waring and Pollack 1987; DiBartolomeis et al. 1992; Kuhn et al. 2012; Gallach 2014). Some euchromatic X-linked repeats have sequence similarity to the large blocks of heterochromatic satDNAs (Waring and Pollack 1987; DiBartolomeis et al. 1992; Kuhn et al. 2012) suggesting they could be a continual source of euchromatic repeats. Studies suggest these euchromatic repeats may play roles in gene regulation by acting as “evolutionary tuning knobs” (King et al. 1997), regulating chromatin (Brajkovic et al. 2012; Feliciello, Akrap, and Ugarkovic 2015), and facilitating X chromosome recognition/dosage compensation (Waring and Pollack 1987; Kuhn et al. 2012; Lundberg et al. 2013; Menon et al. 2014; Lucchesi and Kuroda 2015; Joshi and Meller 2017; Deshpande and Meller 2018; Kim et al. 2018).
Much of the species-level variation in satDNA arises through movement and divergence of an ancestral “library” of satellites inherited through common decent (Fry and Salser 1977). Unequal exchange between different repeats within a tandem array leads to expansions and contractions of repeats at a locus (Smith 1976) and, along with gene conversion, causes the homogenization of repeated sequences. This homogenization can occur both within repeat arrays (Schlotterer and Tautz 1994) and between repeats on different chromosomes, causing repeat divergence between species (reviewed in Dover 1982). These processes result in the concerted evolution (Dover 1994) of satDNAs (Strachan et al. 1982) and multicopy gene families like rDNA and histones (Coen et al. 1982), leading to species-specific repeat profiles. Novel satDNAs can arise within a species from the amplification of unique sequences through replication slippage (Levinson and Gutman 1987; Schlotterer and Tautz 1992), unequal exchange, rolling-circle replication (Britten and Kohne 1968; Southern 1970; Lohe and Brutlag 1987; Walsh 1987), and transposable element (TE) activity (Dias et al. 2014; McGurk and Barbash 2018; Vondrak et al. 2020). Recombination involving satDNA can cause local rearrangements or large-scale structural rearrangements such as chromosomal translocations (Richardson and Jasin 2000; Lieber et al. 2006). Intrachromatid recombination events give rise to extrachromosomal circular DNAs (eccDNAs) that are common across eukaryotic organisms (Cohen et al. 1999, 2003, 2006; Zellinger and Riha 2007; Navratilova et al. 2008; Cohen and Segal 2009; Paulsen et al. 2018) and may contribute to the rapidly changing repeat landscape across genomes.
We have limited resolution on the evolutionary dynamics and molecular mechanisms that drive the rapid turnover of satDNA and its distribution in genomes. This lack of resolution is, in part, due to the challenges that repetitive DNA presents to sequence-based and molecular biology approaches. Here, we characterize patterns and mechanisms underlying the evolution of complex satellites over short evolutionary time scales in D. melanogaster and the closely related species in the simulans clade, D. mauritiana, D. sechellia, and D. simulans. We focus on the two abundant satellite repeat families that are present in the euchromatin of all four study species: 1.688 g/cm3 and Rsp-like. 1.688 g/cm3 (hereafter called 1.688) is a family of several related repeats named after their monomer lengths, including 260, 353, 356, 359, and 360 bp (Losada and Villasante 1996; Abad et al. 2000). Rsp-like is a 160-bp repeat named for its similarity to the 120-bp Responder (Rsp) satellite (Larracuente 2014). We studied broad-scale patterns using cytological and genomic approaches. By leveraging new reference genomes based on long single-molecule sequence reads (Chakraborty et al. 2020), we study the dynamics of these repeats at base-pair resolution across the X chromosome. We discovered the rapid spread of Rsp-like repeats to new locations across the X chromosome in D. simulans and D. mauritiana. We explored the mechanism of satDNA movement, including the potential role of interlocus gene conversion and eccDNA in facilitating the spread of satellites across long physical distances on the X chromosome. Revealing the processes that shape satDNA evolution over short time scales is a critical step toward understanding the functional and evolutionary consequences of repeat turnover.
Results
Heterochromatic and Euchromatic satDNA Composition Varies across Species
Our analysis of mitotic chromosomes with fluorescence in situ hybridization shows that large heterochromatic blocks of 1.688 repeats are primarily X-linked in D. melanogaster and D. sechellia but are autosomal in D. simulans and D. mauritiana (supplementary fig. S1, Supplementary Material online). Drosophila melanogaster also has two smaller blocks of 1.688 family repeats in the heterochromatin of chromosome 3 (Abad et al. 2000). The distribution of the Rsp-like family is similarly dynamic in the heterochromatin: large blocks are X-linked in D. simulans, autosomal in D. sechellia (chromosomes 2 and 3), and lacking in the heterochromatin of D. mauritiana and D. melanogaster (Larracuente 2014; supplementary fig. S1, Supplementary Material online). The 1.688 repeat family also exists in the euchromatin (Waring and Pollack 1987; DiBartolomeis et al. 1992; Kuhn et al. 2012; Gallach 2014), where they are overrepresented on the X chromosome relative to the autosomes in these Drosophila species (Chakraborty et al. 2020).
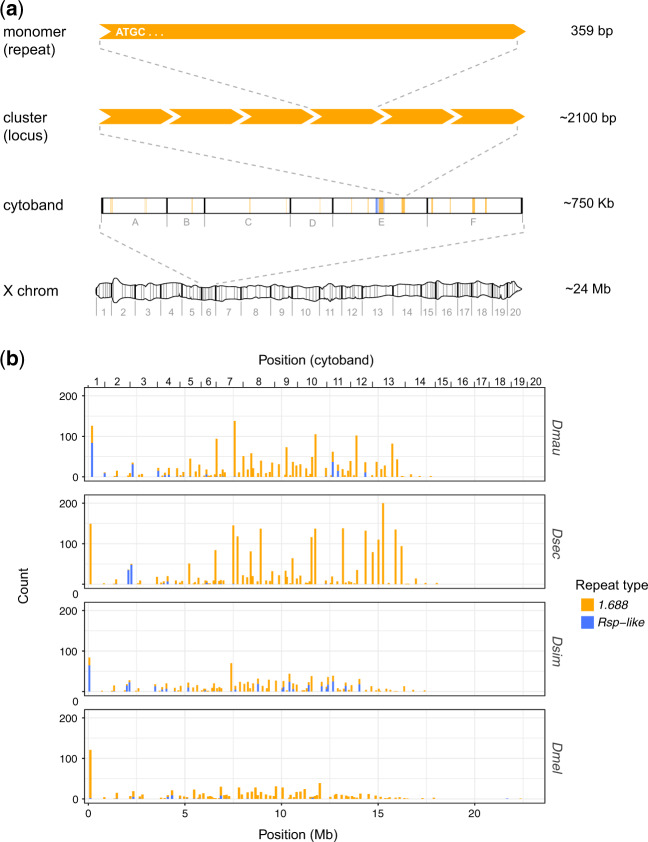
We mapped euchromatic satDNA repeats at a fine scale across the X chromosome. We find that similar to 1.688, Rsp-like repeats are also present in the X euchromatin (fig. 1 and supplementary figs. S2 and S3, Supplementary Material online). We describe the location of these repeats relative to their cytological divisions (i.e., cytobands) on D. melanogaster polytene chromosomes and hereafter use the terms “cytobands,” “clusters,” and “monomers” as illustrated in figure 1a. Both satellites accumulate near the telomere (cytoband 1) and in the middle of the X chromosome but are uncommon from cytoband 15 to the centromere (fig. 1 and supplementary fig. S3, Supplementary Material online). We confirmed the euchromatic enrichment of these repeats using FISH on polytene chromosomes, where we see a high density of bands on the polytenized arm of the X chromosome in the simulans clade species (e.g., representative FISH image; supplementary fig. S2, Supplementary Material online).
Fig. 1.
Euchromatic X-linked satellites are unevenly distributed across the X chromosome. (a) A schematic illustrating terms frequently used in the text. We use “cytoband” to reference large regions of the X chromosome that are defined by banding patterns in polytene chromosomes. The X Chromosome polytene graphic was redrawn based onBridges (1938) as a reference image. We use “cluster” to mean any distinct genomic locus containing the repeat of interest; typically >1 repeat. “Monomer” refers to a single repeat unit; the example shown represents a 1.688 monomer. (b) The x axis shows position of 1.688 and Rsp-like satDNA clusters along the X chromosome for each species. Each bar on the chart represents a cytological subdivision (e.g., 1A, 1B, etc.) in which counts of all repeats are pooled. The y axis indicates the number of repeat copies (i.e., monomers) within a subdivision.
The abundance of euchromatic complex satellite repeats shows a 3-fold variation among species. Drosophila sechellia has the most euchromatic X-linked repeats (2,588 annotations), followed by D. mauritiana (1,390), D. simulans (1,112), and D. melanogaster (849) (table 1). The D. sechellia X chromosome assembly contains 19 gaps, six of which occur within satellite loci (Chakraborty et al. 2020); therefore, the X-linked copy number represents a minimum estimate for this species. The other species have fewer gaps in the X chromosome assembly (11, 5, and 9 gaps in D. melanogaster, D. simulans, and D. mauritiana, respectively) and none that occur at satellite loci.
