Abstract
Understanding how organisms adapt to extreme environments is fundamental and can provide insightful case studies for both evolutionary biology and climate-change biology. Here, we take advantage of the vast diversity of lifestyles in ants to identify genomic signatures of adaptation to extreme habitats such as high altitude. We hypothesized two parallel patterns would occur in a genome adapting to an extreme habitat: 1) strong positive selection on genes related to adaptation and 2) a relaxation of previous purifying selection. We tested this hypothesis by sequencing the high-elevation specialist Tetramorium alpestre and four other phylogenetically related species. In support of our hypothesis, we recorded a strong shift of selective forces in T. alpestre, in particular a stronger magnitude of diversifying and relaxed selection when compared with all other ants. We further disentangled candidate molecular adaptations in both gene expression and protein-coding sequence that were identified by our genome-wide analyses. In particular, we demonstrate that T. alpestre has 1) a higher level of expression for stv and other heat-shock proteins in chill-shock tests and 2) enzymatic enhancement of Hex-T1, a rate-limiting regulatory enzyme that controls the entry of glucose into the glycolytic pathway. Together, our analyses highlight the adaptive molecular changes that support colonization of high-altitude environments.
Keywords: genomics, adaptation, molecular evolution, cold adaptation; relaxation
Introduction
Adaptation of organisms to climate drives variation in diversification rates and species richness among clades. Their climatic niche affects a species’ distribution in space and time (Soberón 2007), critically affecting both speciation and extinction. Therefore, understanding how organisms manage thermal adaptation is important in terms of both evolutionary biology and climate-change biology, considering the potential world-wide loss of ecological niches (Lamprecht et al. 2018; Rogora et al. 2018). In this context, high elevations, characterized by a short growing season and low annual minimum and mean temperatures, with high daily fluctuation temperatures (Körner et al. 2011), are an important open-air laboratory to study speciation and adaptation to cold habitats; and compared with the great effort in gene-based studies on cold tolerance of model organisms (Clark and Worland 2008), a lesser effort has been put toward understanding truly cold-tolerant animals at the genomic level (Clark and Worland 2008; Parker et al. 2018). This is even more evident when considering that among the ∼600 sequenced insect genomes available today only four belong to species ecologically restricted to high altitudes or Antarctic habitats (Keeling et al. 2013; Kelley et al. 2014; Macdonald et al. 2016; Cicconardi, Di Marino, et al. 2017).
As yet, potential patterns of genomic signatures in arthropods adapting to more extreme habitats, such as high elevations, have been sparse and, more generally, there is no theory that predicts the rates of genomic change for extreme habitats. Here, we hypothesize two parallel patterns to occur in a genome adapting to an extreme habitat. 1) Strong positive selection on genes related to adaptation, such as genes involved in metabolic pathways, and 2) relaxation of previous selecting forces, as the conditions of the previous niche are lacking in the new niche. The latter should lead to a reduced number and/or a different set of genes under purifying selection. Specifically, in the case of high-elevation habitats, heat-shock proteins (HSPs) related to extreme heat response could be under relaxed selection. A problem with the hypothesis of these two parallel patterns is that there is no direct test available for the functional consequences of the loss or gain of a specific gene. There are often many pleiotropic effects for genes that make interpretations difficult, and HSPs, expressed upon exposure to stress or during development and growth, are likely to fall into this category (King and MacRae 2015). Therefore, we expect strong positive selection in genes involved in energetic metabolism, and an increase in the relaxation rates in other genes, without being able to specify which these might be. Identifying such genes will help to set up hypotheses to test in the future. A comparative approach offers the strongest method for testing our hypothesis by comparing genomes of species closely related to each other but divergent in terms of adaptation to different environments. However, this approach can be limited by insufficient niche divergence within a group or the number of genomes sequenced. Ants are emerging as a leading system for comparative genomics due to the constantly increasing number of available genomes. This resource offers the opportunity to improve the accuracy of orthology detection (Nygaard et al. 2016), to scan for specific mutations, candidate genes, and patterns of acceleration and relaxation in the ant genomes associated with adaptation to cold habitats.
Ants are key species in the Earth’s terrestrial ecosystems. They comprise more than 15,442 described species and display an impressive diversity of lifestyles (Hölldobler and Wilson 1990; Bolton 2018; Seifert 2018). Ants are especially notable among insects for their ecological dominance as predators, scavengers, and indirect herbivores. They compose at least one-third of the entire insect biomass (Wilson 1990) and colonize all kinds of habitats, including thermobiologically challenging environments. On the one hand, he formicine ant Melophorus bagoti, for instance, is active during the hottest periods of the summer day, when air temperatures at ant height exceed 50 °C (Christian and Morton 1992). On the other hand, for example, the myrmicine ant Tetramorium alpestre inhabits the montane and subalpine belt of the Central and South European mountain systems, with the Alps as its main distribution area (Steiner et al. 2010; Wagner et al. 2017). This species lives mainly between 1,300 and 2,300 m above sea level (a.s.l.), forages below the ground, and nests are established in cool grassland under stones, in moss, rootage, and dead wood, especially subalpine and alpine grass mats (Seifert 2018). Tetramorium alpestre is thus a system well suited for studying both genomic adaptation to high elevation and sociobehavioral evolution.
Here, we tested our hypothesis of both increased diversifying and reduced purifying (relaxing) selection resulting from adaptation to an extreme niche using T. alpestre. We newly sequenced its genome and those of four related Tetramorium species with diverging ecological niches, T. immigrans, T. parvispinum, T. bicarinatum, and T. simillimum. Tetramorium immigrans has a palearctic distribution (Europe, Anatolia, Caucasus, both Americas) with a broad ecological occurrence and is an invasive species in the Americas (Steiner et al. 2008). Compared with T. alpestre, it occurs at significantly lower altitude (mean elevation occurrence at 285 vs. 1,856 m a.s.l.) (Wagner et al. 2018). They are not sister species but belong to the Tetramorium caespitum complex, which include at least 11 cryptic species. The complex comprises two clusters, and the two species are the members of those two, hat is, they are not directly related (Wagner et al. 2017, 2018). Tetramorium immigrans should withstand lower temperatures compared with many other ant species and therefore can be considered as a tolerant species and not a specialist to cold habitats as T. alpestre. Tetramorium parvispinum is restricted to mountain forest habitats of the Austral-Asian and Indo-Malayan subregions (Liu et al. 2015), and T. bicarinatum and T. simillimum are as well generalist species, which occur in warmer habitats with the potential to become invasive (Bertelsmeier et al. 2017; Guénard et al. 2017). Our goals were to define the ecological niche of these ant species based on environmental data and subsequently to perform gene family expansion/contraction and protein-coding scans for signatures of diversifying and relaxing selection. We also annotated the five HSP subfamilies for 19 ant species to test for possible shifts in selection acting on these gene families, and we performed two experiments in T. alpestre and its relative, T. immigrans: We assessed 1) chill-shock triggered gene-expression patterns of starvin (stv), a modulator of the activity of the Hsp70 chaperone machinery during recovery from cold stress, and certain HSPs involved in recovering from extreme cold and 2) the temperature dependence of the enzyme activity of Hexokinase type 1 (Hex-T1), a rate-limiting and regulatory enzyme that controls the entry of glucose into the glycolytic pathway, one of the most conserved and essential hexokinase isoenzymes (Jayakumar et al. 2007). The genomic data presented here contribute to a foundation for studying insect genome evolution, particularly also in the light of climate change.
Results
Climatic Niches
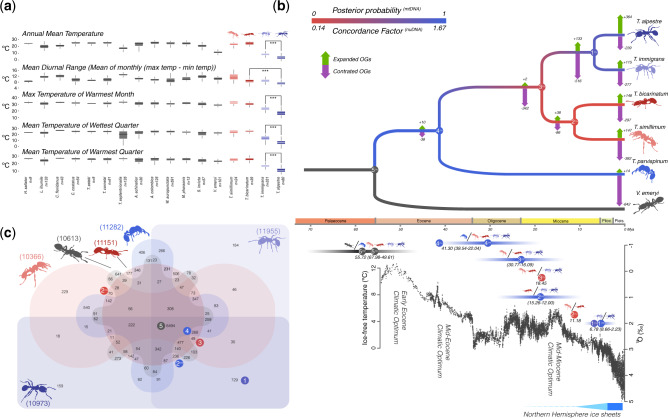
To define the environment of T. alpestre and the other four Tetramorium species, a data set containing 1,835 localities and representing 17 species was assembled, using georeferenced occurrence data compiled from the Global Ant Biodiversity Informatics database (see Materials and Methods section), with the number of localities per species ranging from nine to 591 (supplementary table S1, Supplementary Material online). Of the 19 bioclimatic variables, four (bio1: Annual Mean Temperature, bio5: Max Temperature of Warmest Month, bio8: Mean Temperature of Wettest Quarter, and bio10: Mean Temperature of Warmest Quarter) differentiated between T. alpestre and the other ants (adjusted P values < 0.001) (fig. 1a). In detail, T. immigrans and T. alpestre occupy habitats that are colder in the growing season than those of the remaining species, with T. alpestre revealing even colder habitats compared with T. immigrans.
Fig. 1.
(a) Boxplots of the five bioclimatic variables significantly different for Tetramorium alpestre and 16 other ant species. (b) Dated Crematogastrini nuclear phylogeny (nuDNA) (above). Branch colors are based on the concordance factors of the nuDNA and the BI of the mtDNA. On each branch, the numbers of expanded (green arrows) or contracted (purple arrows) OGs are shown. Below the phylogeny, the global δ18O (‰), derived from analyses of two common and long-lived benthic taxa are given, Cibicidoides and Nuttallides, which reflect the global deep-sea oxygen and carbon isotope and thus the temperature (from Zachos et al. [2001]). Bars represent the 95% confidence interval (CI) of the BI analysis. The circles on top of the CI represent the timing of the phylogenetic splits for the nuDNA (Nnu) and mtDNA (Nmt) phylogenies, whereas the numbers within the circles represent the nodes in the phylogenetic tree (above). The same splits (i.e., nodes) are also represented by the ant diagrams and indicated bya /. (c) Venn diagram displaying overlap in orthologous genes in the five Tetramorium spp. + Vollenhovia emeryi (Crematogastrini). Numbers close to the species diagrams represent the total number of orthologous groups. Colored circles with numbers represent phylogenetic splits in the nuDNA (Nnu) and mitochondrial phylogenies in (b).
