Abstract
For most sequenced flowering plants, multiple whole-genome duplications (WGDs) are found. Duplicated genes following WGD often have different fates that can quickly disappear again, be retained for long(er) periods, or subsequently undergo small-scale duplications. However, how different expression, epigenetic regulation, and functional constraints are associated with these different gene fates following a WGD still requires further investigation due to successive WGDs in angiosperms complicating the gene trajectories. In this study, we investigate lotus (Nelumbo nucifera), an angiosperm with a single WGD during the K–pg boundary. Based on improved intraspecific-synteny identification by a chromosome-level assembly, transcriptome, and bisulfite sequencing, we explore not only the fundamental distinctions in genomic features, expression, and methylation patterns of genes with different fates after a WGD but also the factors that shape post-WGD expression divergence and expression bias between duplicates. We found that after a WGD genes that returned to single copies show the highest levels and breadth of expression, gene body methylation, and intron numbers, whereas the long-retained duplicates exhibit the highest degrees of protein–protein interactions and protein lengths and the lowest methylation in gene flanking regions. For those long-retained duplicate pairs, the degree of expression divergence correlates with their sequence divergence, degree in protein–protein interactions, and expression level, whereas their biases in expression level reflecting subgenome dominance are associated with the bias of subgenome fractionation. Overall, our study on the paleopolyploid nature of lotus highlights the impact of different functional constraints on gene fate and duplicate divergence following a single WGD in plant.
Keywords: whole-genome duplication, gene expression, methylation, gene balance, subgenome dominance
Introduction
Gene duplication is one of the most important drivers of eukaryotic evolution. Indeed, by increasing the amount of raw genetic material on which evolution can work, gene duplication generates the genetic redundancy through which processes such as subfunctionalization and neofunctionalization can create functional novelty (Ohno 1970; Shiu and Bleecker 2001; Zhang 2003; Blanc and Wolfe 2004; Gout and Lynch 2015; Sandve et al. 2018). Apart from small-scale gene duplication (SSD), also whole-genome duplication (WGD), whereby thousands of novel genes are created at once, has been frequently observed during evolution, especially in flowering plants (Cui et al. 2006; Fawcett et al. 2009; Jiao et al. 2011; Vanneste et al. 2014; Li, Baniaga, et al. 2015; Ruprecht et al. 2017; Van De Peer et al. 2017). Interestingly, the fate of genes duplicated through such large-scale duplication (LSD) events often seems to be different from that of genes duplicated in small-scale events, and previous studies have shown that the chance of survival and maintenance of genes duplicated in a WGD is very much dependent on their function. On the one hand, despite repeated WGDs in angiosperms, many genes were found that convergently revert to single-copy status, and in Arabidopsis, they exhibit more constitutive and higher expression than duplicate genes in general and are enriched in house-keeping functions (Paterson et al. 2006; De Smet et al. 2013). One explanation is that the deletion of duplicates is needed to prevent copies with dominant-negative mutations, which might interfere with the correct functioning of the wild type copy (Paterson et al. 2006; De Smet et al. 2013). On the other hand, there are those genes that are retained in excess following WGD for a longer time. For these retained duplicate genes, gene balance hypothesis (GBH) states that maintaining stoichiometric balance is crucial, and genes can only be deleted together with their “interactors” where losing or further duplication of part of the network or complex is detrimental because the stoichiometry is challenged (Birchler et al. 2005; Freeling 2009; Bekaert et al. 2011; Birchler and Veitia 2012; De Smet and Van de Peer 2012; Tasdighian et al. 2017). Genes that underwent SSDs, such as tandemly duplicated genes, in contrast were found to be selected for either increased gene dosage or rapid gene turnover in order to confer lineage-specific adaptation because they are mostly insensitive to dosage imbalance (Coate et al. 2016; Lan et al. 2017). Although these theories explain how different mechanisms that potentially affect gene fate after WGD, we still do not know the difference in functional constraints including quantifiable features such as expression, epigenetic regulation, and protein–protein interactions (PPIs) imposed on those genes with different fates after a WGD (single-copy, WGD, and SSD genes).
Studies including a recent investigation on WGDs across plants including 134 sequenced angiosperms suggest that after diverging from the extant basal-most angiosperm (Amborella), only lotus and seagrass (Zostera marina) experienced a single WGD (4×), whereas the other angiosperms experienced at least a genome triplication (6×) or sequential WGDs (Qiao et al. 2019). However, the scaffold-level genome assembly of seagrass provides limited information on synteny to study the gene fates after its WGD (Olsen et al. 2016). Case studies of recently released genomes also show that columbine, Liriodendron and water lily experienced a single WGD (Aköz and Nordborg 2019; Chen et al. 2019; Zhang et al. 2020). Therefore, the genome of sacred lotus (Nelumbo nucifera Gaertn.) is one of the few angiosperms carrying a well-retained intraspecific synteny reflecting only a single ancient WGD coincided with the K–pg boundary (Ming et al. 2013; Wang, Fan, et al. 2013; Vanneste et al. 2014; Shi et al. 2017). Because of its relatively simple and ancient WGD history, lotus genome facilitates comparing genes with different fates (duplication status) following a single WGD. In addition, because long-retained duplicate pairs descending from the same WGD event can be easily tracked in species such as lotus, the (functional) factors, including dosage-balance constraint, that shape the expression pattern divergence of duplicate gene pairs can also be well investigated. Yet, in Arabidopsis, poplar, soybean, tomato, or maize, the fact that multiple different rounds of WGDs occurred makes it difficult to study the fate of the most ancient duplicates (Rodgers-Melnick et al. 2012; Jiang et al. 2013; Defoort et al. 2019). Other than divergence in expression pattern, many duplicate pairs might have bias in expression level (Lehti-Shiu et al. 2015). Often, this expression bias between the two copies is associated with subgenome dominance which is a phenomenon that was initially defined in allopolyploid cotton and later in other (presumed) paleoallopolyploids: copies residing in one less fractionated (LF; parental) subgenome tend to show higher expression than those in the other (parental) subgenome (Langham et al. 2004; Rapp et al. 2009; Flagel and Wendel 2010; Woodhouse et al. 2014; Cheng et al. 2016, 2018; Edger et al. 2017; Vicient and Casacuberta 2017; Bottani et al. 2018).
Therefore, understanding the mechanisms such as epigenetic regulation and subgenome dominance underlying the divergence in expression pattern and level after a WGD in lotus will improve our understanding of how a duplicate pair diverges in function. To better address the questions as mentioned above, we build an improved genome assembly of the lotus var. “China Antique” by PacBio long-read sequencing and scaffolding using high-throughput chromosome conformation capture (Hi-C). This can optimally identify the genomic relics from both ancient SSD and WGD events. Complementing this chromosome-level assembly with further whole-genome bisulfite (methylation) sequencing, RNA-seq, and genome resequencing data, not only allow us to study the mechanisms, such as expression and epigenetic regulation that coordinate and maintain the functional integrity of genes displaying different evolutionary fates, but also provide further insight into the genetic mechanisms that create functional divergence of duplicates retained after a WGD.
Results
A Chromosome-Level Assembly of Lotus
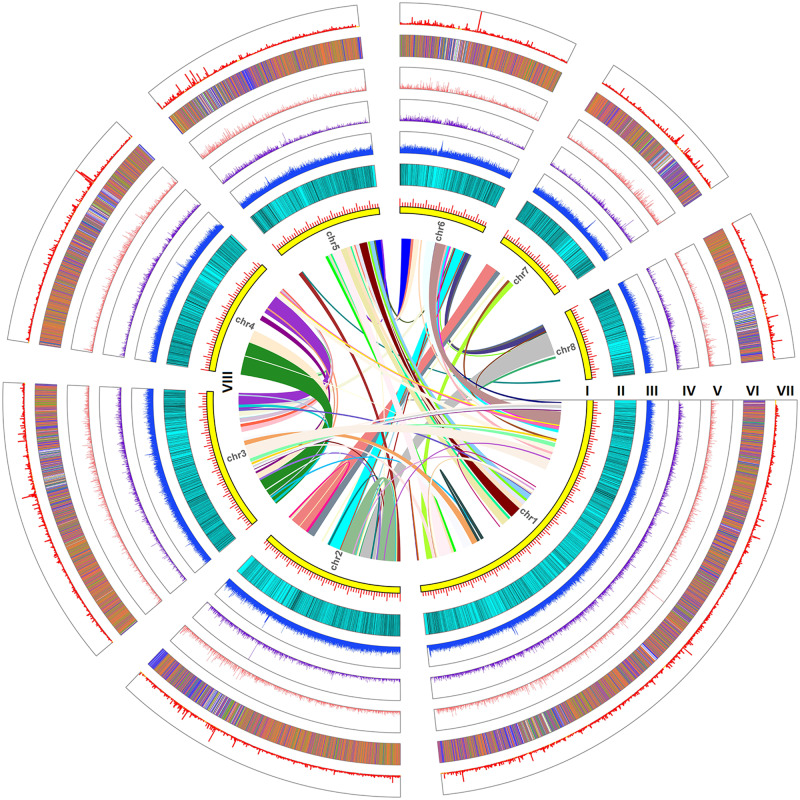
Based on newly generated data, we obtained an improved assembly and annotation of the lotus genome. Combining PacBio Sequel subreads (11.9 G; 1,330,739 subreads with a mean length of 8.8 kb and N50 of 12.7 kb) with previously published Illumina paired-end (PE) reads (94.2 Gb) (Ming et al. 2013), resulted in a hybrid assembly, containing contigs with an N50 length of 484.3 kb. This assembly is about 12.5 times the length of previously assembled contigs (v2013) (N50 = 38.8 kb) (supplementary fig. S1, Supplementary Material online). The final 4,709 contigs cover about 807.6 Mb. Using genome-wide HI-C, overall, 4,248 contigs (799.7 Mb) were anchored and ordered into eight different pseudomolecules (chromosomes) (supplementary fig. S2, Supplementary Material online). Further optimization of the assembly by gap filling and polishing (error correction using accurate Illumina reads) resulted in a final assembly consisting of eight pseudochromosomes (813.2 Mb) and 456 unanchored contigs (8.0 Mb) (fig. 1 and supplementary table S1, Supplementary Material online).
Fig. 1.
Circos plot of lotus genome assembly. From outside to inside rings: (I) size (Mb) of the assembly for each chromosome; (II) density distribution of genes; (III) density distribution of sRNA − TEs; (IV) density distribution of sRNA + TEs; (V) dot plot of nucleotide diversity of CDS for each gene; (VI) methylation level of genes and flanking regions; (VII) gene expression level (log-transformed FPKM value); and (VIII) syntenic paralogs are linked by colored lines.
The newly assembled genome contains 58.5% repetitive sequences, of which 48.7% of the total assembly consists of known transposable elements (TEs) and 9.1% of unknown repeats (fig. 1 and supplementary table S2, Supplementary Material online). Gene annotation based on a repeat-masked genome yielded a total of 32,124 protein-coding genes (fig. 1). The accuracy of the new assembly was assessed by a previous single-nucleotide polymorphism (SNP)-based linkage map of lotus (Liu et al. 2016). The majority of uniquely mapped SNP markers from a given linkage group aligned within the same pseudochromosome in the new assembly, whereas in the old assembly these markers showed a partitioned and mosaic distribution over different megascaffolds (v2013) (supplementary fig. S3, Supplementary Material online). To assess the completeness of the assembly, we investigated to what extent the 1,440 plant conserved gene set of BUSCO was recovered: 94.6% (1,362) of the gene set was completely retrieved, 3.1% (44) was partially retrieved, and 2.3% (34) was “missing.” This shows that our assembly is the most complete lotus assembly to date when comparing to the other lotus assemblies (supplementary table S3, Supplementary Material online) (Gui et al. 2018). This is supported by the fact that the number of syntenic orthologs, for instance in relation to monocots, is substantially higher in our new assembly than in an older version: 5,421 Brachypodium distachyon genes and 5,922 rice genes showed a collinear relationship in the new assembly, whereas in the old assembly the numbers were 3,690 and 4,040, respectively (v2013) (supplementary fig. S4, Supplementary Material online). Comparing eudicot genomes from the Plant Genome Duplication Database (PGDD) and our lotus assembly to both B. distachyon and rice learns that both the new and old assemblies of lotus share more collinear orthologs with the two monocot genomes than the other eudicots (supplementary fig. S4, Supplementary Material online). Although lotus and the other eudicots in the PGDD together form a sister group to monocots, the genome architecture (at least considering synteny) of lotus seems to resemble that of monocots most, probably because most eudicots present in the PGDD have undergone at least one triplication or further rounds of WGDs subsequent to eudicot radiation (Ming et al. 2013).
