Abstract
Background
LCL161, a SMAC’S small molecule mimetic, can bind to a variety of IAPs and activate Caspases. We found that on its own, LCL161induces apoptosis of drug-resistant breast cancer cells by binding to a variety of IAPs and activating Caspases. However, when LCL161 is used in combination with Caspase Inhibitors (CI), its capacity to induce apoptosis of breast cancer cells is enhanced.
Objective
To carry out proteomic and bioinformatics analysis of LCL161 in combination with CI. We aim to identify the key proteins and mechanisms of breast cancer drug-resistant apoptosis, thereby aiding in the breast cancer drug resistance treatment and identification of drug targeting markers.
Methods
Cell culture experiments were carried out to explore the effect of LCL161 combined with CI on the proliferation of breast cancer drug-resistant cells. Proteomic analysis was carried out to determine the protein expression differences between breast cancer drug-resistant cells and LCL161 combined with CI treated cells. Bioinformatics analysis was carried out to determine its mechanism of action. Validation of proteomics results was done using Parallel Reaction Monitoring (PRM).
Results
Cell culture experiments showed that LCL161 in combination with CI can significantly promote the apoptosis of breast cancer drug-resistant cells. Up-regulation of 92 proteins and down-regulation of 114 proteins protein were noted, of which 4 were selected for further validation.
Conclusion
Our results show that LCL161 combined with CI can promote the apoptosis of drug-resistant breast cancer cells by down-regulation of RRM2, CDK4, and ITGB1 expression through Cancer pathways, p53 or PI3K-AKT signaling pathway. In addition, the expression of CDK4, RRM2, and CDC20 can be down-regulated by the nuclear receptor pathway to affect DNA transcription and replication, thereby promoting apoptosis of breast cancer drug-resistant cells.
Keywords: Breast cancer, proteomics, smac mimetics, Caspase Inhibitors (CI), p53, PI3k-AKT
1. INTRODUCTION
According to the 2018 global cancer statistics, the diagnosis of lung cancer is most commonly accompanied by the highest mortality rate among men and women. With regard to women, in particular, the prevalent cause of death is Breast Cancer (BC), followed by lung and colorectal cancer [1]. The most effective first-line chemotherapy drugs for the treatment of BC are: cisplatin, paclitaxel, and cyclophosphamide [2]. However, its efficacy is limited and its side effects are severe. Multidrug resistance of tumor cells is the primary reason chemotherapy fails in clinical BC cases [3]. In addition, drug-resistant cells can also lead to tumor treatment failure by promoting tumor progression [4]. Therefore, it is imperative to develop a highly effective strategy for multi-drug resistant BC treatment.
Inhibitor of Apoptosis Proteins (IAPs) are proteins that are vital in controlling Programmed Cell Death (PCD) [5]. The expression of IAPs is significantly elevated in cancer cells, thereby increasing cell viability, enhancing tumor growth and subsequent metastasis. Uninhibited IAPs can inhibit apoptosis and promote cell cycle progression [6]. The activity of pro-apoptotic factors against anti-apoptotic proteins are hindered in cancer cells, e.g., there is an inhibition in the up-regulation of apoptosis protein, resulting in uncontrolled cell division [7]. In a variety of tumor cells, the high expression level of apoptosis inhibitory factor is positively correlated with poor prognosis, metastasis, and tumor resistance. Studies have shown that interference of siRNA can reduce the expression of IAPs, promote tumor cell apoptosis, and can make tumor-resistant cells regain sensitivity to chemotherapy drugs [8]. These studies suggest that apoptosis inhibitors are a novel approach to treat tumor resistance. Despite the novelty of these inhibitors, studies have shown their effectiveness in promoting apoptosis [9]. The ability of tumor cells to acquire the ability to undergo apoptosis resistance is among the main markers of cancer. Therefore, endogenous IAPs are promising targets for anticancer therapy.
Apart from their anti-apoptotic activity, IAPs are capable of regulating many other cellular functions, like differentiation, proliferation, and migration. They also contribute to pro-inflammatory and immune responses. The potential therapeutic agents, Second Mitochondria-derived Activator of Caspases (SMAC) mimetics have recently been in the spotlight as synthetic IAP antagonists. The (SMAC) mimetic LCL161 acts as a synthetic IAPs antagonist, which binds and participates in the down-regulation of various IAPs, and then activates caspase to induce apoptosis [10]. Studies have shown that LCL161 significantly inhibits proliferation and promotes apoptosis in multiple solid tumors and cell lines such as multiple myeloma [11], hepatocellular carcinoma [12], and leukemia [13]. Synergism in radiotherapy or targeted therapy may enhance their therapeutic effect [14]. Studies have shown that when CI are combined with LCL161, they can further promote BC cell apoptosis [15], which is contradictory as theoretically the apoptosis should be reduced during caspase enzyme inhibition. Therefore, we investigated the efficacy of LCL161-CI combination on the proliferation of BC resistant cell line MCF-7/DDR. Comparison of the protein expression differences in the control group and the LCL161-CI combination experimental group was done by proteomics. The possible mechanism action was predicted to provide a theoretical basis and support for future studies with LCL161.