Sequencing Data and Phylogenetic Analyses
All five de novo Tetramorium draft assemblies were ∼240 Mb long, comparable in size with the average of all ant genomes available (∼278 Mb). The total T. alpestre genome size was 245.72 Mb, slightly smaller than the estimated genome size as estimated by flow cytometry of 291.84 ± 1.76 Mb (n = 12). The genome fractions of the repetitive elements were similar across all five species (see supplementary tables S1–S3 and figs. S1–S6, Supplementary Material online). The combinatorial approach of unsupervised RNA-seq-based, homology-based, ab initio, and de novo gene annotations gave a number of genes comparable with those in the phylogenetically closest annotated species (see supplementary tables S1 and S3 and figs. S7 and S8, Supplementary Material online). The orthology-search analysis produced a total of 7,195 orthologous groups (OGs) between Drosophila melanogaster and Hymenoptera; 3,261 of these represented single-copy orthologous groups (scOGs) present in all 22 species analyzed. Restricting the orthology analysis to Tetramorium spp. and Vollenhovia emeryi resulted in 6,494 OGs (fig. 1c and Supplementary Material online). The estimated divergence ages of the most recent common ancestors between T. alpestre and T. immigrans was dated at 6.8 Ma (median: 5.15 Ma; 95% Highest Posterior Density: 2.66, 8.23). The phylogenetic analyses gave generally strongly supported and highly congruent topologies (see fig. 1b and supplementary figs. S11–S14, Supplementary Material online). The relation of T. bicarinatum and T. simillimum was uncertain: Our nuDNA phylogeny recovered them as sister species with low Coalescent Units length (0.11), whereas our mitochondrial DNA (mtDNA) phylogeny placed T. bicarinatum as sister to the T. alpestre–T. immigrans cluster (Supplementary Material online). The evolutionary dynamics of OGs had a more dynamic range corresponding with terminal branches; T. alpestre showed the highest number of expanded OGs (+364) (fig. 1b). Among others, there seems to be an expansion of hemolymph juvenile hormone-binding proteins, zinc fingers, and sugar transporters (see Supplementary Material online).
Dual Signature of Evolutionary Pressures in Single-Copy OGs in the T. alpestre Genome
We investigated the impact of evolutionary pressures on one-to-one orthology in the five Tetramorium spp. and 14 other ant species and three outgroups (Apis mellifera, Nasonia vitripennis, and D. melanogaster) by computing both mean ω (dN/dS) for terminal branches and relaxing selection (k) within the Crematogastrini.
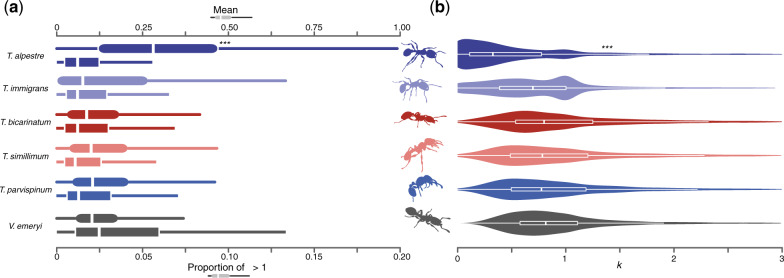
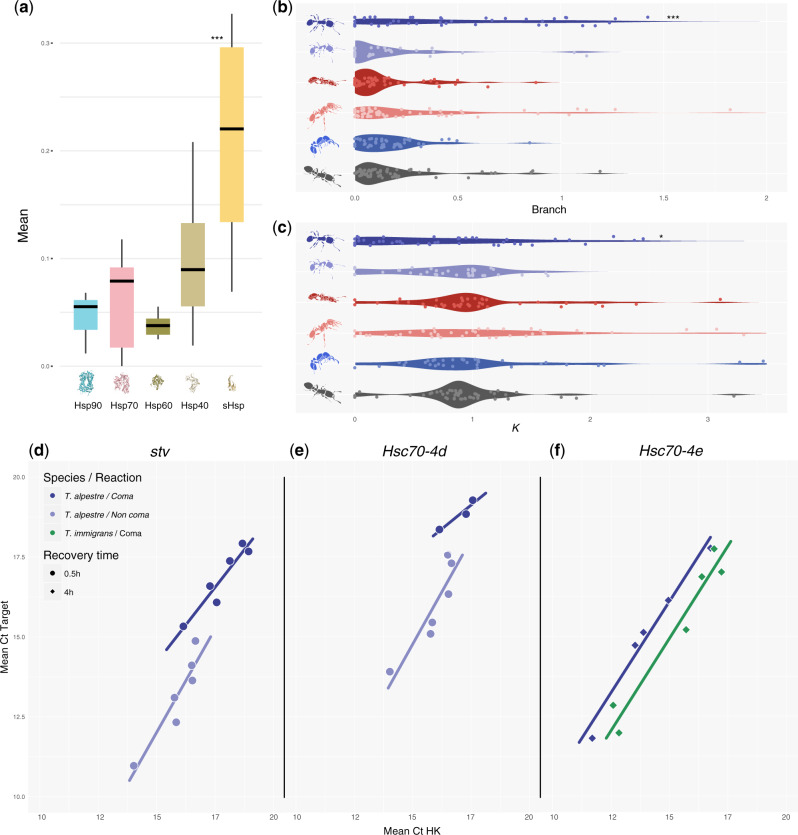
The measure of the mean-branch ω showed an overall high level of purifying selection along the whole ant phylogeny, including for short branches (Roux et al. 2014; Cicconardi, Marcatili, et al. 2017), with median values of 0.10. However, this distribution was significantly shifted in T. alpestre, which showed a median value of 0.38 (Wilcoxon rank-sum test “greater,” P value < 2.2e−16), while keeping the proportion of ω > 1 in each branch unchanged (fig. 2a and supplementary fig. S13, Supplementary Material online). The evaluation of k in the five Tetramorium spp. and V. emeryi had a mean of median values of 0.99, that is, twice the value in T. alpestre (median k = 0.47; Wilcoxon rank-sum test “less,” P value < 2.2e−16) (fig. 2b). Two evolutionary trajectories appeared to be acting simultaneously: One trajectory seems to have led to episodic diversifying selection, promoting the fixation of nonsynonymous mutations with presumably advantageous fitness effects for genes with increased values of ω and k, also present in the other species. The other trajectory showed a relaxation of the overall purifying selection in genes with increased values of ω and decreased values of k, only mildly present in T. immigrans.
Fig. 2.
(a) Boxplots showing the distribution and median (vertical line) of mean ω (dN/dS) rates (square shapes) in terminal branches of Tetramorium spp. and Vollenhovia emeryi (Crematogastrini) in scOGs, and the proportion of genes for which ω is higher than one. (b) Boxplots overlapped by violin plots showing the distribution of k in Crematogastrini. The distribution in Tetramorium alpestre is bimodal as in Tetramorium immigrans, but with a much more skewed distribution toward 0 values. In both sections, asterisks indicate the degree of significance between T. alpestre and the other species (Wilcoxon rank-sum tests).
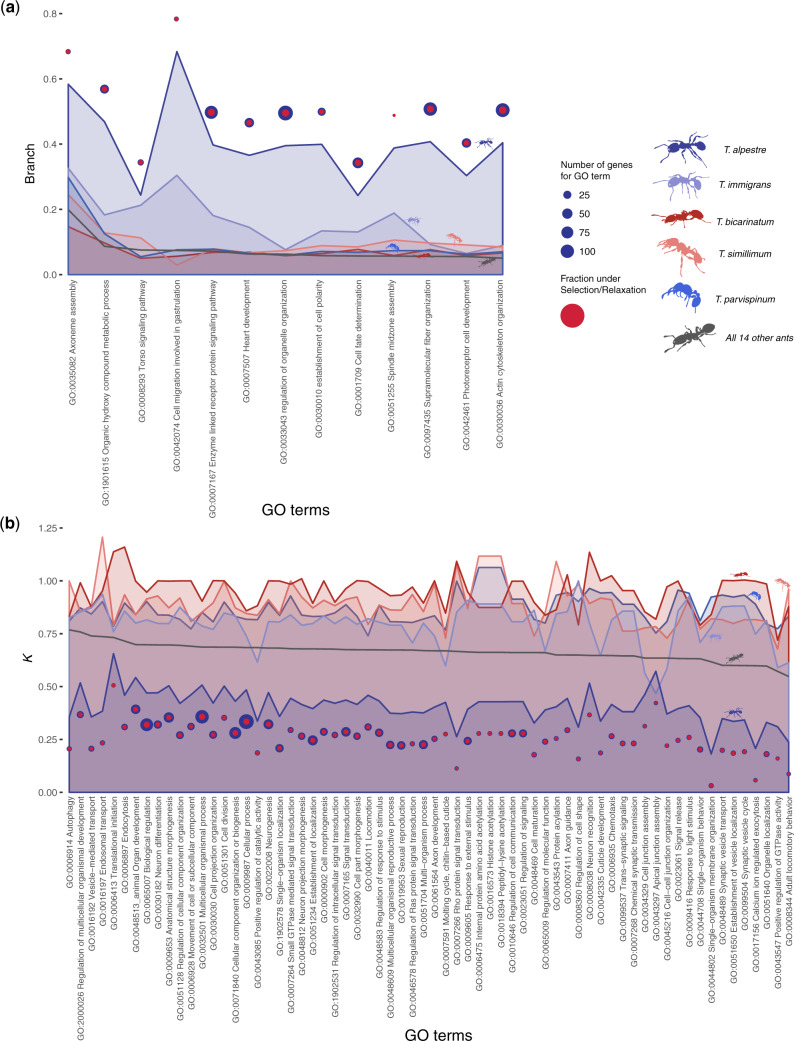
After a stringent correction for multiple testing, we found 175 scOGs with a putative signature of diversifying selection (adjusted P values < 0.005; supplementary table S5, Supplementary Material online), enriching 13 biological process terms (P values < 0.005; fig. 3a and supplementary table S6, Supplementary Material online), nine related to cell development (e.g., cell migration involved in gastrulation, photoreceptor cell development, and cell fate determination) and organization (e.g., regulation of organelle organization), two related to signaling pathways (enzyme-linked receptor protein signaling pathway and torso signaling pathway), one to heart development, and one to organic hydroxy-compound metabolic processes. The most representative genes were Pten and Gbeta13F, involved in six biological processes, DCTN1-p150 and dsh, involved in five of them; all of them involved in the proliferation and division of cells, especially neurological stem cells (Gbeta13F), respiratory system development (Pten and dsh), and the serine/threonine-protein kinase polo, present in four enriched Gene Ontology (GO) terms. In the catalytic domain of this protein (255 aa), we found five amino acid substitutions compared with T. immigrans, three of them corresponding to active sites (TalpPoloG29V, TalpPoloG30R, and TalpPoloS104R). We also tested for possible enrichment in KEGG pathways and found three significant pathways (P values < 0.02; supplementary table S7, Supplementary Material online), all involved in the metabolism of sugars, specifically: glycolysis, pentose phosphate pathway, and galactose metabolism. Within these pathways, five enzymes were found under diversifying selection: a glucose-6-phosphate (CG9008), the hexokinase type 1 (Hex-T1), a phosphofructokinase (Pfk), a transketolase (CG8036), and a phosphoglucosemutase (pgm).