Classification of Single-Copy and Duplicated Lotus Genes
To define different classes of lotus duplicates (Wang Y, Wang X, et al. 2013; Wang, Zhang, et al. 2017), first, within-species syntenic blocks were identified (see Materials and Methods). Such blocks, showing conservation in gene content and order, and thus potentially representing remnants of a WGD, were found across all chromosomes (fig. 1 and supplementary table S4, Supplementary Material online). Comparison of peaks in 4dTv (4-fold degenerate site transversion) distances which represent age distributions formed by the divergence of syntenic duplicates (4dTv median = 0.158) and divergence of orthologs between lotus and Macadamia ternifolia (the other sequenced Proteales species) (4dTv median = 0.405) suggests that most syntenic duplicates (WGD) have been derived from a duplication event after the split between Macadamia and lotus (Mann–Whitney U test, P < 0.01) (supplementary fig. S5, Supplementary Material online).
Next to 2,353 orphan genes (defined as genes in lotus that have no homolog in any other considered plant species), we identified 29,771 genes with homologs in other species (nonorphan genes) (supplementary table S5, Supplementary Material online). Among these lotus genes, so-called dispersed duplicates are the most abundant (13,235), followed by duplicates resulting from WGD (referred to WGD) (9,482), tandemly duplicated genes (2,622), single-copy genes (2,261), proximal duplicated genes (1,566), and finally duplicates that underwent both WGD and tandem duplication (WGD&TD) (605), as classified by MCscanX (supplementary fig. S6A and table S5, Supplementary Material online). Orphan genes are mostly either single-copy (62.14%) dispersed duplicates (33.81%) (supplementary fig. S6B and table S5, Supplementary Material online). The above-defined gene groups were used to further study how the fate of genes, for instance after WGD, correlates with functional constraints, reflected by PPIs, gene expression, and epigenetic and sequence properties. Lotus-specific orphan genes were analyzed separately.
Single-Copy Genes and WGD-Derived Duplicates of Lotus Show Conservation in Copy Number in Related Taxa
Here, we estimated the extent to which dosage sensitivity (copy number conservation) of lotus genes depends on their duplication status. Hereto, we first grouped lotus genes according to their duplication status in lotus (as defined above, “single-copy genes,” “WGD,” “tandem duplicates,” and others) and subsequently assessed whether the orthologs of these lotus genes retained the same copy number status in two related eudicot species, namely M. ternifolia and Vitis vinifera. Macadamia was chosen because it is the sequenced Proteales species that is closest to lotus, whereas Vitis, with only one eudicot genome triplication, was also chosen because of its relatively conserved genome architecture compared with the other core eudicots (Jaillon et al. 2007). To assess the variation in copy number across the studied species, we used the coefficient of variation (CV). The average copy number among the three species (as shown in the violin plot) varies largely among the genes of different duplication status, and therefore standard deviation cannot serve to assess the variation in this case (fig. 2A). Single-copy genes (grouped according to their single-copy status in lotus) have a median of the average copy number among the three species close to one, indicating that, for genes grouped as single-copy in lotus, there is a general strong selection against gene redundancy in the related species as well (fig. 2A). For genes classified as lotus WGD-derived duplicates, a median of the average copy number between 1 and 2 was found, suggesting that genes belonging to this group also tend to display a limited level of gene redundancy in the three studied taxa (fig. 2A). Interestingly, dispersed and WGD-derived duplicates show, after single-copy genes, respectively the second and the third-lowest CV for variation in copy number, and therefore presumably exhibit higher dosage sensitivity than local duplicates (tandem, proximal, and WGD&TD) (Kruskal–Wallis test, all P values < 0.01) (fig. 2B). This is in line with the GBH, which states that WGD-derived duplicates are more dosage-sensitive or more strict in preserving their copy numbers than local duplicates (Coate et al. 2016; Lan et al. 2017). For the group of the dispersed duplicates, the interpretation is less trivial as these genes contain WGD-derived duplicates that lost collinearity, local duplicates that lost “proximity” to other duplicates, transposed duplicates, or “angiosperm-conserved single-copy genes” (“angio-singles”) that were created by earlier preangiosperm duplications but stopped duplicating during angiosperm radiation. By examining the proportion of “angio-singles” in each of the studied gene groups using annotations described in a previous publication (De Smet et al. 2013), we found that next to the group of single-copy genes, the group of dispersed duplicates contains the second-highest enrichment of “angio-singles” (fig. 2C). Greater 4dTv distances between the most similar dispersed duplicates than between corresponding orthologs (Nelumbo vs. Amborella) (Kruskal–Wallis test, all P values < 0.01) (supplementary fig. S7, Supplementary Material online) suggest that “angio-singles” in dispersed duplicates were mostly created by early duplications prior to angiosperm radiation. As those early duplicates stopped duplicating during angiosperm radiation, they were classified as so-called single-copy genes in angiosperms. This explains why the group of dispersed duplicates also shows a low CV in copy number.
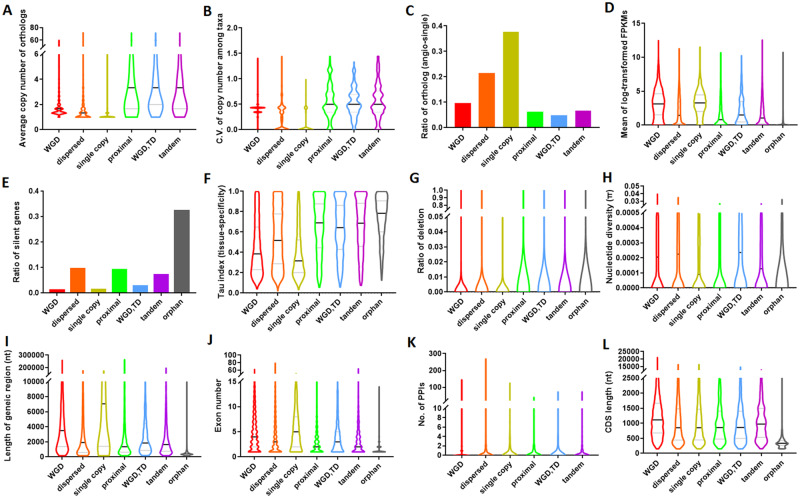
Fig. 2.
Violin plots of expression, functional, and genomic features of genes from different gene groups (based on duplication status). (A) The average copy number of orthologs. (B) Coefficient of variation (CV) of copy number among taxa. (C) Ratio of orthologs as “angio-singles.” (D) The mean of log-transformed FPKM. (E) The ratio of silent genes. (F) Tissue specificity index (based on Tau index). (G) The average portion of the deleted genic sequence in tropical lotus comparing to the reference genome (ratio of deletion). (H) Nucleotide diversity (π). (I) Length of the genic region. (J) Exon number. (K) The number of protein–protein interactions inferred from the closest orthologs in Arabidopsis. (L) CDS length. Black line: median; gray line: quantile.
Single-Copy Genes and WGD-Derived Duplicate Genes Have High Expression Level and Breadth
To understand why single-copy genes and WGD-derived duplicates are more highly constrained in copy number, we compared the level and breadth of gene expression for the above-defined gene groups. This is because genes expressed at higher levels tend to be under stronger selective pressure (Akashi 2001; Drummond et al. 2005; Jovelin and Phillips 2011; Song et al. 2017). Average gene expression levels (log-transformed FPKMs), observed in 41 samples representing a variety of tissue-types, varied substantially among the studied gene groups. Single-copy genes showed on average the highest expression level (Kruskal–Wallis test, all P values < 0.01) (fig. 2D and supplementary table S6, Supplementary Material online). This result is consistent with a previous finding in Arabidopsis showing that the angiosperm-conserved single-copy genes generally show higher expression than duplicated genes (De Smet et al. 2013). This larger expression ubiquity also implies that single-copy genes are more likely involved in house-keeping functions than genes belonging to the other groups. When focusing on the duplicated genes, genes retained after WGD show on average a significantly higher expression level than genes from groups representing other types of duplicates (Kruskal–Wallis test, all P values < 0.01) (fig. 2D). Because essential genes are found to be highly expressed in Arabidopsis and other plants (Lloyd et al. 2015), this suggests that both single-copy and WGD-derived duplicates might constitute the more essential genes in lotus. Therefore, the strong purifying selection from gene essentiality of these two groups of genes might play an important role in constraining their dosage sensitivity (copy number change among taxa).
Further, we found that in lotus the largest gene group, namely the dispersed duplicates, possesses the highest ratio of silent genes (genes that are not expressed in any of the investigated samples) (9.61%), followed by proximal duplicates (9.20%) and tandem duplicates (7.29%), whereas genes resulting from WGD&TD (2.81%), from WGD (1.15%) and single-copy genes (1.42%) display much lower ratios of silent genes (fig. 2E). This explains that even though dispersed duplicates contain a large portion of “angiosperm-conserved single-copy genes,” they do not show a higher expression level than duplicates retained from WGD because they also contain a substantial number of silent (likely pseudogenized) duplicate genes. We further showed that compared with the expressed dispersed duplicates, the silent dispersed duplicates generally have younger ages (measured by 4dTv), lower number of introns, smaller protein length, and lower selective pressure, suggesting that they might be recent retrotransposed duplicates (supplementary fig. S8, Supplementary Material online). Overall, these comparisons further confirm that losing function by gene silencing is not a random phenomenon and that single-copy genes and duplicates retained after a WGD are the least likely to be silenced.
Moreover, using the Tau index to measure expression specificity across different lotus tissues, we revealed that single-copy genes (mean Tau index of 0.38) show the lowest expression specificity of all gene groups (Kruskal–Wallis test, all P values < 0.01). In addition, WGD duplicates (mean Tau index = 0.45) exhibit significantly lower expression specificity than other types of duplicates (Kruskal–Wallis test, all P values < 0.01) (fig. 2F). Both single-copy genes and genes retained from a WGD tend to have a wider “expression breadth” than small-scale duplicates, and hence their expression might be essential in most tissues as is supported by findings in Arabidopsis (Lloyd et al. 2015). By showing higher expression level and breadth, both single-copy genes and WGD-derived genes might expose themselves to stronger purifying selection. This is supported by lotus genome resequencing data that show significantly lower ratios of sequence deletion and nucleotide diversity (π) for single-copy genes and WGD-derived duplicates than for small-scale duplicates (Kruskal–Wallis test, all P values < 0.01) (fig. 2G and H).