Table 1.
Summary of Euchromatic satDNA Cluster Sizes on X Chromosome.
| Species | Total No. 1.688 | No. 1.688 Clust | % N = 1 1.688 | % N < 4 1.688 | No. Rsp-like | No. Rsp-like Clust | % N = 1 Rsp-like | % N < 4 Rsp-like |
|---|---|---|---|---|---|---|---|---|
| Drosophila mauritiana | 1,165 | 325 | 24.00 | 68.31 | 225 | 26 | 30.77 | 34.62 |
| Drosophila sechellia | 2,486 | 308 | 33.44 | 82.14 | 102 | 12 | 50.00 | 58.33 |
| Drosophila simulans | 786 | 324 | 31.17 | 89.20 | 326 | 38 | 18.42 | 34.21 |
| Drosophila melanogaster | 808 | 274 | 33.94 | 83.94 | 41 | 19 | 73.68 | 78.95 |
Note.—Total No., number of total repeats; No. Clust, total number of clusters at distinct loci; % N = 1, percentage of singletons (clusters of a single repeat); % N < 4, percentage of small clusters (<4 repeats).
Within each species, 1.688 is more abundant on the X chromosome than Rsp-like, both in terms of total repeats (i.e., the number of euchromatic repeat monomers annotated in our assemblies) and the number of clusters (i.e., the number of distinct genomic loci containing repeats) (fig. 1b). Single-monomer clusters exist in both satDNA types; they represent ∼30% of all 1.688 clusters and ∼43% (∼33% if D. melanogaster is excluded) of all Rsp-like clusters (table 1 and supplementary fig. S4, Supplementary Material online). These single-monomer clusters are considered “dead” as they cannot undergo unequal exchange and expand (Dover 1982; Langley et al. 1988; Charlesworth et al. 1994). The majority of the remaining 1.688 clusters are also small (i.e., contain 2–3 repeats), whereas the majority of the remaining Rsp-like clusters are larger (i.e., contain ≥4 repeats; table 1 and supplementary fig. S4, Supplementary Material online).
Both the number of total repeats and the number of clusters for each satellite also vary among species in the X chromosome euchromatin. Rsp-like shows an 8-fold difference in total repeat number and a 3-fold difference in number of clusters across species, with D. simulans and D. mauritiana having more total repeats as well as more clusters than D. sechellia and D. melanogaster (table 1 and supplementary fig. S4, Supplementary Material online). In D. simulans and D. mauritiana, Rsp-like clusters have apparently spread to cytobands that lack such clusters in one, or both of the other species (e.g., clusters at cytobands 7–12 in D. simulans and cytobands 11–12 in D. mauritiana; fig. 1 and supplementary fig. S3, Supplementary Material online). A relatively recent spread is consistent with D. simulans and D. mauritiana having a lower proportion of single repeat, or “dead” clusters (18.4% and 30.8%, respectively) than the other species (table 1). In 1.688, D. sechellia shows as much as a 3-fold increase in total repeats despite having fewer 1.688 loci than the other simulans clade species, a pattern driven by a high number of large clusters in D. sechellia (16 clusters with ≥50 monomers), which are less common in other species (six clusters in D. mauritiana, one in both D. simulans and D. melanogaster; table 1).
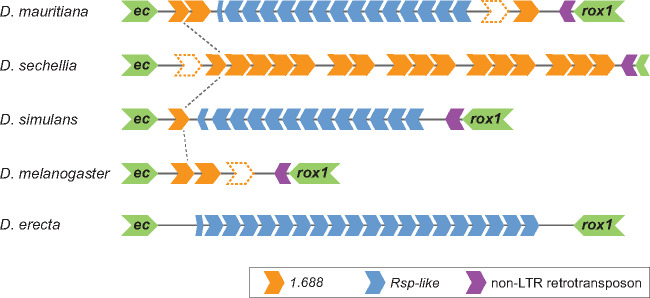
The collective differences in abundance and location of these satellites suggest dynamic turnover of satDNA repeat composition across the X chromosome euchromatin over short evolutionary time scales. The repetitive nature of these loci makes it difficult to systematically establish orthology on a locus-by-locus basis to accurately quantify the rate of turnover across the X chromosome. However, we can explore the dynamics of specific clusters for which synteny of unique flanking sequences strongly suggests orthology across species. One such representative cluster is embedded between two genes—echinus and roX1—at cytoband 3F (fig. 2). In D. melanogaster, this cluster has only two 1.688 repeats, the first of which is truncated, plus an unannotated adjacent region that contains degenerated 1.688 sequence. Drosophila sechellia also has 1.688 at this location, but the cluster is expanded relative to D. melanogaster. In contrast, both Rsp-like and 1.688 repeats are present at this locus in D. mauritiana and D. simulans; however, each species shows differences in repeat number of the respective satellites (fig. 2). The Rsp-like repeats in D. mauritiana and D. simulans are homogenized within each locus but are highly divergent between species. We see similar shifts in repeat composition at 12 other loci that we are able to confidently identify as orthologous (supplementary table S1, Supplementary Material online), suggesting that this is a general pattern. The major differences in X-linked satellite composition among species at specific loci further suggest that euchromatic satellites, like heterochromatic satellites, evolve dynamically over short evolutionary time scales.
Fig. 2.
Schematic of 3F cluster in Drosophila melanogaster, the simulans clade species, and D. erecta. Cluster is flanked by two genes, echinus (ec) and roX1 (last green chevrons), with a TE insertion at the distal side of the locus (purple chevrons adjacent to roX1). Complex satellite monomers are indicated by blue (Rsp-like) or orange (1.688) chevrons located between the genes. Chevrons with dotted outline indicate sequences that were not annotated but were determined manually by BLAST to be highly degenerated satellite monomers. Black dotted lines between species indicate shared repeats. Although we include D. erecta as an outgroup taxon, we consider the ancestral state of this locus to be unknown given the fast turnover rate of these repeats and the divergence time between the study species and D. erecta (∼9 Ma).
Recent Proliferation of satDNA across the X Euchromatin
Analysis of the nearest upstream and downstream genomic features relative to 1.688 and Rsp-like satellites showed that Rsp-like clusters have a nonrandom distribution, particularly in D. simulans and D. mauritiana. Rsp-like clusters are directly adjacent to, or interspersed with, 1.688 clusters in 82% of euchromatic X-linked clusters in D. simulans and in 62% of clusters in D. mauritiana (table 2 and supplementary figs. S5 and S6, Supplementary Material online). Conversely, the 1.688 clusters do not seem to preferentially associate with Rsp-like, though they are often located near genes, consistent with previous findings (Kuhn et al. 2012; supplementary figs. S5 and S6, Supplementary Material online).
Table 2.
Rsp-like Clusters Associate with 1.688.
| Species | No. Rsp-like | No. Rsp-like/1.688 | % Rsp-like/1.688 |
|---|---|---|---|
| Drosophila mauritiana | 26 | 16 | 62 |
| Drosophila sechellia | 12 | 3 | 25 |
| Drosophila simulans | 38 | 31 | 82 |
| Drosophila melanogaster | 19 | 7 | 37 |
Note.—No. Rsp-like, number of Rsp-like clusters on X chromosome; No. Rsp-like/1.688, number of Rsp-like clusters (including singletons) that have 1.688 repeats within 100 bp either upstream or downstream.
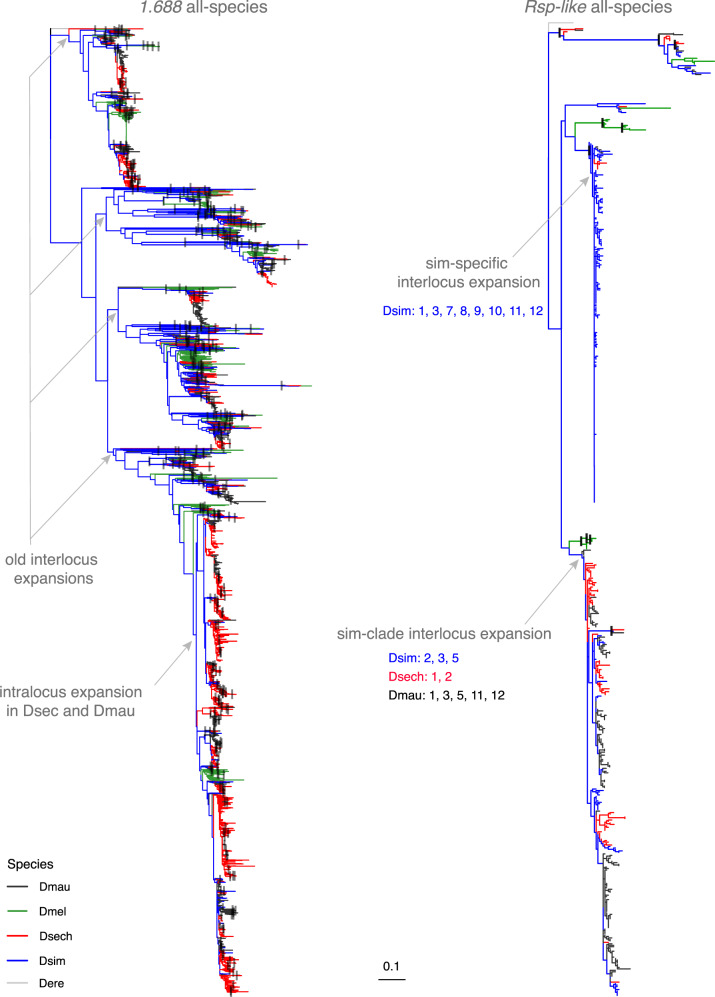
Examination of within-species and all-species phylogenetic trees of satellite repeats led to four major findings: 1) Heterochromatic repeats form clades that are generally separate from euchromatic repeats for both satellites in all species except D. sechellia, for which euchromatic and heterochromatic repeats are interspersed in both 1.688 and Rsp-like (supplementary figs. S7–S14, Supplementary Material online). 2) Drosophila sechellia and D. mauritiana (especially the former) show repeated evidence of intralocus expansion of repeats (supplementary figs. S15 and S16, Supplementary Material online). 3) 1.688 euchromatic repeats have a relatively old diversification history that largely predates the speciation events that gave rise to the study species (figs. 3and 4 and supplementary figs. S7, S9, S11, S13, S15, and S16, Supplementary Material online). This contrasts with Rsp-like, which shows evidence of relatively recent diversification, particularly in the simulans clade species (figs. 3and 4 and supplementary figs. S8, S10, S12, S14, and S17–S20, Supplementary Material online). 4) Rsp-like repeats show evidence of two major expansions (figs. 3and 4 and supplementary figs. S8, S10, S12, S17, and S18, Supplementary Material online), which encompass large physical distances across the X chromosome (i.e., “interlocus” expansions) and mainly occurred independently in D. simulans and D. mauritiana. The latter two findings are discussed in additional detail in the Supplementary Material online.