Fig. 3.
For each enriched GO term category, (a) the median values of ω and (b) k for genes enriching the specific category. Tetramorium spp. are color coded, the other 14 species, merged to obtain a background signal, are in gray. Blue circles are scOGs tested for each GO term, and red circles are scOGs returned as significant. Circle sizes are proportional to the number of genes. To be noted are the overall extremely higher values of ω and lower values of k in Tetramorium alpestre branches compared with those of the other species. Seldomly, also Tetramorium immigrans shows a few GO terms with high ω and low k values in particular, the Apical junction assembly (GO:0043297) shows an inverted trend between T. alpestre and T. immigrans.
One hundred and thirty-two scOGs had a signature of putative relaxation (P values < 0.005; supplementary table S8, Supplementary Material online). Although the absolute number of scOGs under relaxation was slightly lower than the number of genes under diversifying selection, the gene-set enrichment tests revealed many more biological processes significantly enriched: 70 terms (P values < 0.005; fig. 3b and supplementary table S9, Supplementary Material online) enriched by 118 genes. Fifteen terms were related to the regulation of cellular development, signal transduction, and cell communication (e.g., regulation of response to stimuli, regulation of cell communication, and regulation of Ras signal transduction), 12 terms related to neurogenesis, axon development, and synaptic transmission (e.g., neuron differentiation, trans-synaptic signaling, and axon guidance), seven terms were related to cell morphogenesis and junction organization, and many other related to response to stimuli (e.g., chemotaxis, response to light) or related to adult development and morphogenesis (e.g., anatomical structure morphogenesis, molting cycle, and chitin-based cuticle). The enrichment of KEGG pathways revealed three pathways: mRNA surveillance pathway, spliceosome, and protein processing in endoplasmic reticulum (P values < 0.05; supplementary table S10, Supplementary Material online).
The Hex-T1 Activity, Its Model Structure, and Molecular Dynamics Simulations in T. alpestre and T. immigrans
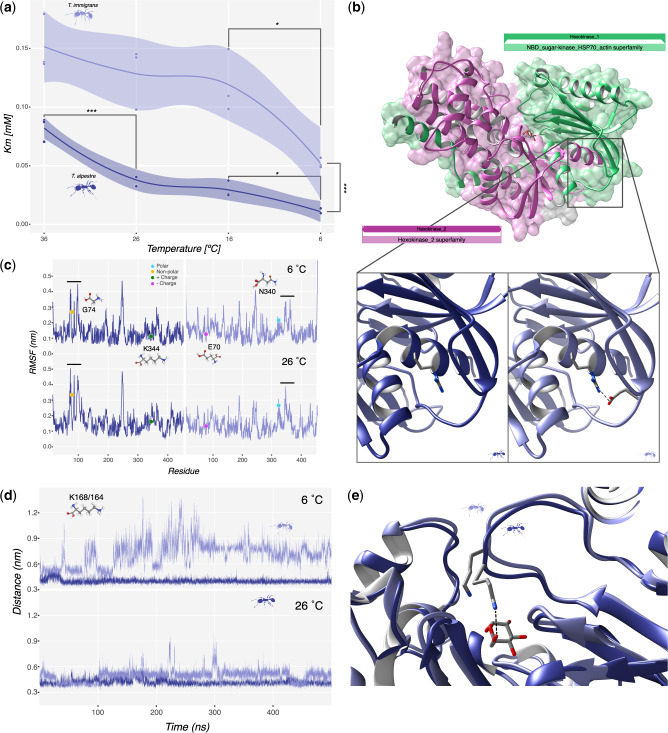
As we describe in the previous section, the enrichment for KEGG pathways led to the identification of three metabolic pathways, all involved in the metabolism of sugars, with five genes under selection. To validate experimentally these in-silico results, we cloned and expressed the Hex-T1 forms for both T. alpestre and T. immigrans into D. melanogaster cell lines for comparing their activities (the aim was to include additional genes, but for these, the process of establishing failed, at different stages of the process; for details, see Synthesis of Hex-t1 Gene and Its Enzyme Activity Assays section). The purified enzyme of both ants was assayed at 6, 16, 26, and 36 °C to establish their efficiency under different thermic conditions and identify possible metabolic adaptations to cold environments. The assays showed that the Km (substrate concentration necessary to reach maximum reaction speed) of Hex-T1 of T. alpestre was lower than the enzyme of T. immigrans at all temperatures (Wilcoxon rank-sum test “less,” P value < 0.00004, fig. 4a), with values ranging from 0.08 to 0.01 mM in T. alpestre versus 0.15 to 0.05 mM in T. immigrans, from the highest to the lowest temperature. These results demonstrate that Hex-T1 of T. alpestre was always more efficient than T. immigrans, requiring less substrate to reach maximum activity. Although the number of replicates per condition and species was small (n = 3), we performed statistical tests to attempt obtaining insight in the enzymatic activity differences within species at different temperatures and in their variance. Interestingly, it seems that for both species there is an improvement in the Km values shifting toward the lowest temperatures (6 °C vs. 16 °C; two-way ANOVA followed by a Tukey’s multiple comparison; adjusted P values < 0.05), and a possible deterioration at higher temperatures in T. alpestre, from 0.04 ± 0.01 mM (26 °C) to 0.08 ± 0.01 mM (36 °C) (two-way ANOVA followed by a Tukey’s multiple comparison; adjusted P value < 0.0002). An F test (var.test() implemented in R) to compare the variances of the enzymatic activities (Km values) between the two species—by pooling the replicates at 16 and 26 °C—suggested that there is a possible significant difference in their variance (P value = 0.015), with the confidence interval of Hex-T1 activity in T. immigrans twice wider than in T. alpestre.
Fig. 4.
(a) Scatter plot of Km score across the temperature gradient in the two species, Tetramorium immigrans on top and Tetramorium alpestre on the bottom. Lines represent average values with confidence intervals. Significances: ***<0.001, **<0.001, and *<0.05. (b) Overview of the homology-modeled structure of Hex-T1 in T. alpestre showing the two Hexokinase domains, pfam00349 (green) and pfam03727 (pink). The lower panels show the location of TalpHex-T1G74 (left) and TimmHex-T1E70 (right) as well as the arginine residue that forms a salt bridge with E70 (dashed line). (c) Per-Residue Root Mean Square Fluctuation calculated along 500 ns trajectories carried out at 6 °C (upper panel) and 26 °C (lower panel) for TalpHex-T1 (left panel, blue line) and TimmHex-T1 (right panel, light blue line). The location of the residues found mutated between the two sequences are indicated. (d) Time evolution of the atomic distance between the center of mass of glucose and the lateral chain nitrogen atom of TalpHex-T1K168 (blue line) and TimmHex-T1K164 (light blue line) in the binding pocket calculated along the 500 ns trajectories carried out at 6°NVTC (upper panel) and 26 °C (lower panel). (e) Representative configurations extracted from the simulations at 26 °C showing the different orientation of TalpHex-T1K168 and TimmHex-T1K164, with the first one being the only one able to bind the glucose (dashed line).
The Hex-T1 enzyme is made of two structurally similar domains, Hexokinase 1 and 2 (pfam00349 and pfam03727), and an N-terminus that is highly variable both in terms of length and amino acidic composition (fig. 4b). As can be discerned from the alignment, the two domains are relatively well conserved across all ants, other Hymenoptera, and even D. melanogaster. Excluding the hypervariable N-terminus, and considering the two functional domains, the amino acid sequences of T. alpestre and T. immigrans only differ by two amino acidic substitutions: a glutamic acid (E) replaced by a glycine (G) in position 74 (TalpHex-T1E74G) (fig. 4b inserts) and an asparagine (N) replaced by a lysine (K) in position 344 (TalpHex-T1N344K). Although the second change probably does not entail a significant functionality effect, the TalpHex-T1E74G instead may be significant. Because of the lack of the long chain and the negative charge of glutamic acid, this substitution could give the protein a higher flexibility and therefore an overall higher kinetics. We tested this hypothesis using the classical molecular dynamics (MD) simulation to understand the molecular mechanism responsible for the different enzymatic activities here described. In the last decades, the use of MD simulation was demonstrated to be a powerful and solid tool to study complex molecular mechanisms (such as the role of pathological mutations affecting the structure and dynamics of different enzymes and proteins’ interactions [Dror et al. 2015; Hollingsworth and Dror 2018]). We therefore chose to proceed with this strategy to simulate the two enzymes in the presence of the glucose substrate at different temperatures, that is, 6 and 26 °C, mirroring the condition in the in vitro assay. In the simulations, we can see a dual effect. For TalpHex-T1N344K, located at the C-terminal region of an α-helix forming a helix-turn-helix motif (VSETEKDPKG), the Lysine to Asparagine transition results in a lowered flexibility of the TalpHex-T1 enzyme, as revealed by the profile of the Root Mean Square Fluctuation (fig. 4c). This is possibly due to the alternation of positive and negative charges that creates a salt-bridges network that stabilizes the whole motif (data not shown). In particular, TalpHex-T1K344 is involved in a stable interaction with TalpHex-T1E341 (data not shown), whereas the corresponding TimmHex-T1N340 is not available for the formation of salt bridges and mainly establishes hydrogen bonds with TimmHex-T1S336 or TimmHex-T1E337 at 6 or 26 °C, respectively. Although the T. alpestre salt bridge brings a higher flexibility of the helix-turn-helix motive, it does not interfere with the catalytic site, given that this motif is far away in the C-terminal region of the protein. In contrast, at the level of the second region (TalpHex-T1E74G), the mutation-caused amino acid change allows the loop to explore a broader conformational space equipping it with a higher degree of flexibility already at 6 °C (fig. 4c). Nevertheless, by increasing the temperature, TalpHex-T1 was subjected to an even higher degree of fluctuation, once again confirming the intrinsic flexibility led by the glycine. Notably, the increased flexibility is also transferred to residues relatively far from the mutation, in particular in regions near the active site (168–169), causing a different pattern of interaction with the substrate (fig. 4d and e). Indeed, TalpHex-T1 established a stronger interaction with glucose at both temperatures with respect to TimmHex-T1 (fig. 4e).