Differences in Expression Might Be Associated with Differences in Methylation Level and TE Distribution
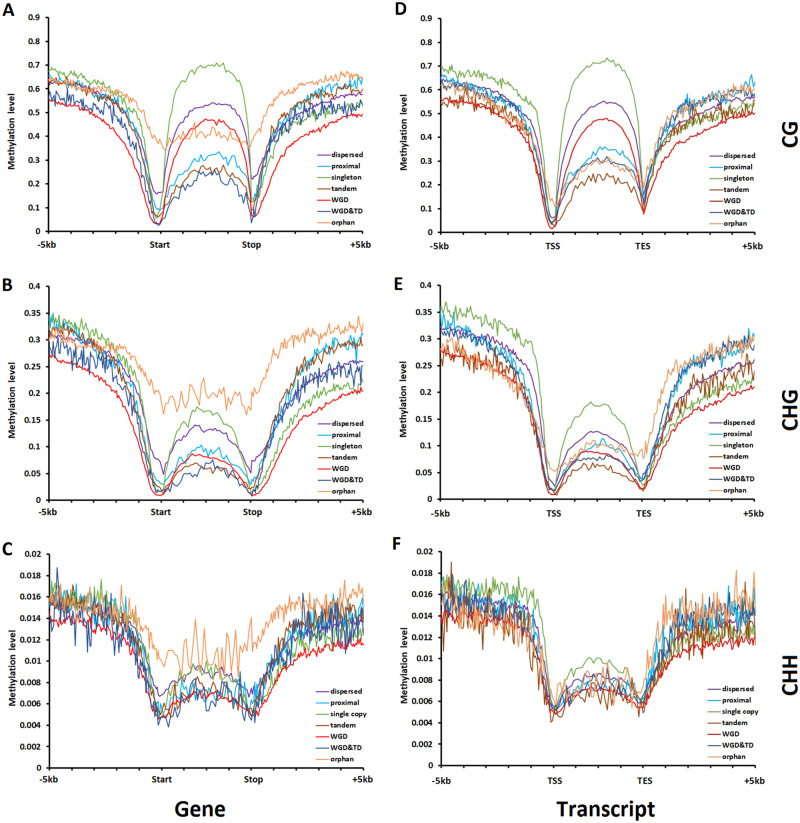
Most cis-regulatory elements reside in gene flanking regions, which play profound roles in gene regulation. Given the impact of epigenetic regulation on gene expression, we assessed whether the above-mentioned differences in expression among different gene groups could be associated with differences in methylation level on gene flanking regions (Lorincz et al. 2004; Luco et al. 2010; Zhang et al. 2015). Hereto, we used methylation data obtained from leaf, petal, stamen petaloid, and stamen. Cytosine methylation levels at CG, CHG, and CHH sites along the gene (upstream, genic and downstream region) generally display a curved “W” shape with the lowest methylation level being observed close to the gene start and stop sites; Note that similar “W”-like shapes were observed when using an alternative definition of flanking regions (see Materials and Methods) (fig. 3 and supplementary figs. S9 and S10, Supplementary Material online). These patterns in which the lowest methylation level is observed near the flanking regions agree with the finding that methylation can inhibit the binding of RNA polymerase II and transcriptional initiation (Lorincz et al. 2004). Among CG, CHG, and CHH sites, the methylation level is the strongest at CG (mean ML = 0.458) (fig. 3A). The average methylation level in flanking regions (promoters and downstream regions) of genes retained after a WGD is significantly lower than the methylation levels of genes belonging to other groups, indicating that duplicates retained after a WGD are transcriptionally less repressed by methylation in flanking regions. This is displayed in figure 3 for methylation levels observed in leaf. Similar figures were obtained for the methylation data obtained from other tissues (supplementary figs. S9 and S10, Supplementary Material online). This average lower methylation level in flanking regions for genes that were retained after a WGD is in line with their relatively higher expression level and breadth. In contrast, the higher expression level and breadth of single-copy genes as compared with genes from other groups seem not to be associated with relatively lower methylation levels of flanking regions: single-copy genes display a higher methylation level in their promoters than genes belonging to the other groups (Kruskal–Wallis test, P values < 0.01).
Fig. 3.
Differences in average CG, CHG, and CHH methylation level (ML) in lotus leaf along the gene and flanking regions among different gene groups based on the duplication status. (A–C) Methylation of all annotated genes. (D–F) Methylation of the genes with RNA-seq evidence.
In plants, (24-nt) RNA-directed DNA methylation is frequent in regions containing TEs, likely because most TEs need to be silenced to reduce TE activity and maintain genome stability. Hence, we assessed the degree to which differences in methylation level in gene flanking regions can be associated with the presence of TEs, including both TEs with 24-nt small (interfering) RNA (sRNA + TE) and those without (sRNA − TE) (Zhai et al. 2008) (see Materials and Methods). Interestingly, the differences in TE density, especially of sRNA − TEs, between the different gene groups resembles the distribution pattern of the overall CG and CHG methylation levels, where the gene group representing duplicates retained after a WGD shows the lowest average TE density in gene flanking regions and concomitantly also the lowest average methylation levels in these flanking regions (supplementary fig. S11, Supplementary Material online).
Unlike gene flanking regions, the methylation level along the gene body (gene region) seems to be more related to differences in gene expression among the different gene groups. Whereas DNA methylation is generally believed to repress gene expression (Weber et al. 2007; Stroud et al. 2013; Hirsch and Springer 2017), we found that higher gene body methylation level tends to occur in the gene groups with higher expression level and breadth, that is, single-copy and WGD duplicates. Interestingly, we found that for the group of single-copy genes, on average, the higher methylation level in the gene body seems to correlate with their greater gene length and exon number (Kruskal–Wallis test, all P values < 0.01) (fig. 2I and J). The fact that introns often contain TEs which are often associated with higher methylation levels might explain why single-copy genes also display the highest TE density in their gene body (Kruskal–Wallis test, all P values < 0.01) (fig. 3 and supplementary table S6, Supplementary Material online) (Swinburne and Silver 2008; Lisch and Bennetzen 2011).
WGD-Derived Duplicates Are Constrained by Gene Dosage Balance
The evolutionary fate of duplicates is often explained employing the GBH: Genes with regulatory or signaling functions such as transcription factors or kinases will largely impact the regulatory network after a duplication because of their hub-like properties. Such duplicates are preferentially retained because the loss of one copy might disrupt many genes to which they directly or indirectly connect (Rody et al. 2017; Tasdighian et al. 2017). If gene balance plays a role in the preferential retention of duplicates after a WGD, this should be reflected in the topological properties of these WGD duplicates (Freeling and Thomas 2006). To assess the effect of gene balance, we analyzed the topological properties of genes belonging to each of the studied gene groups in the physical interaction network. As 27,458 out of 32,124 lotus genes (85.5%) can have the closest ortholog to corresponding Arabidopsis genes, the protein–protein interactome map from the “Arabidopsis Interactome Map” was used as a scaffold for the lotus (PPI) network (see Materials and Methods) (Arabidopsis Interactome Mapping Consortium 2011). We found that indeed genes retained after a WGD show the highest average number of PPIs (mean PPIs = 1.31) (Kruskal–Wallis test, all P values < 0.01), whereas genes belonging to the other groups only differ marginally in the number of PPIs in which they tend to be involved (fig. 2K). Even though the analyses above suggest that, based on their relatively high expression level and breadth, single-copy genes are likely to be the more essential genes, these single-copy genes are not involved in more PPIs than genes from other groups. It appears that single-copy genes tend to immediately return to their single-copy status after a WGD because little dosage-balance constraint is imposed by the interaction network and a strong selection against gene redundancy is present (De Smet et al. 2013). Larger protein length for genes is often found to be associated with the possibility of increased interfacing with different interactors (Jones and Thornton 1996; Caffrey 2004). Intriguingly, we also found that genes retained from a WGD have the largest average coding sequences (CDS) or protein length (Kruskal–Wallis test, all P values < 0.01), whereas genes retained after SSDs show a comparably smaller protein length, which further supports the stronger constraint of dosage balance on genes retained from a WGD (fig. 2L).
For the different groups of genes, we also assessed the bias in which genes are retained following duplication by calculating their Gene Ontology (GO) enrichment (K–S test with P value < 0.01). We showed that the top 30 most significantly enriched GO terms for gene groups with different duplication status have no overlapping functionalities (GO terms) (supplementary fig. S10, Supplementary Material online). In line with the GBH, we observed that genes retained after a WGD are mostly enriched in biological terms relating to protein phosphorylation and regulation of transcription (supplementary fig. S12, Supplementary Material online). In addition, we found that duplicates from the lotus WGD were significantly enriched in genes related to trehalose biosynthesis, polyamine biosynthesis, xylem, and phloem development (supplementary fig. S12, Supplementary Material online). These duplications might have contributed to unique features of lotus: Because both trehalose and polyamine (metabolites) help plants to survive in stresses such as drought and cold (Zentella et al. 1999; Montilla-Bascón et al. 2017; Zhao et al. 2019), the unique longevity of lotus seeds and their survival during K–pg boundary might have benefited from the duplication of these biosynthesis genes. Also, the well-developed aerenchyma in stem and rhizome of lotus might have benefited from the duplication of genes related to xylem and phloem (Casto et al. 2018). In contrast, small-scale duplicates (groups of tandem and proximal duplicates) are mostly enriched in metabolic processes, whereas genes resulting from a combination of WGD&TD are enriched in transport processes (supplementary fig. S12, Supplementary Material online). Thus, both the PPI network and GO functional enrichment analyses suggest that gene-balance-driven selection determines the retention of duplicates after a WGD.
Orphan Genes in Lotus Display Unique Properties
Orphan genes, comprising 7.32% of all lotus genes, are either single-copy genes or form dispersed duplicates, suggesting they are either not retained after lotus WGD or appeared after the lotus WGD (supplementary fig. S6A and B, Supplementary Material online). They show a much lower average expression level, an elevated ratio of silent genes, and a higher expression specificity than genes with homology to known proteins (nonorphan genes) (Kruskal–Wallis test, P values of all pairwise comparisons <0.01) (fig. 2D–F). The relatively higher average π and the ratio of sequence deletion of orphan genes suggest that they are under more relaxed selection than genes from other groups (fig. 2G and H). Moreover, they have on average a shorter CDS, a shorter gene length and the lowest number of exons, implying that they are shorter and have a less complex gene structure (fig. 2I, J, and L). Additionally, orphan genes only display small differences in ML and TE density between their flanking regions and gene bodies (fig. 3 and supplementary fig. S9, Supplementary Material online). Meanwhile, with much higher ML and TE density in gene flanking regions than nonorphan genes, it is more likely that most dispersed orphan genes were created by transposed duplications mediated by TEs (fig. 3 and supplementary fig. S11, Supplementary Material online). Hence, as orphan genes exhibit features that reflect their relatively weaker functional relevancy, especially weak expression and rapid sequence turnover within lotus populations, than all nonorphan genes, they were not used to study the fate of genes after a WGD.
WGD-Derived Duplicates That Have Diverged in Function
WGD-derived duplicates can subfunctionalize and/or neofunctionalize due to changes in the protein-coding domain, or because of regulatory changes causing divergence of expression. Here, we focused on the latter phenomenon and assessed the degree to which duplicate pairs retained from a WGD diverged in gene expression behavior. Hereto, we relied on the interconnectivity score calculated based on the coexpression network (HSu et al. 2011) (fig. 4A). Based on the interconnectivity score, duplicates retained after a WGD were subdivided into five groups: Gene duplicates belonging to group A (connectivity >0.5 with a P value <0.01) tend to share many neighbors in the coexpression network and are unlikely to have subfunctionalized or neofunctionalized. The degree of connectivity gradually decreases for duplicates belonging to groups B and C but still is larger than what can be expected by chance, given the local connectivity of the duplicate pairs under study. In contrast, duplicate pairs belonging to group D share no coexpressed neighbors and the absence of shared neighbors is significant given the local connectivity of the genes in a pair (connectivity <0.15 and P value >0.99). These genes diverged in expression pattern are more likely to have subfunctionalized or neofunctionalized (fig. 4A). Genes belonging to group E (with connectivity <0.15 and 0.99 > x > 0.1) show detectable connectivity in the coexpression network but this connectivity is not higher than what can be expected by chance. As for these gene pairs, it is difficult to decide whether they share coexpression neighbors, they were not considered for further analyses.