Our aim was to elucidate the effect of LCL161 combined with CI on the proteome of MCF-7 BC resistant cells, to explore its possible mechanism of action at a molecular level. We also wished to explore the therapeutic potential of LCL161 in combination with CIs. This study first demonstrated the effect of LCL161 combined with CI on the proliferation of drug-resistant BC cells by cell experiments. Proteomic analysis to select potential target proteins was then carried out followed by the selection of differential proteins by PRM (Parallel Reaction Monitoring) verification. We found 206 significantly differentially expressed proteins that play a role in various cellular signaling pathways, as well as cancer pathways: PI3-Akt and p53 signaling pathways. CDC20, IGTB1, RRM2, and CDK4 were identified as key proteins that may be associated with LCL161 in combination with CI to promote drug-resistant cells death in breast cancer. With the help of proteomic analysis, we wish to explore the molecular mechanism involved when LCL161 combined with CI promotes the apoptosis of BC drug-resistant cells. We also wish to provide new ideas to subsequently treat BC resistance.
2. MATERIALS AND METHODS
2.1. Chemicals and Reagents
LCL161, as well as z-VAD-fmk, were purchased from MCE Corporation (US). RMPI 1640 medium was purchased from Hyclone Company (US). Fetal bovine serum was produced by Pan Company (Germany). Cell Counting Kit-8 Cell Proliferation Assay Kit and ATP Assay Kit were purchased from Biosharp Company, (China). 96 and 6-well cell culture plates were obtained from Sigma-Aldrich, USA. 60mm, 75, 100mm cell culture dishes were purchased from Corning (USA).
2.2. Cell Lines and Cell Culture
Human BC cell line MCF-7 cells were obtained from Nanjing Kaiji cell bank and gradually induced into cisplatin-resistant MCF-7/DDP by low concentration cisplatin culture. MCF-7/DDR cells were cultured in an RMPI 1640 medium that contained 10% fetal bovine serum, 100 U/ml penicillin, 100μg/ml streptomycin, and placed in a cell incubator at 37°C, 5% CO2 saturated humidity.
2.3. Detection of Drug Resistance Index in Cisplatin-Resistant MCF-7/DDP Cells of Breast Cancer
The low-concentration gradual induction method allows BC resistant strains to grow stably in high concentrations of cisplatin. BC cisplatin-resistant cell line was cultured in complete medium in the absence of cisplatin for 2 weeks, and the parental cells and drug-resistant cells grown in log phase were inoculated into 96-well culture plates, respectively. 5x103 cells were seeded into each well, and after 24 hours of incubation, different concentrations of cisplatin drugs were added to the corresponding 96-well plates in the parental cells and drug-resistant cells. 5 parallel holes were set for each concentration. The half-inhibitory concentration (IC50) of cisplatin in various parental and drug-resistant cells was calculated using SPSS22.0. Drug Resistance Index (RI) was calculated using the formula below. The experiment was done in triplicates.
RI = semi-lethal concentration of drug-resistant cells / semi-lethal concentration of parental cells
2.4. CCK-8 Assays Detects Cell Viability
Cells plating (density 6×103 cells/well) in 96-well plates were carried out and incubated for 24h, following treatment with varying concentrations of LCL161 (0 to 5μM) and different concentrations of docetaxel (0 to 2.4mg/L). 5 duplicate wells per group were maintained and the drug was allowed to act for 24 to 72h. In addition, the experiment was divided into 4 groups: docetaxel group, LCL161 group (2μM), LCL161 with docetaxel group and blank control group. The drug was allowed to act for 48h followed by the addition of 10μL CCK-8. After incubation for one hour at 37°C, absorbance was measured at 450nm with a plate reader.
2.5. Detection of Mitochondrial Membrane Potential
Seeding of cells was done into the wells of a 6-well plate (2×105 cells/well). After the cells were attached to the plates, different treatments were added to the medium. The cells were then collected after 5h of incubation. A luminometric-based ATP assay kit was used to detect intracellular ATP levels for the different treatments, as per manufacturer’s protocol. Measurement of absorbance was done using a microplate reader and ATP levels were calculated as a percentage of the blank group.
2.6. Methods of Proteomics Experiments
2.6.1. Sample Preparation for 2-DE and iTRAQ Analysis
Seeding of MCF-7/DDP cells in 75cm2 flasks was done and placed in a humidified incubator at 37°C with 5% CO2. At 70% confluency, treatment with 2μM LCL161 and 10μM z-VAD-zmk or DMSO (control cells) was carried out. Following 24 hours of incubation, the drug group and the control group underwent three washings with pre-cooled PBS, centrifuged to obtain a cell pellet. Storage at -80°C was maintained till required for further analysis by mass spectrometry based on TMT/iTRAQ.
2.6.2. Protein Extraction
Sonication was carried out thrice on ice with a high-intensity ultrasonic processor (Scientz) in lysis buffer (8M urea, 1% Protease Inhibitor Cocktail). (Note: For PTM experiments, the addition of inhibitors to lysis buffer was made, e.g. 3μM TSA and 50mM NAM for acetylation) It was centrifuged to remove the debris at 12,000g for 10 minutes at 4°C. This was followed by collection of the supernatant and determination of protein concentration was carried out with the BCA kit as per the manufacturer’s protocol.
2.6.3. Trypsin Digestion
Reduction of protein solution was done by adding 5mM dithiothreitol for 30 minutes at 56°C. This was followed by alkylation with 11mM iodoacetamide for 15min at RT (room temp) in darkness. Dilution of the sample by addition of 100mM TEAB to urea concentration less than 2M was then done. For the first digestion overnight, the addition of trypsin at 1:50 trypsin-to-protein mass ratio was maintained and for the second 4 h-digestion 1:100 trypsin-to-protein mass ratio was maintained.