The Evolution of HSPs in T. alpestre and Their Expression Pattern under Chill Shock
We investigated the adaptive value of critical stress proteins known as HSPs (Hsp90s, Hsp70s, Hsp60s, Hsp40s, and sHsps) to determine their contribution to possible mechanisms of adaptation to cold environments. To do this, we first annotated all the members of the five subfamilies in 19 ant species and searched for signals of diversifying and relaxed selection, recovering strong purifying selection across all the subfamilies (fig. 5a and supplementary table S11 and figs. S14–S18, Supplementary Material online). We computed the mean values of ω and k for all branches leading to Tetramorium spp. and V. emeryi, integrating 13 ant species, and scanned for diversifying and relaxed selection specifically in T. alpestre. The first part of the analysis showed a similar landscape as observed in scOGs. Among the five Tetramorium species and V. emeryi, five species displayed a distribution of the mean-branch ω ranging between 0.07 (T. immigrans) and 0.13 (T. parvispinum); T. alpestre instead showed a significant 2.7-fold higher value of the median mean-branch ω (ω = 0.36, Wilcoxon rank-sum test “greater,” adjusted P values < 0.004, fig. 5b). The distribution of k among HSPs in the six species showed median values of k all close to the expected (k = 1), between 0.90 in V. emeryi and 1.05 in T. simillimum, again with the exception of T. alpestre, where the median k = 0.63, 1.4-fold lower than the lowest median k (Wilcoxon rank-sum test “less,” adjusted P values < 0.05, fig. 5c). The scan for diversifying positive and relaxed selection in branches of the HSPs of T. alpestre showed no gene under putative diversifying positive selection, whereas six loci were identified to be under putative relaxing selection: one Hsp90 (Trap1), two Hsp60 (CCT4, CCT5), and three Hsp40s (Sec63, DnajC8, DnajC11) (k < 0.14, adjusted P values < 0.005). Using these genes in interaction prediction networks, we found that the 2 Hsp60s interacted with other 21 genes, giving 17 significant enriched functions (FDR < 0.05) mainly related to protein folding, microtubule, and spindle organization; the 3 Hsp40s interacted with 21 genes, giving 2 enriched functions (FDR < 0.05) related to response to heat and response to temperature stimulus. (supplementary fig. S19, Supplementary Material online).
Fig. 5.
(a) Boxplots showing the distribution and median (horizontal black line) of mean ω (dN/dS) rates for each of the HSP subfamily. (b) Violin plot showing the distribution of the mean ω values and (c) k in the terminal branches of the Crematogastrini (from up to bottom: Tetramorium alpestre, T. immigrans, T. bicarinatum, T. simillimum, T. parvispinum, and Vollenhovia emeryi). (d–f) Scatter plots of target genes (stv, Hsc70-4d, and Hsc70-4e) versus housekeeping gene (HK) concentrations; concentrations given as total number of cycles minus cycle threshold (Ct) values, that is, higher values represent higher concentrations. Straight lines are linear regressions for the target gene against HK using the different recovery times separately. Only data and regression lines shown for which significant differences (P adjusted < 0.05) were identified using one-way ANCOVA within and among species (four comparisons per recovery time and gene) followed by correction for multiple test corrections (see supplementary figs. S20–S22, Supplementary Material online, for all data and regression lines).
Among all the scOGs with a putative signature of diversifying selection (see previous sections), we also found starvin (stv), a Bcl-2-associated athanogene (BAG)-family member, which seems to be implicated in Hsp70 ATPase activity during chill-shock recovery in D. melanogaster (Colinet and Hoffmann 2010). This might be a clear indication that there could be selection toward a better recovery from cold and that it can be critical to the survival in harsh cold environments. Unfortunately, so far there are no data indicating that stv may be implicated in the same physiological mechanisms in Hymenoptera as in Diptera, also because Hsp70s orthologs are lacking in Hymenoptera. Therefore, we decided to perform pilot tests measuring the different expression patterns of stv, two paralogs of D. melanogaster Hsc70-4 (Hsc70-4d and Hsc70-4e), a member of the Hsp90 subfamily (Hsp83), and a sHsp (l(2)efl4) in the two species; to test 1) if the modulation of stv expression may play a role in Hymenoptera as well, by focusing on the adaptation of T. alpestre compared with the related species T. immigrans and 2) if along with stv the two Hsc70 paralogs possess the same modulatory mechanisms.
We measured the transcription activity of three recovery times (0.5, 2.0, and 4.0 h) and compared coma and noncoma individuals, within and between species, and found a significant difference in the expression patterns of stv, Hsc70-4d, Hsc70-4e, and Hsp83, but not of l(2)efl4 (one-way analysis of covariance [ANCOVA] followed by Bonferroni–Holm correction). In particular, looking at the different recovery times, stv and Hsc70-4d responded quickly in T. alpestre but not in T. immigrans, being significantly expressed only in workers of T. alpestre after 0.5 h recovery, but not in T. immigrans (fig. 5d and e). Comparing the expression levels between species, we found higher mRNA levels for Hsc70-4e in T. alpestre after 4 h recovery (fig. 5f).
Discussion
Tetramorium alpestre represents a critical example of evolutionary adaptation to cold environments such as alpine habitats (fig. 1a). By comparing its genome with those of 18 other ant species, we shed new light on how T. alpestre likely evolved to colonize subalpine habitats and propose it as a model for studies on arthropod adaptation to cold. We found significant concordance between the nuclear and the mitochondrial dated phylogenies concerning the dating of the speciation of T. alpestre, which seems to have diverged from its closest relative in these phylogenies, T. immigrans, between 2 and 9 Ma. This period overlaps with the beginning of a new climatic zonation of the European continent during the Middle and earliest Late Miocene (Zachos et al. 2001). At the end of this period, a rapid uplift (∼5 Ma) fundamentally changed the paleogeographic and topographic setting of central and southern Europe and triggered Alpine glaciation (Kuhlemann 2007). Both paleoclimatic and paleogeographic changes possibly co-occurred with the origin of the lineage leading to T. alpestre (fig. 1b). Although these glaciations dramatically reshaped global biodiversity patterns, eliminating terrestrial biota from many mid- to high-latitude areas, they may have been the main driver of diversification on highlands by reducing gene flow among populations (Wallis et al. 2016).
The present results support our prediction that two parallel patterns will occur in the genome of species adapting to extreme habitats. The genome of T. alpestre appears to be under the influence of two strong evolutionary trajectories: on the one hand, a diversifying selective force, 2-fold higher compared with all other ants, and on the other hand, a relaxation of the overall purifying selection present in other ants, identified by the skewed distribution of k toward 0. Specifically, the diversifying selective force affects more than 100 genes in many key biological processes, such as those related to development, cell migration, and gastrulation. Genes under the category of biological processes are also under selection in the genome of the Antarctic midge Belgica antarctica (Kelley et al. 2014). In fact, it has been shown in vertebrates that early stages are the most fragile ones, having the highest rate of lethal phenotypes during heat shock and UV irradiation, particularly around gastrulation (Uchida et al. 2018). The same may pertain to genes related to cellular-level organization (e.g., spindle midzone assembly, supramolecular fiber organization, and actin cytoskeleton organization), where environmental stress may jeopardize a well-working cell division machinery, as single-nucleotide polymorphisms associated to long-term cold-adaptation plasticity were found to be present in genes related to cytoskeletal and membrane structural components in D. melanogaster (Gerken et al. 2015). Among 25 genes involved in these biological processes, polo, a polo-like kinase that plays a central role as regulator of cell division and is required for several events of mitosis and cytokinesis (Archambault et al. 2015), shows three amino acid substitutions in positions corresponding to active binding sites and thus could be a good candidate for further studies.
The two evolutionary trajectories were predicted in our hypothesis, and although diversifying selection is somewhat expected in species adapting to a new environmental niche, the magnitude of relaxed selection found is surprising. Although both ω and k distributions in T. alpestre are highly skewed, the absolute number of genes under diversifying and relaxed selection is not markedly greater than in other species in similar studies (Harpur et al. 2014; Roux et al. 2014; Cicconardi, Marcatili, et al. 2017). This is unlikely to be due to the type I error rate because the same rate should be equally randomly present in all the other branches of the phylogeny. Nevertheless, T. alpestre, as part of a species complex (Steiner et al. 2010; Wagner et al. 2017), may have more recently diverged from more closely related species not yet adapted to the alpine habitat. The magnitude of this relaxed selection is evidenced by the 70 enriched biological processes and can be seen as the consequence of a shift and/or decreased magnitude in the purifying selection. The direct implication of this relaxation is not clear, especially its effect on genes and their biological processes, such as the regulation of cellular development, signal transduction, cell communication, neurogenesis, axon development, and many others. If the physiology of cold adaptation utilizes different metabolic and developmental strategies to minimize and optimize energy consumption, a new balance needs to be reached. The two patterns here observed (diversifying/relaxing selection) might represent this new shift. Interestingly, T. immigrans, which inhabits intermediate environmental conditions (fig. 1a), shows also an intermediate pattern of k, with an intriguing bimodal distribution (fig. 2b). Therefore, we can speculate that the highly skewed distributions in T. alpestre point to ongoing adaptation to a colder climate and a relaxation of selective forces present in warmer habitats. To our knowledge, this is the first time that this effect is found in an organism, most likely due to the underrepresentation of genomic studies on alpine and cold-adapted species, and, importantly, the lack of application of the RELAX test in a genome-wide manner.
Examining the effect of these evolutionary trajectories, energetic metabolism seems to be involved in the subalpine ant’s adaptation. Several clues may point in this direction, such as a more specialized diet underlined by the presence of commensal underground aphids in T. alpestre nests but not in those of T. immigrans (unpublished data), the expansion of OGs related to sugar transporters, and five enzymes under diversifying selection related to metabolism of sugars. We explored and validated one of these enzymes, Hex-T1, a key regulator and rate-limiting enzyme for energy (sugars) metabolism and reactive oxygen species activity in insects (Xian‐Wu and Wei‐Hua 2016), by contrasting its enzymatic activity between T. alpestre and T. immigrans. We showed that the mutated form in T. alpestre is indeed more efficient over all temperatures, at least in our in vitro assays, with less substrate needed to reach the highest enzymatic activity, which may be key to an organism living at high elevation, where daily temperature amplitudes at the microhabitat level can be larger than at lower elevation (Franz 1979). Also, the protein-modeling simulations demonstrated that the single amino acid mutation enhances the catalytic efficiency of the enzyme due to the formation of a more functional network of hydrogen bonds with its substrate. In fact, the path of the hydrogen bonds found in TalpHex-T1 resembled the one depicted by the X-ray solved structure of Hex-T1 from Mulichak et al. (1998), used as a template to obtain the three-dimensional coordinates of both TimmHex-T1 and TalpHex-T1 (see Materials and Methods). Mulichak et al. (1998) identified their model residues Lys621 and Asp657 as crucial for the catalytic activity of the protein; the corresponding residues TalpHex-T1K168 and TalpHex-T1D204 are indeed interacting, together with other residues, with glucose in TalpHex-T1 at both 6 and 26 °C. Although the interaction with TalpHex-T1D204 is conserved, the one with the lysine is lost in TimmHex-T1 and a looser protein-sugar network is formed, explaining the higher catalytic efficiency of TalpHex-T1 versus that of TimmHex-T1. We infer that the higher flexibility conferred by TalpHex-T1G74 increases the shift of the residue TalpHex-T1K168, belonging to the catalytic site (fig. 4d and e), allowing it to move closer and establish a stronger interaction with glucose, possibly contributing to lower the Km of the catalytic reaction. Although sugar intake is a basic metabolic process across animals, sugar-containing food is very relevant to ants and possibly in particular to T. alpestre. Ants and plant-sap-sucking insects have evolved mutualistic relationships in which one partner provides sugar-rich honeydew and the other in return offers protection from and adverse conditions. Usually, this interaction is above ground level, but there are cases in which ants are specialized to farm subterranean honeydew producers and rely on them as food source (Ivens et al. 2012). Field observations (Krapf P, Contala M-L, Steiner FM, Schlick-Steiner BC, unpublished data) indicate that this may also apply to T. alpestre. Thus, we hypothesize that the survival of T. alpestre may be improved considerably by sugar availability through subterranean aphid farming in an environment otherwise comparatively poor in food sources, and the relevance of this relationship might also be one of the reasons why this ant has to forage less time above ground and often behaves outstandingly peaceful when confronted with conspecifics from another nest (Krapf et al. 2018, 2019).