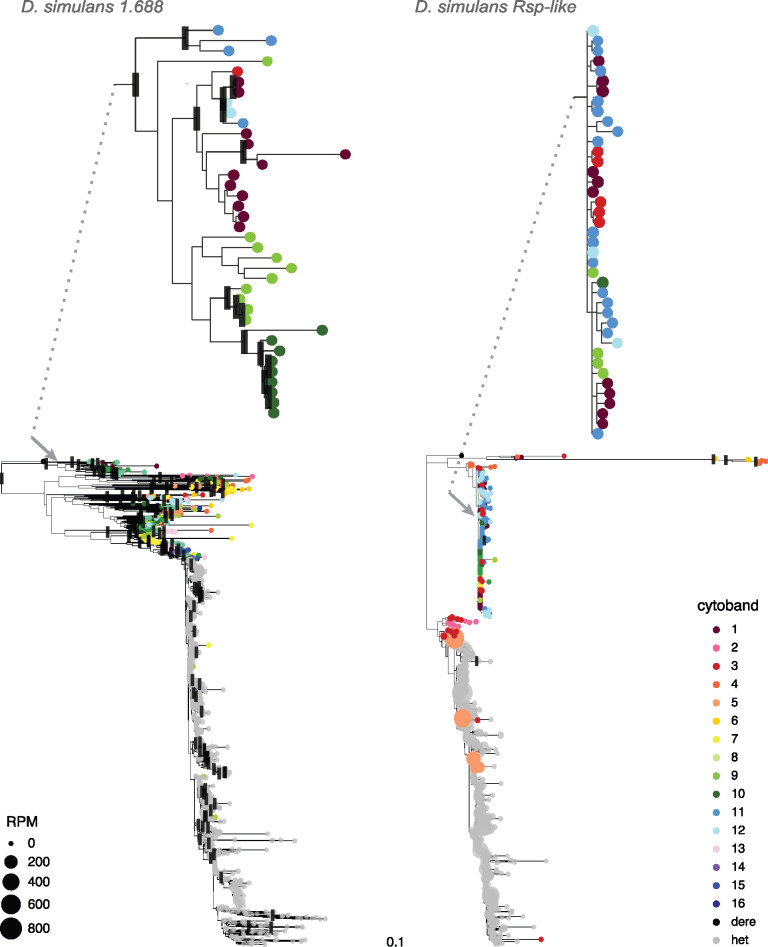
Fig. 3.
Comparison of phylogenetic patterns between 1.688 and Rsp-like for Drosophila simulans. Each terminal represents an individual repeat monomer from the X chromosome. Colored tip terminals indicate euchromatic repeats; gray tip terminals represent repeats from heterochromatic loci (defined as unassigned scaffolds in the assembly). Black rectangles indicate nodes with bootstrap support ≥90. Two regions in each tree are shown in greater detail to highlight differential phylogenetic patterns observed in euchromatic repeats of 1.688 and Rsp-like; arrows and dotted lines indicate relative position of enlarged regions in the tree. Branch lengths shown are proportional to divergence with both trees shown on the same relative scale. Sizes of the tips are scaled to reflect proportion of eccDNA reads mapping to a given variant, expressed as reads per million (RPM) (see eccDNA analysis). Maximum likelihood trees were inferred in RAxML with nodal support calculated following 100 bootstrap replicates.
Fig. 4.
All-species maximum likelihood trees of euchromatic 1.688 and Rsp-like. Each terminal represents an individual repeat monomer. All monomers from clusters with ≥3 repeats were included in the analysis. Species identity is indicated by branch color. Major inter- and intralocus expansions of satellites discussed in the text are labeled with gray arrows. For interlocus expansions in Rsp-like, the species involved are listed along with cytological bands that are represented by monomers within the expansion. The outgroup (Drosophila erecta) is indicated by gray branches. Black rectangles indicate nodes with bootstrap support ≥90. Maximum likelihood trees were inferred in RAxML with nodal support calculated following 100 bootstrap replicates. Branch length is shown proportional to relative divergence with both trees on the same relative scale. (See figures S15–S18, Supplementary Material online, for added detail as to genomic location of terminals.)
Mechanisms Driving satDNA Turnover in the Euchromatin
How did the new Rsp-like clusters observed in D. simulans and D. mauritiana (i.e., finding four in the previous section) arise? We found frequent colocalization of Rsp-like and 1.688 repeats in these species, which was surprising because these two repeats are unrelated at the sequence level. We therefore looked for sequence motifs at these junctions that could facilitate insertion of new Rsp-like repeats into pre-existing 1.688 clusters.
Our analysis of the 1.688/Rsp-like junctions on each end of newly inserted Rsp-like clusters in D. simulans and D. mauritiana revealed multiple independent insertion events with shared signatures (fig. 5). One prominent signature is that junctions between the Rsp-like and 1.688 sequences commonly occur at positions of microhomology. The same junction sequence is often shared between clusters at different locations across the X chromosome. We use the sequence of these microhomologies to define clusters of the same “type”: type 1 was found in D. simulans and types 2 and 3 were found in D. mauritiana. Because there are different 1.688 variants adjacent to both type 1 and 2 junctions (e.g., compare Dsim10A and Dsim11E1, fig. 5), we infer that five or more independent events have created the three junction types.
Fig. 5.
Junctions at new Rsp-like insertions in Drosophila simulans and D. mauritiana. Junctions from a subset of the newer Rsp-like clusters (blue text/lines/bars in the center) are aligned and grouped into three types (gray “Type” boxes) based on common signatures with nearby 1.688 monomers (flanking orange text/lines/bars). Type 1 junction is found in D. simulans, whereas types 2 and 3 are found in D. mauritiana (cytoband location of each cluster is indicated in the names at far left). Within each type, identical truncated Rsp-like monomers abut 1.688 at the same position in the 1.688 repeat monomer. In all three junction types, there is overlap between the two satellite sequences (black text) which, for at least the longer overlaps, potentially represents microhomology involved in the original insertion event. The second junction associated within and among these types is more variable (gray “variable” boxes) with Rsp-like sequences abutting different positions of the 1.688 repeat or different unannotated sequences (gray bars). The number of full length Rsp-like monomers as well as the lengths of truncated Rsp-like monomers, unannotated regions, and 1.688 sequences in these variable regions are indicated for each cluster. Note that while lacking a common signature, subsets of clusters within each variable region are nearly identical (e.g., Dsim7D and Dsim12F). The 1.688 sequences for an uninserted monomer (dark gray text) as well as the base positions relative to the 1.688 consensus sequence are indicated above each junction type. *Indicates the duplicated 1.688 sequences at the two junctions in the first four D. simulans clusters presented.
In D. simulans, type 1 is the predominant junction and is observed in 19/31 Rsp-like clusters located near 1.688 repeats, 12 of which are diagramed in figure 5. The type 1 junction is associated with a 42-bp truncated Rsp-like monomer abutting 1.688 sequences. The transition between the two satellite types includes a 7-bp region of microhomology (TGGTACC). Among these 12 Rsp-like clusters there are, however, at least 6 different junction sequences at the other end of the cluster. One of these variable junctions includes four clusters in which the sequences adjacent to Rsp-like are a duplication of the 32 bp (including the microhomology) of 1.688 sequences found at the type 1 junction. The remaining clusters have varying lengths of unannotated (5–397 bp) and 1.688 sequences (1–310 bp) in the variable region. All 19 Rsp-like insertions, which includes the clusters at 3F, 9D, 9F, 11C, 11D, 12C, and 12F-1 not diagramed in figure 5, are associated with a minor subset of 1.688 repeat variants comprising ∼15% of the 787 monomers examined.