2.6.4. TMT/iTRAQ Labeling
Peptide desalting was done with the Strata X C18 SPE column (Phenomenex) followed by vacuum-drying. Reconstitution of peptides in 0.5M TEAB was done as per the manufacturer’s instruction for TMT /iTRAQ kit. One unit of TMT/iTRAQ reagent was allowed to thaw followed by reconstitution in acetonitrile. This was followed by incubation for 2 hours at RT. Following this, pooling, desalting, and drying by vacuum centrifugation were carried out.
2.6.5. LC-MS/MS Analysis
The tryptic peptides were allowed to dissolve in 0.1% formic acid (solvent A), followed by loading onto a home-made reversed-phase analytical column (15cm length, 75μm i.d.). The gradient was made of an increase from 6% to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26min, 23% to 35% in 8min and climbing to 80% in 3min then holding at 80% for the last 3min, all at a constant flow rate of 400nL/min on an EASY-nLC 1000 UPLC system.
The peptides were exposed to NSI source and then tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC. The application of 2.0kV of electrospray voltage was maintained. The m/z scan range was 350 to 1800 for a full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were then selected for MS/MS using the NCE setting as 28 and the fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15.0s dynamic exclusion was carried out. Automatic Gain Control (AGC) was set at 5E4. The fixed first mass was set as 100 m/z.
2.6.6. Database Search
Processing of MS/MS data was done by the Maxquant search engine (v.1.5.2.8). Tandem mass spectra were searched against the human UniProt database concatenated with the reverse decoy database. Trypsin/P was indicated as a cleavage enzyme that allowed up to 4 missing cleavages. The setting for the mass tolerance for precursor ions was 20 ppm in the primary search followed by 5 ppm for the main search. The setting for the mass tolerance for fragment ions was 0.02 Da. Carbamidomethyl on Cys was stated as fixed modification and acetylation modification and oxidation on Met were set as variable modifications. FDR was changed to <1% with the minimum score for modified peptides set at > 40.
2.7. Bioinformatics Methods
2.7.1. Annotation Methods
2.7.1.1. GO Annotation
The Gene Ontology, or GO, is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to cover three domains: Cellular component, Molecular function, and Biological process. Gene Ontology (GO) annotation proteome was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). The first step was to convert the identified protein ID to UniProt ID and then map it to GO IDs by protein ID. The identified proteins that the UniProt-GOA database was unable to identify were annotated using the InterProScan soft based on the protein sequence alignment method. Classification of proteins was then done by Gene Ontology annotation on the basis of three categories: biological process, cellular component, and molecular function.
2.7.1.2. KEGG Pathway Annotation
KEGG has the ability to collect known information on molecular interaction networks, such as pathways and complexes (the “Pathway” database), gene and protein information generated by genome projects (including the gene database) and information about biochemical compounds and reactions (including compound and reaction databases). These databases are different networks, known as the “protein network” and the “chemical universe”,
respectively. There are efforts in progress to add to the knowledge of KEGG, including information regarding ortholog clusters in the KEGG Orthology database. Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used for annotation of the protein pathways. KEGG online service tools KAAS was used for annotation of protein’s KEGG database description. Then the annotated result was mapped on the KEGG pathway database by KEGG online service tools KEGG mapper.
2.7.2. Functional Enrichment
2.7.2.1. Enrichment of Gene Ontology Analysis
Using GO annotation, classification of proteins in the following three categories was done: biological process, cellular compartment, and molecular function. For each, a two-tailed Fisher’s exact test was carried out for testing the enrichment of the differentially expressed protein against all identified proteins. The GO with a corrected p-value <0.05 is considered significant.
2.7.2.2. Enrichment of Pathway Analysis
Encyclopedia of Genes and Genomes (KEGG) database was utilized for the identification of enriched pathways using two-tailed Fisher’s exact test to test the enrichment of the differentially expressed proteins against all identified proteins. The pathway with a corrected p-value <0.05 was considered significant. Classification of these pathways into hierarchical categories was done based on the KEGG website.
2.7.3. Protein-Protein Interaction Network
Differential protein database numbers or protein sequences screened in different comparison groups were compared with the STRING (v.10.5) protein network database, and the differential protein interaction relationship was extracted according to the confidence score >0.7 (high confidence). The differential protein interaction network was visualized using the R package “networkD3” tool.
2.8. PRM Analysis
Confirmation of the protein expression levels by LC-MS/MS analysis was done by quantification of the expression levels of four selected proteins by PRM analysis carried out at the “Hangzhou Jinjie Bioscience Co., Ltd (Hangzhou, China)”. Signature peptides for the target proteins were defined according to the LC-MS/MS data, and only unique peptide sequences were selected for PRM analysis. The tryptic peptides were first dissolved and then directly loaded onto a home-made reversed-phase analytical column. The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC. A data-independent procedure that consisted of alternation between one MS scan followed by 20 MS/MS scans was run. Automatic Gain Control (AGC): 3E6 (full MS) and 1E5 (MS/MS) and maximum IT: 20 ms (full MS) and auto (MS/MS) were maintained. The setting for the isolation window for MS/MS was 2.0m/z.
The processing of MS data was done with Skyline (v.3.6). The following peptide settings were maintained: enzyme- Trypsin [KR/P], Max missed cleavage- 2, Peptide length: 8-25, Variable modification: Carbamidomethyl on Cys and oxidation on Met, max variable modifications: 3.
2.9. Statistical Analysis
Student's t-test was used to compare the two groups and One-way analysis of variance was used for comparison between groups. SPSSv.22.0 software was used for data Process. All analyses were statistically significant with p values < 0.05. *P <0.05, **P <0.01, ***P <0.001.