We make plausible that adaptation to cold environments can be related to changes in protein-coding sequences, but it has long been postulated that phenotypic divergence between closely related species, such as T. alpestre and T. immigrans, is primarily driven by quantitative and spatiotemporal changes in gene expression, mediated by alterations in regulatory elements (e.g., Prescott et al. 2015; Danko et al. 2018). We validated this for T. alpestre and T. immigrans by looking at the expression patterns of stv and four other HSPs (Hsc70-4d, Hsc70-4e, Hsp83, and l(2)efl4). Stv is a member of BAG-family proteins, which interact with Hsc70s and Hsp70s, and can modulate, either positively or negatively, the functions of these chaperones (Doong et al. 2002). It is implicated in the recovery from chill shock in D. melanogaster during cold recovery (Colinet and Hoffmann 2010, 2012). The concerted synthesis of D. melanogaster stv and Hsp70s suggests cooperation to offset cold injury, possibly by preserving the folding/degradation and by regulating apoptosis (Colinet and Hoffmann 2010). In our experiment, an acute cold stress (−6 °C) was used to knock down individuals. By looking at the differential expression of these genes in coma and noncoma individuals within and between species, we were not only able to show that stv may interact with Hsc70s but also that T. alpestre has a quicker response in these genes compared with T. immigrans, the related species not cold adapted. Given these results, which are in line with the selection signature we identified, stv may play a similar role in the physiology of ants. As we have shown in this study, ants completely lack Hsp70s (supplementary fig. S14, Supplementary Material online) and retained only the heat-shock cognates (Hsc70s) with mainly two conserved paralogs (Hsc70-4d and Hsc70-4e), which may have evolved to function as the dipteran Hsp70s. The correlation we found among chill shock, stv, and Hsc70s, is an interesting and promising starting point, but independent, hypothesis-free analysis using RNA sequencing will be important to evaluate how many genes under positive versus relaxed selection are involved in cold response. Likewise, it will be important to assess whether stv has tissue-specific expression patterns, as was found in D. melanogaster (Coulson et al. 2005). All these approaches should be applied to a wider range of species with different ecological adaptations.
Our comparative genomic analysis shows how natural selection could trigger complicated patterns of change in genomes, especially protein-coding sequences, both in terms of diversifying selection and relaxation of purifying selection. Many of these changes are in genes that may be associated with aspects of development, either directly or through the associated complex changes in ecology and natural history. Furthermore, this work characterized some candidate gene activities, both in vitro and in silico, which confirms our hypothesis and may become a starting point in identifying other genes possibly important in adaptation to new ecological niches. Overall, this study represents a systematic attempt to provide a framework for the genomic analysis of adaptation to extreme environments and underlines the importance of studying organisms with different ecological niches in understanding the genetic basis of ecological adaptation.
Materials and Methods
Species Locality and Climatic Data
To define the environment of T. alpestre and the other four Tetramorium species, georeferenced occurrence data were compiled and complemented with those for the native ranges of other ant species with available genomic data. Occurrence records for 17 species (supplementary table S1, Supplementary Material online) were compiled from the Global Ant Biodiversity Informatics database (Guénard et al., 2017, accessible via www.antmaps.org, last accessed April 6, 2020, Janicki et al., 2016). The species determinations in the records were evaluated by specialists in ant taxonomy, and erroneous data were excluded. Slightly incorrect coordinates (i.e., in the ocean within 8.5 km of land) were assigned to the nearest land point. Additionally, georeferenced occurrence data of two species (T. alpestre, T. immigrans) were compiled from the latest taxonomic revision (Wagner et al. 2017). The occurrence data of each species were spatially rarefied at a 5-km level using the R package spThin v. 0.1.0 (Aiello-Lammens et al. 2015), to counter spatial autocorrelation and to provide some protection against sampling bias issues (Merow et al. 2013). The Bioclim variables bio1-bio19 from the WorldClim database v. 1.4 (Hijmans et al. 2005) were extracted for each unique species locality at a 2.5-arc min resolution (4.65 × 4.65 = 21.6225 km2 at the equator) using ArcGIS v. 10.4 (ESRI, Redland, CA). A Wilcoxon–Mann–Whitney rank-sum test as implemented in the R function wilcox.test (http://www.r-project.org) v. 3 (R Development Core Team 2017) was adopted to test for differences between T. alpestre and the other species individually for each of the Bioclim variables. The Bonferroni–Holm sequential rejection procedure (Abdi 2010) was used to control for multiple testing.
Biological Sample Processing, Genome Size Estimation, and Genome Sequencing
We collected T. alpestre from six colonies from different geographical locations for flow cytometry analysis and DNA and RNA extractions (Supplementary Material online).
All T. alpestre DNA-seq and RNA-seq Illumina libraries were prepared by IGATech (http://igatechnology.com/, last accessed April 6, 2020). Four libraries were produced for the T. alpestre genome: one overlapping-reads library, one regular PE library, and two mate pair libraries (see supplementary table S2, Supplementary Material online). For RNA-seq, six strand-specific libraries were constructed from three stages of workers, two stages of gynes, and adult males (Supplementary Material online). Each of the other Tetramorium species was collected and identified by Franscisco Hita Garcia, and TruSeq libraries were prepared and sequenced on an Illumina platform (Supplementary Material online).
Genomic and Mitogenomic Assembly and Gene Prediction
Genomic sequencing reads of T. alpestre were filtered to remove low-quality reads, corrected, and assembled using Allpaths-LG v. 52488 (Gnerre et al. 2011). For the other species, Trimmomatic was also used to filter out reads, and SPAdes v. 3.10.0 (Bankevich et al. 2012) was used to run error correction and the genome assemblies (Supplementary Material online).
A bioinformatic pipeline was used that included combinatorial approaches: Homology-based, ab initio, and de novo methods were implemented to predict gene models for T. alpestre, using the BRAKER1 procedure (Hoff et al. 2016), AUGUSTUS v. 3.2.1. (Stanke et al. 2006), Trinity v. 2.2.0 (Grabherr et al. 2011), and other tools. The results were then evaluated using Benchmarking Universal Single Copy Orthologs (BUSCO v. 1.2) (Simão et al. 2015) (Supplementary Material online).
All available complete ant mitochondrial genomes (mtDNA) were downloaded from GenBank (Benson et al. 2015) (supplementary table S1, Supplementary Material online). Illumina raw SRA data of other ant species were downloaded and processed following Cicconardi, Di Marino, et al. (2017). In brief, reads were quality filtered and assembled using IDBA-UD v. 1.1.1 (Peng et al. 2012) and SPAdes v. 3.10.0 (Bankevich et al. 2012) and annotated using MITOS v. 2 web server (Bernt et al. 2013).
Phylogenetic Analysis
Nuclear scOGs and complete mitochondrial genomes were used to compute the ant species tree. Once scOGs were determined (see below), single-locus trees and a species tree of concatenated scOGs were estimated using maximum-likelihood (ML) search as implemented in FastTree v. 2.1.8 SSE3 (Price et al. 2010). Gene trees were summarized with MP-EST (Liu et al. 2010) using the STRAW web server (Shaw et al. 2013).
For the mtDNA phylogeny, protein-coding genes were aligned and concatenated. MtDNA trees were searched with ML and Bayesian Inference (BI) algorithms (see Supplementary Material online for more detailed description). BEAST v. 2 (Bouckaert et al. 2014) was adopted to date the mtDNA tree and to estimate the divergence of the most recent common ancestors (template: BEAST [Heled and Drummond 2010]). Fossil data were taken mainly from Moreau and Bell (2013) and PaleoBio DB (paleobiodb.org) (see supplementary table S4, Supplementary Material online, for detailed methods and the list of references). Divergence dates for the ML nuclear DNA (nuDNA) tree (only ants) were also inferred using the penalized likelihood approach (Sanderson 2002) implemented in the R package APE v. 5.1 (Paradis et al. 2004) by calibrating four nodes: the root, the Tetramorium, the Solenopsis + Monomorium, and the Attini branches (supplementary table S4, Supplementary Material online).
Functional Annotation and Orthologous-Group Dynamic Evolution
Genes from 19 ant and three outgroup species (supplementary table S3, Supplementary Material online) were clustered into OGs with Hieranoid v. 2 (Kaduk and Sonnhammer 2017), which implements InParanoid v. 8 (Sonnhammer and Östlund 2015) using a guide tree topology based on Moreau and Bell (2013) and BlastP (e-value cutoff of 10−5) to improve the reciprocal hit accuracy (Edgar 2010), whereas homology relationships were searched with deltaBLAST (Boratyn et al. 2012). The functional annotation of each putative protein-coding sequence was performed by identifying both the protein domain architecture and GO terms. For each sequence, first HMMER v. 3.1b2 (hmmscan) (Eddy 2011) was used to predict Pfam-A v. 31.0 domains, then Domain Annotation by a Multi-objective Approach v. 2 (Bernardes et al. 2016) was applied to identify architectures combining scores of domain matches, previously observed multidomain co-occurrence, and domain overlapping. Annotation of GO terms was performed as implemented in the CATH assignments for large sequence data sets (Das et al. 2015). Briefly, each input sequence was scanned against the library of CATH functional families (FunFams) HMMs (Sillitoe et al. 2015) using HMMER3 (Eddy 2011) to assign FunFams to regions on the query sequence (with conditional e-value < 0.005). Then, the GO annotations for a matching FunFam were transferred to the query sequence with its confidence scores, calculated by considering the GO term frequency among the annotated sequences. Finally, a nonredundant set of GO annotations was retained, making up the GO annotations for the query protein sequence (Sillitoe et al. 2015).