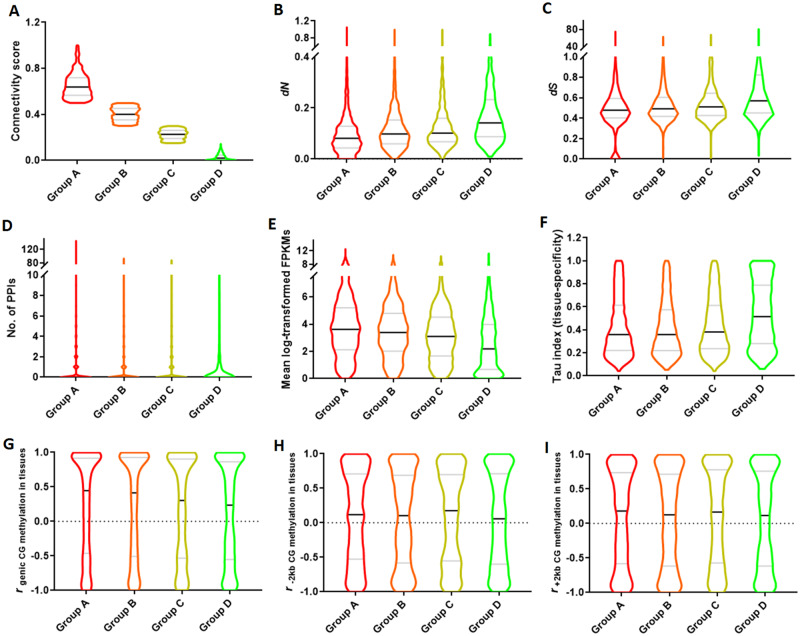
Fig. 4.
Violin plots of expression, functional, methylation, and evolutionary features of WGD-derived duplicate genes with different level of expression divergence (group A, group B, group C, and group D). (A) Connectivity score. (B) dN, nonsynonymous mutation. (C) dS, synonymous mutation. (D) The number of protein–protein interaction inferred from the closest orthologs in Arabidopsis. (E) The mean of log-transformed FPKM. (F) Tissue specificity index (based on Tau index). (G–I) r (correlation coefficient) of CG methylation levels in tissues between duplicates for gene body (G), upstream (H), and downstream region (I). Black line: median; gray line: quantile.
To compare the degree of functional constraint on duplicates with different levels of expression divergence, we further assessed sequence and expression related characteristics for gene pairs belonging to each of the different groups (excluding group E). In line with the observed increase in expression divergence, also both the number of nonsynonymous substitutions (dN) and the number of synonymous substitutions (dS) in group A (the group with duplicates that display the most conserved expression behavior) are significantly lower than those in group D (the group most diverged in expression behavior) (Kruskal–Wallis test, all P values < 0.05), which further shows a gradual increase from group A to group D (fig. 4B and C). Thus, duplicate pairs that show little expression divergence tend to retain their sequence similarity (especially groups A and B). This indicates that these genes are conserved and under higher functional constraint which might be related to a relatively stronger dosage balance. We indeed also observed that duplicates that displayed the largest sequence and expression conservation (group A) are also more frequently interacting in the PPI network than duplicates that display the most divergent expression behavior (group D) (as assessed by the degree of the duplicate genes in the PPI network) (Kruskal–Wallis test, all P values < 0.01), and accordingly a gradual decrease from group A to group D was observed, which seems in line with a previous study on WGD-derived duplicates and small-scale duplicates in Arabidopsis, tomato, and maize (fig. 4D) (Defoort et al. 2019). Moreover, both the average gene expression level and expression breadth (expressed as the opposite of the Tau index) in group A are significantly higher than group D (Kruskal–Wallis test, all P value < 0.01), which also exhibit a gradual change from group A to group D (fig. 4E and F). This indicates that duplicate pairs more conserved in their expression behavior are involved in more generic functions, whereas as expected, the duplicates more divergent in expression behavior tend to have more specialized functions. The small difference of tissue specificity (Tau index) between group A and group B might indicate they are both still under strong functional constraints (Kruskal–Wallis test, all P value = 0.141). However, we did not observe that the degree of expression divergence between duplicated gene pairs belonging to different groups exhibits any significant association with overall methylation level (in tissues) or TE density (supplementary figs. S13–S15, Supplementary Material online). This suggests that the gradual increase in gene expression level of duplicates from group D (less conserved in expression behavior) to group A (most conserved in expression behavior) is not related to a decline in methylation level. Because the methylation level of a gene can change in different tissues, we also calculated how the methylation pattern between duplicates is different in a well-defined region of the gene (gene body, upstream or downstream) by using correlation coefficient (r). A gene’s methylation pattern is here defined as the variable of methylation levels in the four tissues on a defined region of the gene (see Materials and Methods). This analysis was performed for CG, CHG, and CHH methylation, and for each genic region separately. We found that duplicates belonging to group A (the group most conserved in expression behavior) display significantly more correlated CG methylation patterns in their genic region (with the highest r) than those of group D (Kruskal–Wallis test, all P value < 0.01), with a gradual decline from group A to group D (fig. 4G). This trend was not visible for the CHG nor CHH sites in upstream and downstream regions of duplicates (fig. 4H and I and supplementary fig. S16A–F, Supplementary Material online). This suggests that the level to which CG methylation occurs in different tissues tends to be more conserved for duplicates that are more conserved in expression behavior. Subfunctionalized genes tend to display more differences in CG methylation level across tissues in their genic regions.
The duplicates with the most conserved expression behavior (group A) are enriched in GO terms related to protein translation (ribosome) and regulation of transcription, both functions which are known to be dosage-sensitive (supplementary fig. S17, Supplementary Material online) (Edger and Pires 2009; Jiang et al. 2013). In contrast, the duplicates that are most diverged in expression (group D) are mainly enriched in transport mechanisms (e.g., transmembrane transport, spermine biosynthetic process, and anion transport), which are not typical dosage-sensitive functions. As a reference, we also analyzed duplicates from the Arabidopsis K–pg boundary WGD (At-β) and the recent WGD (At-α) with a similar strategy (using a similar grouping based on their degree of expression divergence) (supplementary fig. S18, Supplementary Material online). In line with our results in lotus, also here GO terms related to ribosome synthesis and regulation of transcription and biological processes are enriched in the groups representing the genes that displayed the least expression divergence after duplication (respectively group A of At-β and At-α) (supplementary figs. S19 and S20, Supplementary Material online). For the duplicates from At-β (group D) that diverged most in expression, GO terms related to response to chemicals, hormone, and stimulus were most enriched, whereas for the diverged genes of At-α (group D) enriched, GO terms related to membrane, transferase activity, and oligopeptide transporter activity (supplementary figs. S19 and S20, Supplementary Material online). This analysis shows that both in lotus and Arabidopsis duplicates that display the least expression divergent are related to dosage-sensitive functions, whereas the duplicated most divergent in expression (subfunctionalized) tend to have lineage-specific functions. For example, group D in lotus was enriched in “circadian regulation of calcium ion oscillation.” This enrichment could be associated with the presence of four lotus genes (namely, Nn-CRY1a, b and Nn-CRY2a, b) being homologous to respectively Arabidopsis Cryptochrome 1 (CRY1) and Cryptochrome 2 (CRY2) (fig. 4A and supplementary figs. S17 and S21, Supplementary Material online). Although CRY1 is a flavin-type blue-light photoreceptor, participating in blue-light induced stomatal opening and thermomorphogenesis, CRY2 is a blue/UV-A photoreceptor controlling flowering time and cotyledon expansion (Endo et al. 2007; Wang et al. 2016; Zhou et al. 2019). Therefore, these four circadian rhythm related genes that underwent post-WGD subfunctionalization might be associated with the lineage-specific adoption of lotus-specific characteristics related to the rigorous rhythm of flower opening and closure.
Subgenome Dominance and Fractionation
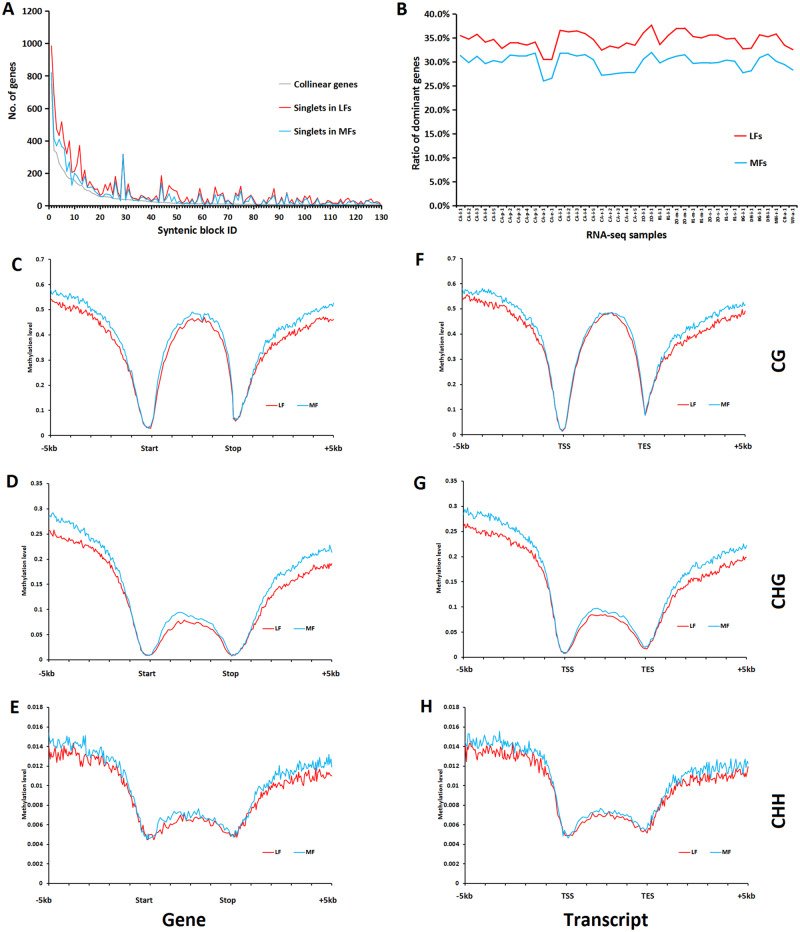
Subgenome dominance is a phenomenon in polyploids, particularly allopolyploids, in which genes are preferentially lost from one parental subgenome and for which the genes that are retained on this parental subgenome are also expressed at lower levels than their corresponding copies on the alternative parental subgenome (Wang, Tu, et al. 2017; Zhao et al. 2017; Liang and Schnable 2018). Here, we wanted to assess whether we could find evidence for subgenome dominance in lotus. For most syntenic blocks, there are many more nonanchor genes (singlets) than anchor genes (collinear genes), suggesting that there has been extensive gene loss and genome rearrangement after the lotus WGD (fig. 5A). Most of the syntenic genome fragments are different in the degree to which gene duplicates are retained (retention of gene numbers), and all pairs of the syntenic regions are different in length (fig. 5A). Only 19 out of the 130 syntenic regions with at least six ancestral genes are significantly biased in gene retention (χ2 test, P < 0.05), rendering it is difficult to partition syntenic genomic fragments based on the significance of gene retention (supplementary table S7, Supplementary Material online). Hence, to study subgenome dominance, we instead grouped the detected syntenic genomic fragments into two groups based on their number of retained ancestral genes and length of the syntenic fragments: We distinguished a group of respectively the LF and the more fractionated (MF) regions (fig. 5A). Duplicated genes of which one copy has an FPKM that is twice as high as that of the alternative copy were identified. The copy with the higher FPKM was referred to as the dominant copy. Interestingly, LF fragments always have a higher ratio of copies with dominant gene expression (mean = 34.49%, SD = 1.16%) than MF fragments (mean = 29.97%, SD = 1.16%). This subgenome dominance can be congruently observed for all 41 surveyed RNA-seq samples obtained from different tissues (fig. 5B). In addition, by investigating the CG, CHG, and CHH methylation and the ratio of sRNA − TE and sRNA + TE in both genic and flanking regions, we found that methylation level and TE density are significantly lower in the LF fragments than in the more fragmented ones (Mann–Whitney U test, all P value < 0.01). This association between subgenome dominance and differential methylation might underly the expression bias between the two copies (fig. 5C–H and supplementary figs. S23 and S24, Supplementary Material online).
Fig. 5.
Subgenome fractionation and dominance in lotus. (A) Differences in the number of singlets (noncollinear) genes across 130 pairs of duplicate syntenic blocks. (B) The ratios of dominant copies in collinear genes between LF blocks and MF blocks across 41 RNA-seq samples. (C–H) Differences in average CG, CHG, and CHH methylation level in leaf along gene and flanking regions between duplicates that belong to LF blocks and MF blocks. (C–E) Methylation of all annotated genes. (F–H) Methylation of the genes with RNA-seq evidence.