In D. mauritiana, type 2 clusters show a similar signature to D. simulans type 1 clusters: one end of the cluster shows a characteristic junction which is associated with a Rsp-like truncated monomer abutting 1.688 sequences, with the other end of the cluster showing more variable patterns. Interestingly, type 2 junctions occur at nearly the same position within the 1.688 monomer and in a similar subset of variants as the D. simulans type 1 junction, however, the position in Rsp-like monomers associated with the junction differs between the two species (i.e., note 27-bp truncated monomers in D. mauritiana and 42-bp truncations in D. simulans; fig. 5). The variable side of the cluster shows four different sequences associated with the junction. The most common variable junction occurs in four of the eight clusters and has a 2-bp deletion before continuing with the interrupted 1.688 repeat sequence. Likewise, the four new clusters in cytoband 11 of D. mauritiana show these junction signatures although unlike the type 1 and type 2 junctions, these type 3 junctions have a larger deletion (36 bp) in the associated 1.688 sequences.
The nature of the variable junctions (unannotated sequences/sequence variation in 1.688 repeat monomers) makes it difficult to determine whether insertion was facilitated by microhomology at these junctions. However, in two cases, short runs of mononucleotides are present at the overlap between 1.688 and Rsp-like sequences. Although nonhomologous end joining does not require, but can use, short stretches of microhomology (<5 bp; Chang et al. 2017), the multiple occurrences of microhomology including the 7 and 4 bp of microhomology observed in the type 1 and type 3 junctions, respectively, suggest that pathways employing microhomology-mediated end joining facilitate Rsp-like insertions (fig. 6a).
Fig. 6.
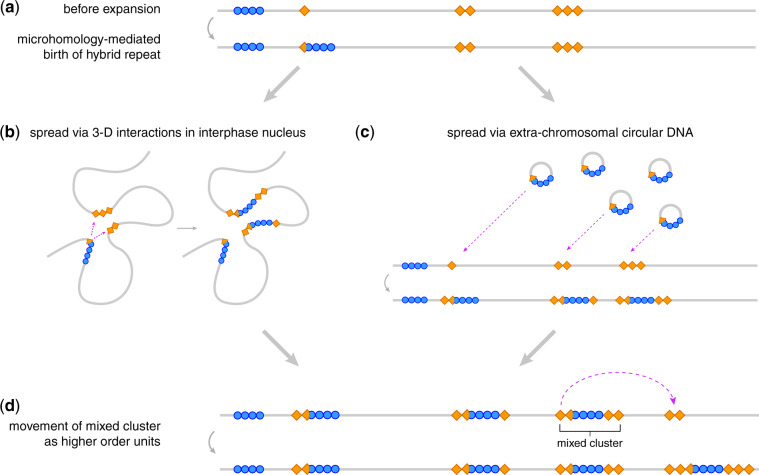
Proposed mechanisms of satDNA dynamics. Blue circles represent an ancestrally rare satellite (i.e., Rsp-like), orange diamonds represent an abundant satellite present at many loci (i.e., 1.688), gray lines represent a fraction of a chromosome that spans many megabases. (a) The microhomology-mediated birth of a hybrid repeat formed from the rare+common satellites facilitates the spread of the rare satellite to loci where the abundant satellite is already present through a process of (b) interlocus gene conversion (loci could be physically distant on a linear X chromosome but in close proximity in 3D space within the interphase nucleus) seeded by the orange (abundant) satellite repeats, or; (c) extrachromosomal circular DNA generation/reintegration, again seeded by the orange satellite repeats. (d) After new insertions of the blue satellite, entire mixed clusters may move as higher order units. The mechanisms illustrated in (b) and (c) could also be responsible for the generation of the hybrid repeat (a) and movement of higher order units (d). Not illustrated is the expansion or contraction of a repeat cluster at a given locus due to unequal exchange with a different cluster of the same repeat type.
As described earlier, the relatively minor 1.688 repeat variants adjacent to the type 1 and type 2 junctions are each shared across multiple 1.688/Rsp-like clusters (fig. 5). This suggests either Rsp-like has repeatedly inserted into a particular subset of variants in both species, or that the multiple 1.688/Rsp-like junctions were not formed independently within either species. In the latter scenario, a relatively rare microhomology-mediated event gives rise to a 1.688/Rsp-like hybrid repeat, which then seeds new Rsp-like clusters at loci where 1.688 clusters were already present, facilitated by homology of the 1.688 portion of the novel hybrid repeat. We made two predictions arising from this model: 1) newly inserted Rsp-like clusters would only occur at genomic loci where 1.688 repeats were already present; 2) any 1.688 sequences moving as a higher order repeat along with Rsp-like sequences may show discordant phylogenetic relationships with 1.688 repeats already present at the new insertion site.
We tested the above predictions using D. simulans Rsp-like clusters with type 1 junctions, focusing on the 12 of 19 clusters that are present at genomic loci where Rsp-like clusters are lacking in one or more of the other three study species (i.e., those clusters at cytobands 7–12). We conducted a synteny analysis across species to establish orthology for the 12 clusters. If a 1.688 cluster was present at a syntenic position in the other species, we inferred that Rsp-like moved into an existing cluster in D. simulans. We found that all 12 new Rsp-like clusters in D. simulans had 1.688 repeats at that same location in each of the other three species with the exception of a single locus in D. melanogaster (supplementary table S1, Supplementary Material online). With the exception of two loci at cytoband 11 in D. mauritiana, none of the syntenic loci in the other species has Rsp-like repeats (supplementary table S1, Supplementary Material online). The fact that 1.688 clusters were already present at the site of new Rsp-like insertions suggests that it is sequence homology (and/or microhomology) with 1.688 repeats that is facilitating new insertions. In 6 of 12 clusters with new insertions, the 1.688 repeat immediately adjacent to the Rsp-like junction shows a strongly discordant relationship with the other 1.688 repeats in the cluster (supplementary table S1, Supplementary Material online), suggesting that at least a partial 1.688 repeat has moved together with Rsp-like repeats.
Our findings from the 1.688/Rsp-like junction and synteny analyses are consistent with a model in which small regions of microhomology can facilitate the integration of Rsp-like into 1.688. Once this association is created, the rapid spread of Rsp-like across the chromosome could be facilitated by hitchhiking with segments of flanking 1.688 repeats (fig. 6b and c), including through the movement of entire mixed clusters to new locations as a higher order unit (fig. 6d).
Mechanisms of satDNA Spread to New Loci
Two mechanisms that can explain the generation of new clusters as well as the spread of nearly identical repeats are: 1) 3D interactions in the nucleus creating opportunities for interlocus gene conversion across long linear distances; and 2) the spread of repeats via extrachromosomal circular DNA (eccDNA) to new loci across the X chromosome (fig. 6).
Our reanalysis of D. melanogaster Hi-C data (Ogiyama et al. 2018) provides some evidence of inter-cytoband interactions, particularly across the middle of the X chromosome (i.e., from cytobands 6 through 14), where we observe sequence blocks flanking satellite repeats that show high interaction values with loci in other cytobands (supplementary fig. S21, Supplementary Material online; see supplementary methods, Supplementary Material online). If long-distance gene conversion is facilitated by 3D interactions in the nucleus, we might expect 1.688 repeats and neighboring Rsp-like repeats to show a similar pattern of gene conversion. Analysis of sequence similarity of the 1.688 repeats adjacent to these Rsp-like clusters showed a mixed pattern, with high-sequence similarity among repeats only at cytobands 1, 11, and 12 (supplementary fig. S22, Supplementary Material online). The majority (64.5%) of 1.688 repeats have <95% sequence similarity with any repeat from another cytoband, whereas the nearest Rsp-like repeat shows >95% similarity with repeats from multiple different cytobands. Thus, we find limited evidence of long-distance gene conversion in 1.688 sequences; however, it is possible that the older age and smaller size of 1.688 clusters relative to Rsp-like clusters may limit interlocus gene conversion.
eccDNA as a Mechanism of satDNA to New Genomic Loci
Reintegration of eccDNA (extrachromosomal circular DNA) is another (nonmutually exclusive) mechanism that could mediate the spread of Rsp-like satellite repeats. We used 2D gel analysis to confirm/show the presence of 1.688 (Cohen et al. 2003) and Rsp eccDNA in D. melanogaster (supplementary fig. S23, Supplementary Material online) and then isolated (supplementary figs. S23 and S24, Supplementary Material online) and sequenced the eccDNA component from all four species. We estimated the abundance of sequences in eccDNA and in the genomic control as reads per million (RPM).