3. RESULTS
3.1. Effects of LCL161 Combined with z-VAD-fmk on the Proliferation of Drug-Resistant Cells
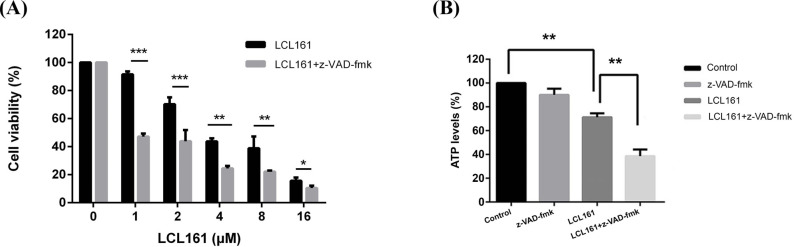
McF-7/DDP cells were exposed to LCL161 for 48h at various concentrations. By cck-8 assay, it was found that LCL161 combined with z-VAD-fmk resulted in significant inhibition of their growth (Fig. 1A).
Fig. (1).
LCL161 in combination with Caspase inhibitors can inhibit growth and induces cell death in human breast cancer MCF-7/DDP cells. (A) MCF-7/DDP cells were treated with various concentrations of LCL161 (from 0 to 16μM) for 24, 48, and 72h. Cell viability was measured using a CCK-8 assay after treatment. (B) Reduction in ATP production after LCL161 in combination with Caspase inhibitors treatment. Cellular levels of ATP were measured after 5h. Results represent means ± SD of three independent experiments. *P<0.05, **P < 0.01, ***P < 0.001. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.2. LCL161 Combined with z-VAD-FMK Decreased Intracellular ATP Levels in Drug Resistant Cells
ATP plays an important role in numerous physiological and pathological processes. During apoptosis and necrosis, cell membrane rupture leads to a decrease in cell membrane potential as well as a decrease in ATP levels. Compared with the control group, LCL161 reduced the ATP level of MCF-7/DDP cells following 5h incubation (P<0.01), but a more significant reduction of ATP was when combined with caspase inhibitors (Fig. 1B). This suggests that when caspase is inhibited, LCL161 interferes with cellular energy metabolism by reducing intracellular ATP levels, leading to cell death.
3.3. Quantitative Intracellular Proteome Analysis and GO analysis of MCF-7/DDP Cells after Treatment with LCL161- z-VAD-FMK Combination
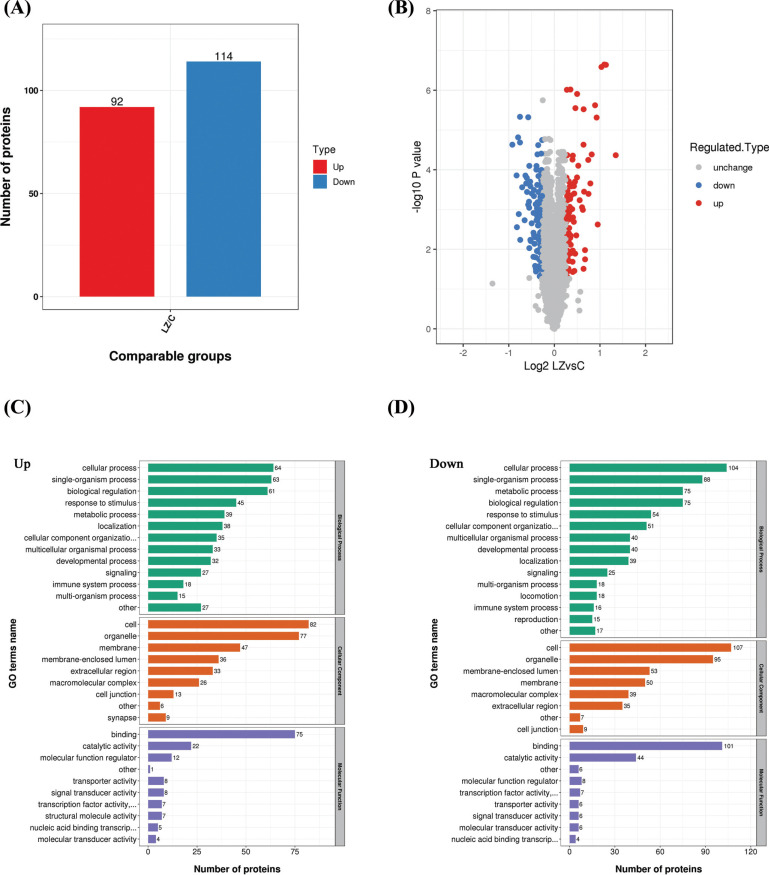
In order to identify the cell biological changes and mechanisms of LCL161 combined with CI, proteomics analysis was done to find the difference in protein expression of McF-7/DDP cells before and after LCL161- z-VAD-zmk combination treatment. From LC-MS/MS, 5220 intracellular proteins were identified between the control (untreated) and LCL161-z-VAD-zmk combination-treated MCF-7/DDP cells across the three sets of biological replicates. It was seen that 206 proteins were at least 1.2 times different (p<0.05) between the control group and before and after the LCL161-z-VAD-zmk combination treatment group. Among them, up-regulation of 92 proteins and down-regulation of 114 proteins were seen (Fig. 2 A, B).