Next, we wondered whether the association between subgenome dominance and differential methylation would still hold if we would focus on the subgroups of genes that are respectively more or less subfunctionalized (where the level of subfunctionalization is proxied by the degree to which the duplicates diverged in expression behavior, see above). We noticed in the analysis performed above that duplicate pairs with more conserved expression behavior across tissues (group A) tend to have mutually more similar patterns of CG methylation levels on gene body across tissues than duplicates with more divergent expression behavior (group D). Because of the aforementioned observation, we would expect that duplicates with more conserved expression behavior would possibly display a smaller difference in methylation level between the MF and LF regions than the duplicates with more divergent expression behavior (group D), that is, group A might be less likely show subgenome dominance. However, the (most) subfunctionalized duplicate pairs (group D) do not show any remarkable differences in methylation level as compared with pairs from the other groups (supplementary figs. S25–S30, Supplementary Material online). This indicates that subgenome dominance is likely a phenomenon that acts independently from subfunctionalization (as defined in this work).
Discussion
Since WGD is frequent and common during plant evolution (Cui et al. 2006; Vanneste et al. 2014; Zwaenepoel et al. 2019; Zwaenepoel and Van de Peer 2019), understanding how different genes evolve after a WGD is important for evolutionary biology. In this study, we updated the assembly and annotation of the lotus var. “China Antique” genome by using long-read sequencing data and HI-C. This updated reference assembly largely improved the detection of collinearity to the other species, as well as within genome collinearity (relics of WGD). Notably, we performed integrative methylation and expression analyses which, when combined with all relevant genomic analyses, provide a unique opportunity to study how functional constraints and dosage balance may determine the fate of genes after a single round of WGD. We observed that single-copy genes display the highest expression level and breadth and do not show a hub-like behavior by having few protein interactors. In line with a previous study, also in lotus single-copy genes maintain their single-copy status regardless of a WGD because there appears to be a strong selection against gene redundancy (Paterson et al. 2006; De Smet et al. 2013; Li et al. 2016). The observed differences in expression behavior and the observed functional bias among duplicates after the WGD in lotus are in line with the GBH (Birchler et al. 2005; Freeling 2009; Bekaert et al. 2011; Birchler and Veitia 2012; De Smet and Van de Peer 2012; Tasdighian et al. 2017). Duplicates retained after a WGD are on average more highly expressed, show a functional bias toward conservative functions shared among plant lineages such as gene transcription and signaling, have the highest number of PPIs, and are the greatest in CDS length by having the longest proteins potentially providing more interface(s) for interacting proteins. However, in keeping with previous studies, local duplicates in lotus show lower and more condition-dependent expression and are enriched in lineage-specific functions such as metabolism, disease-resistance, and other dosage-insensitive functions (Rodgers-Melnick et al. 2012; Wu et al. 2012; Denoeud et al. 2014; Lan et al. 2017).
The above observations are further supported by evolutionary patterns observed at the sequence level (nucleotide diversity and the ratio of sequence deletion in gene coding regions). Single-copy genes show the highest sequence conservation which is consistent with studies in Arabidopsis (De Smet et al. 2013). In addition, WGD duplicates exhibit relatively higher sequence conservation than local duplicates, agreeing with what has been observed in Arabidopsis, rice, and Populus (Rodgers-Melnick et al. 2012; Wang 2013). The degree to which genes display sequence conservation seems to be correlated to their expression breath rather than to their expression level. Genes that have been retained following multiple ways of duplications such as TD and WGD have been suggested to have undergone strong selection for higher dosage (Katju and Bergthorsson 2013). For instance, in lotus, the expansion of the LPR1/2 gene by TD and WGD resulted in adaptation to a low-phosphate aquatic environment (Ming et al. 2013). Other examples of multiple duplication events in certain gene families in Arabidopsis and Brassica have been associated with increased immunity (Hofberger et al. 2015). Interestingly, among all locally duplicated genes detected in our study, genes that underwent both “WGD&TD” show significantly higher average expression levels, lower methylation levels, and lower TE densities in promoters than proximal and tandem duplicates. This suggests that also in lotus, genes that underwent both WGD and tandem duplication are selected for the higher overall gene products not only through multiple duplication events but also by other mechanisms such as transcriptional and epigenetic regulation.
Notably, we could show that the relatively higher expression level of genes retained after WGD might be associated with a differential epigenetic regulation. Cytosine methylation in genic and flanking regions affects gene expression (Hirsch and Springer 2017). We observed that indeed genes that were retained after a WGD showed decreased methylation levels in gene flanking regions as compared with other gene groups explaining their higher expression level. In addition, as was observed in other studies, increased methylation was associated with a higher presence of TEs (Weber et al. 2007; Zemach et al. 2010; He et al. 2011; Park et al. 2012; Stroud et al. 2013; Hirsch and Springer 2017). In contrast to what is generally expected, that is, that gene body methylation generally represses gene expression, we found that lotus single-copy genes which are the most abundantly expressed were also the most abundantly methylated in their gene bodies. This has also been observed in rice (Wang Y, Wang X, et al. 2013). So in lotus, it appears that gene body methylation of single-copy genes seems to induce expression rather than repressing it (Su et al. 2011; Takuno and Gaut 2012; Bewick et al. 2016). In lotus, the observed gene body methylation pattern of single-copy genes is also associated with the presence of TEs. The abundant methylation on the gene bodies (genic regions) for single-copy genes could be associated with a similar TE distribution and the presence of abundant introns, indicating that methylation is involved in silencing TEs. Alternatively, it has been shown that gene body methylation can enhance splicing accuracy by improving the distinction of exon–intron boundaries (Lorincz et al. 2004; Luco et al. 2010). This might be particularly relevant in maintaining the functional integrity of single-copy genes, given their high intron number (Lorincz et al. 2004; Luco et al. 2010). However, future functional and genetic studies on TEs and introns of single-copy genes are necessary to support these hypotheses.
Lotus orphan genes were treated separately in the current study because of their evolutionary transience. The lotus WGD occurred 66 Ma after the split with its closest sequenced relative, Macadamia, about 111 Ma (Ming et al. 2013; Hedges et al. 2015). Their low expression level, high expression specificity, and high methylation level imply that orphan genes tend to be transcriptionally repressed to avoid producing nonfunctional peptides (proteins) and that they are not required in most tissues or organs. Their small protein size, gene length, and exon number are consistent with observations in Drosophila and Arabidopsis (Guo 2013; Neme and Tautz 2013; Palmieri et al. 2014). Although their high nucleotide diversity suggests relatively low functional importance, their functionality cannot be excluded (Li et al. 2009; McLysaght and Hurst 2016).
Given that long-retained duplicates from a WGD are important genetic material for plant innovation and evolution, our current study further focused on how those retained duplicates diverge in expression pattern and level across different lotus tissues. Whereas maintaining gene balance plays right after WGD, subfunctionalization and neofunctionalization explain the long-term evolution of duplicates retained from WGD (Lynch and Force 2000; Duarte et al. 2006; Bekaert et al. 2011; Jiang et al. 2013; Gout and Lynch 2015; Teufel et al. 2016). In lotus, WGD duplicates displayed a continuous spectrum of expression divergence where some duplicates share largely the same coexpression partners, whereas other duplicates display a completely distinct expression pattern. Lotus duplicates that display lower expression divergence tend to correspond to the hubs of PPI networks, have relatively longer protein and gene lengths, display higher average expression levels and breadth, more similar pattern of change of CG methylation in gene bodies across different tissues between duplicate pairs, relatively low pairwise amino acid sequence divergence and low nucleotide diversity, which all support they are under a stronger gene balance constraints (De Smet and Van de Peer 2012). Many of these observations are in accordance with studies in, for instance, Arabidopsis, maize, and tomato (Defoort et al. 2019). Yet, in contrast to lotus, these plants underwent sequential rounds of WGDs which makes it difficult to study the fate of the most ancient duplicates. Hence, the fact that the same findings made in these other species are also observed in lotus indicates that they are truly associated with the fate of ancient duplicates.
Subgenome dominance can be an important source of bias in expression level between duplicated gene pairs retained from a WGD and can result in significant differences in gene retention (content), the intensity of TE insertion, methylation, and population-level polymorphisms between subgenomes (Hughes et al. 2014; Woodhouse et al. 2014; Li, Fan, et al. 2015; Cheng et al. 2016; Zhao et al. 2017). Depending on the studied species, the level of subgenome fractionation that occurs after a WGD can be significantly different, ranging from extensive fractionation in, for example, monkeyflower (WGD estimated at 140 Ma), maize (11.9 Ma), Brassica (13–17 Ma), Arabidopsis (40 Ma), and cotton (60 Ma) (Hughes et al. 2014; Woodhouse et al. 2014; Li, Fan, et al. 2015; Cheng et al. 2016; Zhao et al. 2017) to little subgenome fractionation in, for example, soybean (5–13 Ma), banana (65 Ma), and poplar (8 Ma) fractionation (Garsmeur et al. 2014; Zhao et al. 2017). In our study, about 14.6% of syntenic block pairs in lotus show significant bias in fractionation, a level which is in between the fraction observed in the paleoautopolyploid soybean (5.4%) and the paleoallopolyploid maize (31%) (Zhao et al. 2017). The LF blocks show on average about 4.52% more (expression) dominant copies than the MF blocks, which is a difference that is higher than what is observed in soybean (0–1%) but lower than in maize (∼10%) (Zhao et al. 2017). As the extent of biases in lotus (66 Ma) is between a paleoautopolyploid and a paleoallopolyploid, likely, its two ancestral parental genomes had already diverged to some extent before the formation of ancient polyploid. So far, lotus shows evidence of one of the oldest appearances of subgenome dominance among the above-mentioned plant genomes.
Materials and Methods
Plant Material, PacBio Sequel, and HI-C Sequencing
Sacred lotus “China Antique” was grown and collected from Wuhan Botanical Garden of the Chinese Academy of Sciences. DNA from young leaves of “China Antique” was extracted using Plant DNA Isolation Reagent (Tiangen, China). Two DNA libraries (insert sizes of 10,123 and 10,157 bp, respectively) were constructed according to the PacBio library preparation protocol and sequenced on a PacBio Sequel platform (Pacific Biosciences, USA) at Annoroad Genomics (Beijing, China). Subreads with a quality score under 0.8 were discarded. The HI-C DNA library of “China Antique” was prepared at Annoroad Genomics (Beijing, China) under a previously published protocol (Lieberman-Aiden et al. 2009). Briefly, the nuclear DNA of young lotus leaves was cross-linked inside the tissue cell sample. Then, the extracted DNA was digested with the restriction enzyme (HindIII/MboI). Biotinylation was tagged at the sticky ends of the digested DNA fragments, and then mutually ligated at random after dilution. The library of condensed, sheared, and biotinylated DNA fragments were prepared for PE sequencing with 150-bp reads on Illumina HiSeq platform.
Chromosome-Level Assembly
All contigs were assembled using PacBio and Illumina reads. SparseAssembler was applied to assemble Illumina PE reads of lotus “China Antique” into short but accurate Illumina contigs (Ye et al. 2011, 2012). These Illumina contigs and PacBio Sequel reads were coassembled into longer contigs with the hybrid assembly tool DBG2OLC (Ye et al. 2016). Errors in these hybrid contigs were further polished with all Illumina PE reads using BWA-MEM and Pilon 2.10 (Li and Durbin 2009; Walker et al. 2014). The HI-C sequencing reads were mapped on the “China Antique” hybrid assembly contigs using BWA-MEM (Li and Durbin 2009). Finally, the chromosome-level scaffolding of these contigs was performed with LACHESIS (Burton et al. 2013). Additional gaps in pseudochromosomes were filled with PacBio subreads using Jelly and polished with Illumina reads using Pilon 2.10 (English et al. 2012).