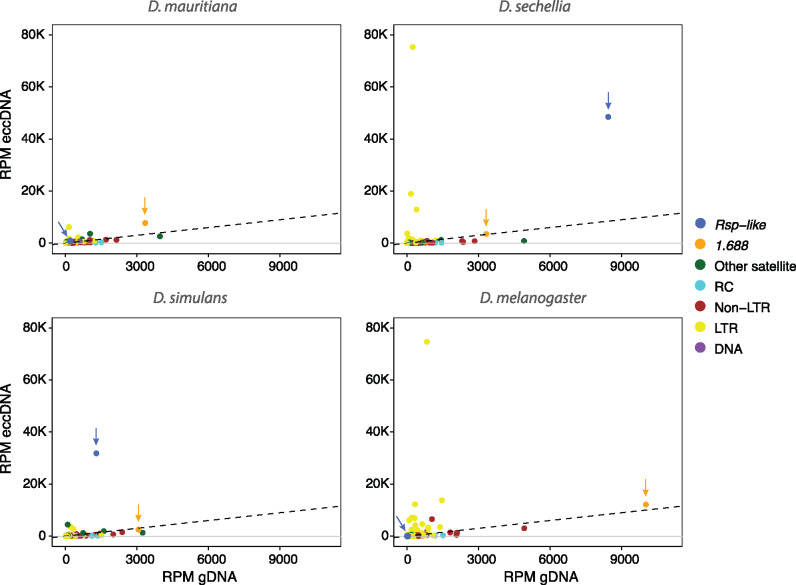
We find long-terminal repeats (LTRs) and complex satellites, including 1.688 and Rsp-like, are abundant on eccDNAs in all four species (supplementary fig. S25, Supplementary Material online and fig. 7). In general, we detect a strong correlation between the abundance of a repetitive element in the genome (estimated by RPM for that element in the nondigested genomic DNA control reads) and the abundance of eccDNA reads derived from that repeat. However, some repeats produce more eccDNA than expected given their genomic abundance (fig. 7). Rsp-like repeats are particularly abundant on eccDNA in D. simulans (fig. 7), where they comprise ∼3% of the total eccDNA-enriched reads (24.5-fold enrichment over the undigested control), and in D. sechellia, where they comprise ∼4.9% of reads (a 5.75 enrichment over the undigested control).
Fig. 7.
Scatter plot of eccDNA RPM and genomic DNA RPM. Repeats in the genome are categorized by color into: other satellite (complex satellites except 1.688 and Rsp-like), LTR retrotransposon, non-LTR retrotransposon, DNA transposon, and rolling-circle (RC) transposon. Rsp-like (shown in blue) and 1.688 (shown in orange) are indicated by arrows. Dotted lines represent the same abundance of eccDNA and genomic DNA such that dots above the dotted line indicate repeats that are enriched in eccDNA libraries relative to genomic controls.
To determine the genomic source of satellite-derived eccDNAs, we estimated abundance of each sequence variant of 1.688 or Rsp-like from euchromatic and heterochromatic loci. We represent the estimated eccDNA abundance on phylogenetic trees by scaling tip labels based on the RPM of each variant (fig. 3 and supplementary fig. S7–S14, Supplementary Material online). With the exception of 1.688 in D. sechellia and D. mauritiana, heterochromatic repeat variants produce more eccDNA than euchromatic variants. Consistent with the lack of heterochromatic Rsp-like repeats (Larracuente 2014), few eccDNAs map to D. mauritiana Rsp-like. Some individual repeats generate more eccDNAs than others, possibly due to sequence composition, chromatin structure, and/or recombination environment. For example, in D. simulans, eight euchromatic Rsp-like variants from cytoband 5A are enriched for eccDNA (RPM ranges from ∼100 to 600, see light orange tips in fig. 3 and supplementary fig. S12, Supplementary Material online). These euchromatic repeats group with the heterochromatic repeats that are also enriched for eccDNA reads (fig. 3 and supplementary fig. S12, Supplementary Material online). It is therefore possible that the repeats at 5 A may be a result of a recent integration of heterochromatic-derived eccDNA carrying Rsp-like repeats.
Discussion
Our comparative analysis of complex satDNA in high-quality genome assemblies reveals that small X-linked euchromatic clusters of 1.688 and Rsp-like repeats evolve rapidly over short evolutionary time scales. Despite diverging from a common ancestor just 240 kya (Garrigan et al. 2012), the simulans clade species differ in the total number of repeats, the number of clusters, and in the composition of clusters across syntenic loci (figs. 1and 2 and supplementary fig. S1, Supplementary Material online; table 1 and supplementary table S1, Supplementary Material online). The dynamic evolution of these repeats within the X chromosome euchromatin is similar to the rapid evolution of large blocks of heterochromatic satDNA across whole chromosomes reported in this (supplementary fig. S1, Supplementary Material online), and other studies (Strachan et al. 1982; Lohe and Brutlag 1987; Lohe and Roberts 1988; Larracuente 2014; Jagannathan et al. 2017; Wei et al. 2018). In the euchromatin, however, the expansion, contraction, sequence turnover, and movement of repeats play out across tens to hundreds of comparatively small loci distributed within a single chromosome. At least some of the differences in repeat abundance between species may be explained by ecology and demographic history. For example, D. sechellia is an island endemic with a historically low-effective population size (Legrand et al. 2009) and natural selection may be less efficacious in this species (McBride 2007). Interestingly, this species has larger euchromatic satDNA clusters suggesting that intralocus expansions of repeats may be weakly deleterious, but it does not have more discrete repeat clusters. In contrast to D. sechellia, we see the birth of new Rsp-like clusters in D. simulans and D. mauritiana across the X chromosome.
We show that euchromatic satDNAs can proliferate rapidly over short evolutionary timescales. Rsp-like repeats recently spread across an ∼14-Mb region of the X chromosomes of D. simulans and D. mauritiana, inserting into existing 1.688 clusters (figs. 1, 3, and 4 and supplementary figs. S3, S8, S10, and S12, Supplementary Material online). Although we find that 1.688 has an old history of diversification, consistent with previous studies (Waring and Pollack 1987; DiBartolomeis et al. 1992), our phylogenetic analysis of 1.688 repeats suggests an evolutionary history characterized by long periods of local differentiation among repeats, punctuated by the occasional proliferation of a particular variant, and subsequent local diversification (fig. 4 and supplementary figs. S15 and S16, Supplementary Material online). Thus, our comparative study of repeat patterns in these species reveals satellite proliferation dynamics that may implicate common processes underlying the evolution of both repeat types. These apparent cycles of proliferation and diversification are somewhat analogous to bursts of TE proliferation, except that rather than spreading by encoding proteins to mediate their movement, satDNAs likely spread through recombination-based mechanisms.
Mechanisms of Rsp-like Movement
We find evidence that microhomology-mediated events generated new hybrid repeats that joined the sequence of a relatively uncommon satellite (i.e., Rsp-like) to that of an abundant satellite with a dense distribution across the X chromosome (i.e., 1.688). The birth of new 1.688/Rsp-like hybrid repeats appears to have occurred independently in D. simulans and D. mauritiana, and likely multiple times within each species (figs. 4and 5 and supplementary figs. S17 and S18 and table S1, Supplementary Material online). Microhomology-mediated repair events are implicated in creating structural rearrangements and chromosomal translocations across organisms (reviewed in McVey and Lee 2008), as well as copy number variations associated with human disease (Hastings et al. 2009), and gap repair after P-element transpositions in Drosophila (Adams et al. 2003; McVey et al. 2004). After the initial microhomology-mediated association of the two repeats, the probability of the Rsp-like repeats being involved in additional repair events at homologous sequences along the chromosome increased because of their association with 1.688, which is abundant across the X chromosome. Our conclusion that this new association with 1.688 facilitated the spread of Rsp-like clusters is supported by both our analysis of junctions and synteny analysis of clusters with new Rsp-like insertions (fig. 5 and supplementary table S1, Supplementary Material online). The movement of these higher order repeats along with intralocus satDNA expansions via unequal exchange further contribute to the fluidity of the repeat landscape (figs. 5 and 6). Our investigation of Rsp-like proliferation provides a nucleotide-scale illustration of the mechanisms that can account for apparently random, differential amplification of ancestral satellites that leads to species-specific satDNA profiles observed by previous studies (Mestrovic et al. 1998; Pons et al. 2004).
Mechanisms Facilitating Long-Distance Spread of New Clusters
Questions remain about the source of the template Rsp-like sequences. We discussed two possibilities here: eccDNA reintegration and interlocus gene conversion. Both exploit DNA breaks which out of necessity must be repaired; the nature/timing of the break is an important factor in determining which of the many repair pathways is involved (Scully et al. 2019). The complexity of the sequences observed in the Rsp-like/1.688 variable junctions could implicate pathways such as FoSteS (fork stalling and template switching; Lee et al. 2007) or MMBIR (microhomology-mediated break-induced replication; Hastings et al. 2009). Both of these repair pathways occur during aberrant DNA replication and can involve multiple template switches facilitated by microhomology. Alternatively, during double-strand break (DSB) repair, synthesis-dependent strand annealing with an interlocus template switch may result in gene conversion events (Smith et al. 2007) that insert Rsp-like sequences into existing 1.688 clusters. Similar events occur at the yeast MAT locus during gene conversion, where interchromosomal template switches occur even between divergent sequences, and these events can proceed based on microhomologies as small as 2 bp (Tsaponina and Haber 2014). DNA prone to forming secondary structures (e.g., non-B form DNA-like hairpins or G quartets) can cause replication fork collapse that leads to DSB formation (reviewed in Mirkin EV and Mirkin SM 2007). Blocks of complex satDNAs may be enriched for sequences that form secondary structures and therefore may have elevated rates of DSBs compared with single-copy sequences. Elevated rates of DSB may make it more likely to observe nonhomologous recombination-mediated repair events resulting in complex rearrangements, differences in repeat copy number and, as we describe here, the colonization of repeats at new genomic positions across large physical distances.