The differentially expressed proteins were categorized according to three aspects: Cellular component, Molecular function, and Biological process by GO analysis based on the UniProt-GOA database. Among the proteins whose expression was up-regulated, the differential proteins of the cellular component are mostly concentrated in cells and organelles. The molecular function was mainly related to binding. In the biological process, the differential proteins are concentrated in the cellular process, the single-organism process, and the biological regulation (Fig. 2C). Among the down-regulated proteins, differential proteins have the same tendency to up-regulate proteins in both the Cellular component and the Molecular function, while in the biological process, the differential proteins are more concentrated in the cellular process and the single-organism process, followed by the metabolic process and biological Regulation (Fig. 2D).
Fig. (2).
(A) The p-value was calculated using the two-sample T-test method. When p-value<0.05, the difference expression amount changed by more than 1.2 as a significantly up-regulated change threshold, and less than 1/1.2 as a significantly down-regulated change threshold. The histogram shows aggregated data for all differential proteins, with red dots indicating significant differential expression of up-regulated proteins and blue dots indicating significant differential expression of down-regulated proteins. (B) The horizontal axis of the volcano plot is the logarithmically converted value of the relative quantitative value of the protein, and the vertical axis is the value of the logarithmically converted p-value after the log-log conversion. The red dot in the figure indicates a significant differential expression of the up-regulated protein, and the blue dot indicates a significant differential expression of the down-regulated protein. (C and D) The biological processes, molecular function and cellular components of the Gene Ontology (GO)‐annotation distribution of the differential proteins in McF-7/DDP cells before and after LCL161 combined with z-vad treatment. The bars represent the numbers of proteins involved in the processes. The two figures respectively represent the up-regulated (C) and down-regulated proteins (D). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.4. Bioinformatic Information for Differentially Expressed Proteins
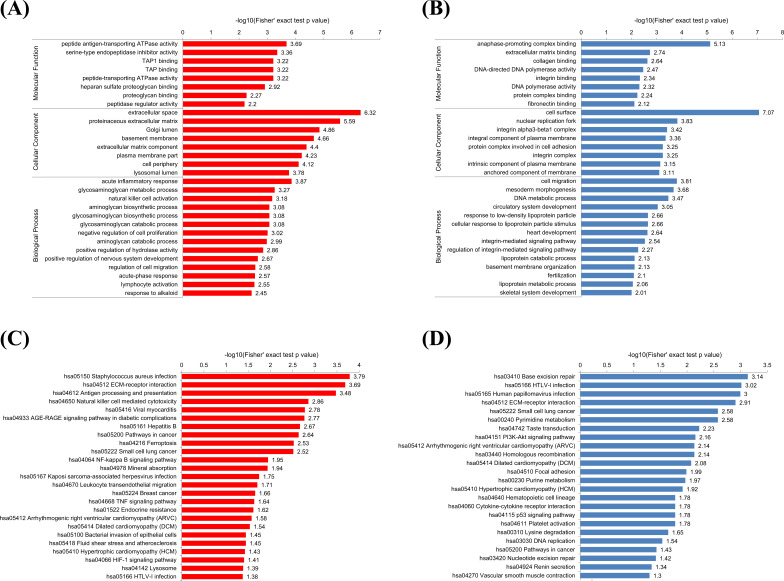
In the GO-based functional enrichment analysis, according to the three categories it was found that the proteins that underwent upregulation were: concentrated in the acute inflammatory response (biological process), had peptide antigen-transporting ATPase activity (molecular function), and was mainly in the extracellular space (cellular component) (Fig. 3A). The down-regulated proteins played a role in migration and DNA metabolic processes (biological processes), played a role in anaphase-promoting complex binding and DNA-directed DNA polymerase activity (molecular function), and were located in cells and nucleus (cellular component) (Fig. 3B). This shows that the proteins that underwent upregulation primarily play a role in the acute stress response of cells while the down-regulation of proteins are involved in cell migration and regulation of nuclear transcription and replication of components.
Fig. (3).
Functional enrichment analysis of up-regulated and down-regulated proteins. A and B, GO-based enrichment analysis of up-regulated (A) and down-regulated (B) proteins. (C and D) KEGG pathway-based enrichment analysis of up-regulated (C) and down-regulated (D) proteins. Red indicates up-regulated differential protein and green indicates down-regulated differential protein. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
KEGG pathway enrichment analysis was then carried out. It was seen that the up-regulated proteins were primarily enriched in disease-related pathway, ECM-receptor interaction, pathways in cancer, and AGE-RAGE signaling pathway (Fig. 3C). The proteins that underwent downregulation were primarily enriched in disease-related pathway, PI3K-Akt signaling pathway, p53 signaling pathway, pathways in cancer, nucleotide excision repair, and DNA replication (Fig. 3D).