Repeat Annotation
Repeats including TEs on the new “China Antique” assembly were annotated following a previously published protocol (Campbell et al. 2014). Generally, MITEs (miniature inverted-repeat transposable elements) were predicted using MITE-Hunter under default settings (Han and Wessler 2010). The most abundant plant TEs, long terminal repeat (LTR) retrotransposons were collected, false-positives were filtered, and redundancy was reduced using LTR-harvest, LTR-digest, and the Perl scripts provided by the protocol “Repeat Library Construction-Advanced” (http://weatherby.genetics.utah.edu/MAKER/wiki/index.php) (Ellinghaus et al. 2008; Steinbiss et al. 2009). Other repeats were collected by RepeatModeler (http://www.repeatmasker.org). Gene fragments in all collected repeats were excluded by searching against all plant protein sequences from Plant Plaza 4.0 (Van Bel et al. 2018). After collecting and building the lotus repeat database, the genome was further annotated using RepeatMasker (http://www.repeatmasker.org).
Gene Annotation
Protein-coding genes on the “China Antique” assembly were annotated based on 1) RNA-seq mapping, 2) protein homology searches, and 3) ab initio prediction. For gene prediction with transcriptional evidence, 41 public available RNA-seq data from leaf, petioles, rhizome, root, and the apical bud of lotus were downloaded from NCBI SRA database and mapped on the genome using HISAT2-StringTie pipeline (Kim et al. 2015; Pertea et al. 2015). Transcript coordinates from different RNA-seq samples were further merged using TACO (Niknafs et al. 2017). Coding regions of transcripts were annotated using Transdecoder (https://github.com/TransDecoder). Homology-based gene prediction was performed using GeMoMa with genome sequences and gene coordinates from Arabidopsis thaliana, Carica papaya, V. vinifera, M. ternifolia (Proteales), and B. distachyon as input (Keilwagen et al. 2016; Nock et al. 2016; Van Bel et al. 2018). Ab initio gene prediction was performed using Braker2 which took in intron hints from transcript coordinates of RNA-seq based assemblies (Hoff et al. 2016). The final consensus gene annotations were produced by EVidenceModeler with weights of “RNA-seq > gene homology > ab initio” (Haas et al. 2008). The longest transcript (isoform) for each gene based on RNA-seq data was retained to represent the expressed lotus genes for methylation analyses. GO annotations were further performed using the “nonredundant” database of plants via BLAST2GO with default settings (Conesa et al. 2005). The lotus interactome was inferred using PPI data from Arabidopsis by the top BLAST hit from orthologs (Arabidopsis Interactome Mapping Consortium 2011; Yang et al. 2013).
Validation of Genome Assembly
Accuracy and structural completeness of the new genome assembly were assessed using 1) previously published SNP markers from genetic linkage groups, 2) ratio of genome collinearity with other species, and 3) conserved single-copy genes of plant from BUSCO. For comparison, SNP markers from a high-density lotus genetic map from a previous study were downloaded and mapped onto the new and old “China Antique” assemblies (Liu et al. 2016) using bowtie allowing no mismatch other than SNP site (Langmead 2010). Collinearity between the genetic map and the genome assembly was anchored by SNP markers. Distributions of SNP markers on genome assemblies were inspected by bar plots, and collinearity was visualized by dot plots. To assess the genome architecture using genome collinearity, we searched homologous genomic blocks in genomes of two monocots, Oryza sativa (rice) and B. distachyon, against the new (v2018) and old (v2013) “China Antique” assemblies using MCScanX (Wang et al. 2012). First, potential anchors between the two genomes were identified using BlastP (E < 1e − 5). Then, MCscanX found all orthologous syntenies with at least six anchor points. For further comparisons, orthologous syntenies between other eudicots species and the two monocot representatives were downloaded from PGDD (Lee et al. 2013). To assess the completeness of the gene regions in the assembly, 1,440 conserved plant single-copy genes as a benchmark were searched using BUSCO v2 (Simão et al. 2015).
Classification of Genes by Duplication Status
Duplicated genes in extant genomes typically originated through different duplication events. Depending on the size of the genomic regions involved in the duplication event, a distinction is made between SSD and LSD. LSD can be maintained as syntenies which likely are retained from WGD. Within SSD, a distinction is made between local (tandem and proximal duplication) versus dispersed duplications (Freeling 2009; Wang, Li, et al. 2013). Tandem duplicates lead to a cluster of two or more consecutive paralogous sequences, whereas proximal having one or a few intervening genes. Dispersed duplicate are mainly unclassified duplicates. Genes that underwent both WGD and tandem duplications often exist, which we refer to as “WGD&TD” (Matus et al. 2008; Liebrand et al. 2014).
To identify ancient genome duplication of lotus, homologs were first identified by all-against-all BlastP for syntenic anchors (E < 1e − 5). Intraspecific syntenic blocks were identified with the same approach as the one used for the identification of orthologous synteny described above using MCscanX (Wang et al. 2012). To identify WGDs, raw 4dTv (the number of transversion 4-fold degenerative sites) of all syntenic paralogous pairs were calculated and further corrected for possible multiple transversions at the same site based on a previous method (Tang et al. 2008). A histogram of 4dTv for all syntenic paralogs was plotted with a bin size of 0.01. To classify syntenic blocks according to WGDs, the median of 4dTv of each syntenic block was used. Syntenic blocks with less than six duplicate pairs with valid 4dTv after correction were classified as syntenies of uncertain origin. Other divergence parameters including dS or Ks (synonymous substitution rate), dN or Ka (nonsynonymous substitution rate), and dN/dS for all syntenic paralogs were calculated using codeML from PAML package (Yang 2007). Further, 4dTv of orthologous divergence were also plotted in histograms. For the fragmented genome assembly M. ternifolia, orthologous pairs were predicted using OrthoMCL (Li et al. 2003; Neale et al. 2014). The chronological order of WGDs and species split (Nelumbo vs. Macademia) were confirmed by the Mann–Whitney U test based on rate calibrated 4dTv.
Single-copy genes and genes of other duplication status including those originating from dispersed duplication, tandem duplication, and proximal duplication events, WGD&TD were also detected by MCscanX (Wang et al. 2012). All lotus genes without homology to other sequenced species were defined as orphan genes, whereas the rest was regarded as nonorphan genes whose ancestral proteins appeared at least before the split of lotus and Macademia (111 Ma). The family Nelombonaceae (in Proteales) is a species-poor clade with only two closely related Nelumbo species. To obtain lotus orphan genes, M. ternifolia (the other only sequenced Proteales genome) and PlantPlaza 3.0 database were used in phylostratigraphic analyses (Arendsee et al. 2014). The groups of genes of different duplication status were used for subsequent comparative analyses. As most orphan genes are evolutionarily transient, they were analyzed separately (Arendsee et al. 2014).
To explore the dosage sensitivity for our studied groups (subdivided as described above), we defined for each lotus gene its orthologs in Macademia and V. vinifera using OrthoMCL (Li et al. 2003; Neale et al. 2014). For each gene, we calculated the average copy number of its orthologs in the three taxa. For each lotus gene, its CV in the number of copies observed in the three species was used to estimate the dosage-sensitive. For each of the studied gene groups, the average copy number and CV were reported.
Nucleotide Diversity and the Ratio of Sequence Deletion of Lotus Genes
Illumina resequencing data from 18 Asian lotus individuals including rhizome lotus, flower lotus, seed lotus, wild lotus, and Thai lotus were downloaded from NCBI (supplementary data, Supplementary Material online) (Huang et al. 2018). Illumina reads were mapped to the new “China Antique” assembly using BWA-mem (Li and Durbin 2009). Mapped files were processed by Picard (https://broadinstitute.github.io/picard/). The SNP variants were called by HaplotypeCaller of GATK 3.7 with further hard filtering of “QD < 2 ‖ FS > 60 ‖ MQ < 30” (McKenna et al. 2010). Nucleotide diversity (π) of each CDS from each annotated gene was estimated using Popgenome (Pfeifer et al. 2014) in R. The ratio of sequence deletion for each gene is calculated by the ratio of InDel length in each CDS. A Mann–Whitney U test or (nonparametric) Kruskal–Wallis test was applied to compare average π and the average ratio of sequence deletion of gene CDS between (among) different gene groups in Graphpad PRISM 7.
Expression Analysis
All 41 RNA-seq samples used for gene annotation were also used for expression analyses. The average expression level per gene for each gene group showing different fates after the lotus WGD was estimated by the average log-transformed FPKM values of the genes in a group. Tissue-specific expression was assessed by the Tau index (Yanai et al. 2005). To define “tissue,” we clustered the 41 samples using the log-transformed FPKM data (Euclidean distance). Samples clustered in eight distinct tissue groups: leaf, petiole, apical, rhizome internode, root, rhizome (later swelling), rhizome (middle swelling), and rhizome (stolon). The Tau index was calculated as follows:
where and is the expression for tissue i.
To build the coexpression network, the genes with an expression value in at least two samples were retained (28,578 are present in the network with 480,849 edges). The “rank of correlation coefficient” (Obayashi and Kinoshita 2009) was used to determine the degree of pairwise coexpression. To calculate the rank-based correlation, the gene–gene Pearson correlation matrix derived from the log 2-transformed FPKM values was transformed into a rank matrix. For every gene–gene combination the Mutual Rank score was calculated using the following formula:
where Rank(A →B) is the rank of the correlation of gene B with gene A as compared with its correlation with all other genes (Obayashi and Kinoshita 2009). Smaller MR scores correspond to a higher degree of pairwise correlation between two genes and can be converted to a network edge weight using the following formula guaranteeing that the range of edge weights in the coexpression network scales between 0 and 1. A small value (one) was added to the FPKM values before log transformation to avoid having undefined values of the rank-based correlation for 0 values.
Grouping WGD Genes Based on Their Expression Behavior
Post-WGD duplicate pairs were subdivided into groups based on their expression divergence. To assess the degree to which duplicate pairs diverged in expression, we used an interconnectivity score (HSu et al. 2011). The interconnectivity between a pair of duplicated genes assesses the degree to which two duplicate genes share neighbors in the coexpression network. The higher the connectivity score, the more the duplicates are assumed to share the same expression profile.
where N(i) and N(j) describe the number of neighbors that are located at most three edges distance of respectively the nodes i and j in the duplicate pair (i, j). The number of shared neighbors between the genes of the duplicate pair is normalized by the total number of neighbors of the two genes in the duplicate pair.
Also, we determined for each duplicate gene pair whether the number of shared neighbors that contributed to the connectivity measure is statistically significant using the hypergeometric test: For every duplicate pair, the number of up to third-order neighbors for one gene NA was determined and used to calculate the chance of a success P = NA/N, where N is the total number of genes in the genome. The number of up to the third-order neighbor for the second gene in the pair (NB) was used as the number of trials and the number of neighbors shared between A and B was considered the number of successes. Using these parameters, the cumulative mass function was calculated to calculate the P value, that is, observing the same number of shared neighbors between two genes just by chance. Based on the combination of the hypergeometric-value and the connectivity score, the duplicates were subdivided in five groups: group A with connectivity >0.5 and P value <0.01, group B with connectivity 0.5 > x > 0.3 and P value <0.01, group C with 0.3 > x > 0.15 and P value <0.01, group D with connectivity <0.15 and P value >0.99, and group E with connectivity <0.15 and 0.99 > x > 0.1. Group E contains the genes that show a certain but insignificant connectivity. This category was not retained for further analysis. Duplicate genes that belong to groups A–C share coexpressed neighbors (more for group A > B > C) and they share more neighbors in the coexpression network than can be expected by chance given the local connectivity of the genes in the pair. Genes belonging to group D show a significant low to no connectivity in the coexpression network.