To identify OGs specifically expanded in the T. alpestre genome, BadiRate v. 1.35 (Librado et al. 2012) was performed twice, once for the nuDNA and one for the mtDNA ultrametric tree topologies, reconstructing ancestral family sizes, gain, death, and innovation, applying stochastic models and allowing estimation of the family turnover rate (λ) by ML. Using this approach, we observed a correlation between the turnover rate (λ) and branch lengths (Spearman correlation: ρ = −0.52, P value = 0), and a bimodal distribution of λ. We interpreted the distribution closer to 0 as background noise and therefore considered only expanded and contracted OGs with λ within the second distribution. The boundary between the two distributions was defined by the valley values between the two, which were computed using optimize and approxfun functions implemented in the stats v. 3.4.0 package in R.
HSP Family Analysis
From the functional annotation analyses of all Hymenoptera, all sequences bearing a valid protein domain matching HSPs were extracted and aligned using the ClustalW v.1.2.1 EBI web server (Larkin et al. 2007); a phylogenetic tree was computed using ML as implemented in FastTree v. 2.1.8 SSE3 (Price et al. 2010) (Supplementary Material online).
Selection on Single-Copy OGs and Gene Families
All selected scOGs and HSP-OGs were scanned to evaluate selection signature on coding regions of T. alpestre. This was done by computing the mean ω (the ratio of nonsynonymous to synonymous substitution rates; dN/dS) and the relaxation of each branch of the phylogeny of Tetramorium spp. + V. emeryi. More specifically, adopting a pipeline similar to that in Cicconardi, Marcatili, et al. (2017) and Cicconardi, Di Marino, et al. (2017), the signatures of diversifying selection were searched in codon-based aligning groups of one-to-one orthologous ant genes with MACSE v. 1.01b (Ranwez et al. 2011), filtering with Gblocks v. 0.91b (Castresana 2000) under a “relaxed” condition (Parker et al. 2013; Cicconardi, Di Marino, et al. 2017; Cicconardi, Marcatili, et al. 2017) and using the adaptive branch-site random effects likelihood (aBSREL) method (Kosakovsky Pond et al. 2011; Smith et al. 2015) as implemented in the HyPhy batch language (Kosakovsky Pond et al. 2005) using a batch script (BranchSiteREL) in HyPhy (http://github.com/veg/hyphy, last accessed April 6, 2020).
In parallel, the same scOGs and HSP-OGs were scanned using the RELAX test (Wertheim et al. 2014) to search for putative signals of relaxation. In brief, RELAX tests the hypothesis of evolutionary-rate relaxation in selected branches of a phylogenetic tree compared with reference branches. A k value is computed to evaluate whether the selective strength ω shifts toward neutrality. The rate of dN/dS (ω) can relax (k < 1), stay stable (k = 1), or intensify (k > 1). For both scOGs and HSP-OGs, and for both the aBSREL and RELAX tests, the Bonferroni–Holm sequential rejection procedure (Abdi 2010) was used to control the false discovery rate with a very stringent cutoff value of 0.005 for the adjusted P values applied (Benjamin et al. 2018). Hsp60s and Hsp40s under relaxed selection were analyzed using the GeneMANIA prediction server (Warde-Farley et al. 2010) to predict their functions by creating an interaction network with genes by including protein and genetic interactions, pathways, coexpression, colocalization, and protein domain similarity.
Finally, the algorithm hyperGTest implemented in the GOstats package for R (Falcon and Gentleman 2007) (settings: annotation org. Dm.eg.db; conditional TRUE; testDirection over) was used to check for GO terms of biological processes and KEGG pathway enrichments of scOGs under selection and relaxation using the whole set of tested scOGs as background. When conditional(p)==TRUE is selected, the test uses a conditional algorithm that uses the structure of the GO graph to estimate for each term whether or not there is evidence beyond that provided by the term’s children to call the term in question statistically overrepresented. The algorithm conditions on all child terms that are themselves significant at the specified P value cutoff. Given a subgraph of one of the three GO ontologies, the terms with no child categories are tested first. Next, the nodes whose children have already been tested are tested. If any of a given node’s children tested significant, the appropriate conditioning is performed. This method is similar to Alexa et al. (2006) which should reduce the false-positive rate, while not missing many true enriched nodes, possibly a better approach of multiple test correction. As a cutoff, we used P values < 0.005 and <0.05 for biological processes GO terms and KEGG pathways, respectively.
Synthesis of Hex-t1 Gene and Its Enzyme Activity Assays
Following identification as under diversifying selection in T. alpestre (see Dual Signature of Evolutionary Pressures in Single-Copy OGs in the T. alpestre Genome section), synthesis of genes and enzymes was attempted for phosphofructokinase, transketolase, phosphoglucosemutase, and Hex-t1, but it was successful only for the Hex-t1. In more detail, the Hex-t1 genes of T. alpestre (1,398 bp) and T. immigrans (1,386 bp) were synthesized by Thermo Fisher Life Technologies (Carlsbad, CA) and Eurofins Genomics (Ebersberg, Germany), respectively. Both constructs carried a 5′ EcoRI and a 3′ HindIII recognition site for subsequent cloning as well as a C-terminal His-tag. The genes were excised from their carrier backbones by enzymatic digestion. The fragments were purified by agarose gel excision and subcloned into the pACEBac1 expression vector (MultiBacTM, Geneva Biotech, Geneva, Switzerland). Correct insertion was verified by Sanger sequencing, and transformation into D. melanogaster cells and protein expression followed the standard protocols of the expression vector manufacturer. After cell harvesting, the presence of the recombinant protein in the cell lysates was verified by a western blot targeting the His-tag. The relative amounts of protein were quantified from the western blot using ImageJ v. 1.3 (https://imagej.net/Welcome, last accessed April 6, 2020). The cell lysates were centrifuged at 4,000 rpm at 4 °C for 5 min, and the supernatant was directly used as protein source.
Enzyme activity assays followed a modified protocol of Crabtree and Newsholme (1972). Briefly, 1,000 µl assay medium containing 75 mM Tris pH = 7.5 (Merck Millipore, Burlington, MA), 7.5 mM MgCl2 (Merck), 0.8 mM EDTA (Sigma-Aldrich, St Louis, MO), 1.5 mM KCl (Merck), 4.0 mM mercaptoethanol (Sigma), 0.4 mM NADP+ (Abcam, Cambridge, UK), 2.5 mM ATP (Abcam), and 10 mM creatine phosphate (Abcam) was placed in a disposable cuvette (Brand, Germany) in a SPECORD 210 PLUS UV/VIS spectrophotometer (Analytic Jena AG, Germany) with an attached 1157P programmable thermostate (VWR, Radnor, PA). Variable amounts of d-glucose (VWR) and synthetic protein were added, and the cuvette was allowed to reach the assay temperature. Then, the reaction was started by adding 15 U creatine phosphokinase (Sigma) and 5 U glucose 6-phospahte dehydrogenase (Sigma).
The Hex-t1 activity was measured in triplicate as the rate of reduction of NADP+ causing an extinction at 340 nm. Each measurement lasted for 60 s, and every second, an extinction value was recorded. For the determination of the Michaelis–Menten constant (Km value) of both enzymes, assays were conducted along a substrate gradient from 0.00167 to 24.30000 mM d-glucose, achieving saturated extinction curves. Assay temperatures ranged from 6 to 36 °C with 10 K increments. The software Prism v. 8 (GraphPad Software, San Diego, CA) was used to calculate the Km values.
Hex-t1 Structure Modeling and MD Simulations
The three-dimensional structures of T. alpestre and T. immigrans Hex-t1 proteins are not available. To study the effect of the amino acid changes on the proteins’ function/dynamics, we modeled the structures of these two proteins via homology modeling. The X-ray structure of the Hex-t1 isoform from Schistosoma mansoni (Mulichak et al. 1998), solved at 2.6 Å resolution, was used as a template and the models were generated using the software Modeller v. 9.21 (Webb and Sali 2014). Once we obtained the models, we built the simulative systems to run MD simulations, using well establish methodologies (D’Annessa et al. 2014, 2019; Di Marino et al. 2014, 2015). Specifically, the two proteins TalpHex-t1 and TimmHex-t1, in complex with the glucose substrate, were immersed in a triclinic box filled with TIP3P water molecules (Jorgensen et al. 1983) and rendered electroneutral by the addition of chloride counterions. The topology of the two systems, consisting of ∼80.000 atoms each, was built using the Amber14 force field (Case et al. 2014), further converted in Gromacs v. 4.6 (Hess et al. 2008) format using Acpype v. 0.1.1 (Sousa Da Silva and Vranken 2012). Simulations were run on a GPU cluster using Gromacs v. 4.6 with the following protocol: 1) 25,000 steps of steepest descent followed by 25,000 steps of conjugate gradient minimization; 2) 5 × 100 ps of equilibration runs in constant volume and temperature (NVT) environment starting at 50 K and performed by increasing the temperature of 50 K after each run until a final value of 250 K; 3) 5 × 100 ps of equilibration runs in constant pressure and temperature (NPT) environment starting at 50 K and performed by increasing the temperature of 50 K after each run until a final value of 250 K; and 4) the equilibrated systems were simulated for 500 ns at 279 or 300 K, that is, at 6 or 26 °C, for a total of four simulations and 2 µs of sampling. Electrostatic interactions were considered by means of the Particle Mesh Ewald method with a cutoff of 1.2nm for the real space and Van der Waals interactions (Darden et al. 1993). Bond lengths and angles were constrained via the LINCS algorithm (Hess et al. 1997). The temperatures were kept constant at 279 or 300K by using the velocity rescale with a coupling constant of 0.1ps, and the pressure was kept at 1bar using the Parrinello–Rahman barostat with a coupling constant of 1.0ps during sampling (Parrinello and Rahman 1981). The four trajectories were collected and comparative analyses performed with the Gromacs suite or with in-house written code (available on request).
Chill-Coma Assay and Quantitative qRT-PCR Assay
The cold hardiness and expression of selected genes were compared between T. alpestre and T. immigrans using chill-coma assays. From four colonies additional to those used for other experiments in this study, two per species, about 500 workers per colony were collected in August 2017; T. alpestre: Jaufenpass (18980: 46.83791°N, 11.29768°E) and Penser Joch (18978: 46.81402°N, 11.44198°E); T. immigrans: Vienna (18977: 48.12448°N, 16.43522°E.; 18983: 48.31341°N, 16.42529°E). Workers were collected alive using aspirators, transported to the laboratory in Innsbruck, and kept in polypropylene boxes (18.5 cm × 11 cm) with chambers of various sizes. The walls of the boxes were Fluon-coated (GP1, De Monchy International BV, Rotterdam, the Netherlands) to prevent workers from escaping. Food (sugar–honey–water and deep-frozen Drosophila flies) and tap water were provided ad libitum three times a week. Workers were kept in a climate chamber (MIR-254, Panasonic, Etten Leur, the Netherlands) at a 12L:12D photoperiod at constant 18 °C. The temperature of 18 °C was the average of the monthly mean temperatures experienced at 200 m a.s.l. (T. immigrans, ∼23.7 °C) and at 2,000 m a.s.l. (T. alpestre, ∼12.3 °C) in July 2017. Workers were acclimatized to 18 °C for at least 3 weeks before the chill-coma assays.