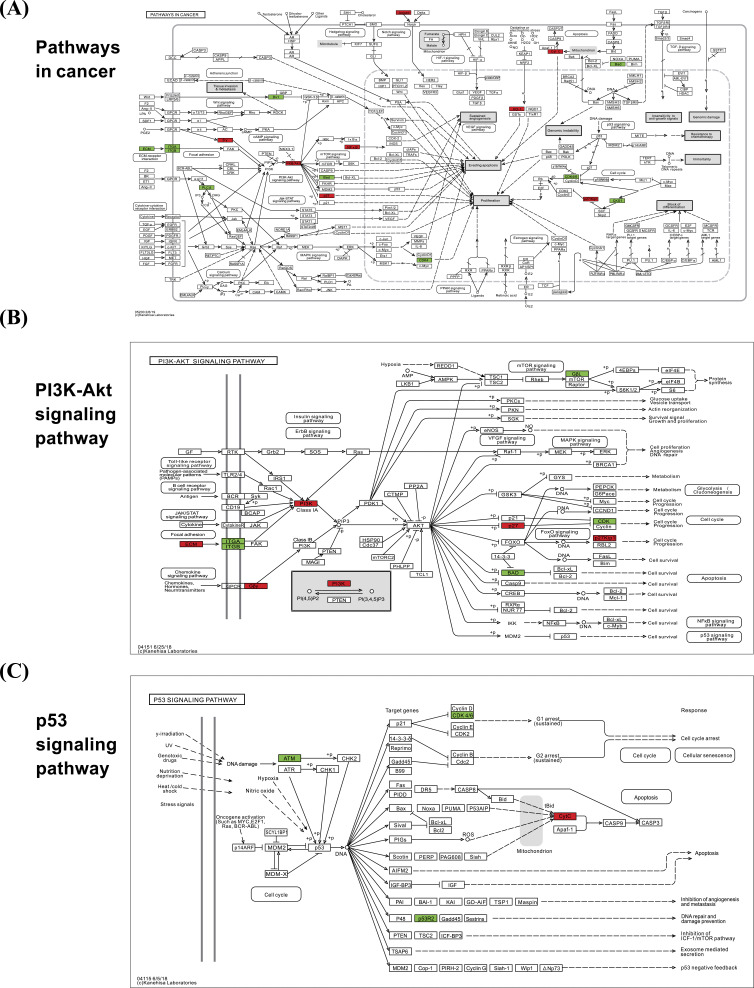
3.5. Cluster Analysis
The KEGG pathway enrichment of the differentially changed proteins disclosed pathways of importance. Specifically, we detected changes in the following signal pathways: Data through the cancer pathway (Fig. 4A), PI3K-AKT signaling pathway (Fig. 4B), and p53 signaling pathway (Fig. 4C). We found that key proteins after treatment with breast cancer-resistant cells via LCL161 in combination with z-VAD-zmk are mainly concentrated in the above
Fig. (4).
Schematic representation of the KEGG pathway with significantly enriched differentially expressed proteins. In the figure, red indicates differentially up-regulated protein; green indicates differentially down-regulated protein; (A) Shown as differentially expressed protein-rich results in pathways in cancer. (B) Schematic representation of enrichment in the PI3K-AKT signaling pathway. (C) Schematic representation of enrichment in the p53 signaling pathway. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
pathways. At the same time, many intranuclear protein expression differences are also seen in the cancer pathway and the PI3K-AKT pathway to regulate cell proliferation.
3.6. PPI Analysis
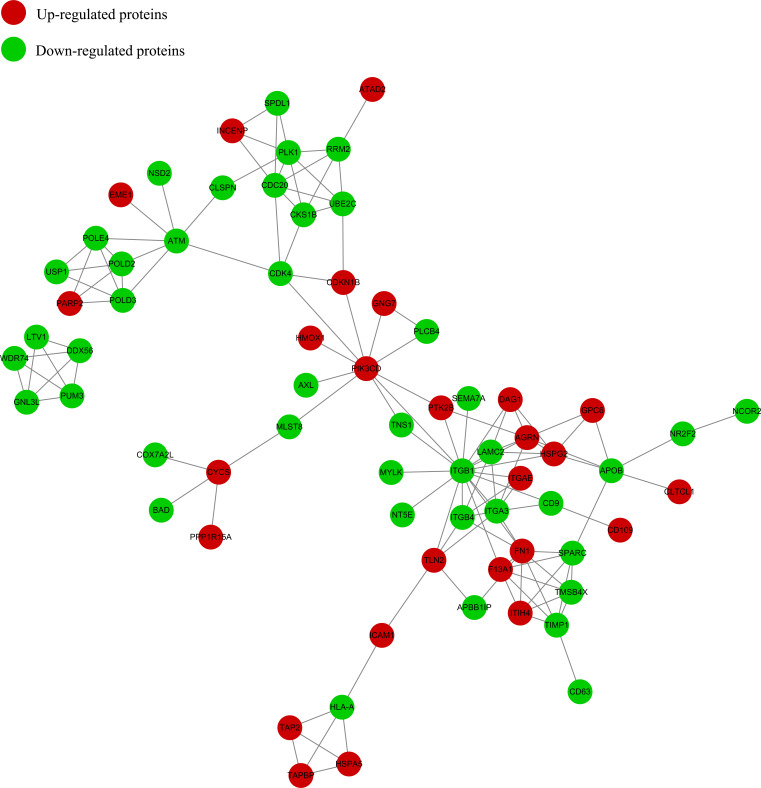
The differential protein interaction network is visually displayed by searching the PPI database to verify the interaction of the identified or differentially expressed protein. In the PPI network, different colors represent a differential expression of proteins (blue are the down-regulated proteins and red are the up-regulated proteins). The circle size denotes the number of proteins interacting with each other. The larger the circle, the more proteins it interacts with, indicating the increased importance of the protein in the network. Through inter-group analysis and comparison, it was found that there are 4 important nodule proteins between the two groups; namely, ITGB1 (connectivity degree of 17), CDC20 (connectivity degree of 6), CDK4, and RRM2 (connectivity degree of 5) (Fig. 5).
Fig. (5).