Comparisons of Different Gene Features
Genomic traits including the length of the CDS, the number of exons and the gene length were directly obtained from the lotus genome annotation. Given that there is currently no protein interactome map for lotus, for those lotus genes displaying homology to Arabidopsis genes, their number of PPIs were inferred from the closest homolog in Arabidopsis (Arabidopsis Interactome Mapping Consortium 2011; Yang et al. 2013). Genomic traits (CDS length, gene length, exon number) and evolutionary parameters (dN, dS, dN/dS, and π) were summarized and compared between different genes of different groups using (nonparametric) Kruskal–Wallis test in Graphpad PRISM 7.
sRNA + TE, sRNA − TE, and Methylation Level Analyses of Gene Duplicates
To test whether TE insertion and methylation level differences might contribute to duplicate gene expression differences, first, sRNAs of “China Antique” were mapped to the genome using bowtie with zero tolerance of mismatch (Shi et al. 2017). Only uniquely mapped sRNAs were used to define TEs (Cheng et al. 2016). TEs were classified into sRNA+ TEs and sRNA − TEs based on whether there was any sRNA aligning to them. Gene flanking regions (defined as the region ± 5 kb the gene body) and gene bodies (defined as the region between the translation start and stop site) were analyzed using a sliding window. Regions in the overlap between the flanking region and the gene body were excluded from the flanking regions. For each 5′ and 3′ flanking region, a 100-bp sliding window with 10-bp step was applied; for each gene body, the 40 evenly divided windows of the gene body were used (Wang et al. 2015). For each sliding window, the proportion of the sequence being composed of sRNA+ TE or sRNA − TEs was calculated. The average proportion in each sliding window was calculated for each gene group under investigation. These averaged proportions were then used to estimate the TE density in the flanking regions and gene bodies of different gene groups. Whole-genome methylation was analyzed based on bisulfite sequencing (BS-seq) on young leaves from a wild lotus (Khabarovsk, Russia) (NCBI accession: SRX4410560), petal (SRX4003561), stamen petaloid (SRX4003562), and stamen (SRX4003563). Flanking regions as defined above were evenly divided into one hundred 50-bp windows, and the gene body was evenly divided into 40 windows (Wang et al. 2015). We included both the exons and introns to the methylation level of gene bodies because pre-mRNAs, being transcribed from DNA, contain introns. Methylation level including CG, CHG, and CHH sites of different gene groups was estimated using BS-Seeker2 and cgmaptools for each window (Guo et al. 2013, 2018). Because the results might be dependent on how the flanking regions and gene bodies are defined, we redid the methylation level assessment with an alternative definition of the gene body and flanking regions in parallel. Here, we used RNA-seq data to define TSS (transcriptional start sites) and TES (transcriptional end sites) of the longest transcript for each gene and defined the flanking regions (fig. 3D–F and supplementary figs. S9 and S10, Supplementary Material online). To measure the similarity of methylation change in the four lotus tissues between a pair of duplicate genes, mean CG, CHG, and CHH methylation levels for 2-kb upstream and downstream regions, and gene bodies of each gene were calculated by cgmaptools. For each gene, a methylation pattern was defined per genic region (upstream, downstream, and gene body). This pattern is represented as a vector with as entries the average methylation level for that genic region per tissue. The similarity in the methylation pattern of duplicates was calculated using the correlation coefficient (r).
Subgenome Fractionation and Dominance
Subgenome fractionation bias was analyzed as outlined previously (Garsmeur et al. 2014). Numbers of collinear genes and noncollinear genes for pairs of syntenic blocks were tested for significant fractionation bias (χ2 test). Differences in TE ratio and methylation between collinear genes in LF and MF syntenic blocks were analyzed with the same approach described above. Subgenome fractionation bias is often associated with subgenome dominance. To test subgenome dominance, all 41 RNA-seq samples were used. For each RNA-seq sample, the dominant copy was defined as the one showing an expression that was more than 2-fold higher than the expression level of the alternative copy (FPKM). Further, for each RNA-seq sample, the ratios of dominant copies in LFs and MFs were summarized and compared.
Supplementary Material
Acknowledgments
We thank Chenxi Ye from University of Maryland (USA) and Katharina Hoff from Ernst Moritz Arndt Universität Greifswald (Germany) for advice on genome analyses, Jia Li from Lanzhou University (China) for discussion, and Pingfang Yang and Yanlin Liu from Wuhan Botanical Garden CAS (China) for conserving plant material. This work was supported by grants from the Strategic Priority Research Program of Chinese Academy of Sciences (No. XDB31000000), the National Natural Science Foundation of China (Nos 31570220, 31870208, and 31700197), Youth Innovation Promotion Association of Chinese Academy of Sciences (No. 2019335), Hubei Provincial Natural Science Foundation of China (No. 2019CFB275), and Hubei Chenguang Talented Youth Development Foundation. Y.V.d.P. acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 833522). R.S.R. is supported by the Ministry of Science, Research and Technology (Iran).
Author Contributions
T.S., K.M., Y.V.d.P., Q.W., and J.C. conceptualized the study; T.S., R.S.R., and K.M. performed the methodology; T.S., H.L., Y.Z., and Z.L. performed the investigation; T.S. and R.S.R. performed the formal analysis; T.S. wrote the original draft; K.M., Y.V.d.P., M.W., P.F.G., and J.C. reviewed and edited the manuscript; Q.W., Y.V.d.P., K.M., and J.C. supervised the study.
All data generated in this study are available from the National Center for Biotechnology Information (NCBI) under BioProject PRJNA481856. The raw PacBio sequences are deposited under SRR7549129 and SRR7549130, HI-C data were deposited under SRR7615553 and SRR7631523, and Bisulfite sequencing data were deposited under SRR7544256. Lotus genome assembly is available at http://nelumbo.biocloud.net.
References
- Akashi H. 2001. Gene expression and molecular evolution. Curr Opin Genet Dev. 11(6):660–666. [DOI] [PubMed] [Google Scholar]
- Aköz G, Nordborg M.. 2019. The Aquilegia genome reveals a hybrid origin of core eudicots. Genome Biol. 20(1):256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333(6042):601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendsee ZW, Li L, Wurtele ES.. 2014. Coming of age: orphan genes in plants. Trends Plant Sci. 19(11):698–708. [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC.. 2011. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell 23(5):1719–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Ji L, Niederhuth CE, Willing E-M, Hofmeister BT, Shi X, Wang L, Lu Z, Rohr NA, Hartwig B, et al. 2016. On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci U S A. 113(32):9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA.. 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21(4):219–226. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA.. 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A. 109(37):14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH.. 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16(7):1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottani S, Zabet NR, Wendel JF, Veitia RA.. 2018. Gene expression dominance in allopolyploids: hypotheses and models. Trends Plant Sci. 23(5):393–402. [DOI] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, Shendure J.. 2013. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 31(12):1119–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey DR. 2004. Are protein–protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci. 13(1):190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Law M, Holt C, Stein JC, Moghe GD, Hufnagel DE, Lei J, Achawanantakun R, Jiao D, Lawrence CJ, et al. 2014. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164(2):513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casto AL, McKinley BA, Yu KMJ, Rooney WL, Mullet JE.. 2018. Sorghum stem aerenchyma formation is regulated by SbNAC_D during internode development. Plant Direct 2(11):e00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hao Z, Guang X, Zhao C, Wang P, Xue L, Zhu Q, Yang L, Sheng Y, Zhou Y, et al. 2019. Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat Plants 5(1):18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Sun C, Wu J, Schnable J, Woodhouse MR, Liang J, Cai C, Freeling M, Wang X.. 2016. Epigenetic regulation of subgenome dominance following whole genome triplication in Brassica rapa. New Phytol. 211(1):288–299. [DOI] [PubMed] [Google Scholar]
- Cheng F, Wu J, Cai X, Liang J, Freeling M, Wang X.. 2018. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants 4(5):258–268. [DOI] [PubMed] [Google Scholar]
- Coate JE, Song MJ, Bombarely A, Doyle JJ.. 2016. Expression-level support for gene dosage sensitivity in three Glycine subgenus Glycine polyploids and their diploid progenitors. New Phytol. 212(4):1083–1093. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M.. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, et al. 2006. Widespread genome duplications throughout the history of flowering plants. Genome Res. 16(6):738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, Adams KL, Vandepoele K, Van Montagu MCE, Maere S, Van de Peer Y.. 2013. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci U S A. 110(8):2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, Van de Peer Y.. 2012. Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr Opin Plant Biol. 15(2):168–176. [DOI] [PubMed] [Google Scholar]
- Defoort J, Van de Peer Y, Carretero-Paulet L.. 2019. The evolution of gene duplicates in angiosperms and the impact of protein–protein interactions and the mechanism of duplication. Genome Biol Evol. 11(8):2292–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, et al. 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345(6201):1181–1184. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH.. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 102(40):14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, DePamphilis CW.. 2006. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 23(2):469–478. [DOI] [PubMed] [Google Scholar]
- Edger PP, Pires JC.. 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17(5):699–717. [DOI] [PubMed] [Google Scholar]
- Edger PP, Smith RD, McKain MR, Cooley AM, Vallejo-Marin M, Yuan Y-W, Bewick AJ, Ji L, Platts AE, Bowman MJ, et al. 2017. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year- old naturally established neo-allopolyploid monkeyflower. Plant Cell 29(9):2150–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U.. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Mochizuki N, Suzuki T, Nagatani A.. 2007. CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19(1):84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS Long-Read Sequencing Technology. PLoS One 7(11):e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y.. 2009. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc Natl Acad Sci U S A. 106(14):5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF.. 2010. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 186(1):184–193. [DOI] [PubMed] [Google Scholar]
- Freeling M. 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 60(1):433–453. [DOI] [PubMed] [Google Scholar]
- Freeling M, Thomas BC.. 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16(7):805–814. [DOI] [PubMed] [Google Scholar]
- Garsmeur O, Schnable JC, Almeida A, Jourda C, D’Hont A, Freeling M.. 2014. Two evolutionarily distinct classes of paleopolyploidy. Mol Biol Evol. 31(2):448–454. [DOI] [PubMed] [Google Scholar]
- Gout JF, Lynch M.. 2015. Maintenance and loss of duplicated genes by dosage subfunctionalization. Mol Biol Evol. 32(8):2141–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui S, Peng J, Wang X, Wu Z, Cao R, Salse J, Zhang H, Zhu Z, Xia Q, Quan Z, et al. 2018. Improving Nelumbo nucifera genome assemblies using high-resolution genetic maps and BioNano genome mapping reveals ancient chromosome rearrangements. Plant J. 94(4):721–734. [DOI] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M.. 2013. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 14(1):774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhu P, Pellegrini M, Zhang MQ, Wang X, Ni Z.. 2018. CGmapTools improves the precision of heterozygous SNV calls and supports allele-specific methylation detection and visualization in bisulfite-sequencing data. Bioinformatics 34(3):381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL. 2013. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 73(6):941–951. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Robin CR, Wortman JR.. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wessler SR.. 2010. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38(22):e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK.. 2011. Regulation and function of DNA methylation in plants and animals. Cell Res. 21(3):442–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S.. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 32(4):835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch CD, Springer NM.. 2017. Transposable element influences on gene expression in plants. Biochim Biophys Acta Gene Regul Mech. 1860(1):157–165. [DOI] [PubMed] [Google Scholar]
- Hofberger JA, Nsibo DL, Govers F, Bouwmeester K, Schranz ME.. 2015. A complex interplay of tandem- and whole-genome duplication drives expansion of the L-type lectin receptor kinase gene family in the Brassicaceae. Genome Biol. Evol. 7(3):720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M.. 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32(5):767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Huang YH, Hsu CT, Yang UC.. 2011. Prioritizing disease candidate genes by a gene interconnectedness-based approach. BMC Genomics. 12(Suppl 3):S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Yang M, Li L, Li H, Yang D, Shi T, Yang P.. 2018. Whole genome re-sequencing reveals evolutionary patterns of sacred lotus (Nelumbo nucifera). J Integr Plant Biol. 60(1):2–15. [DOI] [PubMed] [Google Scholar]
- Hughes TE, Langdale JA, Kelly S.. 2014. The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome Res. 24(8):1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449(7161):463–467. [DOI] [PubMed] [Google Scholar]
- Jiang W, Liu Y, Xia E, Gao L.. 2013. Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol. 161(4):1844–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473(7345):97–100. [DOI] [PubMed] [Google Scholar]