After the acclimation period, from each of the four colonies, 90 workers were randomly chosen and divided into 18 equivalent pools of five workers, each of which were used for a single replicate of either the chill-coma or the control assays. For the chill-coma assay, worker pools were transferred into empty 5-ml glass vials, sealed, and immersed at 2300 h Central European Summer Time (CEST) for 6.5 h in a water:ethane-1,2-diol mix (1:1) bath set at −6 °C; in pilot experiments, this temperature had been determined as the highest temperature at which the ants had fallen into chill coma after 6.5 h exposition; the temperature was identical for both species. Temperature was monitored using an electronic thermometer (TFX 430, ebro Electronic GmbH; Ingolstadt, Germany) with an accuracy of 0.05 °C inserted into an additional, empty vial treated in the same way as the vials with ants. After chill coma, at 0530 h CEST, the vials were transferred to a climate chamber at 18 °C, and the workers were allowed to recover for 0.5, 2.0, and 4.0 h, terminated at 0600, 0730, and 0930 h CEST, respectively. For each recovery time, three replicate pools were used, and each chill-coma treatment and its corresponding control hence were replicated 6-fold per species. After recovery, the workers were instantly transferred to RNAse-free reaction tubes and killed in liquid nitrogen. Tubes were stored at −70 °C. For the control assay, worker pools were transferred to empty glass vials and placed in the climate chamber at 18 °C. After 6.5 h plus respective recovery time, the workers were killed and stored as described above.
For each recovery time and replicate, complete worker pools were used for the molecular analyses. RNA was extracted using the NucleoSpin RNA Kit (Macherey-Nagel, Düren, Germany) following the instructions of the manufacturer. First-strand cDNA was synthesized using 200 U RevertAid reverse transcriptase (all reagents by Thermo Fisher Scientific, Waltham), 40 U RiboLock RNase inhibitor, 5 µM random hexamer primers, 100 µM dNTPs, and 2 µl RNA extract in a total volume of 40 µl. The mixture was incubated for 5 min at 25 °C, 60 min at 42 °C, and 5 min at 70 °C on a UnoCycler 1200 (VWR). Quantitative PCR was conducted on a Rotorgene Q (Qiagen) PCR system. Each reaction contained 1× Rotor-Gene SYBR Green PCR Mastermix (Qiagen), 0.2 μM of primers, specifically designed for target genes (supplementary table S12, Supplementary Material online), and 1 µl cDNA in a total volume of 10 µl. All qRT-PCR reactions were performed as triplicates. RpS20 was used as housekeeping gene as it had been established and used as housekeeping gene for comparable chill-coma assays in Drosophila (Colinet et al. 2010, 2013). Cycling conditions were 95 °C for 5 min, followed by 40 cycles of 94 °C for 15 s, 58 °C for 10 s, and 72 °C for 15 s. Fluorescence was acquired at the end of each elongation step. PCR was followed by a melting curve analysis from 60 to 95 °C with 0.1 °C increments held for 5 s before fluorescence acquisition. Cycle threshold (Ct) values were subtracted from total number of cycles. On these values, linear-regression analyses using the software PAST v. 3.18 (Hammer et al. 2001) were based separately for each target gene, treatment, recovery time, and species (in each regression analysis, two populations and three replicate values were used, i.e., n = 6) using the housekeeping gene as independent variable and the target gene as dependent variable. For each target gene/recovery-time combination, one-way ANCOVA implemented in PAST followed by Bonferroni–Holm correction was then used to identify significant differences between treatments within species and between species within treatment. It is acknowledged that circadian rhythms in metabolic activities (Bloch et al. 2013) may be reflected in the data, in that the three different recovery times were initiated synchronously but terminated asynchronously. However, any such influence was controlled for as far as possible in the frame of this project in that each treatment with its corresponding control was synchronized as were the two species.
Supplementary Material
Acknowledgments
We thank Stefan Gross for valuable work in the laboratory and two anonymous reviewers for the criticism of an earlier version of the manuscript. Research was supported by the Austrian Science Fund (FWF, P23409 and P30861). The computational results presented here have been achieved in part by using the HPC infrastructure of the University of Innsbruck (LEO), the Vienna Scientific Cluster (VSC), the collaborative system of the Universities Innsbruck and Linz (MACH), and the Swiss National Supercomputing Centre (CSCS) hosted in Lugano. Finally, F.C. thanks his mother, who recently died, for her unconditional love and support.
The sequence data from this study have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject numbers PRJNA532334, PRJNA533534, PRJNA533535, PRJNA533536, and PRJNA533537 (https://www.ncbi.nlm.nih.gov/bioproject/). The mtDNA genome assemblies have been submitted to the NCBI (GenBank accessions: MK861047–MK861070). All accession codes of deposited and retrieved data are provided in supplementary table S1, Supplementary Material online.
References
- Abdi H. 2010. Holm’s sequential Bonferroni procedure. Encyclopedia of research design 1(8):1–8. [Google Scholar]
- Aiello-Lammens ME, Boria RA, Radosavljevic A, Vilela B, Anderson RP.. 2015. spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38(5):541–545. [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T.. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22(13):1600–1607. [DOI] [PubMed] [Google Scholar]
- Archambault V, Lépine G, Kachaner D.. 2015. Understanding the Polo Kinase machine. Oncogene 34(37):4799–4807. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DJ, Berger JO, Johannesson M, Nosek BA, Wagenmakers E-J, Berk R, Bollen KA, Brembs B, Brown L, Camerer C, et al. 2018. Redefine statistical significance. Nat Hum Behav. 2(1):6–10. [DOI] [PubMed] [Google Scholar]
- Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW.. 2015. GenBank. Nucleic Acids Res. 43(D1):D30–D35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes JS, Vieira FRJ, Zaverucha G, Carbone A.. 2016. A multi-objective optimization approach accurately resolves protein domain architectures. Bioinformatics 32(3):345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Bertelsmeier C, Ollier S, Liebhold A, Keller L.. 2017. Recent human history governs global ant invasion dynamics. Nat Ecol Evol. 1:0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch G, Hazan E, Rafaeli A.. 2013. Circadian rhythms and endocrine functions in adult insects. J Insect Physiol. 59(1):56–69. [DOI] [PubMed] [Google Scholar]
- Bolton B. 2018. An online catalog of the ants of the world. Available from http://antcat.org.
- Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL.. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct 7(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Gohlke H, et al. 2014. AMBER14. AMBER 14. San Francisco: University of California.
- Christian KA, Morton SR.. 1992. Extreme thermophilia in a central Australian ant, Melophorus bagoti. Physiol Zool. 65(5):885–905. [Google Scholar]
- Cicconardi F, Di Marino D, Olimpieri PP, Arthofer W, Schlick-Steiner BC, Steiner FM.. 2017. Chemosensory adaptations of the mountain fly Drosophila nigrosparsa (Insecta: Diptera) through genomics’ and structural biology’s lenses. Sci Rep. 7:43770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicconardi F, Marcatili P, Arthofer W, Schlick-Steiner BC, Steiner FM.. 2017. Positive diversifying selection is a pervasive adaptive force throughout the Drosophila radiation. Mol Phylogenet Evol. 112:230–243. [DOI] [PubMed] [Google Scholar]
- Clark MS, Worland MR.. 2008. How insects survive the cold: molecular mechanisms—a review. J Comp Physiol B 178(8):917–933. [DOI] [PubMed] [Google Scholar]
- Colinet H, Hoffmann A.. 2010. Gene and protein expression of Drosophila Starvin during cold stress and recovery from chill coma. Insect Biochem Mol Biol. 40(5):425–428. [DOI] [PubMed] [Google Scholar]
- Colinet H, Hoffmann AA.. 2012. Comparing phenotypic effects and molecular correlates of developmental, gradual and rapid cold acclimation responses in Drosophila melanogaster. Funct Ecol. 26(1):84–93. [Google Scholar]
- Colinet H, Lee SF, Hoffmann A.. 2010. Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. FEBS J. 277(1):174–185. [DOI] [PubMed] [Google Scholar]
- Colinet H, Siaussat D, Bozzolan F, Bowler K.. 2013. Rapid decline of cold tolerance at young age is associated with expression of stress genes in Drosophila melanogaster. J Exp Biol. 216(2):253–259. [DOI] [PubMed] [Google Scholar]
- Coulson M, Robert S, Saint R.. 2005. Drosophila starvin encodes a tissue-specific BAG-domain protein required for larval food uptake. Genetics 171(4):1799–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree B, Newsholme EA.. 1972. The activities of lipases and carnitine palmitoyl-transferase in muscles from vertebrates and invertebrates. Biochem J. 130(3):697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Choate LA, Marks BA, Rice EJ, Wang Z, Chu T, Martins AL, Dukler N, Coonrod SA, Tait Wojno ED, et al. 2018. Dynamic evolution of regulatory element ensembles in primate CD4+ T cells. Nat Ecol Evol. 2(3):537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Annessa I, Coletta A, Sutthibutpong T, Mitchell J, Chillemi G, Harris S, Desideri A.. 2014. Simulations of DNA topoisomerase 1B bound to supercoiled DNA reveal changes in the flexibility pattern of the enzyme and a secondary protein–DNA binding site. Nucleic Acids Res. 42:9304–9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Annessa I, Raniolo S, Limongelli V, Di Marino D, Colombo G.. 2019. Ligand binding, unbinding, and allosteric effects: deciphering small-molecule modulation of HSP90. J Chem Theory Comput. 15:6368–6381. [DOI] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L.. 1993. Particle Mesh Ewald: an N·log(N) method for Ewald sums in large systems. J Chem Phys. 98:10089. [Google Scholar]
- Das S, Sillitoe I, Lee D, Lees JG, Dawson NL, Ward J, Orengo CA.. 2015. CATH FunFHMMer web server: protein functional annotations using functional family assignments. Nucleic Acids Res. 43(W1):W148–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marino D, Achsel T, Lacoux C, Falconi M, Bagni C.. 2014. Molecular dynamics simulations show how the FMRP Ile304Asn mutation destabilizes the KH2 domain structure and affects its function. J Biomol Struct Dyn. 32(3):337–350. [DOI] [PubMed] [Google Scholar]
- Di Marino D, Chillemi G, De Rubeis S, Tramontano A, Achsel T, Bagni C.. 2015. MD and docking studies reveal that the functional switch of CYFIP1 is mediated by a butterfly-like motion. J Chem Theory Comput. 11(7):3401–3410. [DOI] [PubMed] [Google Scholar]
- Doong H, Vrailas A, Kohn EC.. 2002. What’s in the “BAG”?—a functional domain analysis of the BAG-family proteins. Cancer Lett. 188(1–2):25–32. [DOI] [PubMed] [Google Scholar]
- Dror RO, Mildorf TJ, Hilger D, Manglik A, Borhani DW, Arlow DH, Philippsen A, Villanueva N, Yang Z, Lerch MT, et al. 2015. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science 348:1361–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Falcon S, Gentleman R.. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23(2):257–258. [DOI] [PubMed] [Google Scholar]
- Franz H. 1979. Ökologie der Hochgebirge. Stuttgart (Germany: ): Eugen Ulmer. [Google Scholar]
- Gerken AR, Eller OC, Hahn DA, Morgan TJ.. 2015. Constraints, independence, and evolution of thermal plasticity: probing genetic architecture of long- and short-term thermal acclimation. Proc Natl Acad Sci U S A. 112(14):4399–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénard B, Weiser MD, Gómez K, Narula N, Economo EP.. 2017. The Global Ant Biodiversity Informatics (GABI) database: synthesizing data on the geographic distribution of ant species (Hymenoptera: Formicidae). Myrmecol News 24:83–89. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD.. 2001. Past: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 4(1):art. 4. [Google Scholar]
- Harpur BA, Kent CF, Molodtsova D, Lebon JMD, Alqarni AS, Owayss AA, Zayed A.. 2014. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc Natl Acad Sci U S A. 111(7):2614–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond AJ.. 2010. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 27(3):570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije J.. 1997. LINCS: a Linear Constraint Solver for molecular simulations. J Comput Chem. 18(12):1463–1472. [Google Scholar]
- Hess B, Kutzner C, Van Der Spoel D, Lindahl E.. 2008. GRGMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 4(3):435–447. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A.. 2005. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 25(15):1965–1978. [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M.. 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32(5):767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölldobler B, Wilson EO.. 1990. The ants. Cambridge (MA): Harvard University Press.