Protein-protein network. The circles in the figure indicate differentially expressed proteins, and different colors represent a differential expression of proteins (blue is the down-regulated protein and red is the up-regulated protein). The number of lines represents the number of different proteins and their interacting proteins. The more proteins that interact with each other, the more important the protein is in the network. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.7. PRM Verification
For verification of the MS results, PRM analysis was carried out for 4 differentially expressed proteins (CDC20, RRM2, ITGB1, and CDK4). They were selected based on the following criteria: 1) they could potentially have biological functions and significance; 2) They had peptide fragments >1 as per LC-MS/MS; and 3) they are associated with cell proliferation and have potential for further research. Based on the relative expression levels of the corresponding peptides of the varied target groups in the different sample groups, PRM quantification was carried out on the selected 4 target proteins of the 6 samples. Table 1 represents the PRM data. The TMT and PRM results were found to be consistent. According to our results, some of the proteins like RMM2, CDK4, ITGB1, and CDC20 undergo down-regulation upon LCL161-z-VAD-zmk combination treatment (Table 1).
Table 1.
PRM protein quantification results.
| Protein Accession | Protein Gene | LZ/C Ratio | LZ/C p-value | LZ/C Ratio(TMT) |
|---|---|---|---|---|
| Q12834 | CDC20 | 0.68 | 0.0148 | 0.69 |
| P31350 | RRM2 | 0.67 | 0.0130 | 0.82 |
| P05556 | ITGB1 | 0.63 | 0.00000273 | 0.81 |
| P11802 | CDK4 | 0.48 | 0.0000588 | 0.78 |
The PRM results in the table are consistent with the TMT quantitative results, and the p-values are less than 0.05.
4. DISCUSSION
For malignant tumors, the resistance of tumor cells to drugs is often the main cause of treatment failure [16]. The mechanism of cancer drug resistance is a complex process with many influencing factors. The abnormal regulation of the apoptosis process is also the cause of tumor cell resistance. Currently, the hot spot for anti-tumor research is to determine how to inhibit tumor growth by promoting tumor cell apoptosis [17-19]. In previous studies, Smac protein is an endogenous regulatory molecule of the mitochondrial apoptotic pathway, which can specifically bind to IAPs to abolish its anti-apoptotic effect and induce apoptosis. As a synthetic IAP antagonist,
Smac mimetic LCL161 binds and participates in the down-regulation of various IAPs due to high affinity with IAPs. It then activates caspase to initiate apoptosis [20]. Vijay et al. (2014) indicated that LCL161 significantly reduced XIAP activity and cIAP levels in multiple myeloma cell lines and their resistant cell lines while promoting TNF-α-mediated apoptosis [21]. In our study, when LCL161 was used alone, it could promote the apoptosis of BC resistant cells. When the downstream caspase was inhibited, contradictory to expected inhibition, it was found that apoptosis was promoted. To further understand the mechanism by which LCL161 in combination with CI promotes apoptosis in BC drug-resistant cells, further experiments were carried out. A TMT-based high-throughput proteomics strategy was used for direct screening of differentially expressed proteins in BC drug-resistant cells before and after exposure to LCL161 and CI. These proteins were analyzed using bioinformatics tools. Moreover, vital proteins were identified.
RRM2 (Ribonucleotide reductase M2) is a protein-translationally modified enzyme in most eukaryotic cells. It is also a key rate-limiting enzyme that promotes DNA synthesis and repair [22]. Studies have shown that RRM2 is not expressed or under-expressed in normal human cells, and is overexpressed in malignant tumors such as cancer of the liver, pancreas, colon, non-small cell lung cancer, melanoma, hydatidiform mole malignant tumor, and trophoblastic tumor [23-29]. The high expression of RRM2 is closely related to the growth, invasion, metastasis, angiogenesis, tumor resistance, and prognosis of malignant tumors [30-35]. Breast cancer cell resistance is the main cause of poor therapeutic effect, and the improvement of DNA repair level is one of the reasons for drug resistance. Chemotherapy drugs mainly kill tumors by destroying or inhibiting tumor cell DNA or RNA. However, tumor cells improve anti-injury ability through multiple pathways during tumor evolution, and RRM2 is associated with DNA synthesis metabolism. Increased expression in cancer drug resistance cells increases resistance to DNA damage as well as anticancer drugs [36-40]. On the other hand, inhibition of RRM2 expression reduces the anti-injury ability of tumor cell DNA and may restore sensitivity to chemotherapeutic drugs [41-44]. Therefore, we speculated that when the caspase enzyme was inhibited, AIF (Apoptosis-Inducing Factor) and Endo located in the middle of the mitochondrial bilayer membrane G (endonuclease G) can be transferred therefrom. Upon entering the nucleus, it can cause chromatin condensation and large fragment DNA degradation (50kb), and these effects of AIF and Endo G are not related to caspase [45-48].