- Jones S, Thornton JM.. 1996. Principles of protein–protein interactions. Proc Natl Acad Sci U S A. 93(1):13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R, Phillips PC.. 2011. Expression level drives the pattern of selective constraints along the insulin/tor signal transduction pathway in Caenorhabditis. Genome Biol Evol. 3:715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V, Bergthorsson U.. 2013. Copy-number changes in evolution: rates, fitness effects and adaptive significance. Front Genet. 4:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, Wenk M, Erickson JL, Schattat MH, Grau J, Hartung F.. 2016. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 44(9):e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T, Renner T, Ibarra-Laclette E, Farr KM, Chang T-H, Cervantes-Pérez SA, Zheng C, Sankoff D, Tang H, Purbojati RW, et al. 2017. Long-read sequencing uncovers the adaptive topography of a carnivorous plant genome. Proc Natl Acad Sci U S A. 114(22):E4435–E4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langham RJ, Walsh J, Dunn M, Ko C, Goff SA, Freeling M.. 2004. Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166(2):935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics 32(1):11.7.1–11.7.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH.. 2013. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41(D1):D1152–D1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Uygun S, Moghe GD, Panchy N, Fang L, Hufnagel DE, Jasicki HL, Feig M, Shiu S-H.. 2015. Molecular evidence for functional divergence and decay of a transcription factor derived from whole-genome Duplication in Arabidopsis thaliana. Plant Physiol. 168(4):1717–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, et al. 2015. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 33(5):524–530. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Foster CM, Gan Q, Nettleton D, James MG, Myers AM, Wurtele ES.. 2009. Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J. 58(3):485–498. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Baniaga AE, Sessa EB, Scascitelli M, Graham SW, Rieseberg LH, Barker MS.. 2015. Early genome duplications in conifers and other seed plants. Sci Adv. 1(10):e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Defoort J, Tasdighian S, Maere S, Van de Peer Y, De Smet R.. 2016. Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell 28(2):326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Schnable JC.. 2018. Functional divergence between subgenomes and gene pairs after whole genome duplications. Mol Plant 11(3):388–397. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326(5950):289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TWH, van den Burg HA, Joosten M.. 2014. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19(2):123–132. [DOI] [PubMed] [Google Scholar]
- Lisch D, Bennetzen JL.. 2011. Transposable element origins of epigenetic gene regulation. Curr Opin Plant Biol. 14(2):156–161. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhu H, Liu Y, Kuang J, Zhou K, Liang F, Liu Z, Wang D, Ke W.. 2016. Construction of a high-density, high-quality genetic map of cultivated lotus (Nelumbo nucifera) using next-generation sequencing. BMC Genomics. 17(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JP, Seddon AE, Moghe GD, Simenc MC, Shiu S-H.. 2015. Characteristics of plant essential genes allow for within- and between-species prediction of lethal mutant phenotypes. Plant Cell 27(8):2133–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M.. 2004. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 11(11):1068–1075. [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T.. 2010. Regulation of alternative splicing by histone modifications. Science 327(5968):996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A.. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154(1):459–473.10629003 [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P.. 2008. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 8(1):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hurst LD.. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet. 17(9):567–578. [DOI] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim MJ, Schatz MC, Campbell M, et al. 2013. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 14(5):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montilla-Bascón G, Rubiales D, Hebelstrup KH, Mandon J, Harren FJM, Cristescu SM, Mur LAJ, Prats E.. 2017. Reduced nitric oxide levels during drought stress promote drought tolerance in barley and is associated with elevated polyamine biosynthesis. Sci Rep. 7(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW, Cardeno C, Koriabine M, Holtz-Morris AE, Liechty JD, et al. 2014. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15(3):R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R, Tautz D.. 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics. 14(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niknafs YS, Pandian B, Iyer HK, Chinnaiyan AM, Iyer MK.. 2017. TACO produces robust multisample transcriptome assemblies from RNA-seq. Nat Methods. 14(1):68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock CJ, Baten A, Barkla BJ, Furtado A, Henry RJ, King GJ.. 2016. Genome and transcriptome sequencing characterises the gene space of Macadamia integrifolia (Proteaceae). BMC Genomics. 17(1):937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K.. 2009. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 16(5):249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Am J Hum Genet. 23(5):541. [Google Scholar]
- Olsen JL, Rouzé P, Verhelst B, Lin YC, Bayer T, Collen J, Dattolo E, De Paoli E, Dittami S, Maumus F, et al. 2016. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530(7590):331–335. [DOI] [PubMed] [Google Scholar]
- Palmieri N, Kosiol C, Schlötterer C.. 2014. The life cycle of Drosophila orphan genes. Elife 3(3):e01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Xu K, Park T, Yi SV.. 2012. What are the determinants of gene expression levels and breadths in the human genome? Hum Mol Genet. 21(1):46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC.. 2006. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 22(11):597–602. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ.. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 31(7):1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH.. 2019. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp RA, Udall JA, Wendel JF.. 2009. Genomic expression dominance in allopolyploids. BMC Biol. 7(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Mane SP, Dharmawardhana P, Slavov GT, Crasta OR, Strauss SH, Brunner AM, DiFazio SP.. 2012. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 22(1):95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rody HVS, Baute GJ, Rieseberg LH, Oliveira LO.. 2017. Both mechanism and age of duplications contribute to biased gene retention patterns in plants. BMC Genomics. 18(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht C, Lohaus R, Vanneste K, Mutwil M, Nikoloski Z, Van De Peer Y, Persson S.. 2017. Revisiting ancestral polyploidy in plants. Sci Adv. 3(7):e1603195–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve SR, Rohlfs RV, Hvidsten TR.. 2018. Subfunctionalization versus neofunctionalization after whole-genome duplication. Nat Genet. 50(7):908–909. [DOI] [PubMed] [Google Scholar]
- Shi T, Wang K, Yang P.. 2017. The evolution of plant microRNAs: insights from a basal eudicot sacred lotus. Plant J. 89(3):442–457. [DOI] [PubMed] [Google Scholar]
- Shiu S-H, Bleecker AB.. 2001. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci U S A. 98(19):10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Song H, Gao H, Liu J, Tian P, Nan Z.. 2017. Comprehensive analysis of correlations among codon usage bias, gene expression, and substitution rate in Arachis duranensis and Arachis ipaënsis orthologs. Sci Rep. 7(1):14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S.. 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37(21):7002–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE.. 2013. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152(1–2):352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Han L, Zhao Z.. 2011. Conservation and divergence of DNA methylation in eukaryotes: new insights from single base-resolution DNA methylomes. Epigenetics 6(2):134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne IA, Silver PA.. 2008. Intron delays and transcriptional timing during development. Dev Cell 14(3):324–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS.. 2012. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol. 29(1):219–227. [DOI] [PubMed] [Google Scholar]
- Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH.. 2008. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18(12):1944–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdighian S, Van Bel M, Li Z, Van de Peer Y, Carretero-Paulet L, Maere S.. 2017. Reciprocally retained genes in the angiosperm lineage show the hallmarks of dosage balance sensitivity. Plant Cell 29(11):2766–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel AI, Liu L, Liberles DA.. 2016. Models for gene duplication when dosage balance works as a transition state to subsequent neo-or sub-functionalization. BMC Evol Biol. 16(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van De Peer Y, Coppens F, Vandepoele K.. 2018. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46(D1):D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Peer Y, Mizrachi E, Marchal K.. 2017. The evolutionary significance of polyploidy. Nat Rev Genet. 18(7):411–424. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van De Peer Y.. 2014. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Pale. Genome Res. 24(8):1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Casacuberta JM.. 2017. Impact of transposable elements on polyploid plant genomes. Ann Bot. 120(2):195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Beyene G, Zhai J, Feng S, Fahlgren N, Taylor NJ, Bart R, Carrington JC, Jacobsen SE, Ausin I.. 2015. CG gene body DNA methylation changes and evolution of duplicated genes in cassava. Proc Natl Acad Sci U S A. 112(44):13729–13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tu L, Lin M, Lin Z, Wang P, Yang Q, Ye Z, Shen C, Li J, Zhang L, et al. 2017. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat Genet. 49(4):579–587. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zuo Z, Wang X, Gu L, Yoshizumi T, Yang Z, Yang L, Liu Q, Liu W, Han YJ, et al. 2016. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354(6310):343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang Z, Fu T, Hu L, Xu C, Gong L, Wendel JF, Liu B.. 2017. Gene-body CG methylation and divergent expression of duplicate genes in rice. Sci Rep. 7(1):2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. 2013. Locally duplicated ohnologs evolve faster than nonlocally duplicated ohnologs in Arabidopsis and rice. Genome Biol Evol. 5(2):362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan G, Liu Y, Sun F, Shi C, Liu X, Peng J, Chen W, Huang X, Cheng S, et al. 2013. The sacred lotus genome provides insights into the evolution of flowering plants. Plant J. 76(4):557–567. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J, Paterson AH.. 2013. MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 29(11):1458–1460. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40(7):e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, Lee TH, Mansoor S, Paterson AH.. 2013. Gene body methylation shows distinct patterns associated with different gene origins and duplication modes and has a heterogeneous relationship with gene expression in Oryza sativa (rice). New Phytol. 198(1):274–283. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D.. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 39(4):457–466. [DOI] [PubMed] [Google Scholar]
- Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X.. 2014. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci U S A. 111(14):5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, et al. 2012. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci U S A. 109(30):12219–12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, et al. 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21(5):650–659. [DOI] [PubMed] [Google Scholar]
- Yang J, Osman K, Iqbal M, Stekel DJ, Luo Z, Armstrong SJ, Franklin F.. 2013. Inferring the Brassica rapa interactome using protein–protein interaction data from Arabidopsis thaliana. Front Plant Sci. 3:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Ye C, Hill CM, Wu S, Ruan J, Ma Z.. 2016. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep. 6:31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Ma ZS, Cannon CH, Pop M, Yu DW.. 2011. SparseAssembler: de novo Assembly with the Sparse de Bruijn Graph. Arxiv Prepr: arXiv1106.2603. [Google Scholar]
- Ye C, Ma ZS, Cannon CH, Pop M, Yu DW.. 2012. Exploiting sparseness in de novo genome assembly. BMC Bioinformatics 13(S6):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D.. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328(5980):916–919. [DOI] [PubMed] [Google Scholar]
- Zentella R, Mascorro-Gallardo JO, Van Dijck P, Folch-Mallol J, Bonini B, Van Vaeck C, Gaxiola R, Covarrubias AA, Nieto-Sotelo J, Thevelein JM, et al. 1999. A Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tps1 mutant. Plant Physiol. 119(4):1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Liu J, Liu B, Li P, Meyers BC, Chen X, Cao X.. 2008. Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet. 4(4):e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18(6):292–298. [Google Scholar]
- Zhang J, Liu Y, Xia E-H, Yao Q-Y, Liu X-D, Gao L-Z.. 2015. Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc Natl Acad Sci U S A. 112(50):E7022–E7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen F, Zhang X, Li Z, Zhao Y, Lohaus R, Chang X, Dong W, Ho SYW, Liu X, et al. 2020. The water lily genome and the early evolution of flowering plants. Nature 577(7788):79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DQ, Li TT, Hao ZJ, Cheng ML, Tao J.. 2019. Exogenous trehalose confers high temperature stress tolerance to herbaceous peony by enhancing antioxidant systems, activating photosynthesis, and protecting cell structure. Cell Stress Chaperones 24(1):247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang B, Lisch D, Ma J.. 2017. Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants. Plant Cell 29(12):2974–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Xun Q, Zhang D, Lv M, Ou Y, Li J.. 2019. TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate thermomorphogenesis in Arabidopsis thaliana. iScience 15:600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A, Li Z, Lohaus R, Peer Y, Van D.. 2019. Finding evidence for whole genome duplications: a reappraisal. Mol Plant 12(2):133–136. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y.. 2019. Inference of ancient whole-genome duplications and the evolution of gene duplication and loss rates. Mol Biol Evol. 36(7):1384–1404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.