We show that complex satellites are abundant on eccDNA (fig. 7 and supplementary figs. S23–S25, Supplementary Material online), and map eccDNA reads to the specific repeat variants from which these circles arise (fig. 3 and supplementary figs. S7–S14, Supplementary Material online). Although the abundance of most eccDNAs correlates with their genomic abundance, some repeats, such as Rsp-like in D. simulans, generate a disproportionate amount of eccDNAs. The formation of eccDNA may depend on DNA sequence, organization (e.g., repetitive vs. unique), chromatin status, and possibly its higher order structure. It is possible that the high abundance of Rsp-like derived eccDNA suggests that this satellite is unstable at the chromatin level, or more prone to DSB. EccDNA formation exploits different methods of DNA damage repair, including homologous recombination (HR) using solo LTRs, (Gresham et al. 2010), microhomology-mediated end joining (Shibata et al. 2012; Moller et al. 2015), and nonhomologous end joining (van Loon et al. 1994). The repetitive nature of 1.688 and Rsp-like makes it difficult to examine junctions in the extrachromosomal circles themselves. We do find evidence suggesting that HR can give rise to Rsp-like circles, however. An eccDNA arising from an intrachromatid exchange event between repeats within the same array, followed by the reintegration of that eccDNA at a new genomic location, could generate new arrays where the first and last repeats are truncated, but together would form a complete monomer. We see this pattern in four of the new Rsp-like arrays in D. simulans (Dsimpre1A-a, Dsimpre1A-b, Dsimpre1A-c, Dsim1A-1; fig. 5) and two arrays in D. mauritiana (Dmau1A-4, Dmau1A-6; fig. 5). It is thus conceivable that eccDNAs are involved in the generation of new Rsp-like clusters. Our finding that satDNAs and LTRs are enriched on circles is consistent with other studies showing that repeats generate eccDNA (Cohen et al. 2003, 2006; Navratilova et al. 2008; Cohen and Segal 2009; Moller et al. 2015; Lanciano et al. 2017; Shoura et al. 2017). EccDNAs may be a source of genomic plasticity within species (Gaubatz 1990); we suspect that they also played a role in the proliferation of satDNAs in the simulans clade, thus contributing to X-linked repeat divergence between these species. Experimental approaches will help explicitly test the hypothesis that satDNA-derived eccDNAs reintegrate in the genome.
Interactions in the 3D nucleus may also contribute to movement of satDNA by facilitating interlocus gene conversion events between loci far apart on a linear chromosome, including through heterochromatin/euchromatin interactions (Lee et al. 2020). Although our data are not suited to directly test this hypothesis, we find indirect evidence that long-distance interactions may occur across the X euchromatin through reanalysis of D. melanogaster Hi-C data and by searching for signatures of recent gene conversion in 1.688 repeats flanking regions with new Rsp-like insertions (supplementary figs. S21 and S22, Supplementary Material online). If these long-distance interactions in the 3D nucleus are conserved between species, this may account for the similar but independent spread of satDNAs to distant loci that we see in D. simulans and D. mauritiana. Data on long-range 3D chromosome interactions in the simulans clade species will be important for testing this hypothesis and for understanding the role of interlocus gene conversion in satDNA movement.
Functional Consequences of Rapid satDNA Evolution
A growing body of research suggests that shifts in satellite abundance and location may have consequences for genome evolution. Large-scale rearrangements or divergence in heterochromatic satDNA may lead to hybrid incompatibilities. In D. melanogaster, a heterochromatic block of 1.688 satDNA is associated with embryonic lethality in D. melanogaster–D. simulans hybrids (Ferree and Barbash 2009; Ferree and Prasad 2012) through mechanisms that we do not yet understand. However, even variation in small euchromatic satDNAs can have measurable effects on gene regulation and thus may be important for genome evolution. Short tandem repeats in vertebrate genomes can affect gene regulation by acting as binding sites for transcription factors (Rockman and Wray 2002; Gemayel et al. 2010). Additionally, repeats can have an impact on local chromatin, which may affect nearby gene expression (Feliciello, Akrap, and Ugarkovic 2015). Novel TE insertions can cause small RNA-mediated changes in chromatin (e.g., H3K9me2) that can spread to nearby regions and alter local gene expression (Lee and Karpen 2017). In D. melanogaster, siRNA-mediated chromatin modifications at 1.688 repeats play a role in X chromosome recognition by helping recruit the male-specific lethal dosage compensation complex (Menon et al. 2014; Joshi and Meller 2017; Deshpande and Meller 2018). Moving specific 1.688 repeats from cytoband 3F on the X chromosome to an autosomal location recruits male-specific lethal to the ectopic autosomal location (Joshi and Meller 2017) and affects both local H3K9 methylation and gene expression, suggesting that these repeats are cis-acting factors for X chromosome recognition (Deshpande and Meller 2018). A subset of 1.688 repeats have similar effects on the targeting of another chromosome-specific protein, Painting of Fourth (Kim et al. 2018), which may be related to an ancient dosage compensation mechanism (Larsson and Meller 2006). The turnover in repeat composition in D. simulans and D. mauritiana that we observe at loci with demonstrated effects on the recruitment of chromosome-binding proteins and chromatin (e.g., fig. 2) raises the possibility that dynamic evolution of euchromatic satDNAs may have functional consequences for dosage compensation.
Understanding of the molecular mechanisms that drive rapid expansion, movement, and rearrangement of satDNAs across the genome is a necessary step in determining the functional and evolutionary consequences of rapid satDNA evolution. In addition to fine-scale mapping of satDNA evolution in a comparative framework, we present initial insights as to the mechanisms that shape the proliferation and movement over short time scales. Future work that includes population data will be important for disentangling species versus population-level variation and addressing whether natural selection plays a role in satDNA evolution within and across loci. We suspect that the rapid satDNA dynamics in one genome compartment (e.g., heterochromatin) may drive corresponding changes in the other genome compartment (e.g., X-linked euchromatin). Future work on the evolutionary forces driving rapid satDNA evolution (e.g., molecular drive [Dover 1982], meiotic drive [Henikoff et al. 2001]), and the molecular and physical interactions between heterochromatin and euchromatin (Lee et al. 2020), will help reveal the broad consequences for rapid satDNA evolution.
Materials and Methods
Repeat Annotation
Repeat annotations were performed as described in Chakraborty et al. (2020). Briefly, we constructed a custom repeat library by downloading the latest repetitive element release for Drosophila from RepBase and added custom satellite annotations. We manually checked our library for redundancies and miscategorizations. We used our custom library with RepeatMasker version 4.0.5 using permissive parameters to annotate the assemblies. We merged our repeat annotations with gene annotations constructed in Maker version 2.31.9 (for the simulans clade species) (Cantarel et al. 2007) or downloaded from Flybase (for D. melanogaster) (Thurmond et al. 2019).
We used custom Perl scripts (Sproul et al. 2020) to define clusters of satellites on the X chromosome and to determine the closest neighboring annotations. We defined clusters as two or more monomers of a given satellite within 500 bp of each other, though in some analyses we also included single monomers. We grouped clusters according to cytoband (FlyBase annotation v6.03; ftp://ftp.flybase.net/releases/FB2014_06/precomputed_files/map_conversion/; last accessed April 7, 2020). We used custom scripts to translate the coordinates of cytoband boundaries from D. melanogaster to the other three species with the following workflow. We extracted 30 kb upstream of the coordinate of each cytoband subdivision in the D. melanogaster assembly and used that sequence as a query in a BLAST search against repeat-masked versions of the simulans clade species genomes. To obtain rough boundaries of D. melanogaster cytobands in each simulans clade species, we defined the proximal-most boundary as the proximal coordinate of the first hit (>1 kb in length) from each cytoband region. We defined the distal boundary arbitrarily as one base less than the proximal coordinate of the next cytoband.
Analysis of 1.688/Rsp-like Junctions
We tested the hypothesis that short regions of microhomology could facilitate the insertion of Rsp-like repeats at new genomic loci using two complementary approaches: 1) We used MEME (Bailey et al. 2015) to computationally detect motifs that are enriched at the edges of new Rsp-like clusters (supplementary fig. S26, Supplementary Material online); and 2) through systematic visual examination of 1.688/Rsp-like junctions in D. simulans and D. mauritiana in the context of multisequence alignments as well as the X chromosome assembly in Geneious v8.1.6. Additional details are provided in the Supplementary Material online.
Analysis of Syntenic 1.688 Clusters with Rsp-like Insertions in D. simulans
We tested the prediction that new Rsp-like clusters would insert only at loci where 1.688 clusters were already present by extracting 5 kb of sequence immediately upstream and downstream of the loci containing a mixed 1.688/Rsp-like cluster in D. simulans. We determined the orthologous position of these flanking sequences in the other three study species by using the flanks as BLAST query sequences which we searched against custom BLAST databases built from the assemblies of the other species. We accepted best hits as orthologous sequences only if they were reciprocal best hits when BLASTed back against the D. simulans genome assembly. We then navigated to the orthologous flanking sequences of each cluster to determine whether a 1.688 cluster was present at that locus in the three other study species.