- Hollingsworth SA, Dror RO.. 2018. Molecular dynamics simulation for all. Neuron 99(6):1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens ABF, Kronauer DJC, Pen I, Weissing FJ, Boomsma JJ.. 2012. Ants farm subterranean aphids mostly in single clone groups—an example of prudent husbandry for carbohydrates and proteins? BMC Evol Biol. 12:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki J, Narula N, Ziegler M, Guénard B, Economo EP.. 2016. Visualizing and interacting with large-volume biodiversity data using client-server web-mapping applications: the design and implementation of antmaps. Org Ecol Inform. 32:185–193. [Google Scholar]
- Jayakumar PC, Shouche YS, Patole MS.. 2007. Functional analysis of Drosophila melanogaster hexokinase Hex-A locus: multiple initiator-like elements enhance DPE containing promoter activity. Insect Mol Biol. 16:3–13. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML.. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926. [Google Scholar]
- Kaduk M, Sonnhammer E.. 2017. Improved orthology inference with Hieranoid 2. Bioinformatics 5:btw774. [DOI] [PubMed] [Google Scholar]
- Keeling CI, Yuen MM, Liao NY, Roderick Docking T, Chan SK, Taylor GA, Palmquist DL, Jackman SD, Nguyen A, Li M, et al. 2013. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 14(3):R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Peyton JT, Fiston-Lavier A-S, Teets NM, Yee M-C, Johnston JS, Bustamante CD, Lee RE, Denlinger DL.. 2014. Compact genome of the Antarctic midge is likely an adaptation to an extreme environment. Nat Commun. 5:4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AM, MacRae TH.. 2015. Insect heat shock proteins during stress and diapause. Annu Rev Entomol. 60(1):59–75. [DOI] [PubMed] [Google Scholar]
- Körner C, Paulsen J, Spehn EM.. 2011. A definition of mountains and their bioclimatic belts for global comparisons of biodiversity data. Alp Bot. 121:73–78. [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5):676–679. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K.. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. 28(11):3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapf P, Hochenegger N, Arthofer W, Schlick-Steiner BC, Steiner FM.. 2019. Comparing ant behaviour indices for fine-scale analyses. Sci Rep. 9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapf P, Russo L, Arthofer W, Möst M, Steiner FM, Schlick-Steiner BC.. 2018. An Alpine ant’s behavioural polymorphism: monogyny with and without internest aggression in Tetramorium alpestre. Ethol Ecol Evol. 30(3):220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlemann J. 2007. Paleogeographic and paleotopographic evolution of the Swiss and Eastern Alps since the Oligocene. Global Planet Change 58(1–4):224–236. [Google Scholar]
- Lamprecht A, Semenchuk PR, Steinbauer K, Winkler M, Pauli H.. 2018. Climate change leads to accelerated transformation of high-elevation vegetation in the central Alps. New Phytol. 220(2):447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Librado P, Vieira FG, Rozas J.. 2012. BadiRate: estimating family turnover rates by likelihood-based methods. Bioinformatics 28(2):279–281. [DOI] [PubMed] [Google Scholar]
- Liu C, Guénard B, Garcia FH, Yamane S, Blanchard B, Yang DR, Economo E.. 2015. New records of ant species from Yunnan, China. Zookeys 78:17–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yu L, Edwards SV.. 2010. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol Biol. 10(1):302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald HC, Cunha L, Bruford MW, Florestas E, Macdonald HC, Sir T, Evans M, Avenue M.. 2016. Development of genomic resources for four potential environmental bioindicator species: Isoperla grammatica, Amphinemura sulcicollis, Oniscus asellus and Baetis rhodani. bioRxivorg:046227.
- Merow C, Smith MJ, Silander JA.. 2013. A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography 36(10):1058–1069. [Google Scholar]
- Moreau CS, Bell CD.. 2013. Testing the museum versus cradle tropical biological diversity hypothesis: phylogeny, diversification, and ancestral biogeographic range evolution of the ants. Evolution (N Y). 67:2240–2257. [DOI] [PubMed] [Google Scholar]
- Mulichak AM, Wilson JE, Padmanabhan K, Garavito RM.. 1998. The structure of mammalian hexokinase-1. Nat Struct Biol. 5(7):555–560. [DOI] [PubMed] [Google Scholar]
- Nygaard S, Hu H, Li C, Schiøtt M, Chen Z, Yang Z, Xie Q, Ma C, Deng Y, Dikow R, et al. 2016. Reciprocal genomic evolution in the ant-fungus agricultural symbiosis. Nat Commun. 7:12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290. [DOI] [PubMed] [Google Scholar]
- Parker DJ, Wiberg RAW, Trivedi U, Tyukmaeva VI, Gharbi K, Butlin RK, Hoikkala A, Kankare M, Ritchie MG.. 2018. Inter and intraspecific genomic divergence in Drosophila montana shows evidence for cold adaptation. Genome Biol Evol. 10(8):2086–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, Stupka E, Rossiter SJ.. 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M, Rahman A.. 1981. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 52:7182. [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL.. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28(11):1420–1428. [DOI] [PubMed] [Google Scholar]
- Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, Gage FH, Swigut T, Wysocka J.. 2015. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163(1):68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5 (3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2017. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery E.. 2011. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One 6 (9):e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogora M, Frate L, Carranza ML, Freppaz M, Stanisci A, Bertani I, Bottarin R, Brambilla A, Canullo R, Carbognani M, et al. 2018. Assessment of climate change effects on mountain ecosystems through a cross-site analysis in the Alps and Apennines. Sci Total Environ. 624:1429–1442. [DOI] [PubMed] [Google Scholar]
- Roux J, Privman E, Moretti S, Daub JT, Robinson-Rechavi M, Keller L.. 2014. Patterns of positive selection in seven ant genomes. Mol Biol Evol. 31(7):1661–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 19(1):101–109. [DOI] [PubMed] [Google Scholar]
- Seifert B. 2018. The ants of Central and North Europe. Tauer (Germany: ): Lutra Verlags- und Vertriebsgesellschaft. [Google Scholar]
- Shaw TI, Ruan Z, Glenn TC, Liu L.. 2013. STRAW: Species TRee Analysis Web server. Nucleic Acids Res. 41:238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillitoe I, Lewis TE, Cuff A, Das S, Ashford P, Dawson NL, Furnham N, Laskowski RA, Lee D, Lees JG, et al. 2015. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 43 (D1):D376–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL.. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32(5):1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón J. 2007. Grinnellian and Eltonian niches and geographic distributions of species. Ecol Lett. 10(12):1115–1123. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Östlund G.. 2015. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43(D1):D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa Da Silva AW, Vranken WF.. 2012. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res Notes 5:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Schöffmann O, Morgenstern B, Waack S.. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner FM, Schlick-Steiner BC, Vanderwal J, Reuther KD, Christian E, Stauffer C, Suarez AV, Williams SE, Crozier RH.. 2008. Combined modelling of distribution and niche in invasion biology: a case study of two invasive Tetramorium ant species. Divers Distrib. 14(3):538–545. [Google Scholar]
- Steiner FM, Seifert B, Moder K, Schlick-Steiner BC.. 2010. A multisource solution for a complex problem in biodiversity research: description of the cryptic ant species Tetramorium alpestre sp.n. (Hymenoptera: Formicidae). Zool Anz. 249(3–4):223–254. [Google Scholar]
- Uchida Y, Uesaka M, Yamamoto T, Takeda H, Irie N.. 2018. Embryonic lethality is not sufficient to explain hourglass-like conservation of vertebrate embryos. Evodevo 9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner HC, Arthofer W, Seifert B, Muster C, Steiner FM, Schlick-Steiner BC.. 2017. Light at the end of the tunnel: integrative taxonomy delimits cryptic species in the Tetramorium caespitum complex (Hymenoptera: Formicidae). Myrmecol News 25:95–129. [Google Scholar]
- Wagner HC, Gamisch A, Arthofer W, Moder K, Steiner FM, Schlick-Steiner BC.. 2018. Evolution of morphological crypsis in the Tetramorium caespitum ant species complex (Hymenoptera: Formicidae). Sci Rep. 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis GP, Waters JM, Upton P, Craw D.. 2016. Transverse Alpine speciation driven by glaciation. Trends Ecol Evol. 31(12):916–926. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38:W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A.. 2014. Protein structure modeling with MODELLER. Methods Mol Biol. 1137:1–15. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2014. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EO. 1990. Success and dominance in ecosystems: the case of social insects. Vol. 2. Oldendorf/Luhe: Ecology Institute.
- Xian‐Wu L, Wei‐Hua X.. 2016. Hexokinase is a key regulator of energy metabolism and ROS activity in insect lifespan extension. Aging 8:245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K.. 2001. Trends, rhythms, and aberration in global climate 65 Ma to present. Science 292(5517):686–693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.