We tested for discordant phylogenetic relationships among 1.688 repeats in clusters with new Rsp-like insertions in D. simulans by extracting 1.688 repeats surrounding the Rsp-like insertion and flagging those sequences in a phylogenetic analysis in which they were included with all 1.688 euchromatic repeats from D. simulans. We extracted flanking sequences, generated custom BLAST databases, conducted BLAST searches, and extracted relevant 1.688 monomers in Geneious v.8.1.9. For both of the above tests, we used as models those Rsp-like clusters that show the dominant junction signature in D. simulans (fig. 5), with a focus on 12 clusters that are present at genomic loci where Rsp-like clusters are lacking in one or more of the other three study species (i.e., those clusters at cytobands 7–12).
Extrachromosomal Circular DNA Isolation and Sequencing
Genomic DNA was isolated from 20 five-day adult females (20–25 mg) from D. melanogaster (strain iso 1), D. mauritiana, (strain 12), D. sechellia (strain C), and D. simulans (strain XD1) using standard phenol–chloroform extractions. The DNAs were ethanol precipitated and resuspended in 10 mM Tris–EDTA, pH 8.0. The concentrations were determined by Qubit fluorometric quantification. About 200 ng of each genomic DNA was subjected to exoV (New England Biolabs) digestion as described by Shoura et al. (2017). In short, after digestion at 37 °C for 24 h, the DNAs were incubated at 70 °C for 30 min. Additional buffer, ATP, and exoV were then added and the samples incubated at 37 °C for another 24 h. The process was repeated for a total of four 24-h incubations with exoV. The concentration of the remaining DNA was determined by Qubit. Following circle isolation, we prepared libraries of circle-enriched and whole genomic control samples using NEBNext FS DNA Ultra II Library Prep Kit (New England Biolabs) using protocol modifications outlined in Sproul and Maddison (2017). Libraries were pooled and sequenced on the same 150 base paired-end lane of an Illumina HiSeq 4000 by GENEWIZ laboratories (South Plainfield, NJ). Additional methods for library preparation and mapping variants of eccDNA to phylogenetic trees are provided in the Supplementary Material online.
Data Availability
All data files and code for analysis and producing plots are deposited at GitHub (https://github.com/LarracuenteLab/simulans_clade_satDNA_evolution; last accessed April 7,2020) and at the Dryad Digital Repository (https://doi.org/10.5061/dryad.2ngf1vhjs)(Sproul et al. 2020).
Supplementary Material
Acknowledgments
We thank Massa Shoura (Stanford University) for advice on eccDNA isolation. We also thank Tom Eickbush, Jack Werren, and Ching-Ho Chang for helpful feedback on the study. This work was supported by National Institutes of Health General Medical Sciences grant (R35GM119515 to A.M.L.), the University of Rochester, and a Stephen Biggar and Elisabeth Asaro Fellowship in Data Science. J.S.S. is supported by a National Science Foundation Postdoctoral Research Fellowship in Biology (Division of Biological Infrastructure [DBI]-1811930).
Illumina genomic DNA and eccDNA raw reads for each species are available in NCBI’s Sequence Read Archive under project accession PRJNA518878.
References
- Abad JP, Agudo M, Molina I, Losada A, Ripoll P, Villasante A.. 2000. Pericentromeric regions containing 1.688 satellite DNA sequences show anti-kinetochore antibody staining in prometaphase chromosomes of Drosophila melanogaster. Mol Gen Genet. 264(4):371–377. [DOI] [PubMed] [Google Scholar]
- Adams MD, McVey M, Sekelsky JJ.. 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299(5604):265–267. [DOI] [PubMed] [Google Scholar]
- Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA.. 2016. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26(10):1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS.. 2015. The MEME suite. Nucleic Acids Res. 43(W1):W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattes R, Monod C, Susbielle G, Cuvier O, Wu JH, Hsieh TS, Laemmli UK, Kas E.. 2006. Displacement of D1, HP1 and topoisomerase II from satellite heterochromatin by a specific polyamide. EMBO J. 25(11):2397–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Campbell P, Leiva-Neto JT, Markow TA.. 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177(3):1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brajkovic J, Feliciello I, Bruvo-Madaric B, Ugarkovic D.. 2012. Satellite DNA-like elements associated with genes within euchromatin of the beetle Tribolium castaneum. G3 (Bethesda) 2:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB. 1938. A revised map of the salivary gland X-chromosome of Drosophila melanogaster. Journal of Heredity. 29(1):11–13. [Google Scholar]
- Britten RJ, Kohne DE.. 1968. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 161(3841):529–540. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M.. 2007. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18(1):188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Chang C-H, Khost DE, Vedanayagam J, Adrion JR, Liao Y, Montooth K, Meiklejohn CD, Larracuente AM, Emerson JJ. 2020. Evolution of genome structure in the Drosophila simulans complex species. bioRxiv 2020.02.27.968743; doi: 10.1101/2020.02.27.968743. [DOI] [PMC free article] [PubMed]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR.. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 18(8):495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH, Stephan W.. 1986. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112:947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W.. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371(6494):215–220. [DOI] [PubMed] [Google Scholar]
- Coen E, Strachan T, Dover G.. 1982. Dynamics of concerted evolution of ribosomal DNA and histone gene families in the melanogaster species subgroup of Drosophila. J Mol Biol. 158(1):17–35. [DOI] [PubMed] [Google Scholar]
- Cohen S, Menut S, Mechali M.. 1999. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol Cell Biol. 19(10):6682–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Segal D.. 2009. Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res. 124(3–4):327–338. [DOI] [PubMed] [Google Scholar]
- Cohen S, Yacobi K, Segal D.. 2003. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 13(6):1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Z, Bacharach E, Lavi S.. 2006. Mouse major satellite DNA is prone to eccDNA formation via DNA ligase IV-dependent pathway. Oncogene 25(33):4515–4524. [DOI] [PubMed] [Google Scholar]
- Courret C, Chang CH, Wei KH, Montchamp-Moreau C, Larracuente AM.. 2019. Meiotic drive mechanisms: lessons from Drosophila. Proc R Soc B. 286(1913):20191430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS.. 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86(1):135–146. [DOI] [PubMed] [Google Scholar]
- Deshpande N, Meller VH.. 2018. Chromatin that guides dosage compensation is modulated by the siRNA pathway in Drosophila melanogaster. Genetics 209(4):1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias GB, Svartman M, Delprat A, Ruiz A, Kuhn GC.. 2014. Tetris is a foldback transposon that provided the building blocks for an emerging satellite DNA of Drosophila virilis. Genome Biol Evol. 6(6):1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBartolomeis SM, Tartof KD, Jackson FR.. 1992. A superfamily of Drosophila satellite related (SR) DNA repeats restricted to the X chromosome euchromatin. Nucleic Acids Res. 20(5):1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. 1982. Molecular drive: a cohesive mode of species evolution. Nature 299(5879):111–117. [DOI] [PubMed] [Google Scholar]
- Dover G. 1994. Concerted evolution, molecular drive and natural selection. Curr Biol. 4(12):1165–1166. [DOI] [PubMed] [Google Scholar]
- Feliciello I, Akrap I, Brajkovic J, Zlatar I, Ugarkovic D.. 2015. Satellite DNA as a driver of population divergence in the red flour beetle Tribolium castaneum. Genome Biol Evol. 7(1):228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciello I, Akrap I, Ugarkovic D.. 2015. Satellite DNA modulates gene expression in the beetle Tribolium castaneum after heat stress. PLoS Genet. 11(8):e1005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA.. 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7(10):e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree PM, Prasad S.. 2012. How can satellite DNA divergence cause reproductive isolation? Let us count the chromosomal ways. Genet Res Int. 2012:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Saunders A.. 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322(5907):1559–1562. [DOI] [PubMed] [Google Scholar]
- Fishman L, Willis JH.. 2005. A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics 169(1):347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry K, Salser W.. 1977. Nucleotide sequences of HS-alpha satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 12(4):1069–1084. [DOI] [PubMed] [Google Scholar]
- Gallach M. 2014. Recurrent turnover of chromosome-specific satellites in Drosophila. Genome Biol Evol. 6(6):1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, Presgraves DC.. 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22(8):1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz JW. 1990. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat Res. 237(5–6):271–292. [DOI] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, Verstrepen KJ.. 2010. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 44(1):445–477. [DOI] [PubMed] [Google Scholar]
- Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B.. 2010. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci U S A. 107(43):18551–18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. 2000. Molecular melodies in high and low C. Nat Rev Genet. 1(2):145–149. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR.. 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5(1):e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS.. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102. [DOI] [PubMed] [Google Scholar]
- Jagannathan M, Warsinger-Pepe N, Watase GJ, Yamashita YM.. 2017. Comparative analysis of satellite DNA in the Drosophila melanogaster species complex. G3 ( Bethesda) 7:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SS, Meller VH.. 2017. Satellite repeats identify X chromatin for dosage compensation in Drosophila melanogaster males. Curr Biol. 27(10):1393–1402.e1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ekhteraei-Tousi S, Lewerentz J, Larsson J.. 2018. The X-linked 1.688 satellite in Drosophila melanogaster promotes specific targeting by painting of fourth. Genetics 208(2):623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DG, Soller M, Kashi Y.. 1997. Evolutionary tuning knobs. Endeavour 21(1):36–40. [Google Scholar]
- Kit S. 1961. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol. 3(6):711–716. [DOI] [PubMed] [Google Scholar]
- Kuhn GC, Kuttler H, Moreira-Filho O, Heslop-Harrison JS.. 2012. The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Mol Biol Evol. 29(1):7–11. [DOI] [PubMed] [Google Scholar]
- Lanciano S, Carpentier MC, Llauro C, Jobet E, Robakowska-Hyzorek D, Lasserre E, Ghesquiere A, Panaud O, Mirouze M.. 2017. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genet. 13(2):e1006630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B.. 1988. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 52(3):223–235. [DOI] [PubMed] [Google Scholar]
- Larracuente AM. 2014. The organization and evolution of the Responder satellite in species of the Drosophila melanogaster group: dynamic evolution of a target of meiotic drive. BMC Evol Biol. 14(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Meller VH.. 2006. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 14(4):417–431. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR.. 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131(7):1235–1247. [DOI] [PubMed] [Google Scholar]
- Lee YCG, Karpen GH.. 2017. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. Elife 6:e25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Ogiyama Y, Martins NMC, Beliveau BJ, Acevedo D, Wu C, Cavalli G, Karpen GH.. 2020. Pericentromeric heterochromatin is hierarchically organized and spatially contacts H3K9me2 islands in euchromatin. PLoS Genet. 16(3):e1008673. [DOI] [PMC free article] [PubMed]
- Legrand D, Tenaillon MI, Matyot P, Gerlach J, Lachaise D, Cariou ML.. 2009. Species-wide genetic variation and demographic history of Drosophila sechellia, a species lacking population structure. Genetics 182(4):1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA.. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 4:203–221. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Yu K, Raghavan SC.. 2006. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst). 5(9–10):1234–1245. [DOI] [PubMed] [Google Scholar]
- Lindholm AK, Dyer KA, Firman RC, Fishman L, Forstmeier W, Holman L, Johannesson H, Knief U, Kokko H, Larracuente AM, et al. 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol Evol. 31(4):315–326. [DOI] [PubMed] [Google Scholar]
- Lohe AR, Brutlag DL.. 1987. Identical satellite DNA sequences in sibling species of Drosophila. J Mol Biol. 194(2):161–170. [DOI] [PubMed] [Google Scholar]
- Lohe AR, Roberts PA.. 1988. Evolution of satellite DNA sequences in Drosophila In: Verma RS, editor. Heterochromatin: molecular and structural aspects. Cambridge (United Kingdom: ): Cambridge University Press. [Google Scholar]
- Losada A, Villasante A.. 1996. Autosomal location of a new subtype of 1.688 satellite DNA of Drosophila melanogaster. Chromosome Res. 4(5):372–383. [DOI] [PubMed] [Google Scholar]
- Lucchesi JC, Kuroda MI.. 2015. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol. 7(5):a019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg LE, Kim M, Johansson AM, Faucillion ML, Josupeit R, Larsson J.. 2013. Targeting of painting of fourth to roX1 and roX2 proximal sites suggests evolutionary links between dosage compensation and the regulation of the fourth chromosome in Drosophila melanogaster. G3 (Bethesda) 3:1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS. 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc Natl Acad Sci U S A. 104(12):4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk MP, Barbash DA.. 2018. Double insertion of transposable elements provides a substrate for the evolution of satellite DNA. Genome Res. 28(5):714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ.. 2004. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167(2):699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Lee SE.. 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24(11):529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon DU, Coarfa C, Xiao W, Gunaratne PH, Meller VH.. 2014. siRNAs from an X-linked satellite repeat promote X-chromosome recognition in Drosophila melanogaster. Proc Natl Acad Sci U S A. 111(46):16460–16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestrovic N, Plohl M, Mravinac B, Ugarkovic D.. 1998. Evolution of satellite DNAs from the genus Palorus–experimental evidence for the “library” hypothesis. Mol Biol Evol. 15(8):1062–1068. [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM.. 2007. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 71(1):13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller HD, Parsons L, Jorgensen TS, Botstein D, Regenberg B.. 2015. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci U S A. 112(24):E3114–E3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova A, Koblizkova A, Macas J.. 2008. Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biol. 8(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang JM, Cavalli G.. 2018. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol Cell. 71(1):73–88.e75. [DOI] [PubMed] [Google Scholar]
- Paulsen T, Kumar P, Koseoglu MM, Dutta A.. 2018. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 34(4):270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons J, Bruvo B, Petitpierre E, Plohl M, Ugarkovic D, Juan C.. 2004. Complex structural features of satellite DNA sequences in the genus Pimelia (Coleoptera: Tenebrionidae): random differential amplification from a common ‘satellite DNA library’. Heredity 92(5):418–427. [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M.. 2000. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405(6787):697–700. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Wray GA.. 2002. Abundant raw material for cis-regulatory evolution in humans. Mol Biol Evol. 19(11):1991–2004. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ruano FJ, Lopez-Leon MD, Cabrero J, Camacho J.. 2016. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci Rep. 6(1):28333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer C, Tautz D.. 1992. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 20(2):211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer C, Tautz D.. 1994. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr Biol. 4(9):777–783. [DOI] [PubMed] [Google Scholar]
- Scully R, Panday A, Elango R, Willis NA.. 2019. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 20(11):698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, Dutta A.. 2012. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 336(6077):82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoura MJ, Gabdank I, Hansen L, Merker J, Gotlib J, Levene SD, Fire AZ.. 2017. Intricate and cell type-specific populations of endogenous circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens. G3 (Bethesda) 7:3295–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS.. 2007. Template switching during break-induced replication. Nature 447(7140):102–105. [DOI] [PubMed] [Google Scholar]
- Smith GP. 1976. Evolution of repeated DNA sequences by unequal crossover. Science 191(4227):528–535. [DOI] [PubMed] [Google Scholar]
- Southern EM. 1970. Base sequence and evolution of guinea-pig alpha-satellite DNA. Nature 227(5260):794–798. [DOI] [PubMed] [Google Scholar]
- Sproul JS, Khost DE, Eickbush DG, Negm S, Wei X, Wong I, Larracuente AM.. 2020. Dynamic evolution of euchromatic satellites on the X chromosome in Drosophila melanogaster and the simulans clade. Dryad. Available from: 10.5061/dryad.2ngf1vhjs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul JS, Maddison DR.. 2017. Sequencing historical specimens: successful preparation of small specimens with low amounts of degraded DNA. Mol Ecol Resour. 17(6):1183–1201. [DOI] [PubMed] [Google Scholar]
- Strachan T, Coen E, Webb D, Dover G.. 1982. Modes and rates of change of complex DNA families of Drosophila. J Mol Biol. 158(1):37–54. [DOI] [PubMed] [Google Scholar]
- Sueoka N. 1961. Variation and heterogeneity of base composition of deoxyribonucleic acids – a compilation of old and new data. J Mol Biol. 3(1):31–40. [Google Scholar]
- Szybalski W. 1968. Use of cesium sulfate for equilibrium density gradient centrifugation. Methods Enzymol. 12B:330–360. [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, et al. 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47(D1):D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaponina O, Haber JE.. 2014. Frequent interchromosomal template switches during gene conversion in S. cerevisiae. Mol Cell. 55(4):615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkovic D, Plohl M.. 2002. Variation in satellite DNA profiles–causes and effects. EMBO J. 21(22):5955–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon N, Miller D, Murnane JP.. 1994. Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res. 22(13):2447–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vondrak T, Avila Robledillo L, Novak P, Koblizkova A, Neumann P, Macas J.. 2020. Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant J. 101(2):484–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JB. 1987. Persistence of tandem arrays: implications for satellite and simple-sequence DNAs. Genetics 115:553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring GL, Pollack JC.. 1987. Cloning and characterization of a dispersed, multicopy, X chromosome sequence in Drosophila melanogaster. Proc Natl Acad Sci U S A. 84(9):2843–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KH, Lower SE, Caldas IV, Sless TJS, Barbash DA, Clark AG.. 2018. Variable rates of simple satellite gains across the Drosophila phylogeny. Mol Biol Evol. 35(4):925–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis JJ, Yasmineh WG.. 1971. Heterochromatin, satellite DNA, and cell function. Structural DNA of eucaryotes may support and protect genes and aid in speciation. Science 174(4015):1200–1209. [DOI] [PubMed] [Google Scholar]
- Zellinger B, Riha K.. 2007. Composition of plant telomeres. Biochim Biophys Acta. 1769(5–6):399–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data files and code for analysis and producing plots are deposited at GitHub (https://github.com/LarracuenteLab/simulans_clade_satDNA_evolution; last accessed April 7,2020) and at the Dryad Digital Repository (https://doi.org/10.5061/dryad.2ngf1vhjs)(Sproul et al. 2020).