In breast cancer, cyclin D1 amplification and CDK4 copy increase are common in luminal and Her2 enriched subtypes, but in basal-like tumors, Rb deletions or mutations and cyclin E1 amplifications [49] are very rare. A large number of preclinical studies have shown that cyclin D1-CDK4/6 is an essential factor in maintaining the tumorigenic potential of breast cancer cells [50-53]. The cell cycle is an important regulator of cell proliferation, growth, and division after DNA damage. It controls the transition from the quiescent state (G0 phase) to cell proliferation and passes the checkpoint [54]. In order to enter DNA synthesis (S phase), all cells must activate Cyclin-Dependent Kinases (CDKs), which need to bind to cyclin subunits to exert catalytic activity. The d-type cyclin and its chaperones, CDK4 and CDK6, play an important role in the cell cycle [55, 56]. In breast cancer resistant cells, the activity of CDK4 is increased, thereby restoring the DNA replication ability of tumor cells, and making breast cancer cells resistant to chemotherapeutic drugs. In addition, studies have shown that the CDK4/6 pathway crosses numerous vital mitotic signaling pathways which provides a powerful theoretical basis for using a combination of CDK4/6 inhibitors along with targeted therapies. A typical example is its combination with endocrine therapy (e.g. tamoxifen, aromatase inhibitor, and fulvast) in er-positive BC [57-59]. The synergy between these drugs can be partially explained as a double-strike mechanism in which endocrine therapy limits CCND1 transcription and CDK4/6 inhibitors directly block kinase activity. Therefore, CDK4 can be considered as a target protein for BC resistant cells, and combined with LCL161 to treat BC resistance. This indicates that when the apoptosis pathway induced by LCL161 is blocked by caspase, the expression of CDK4 protein can be down-regulated to promote drug-resistant cell death, indicating that CDK4 is a target protein that promotes drug-resistant cell death in breast cancer.
The B-cell lymphoma 2 (Bcl-2) family plays a key role in controlling cell apoptosis, which can be divided into two functionally distinct types: the anti-apoptotic protein group and the pro-apoptotic protein group [60]. It is a key protein in the mitochondrial pathway during programmed cell death [61, 62]. The Bcl-2 protein is a family of anti-apoptotic proteins that were originally identified as a primitive in cell follicular lymphoma. The oncogene Bax protein acts as a pro-apoptotic protein during apoptosis and death [63, 64]. In many tissues, the expression ratio of Bcl-2 and Bax protein is used to study cell apoptosis [65]. The balance between Bcl-2 and Bax protein expression determines cell survival and death. Our results showed that the down-regulation of ITGB1 promoted the decrease of anti-apoptotic protein Bcl-2 expression, while the pro-apoptotic protein Bax increased and activated Caspase protein [66-69]. These results suggest that combating the high expression of ITGB1 in breast cancer resistant cells could be a potential method of treatment. It is possible to inhibit the expression of Bax through the mitochondrial signaling pathway, thereby inhibiting the apoptosis of tumor cells.
CDC20 (Cell-Division Cycle 20) is an important regulatory molecule of cell cycle checkpoints that has a vital role in the regulation of cell mitosis by direct binding and activation of APC (Anaphase Promoting Complex) [70]. Previous research has shown that the high expression of CDC20 is likely to be an important factor in promoting the abnormal proliferation of tumor cells and resisting apoptosis [71]. Bim is a pro-apoptotic molecule in the Bcl-2 family [72], and CDC20 disrupts its protein stability by ubiquitination of Bim, which in turn inhibits apoptosis [73]. Studies have found that down-regulating the expression level of CDC20 can significantly increase the expression of Bim protein in BC cells, indicating that the E3 ubiquitin ligase activity targeting CDC20 increases the apoptosis sensitivity of tumor cells by up-regulating the expression level of Bim [74-76]. In summary, we believe that CDC20 is highly expressed in breast cancer resistant cells, which in turn inhibits the expression level of Bim, thereby reducing the apoptosis sensitivity of tumor cells. This suggests that drug resistance in the BC cells may be due to increased expression of CDC20. LCL161- z-VAD-zmk combination can induce the apoptosis of BC drug-resistant cells by inhibiting the expression of CDC20 protein, which may be caused by mitosis and apoptosis signals. Therefore, CDC20 can be used as a target protein for promoting apoptosis of BC resistant cells.
CONCLUSION
In summary, our research was the first to use proteomics to analyze the protein changes of Smac mimetic combined with CI on drug-resistant BC cells. The GO analysis of differentially expressed proteins in our study indicates that they could play a role in the cellular and the single-organism process and are mostly located on the extracellular space, organelles, and nucleus. This indicates that these differential proteins are transmitted intracellularly through signaling pathways, and then induce nuclear replication and transcriptional disorders which lead to cell death. This result is in accordance with the enrichment of KEGG pathway. Through the KEGG pathway, a variety of classical pathways related to cell development, proliferation, and metabolism can be found. The pathways in cancer, PI3-Akt and p53 signaling pathways are the most noteworthy changes among numerous signal pathways. Through PPI analysis, 4 important differentially expressed proteins were identified, which played a role in exploring the mechanism of action of LCL161 in combination with CI and drug targeting for the subsequent treatment of BC drug-resistance. They were highly consistent with TMT results. CDC20 is mainly located in the nucleus, and IGTB1, RRM2, and CDK4 are mainly localized in the cytoplasm and organelles. IGTB1 is localized extracellular, and they help regulate tumor cell apoptosis through different pathways. The results of this research may be helpful in understanding the molecular mechanisms by which Smac mimics induce apoptosis in BC drug-resistant cells when caspase is inhibited. It can help to identify its potential as a target protein for promoting drug-resistant apoptosis and even reverse the possibility of drug resistance, thereby providing a new method for clinical treatment of BC drug-resistance. More importantly, our study provides a novel methodology that can be used for future research to reverse BC drug-resistance. Further studies are required to identify the association of differentially expressed proteins with Smac mimics in combination with CI, and the exact molecular mechanisms of action like post-translational modification.
Acknowledgements
We thank Jingjie PTM Biolabs for the help of mass spectrometry analysis.
list of ABBREVIATIONS
- BC
Breast Cancer
- CI
Caspase Inhibitors
- GO
Gene Ontology
- IAPs
Inhibitor of Apoptosis Proteins
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MS
Mass Spectrometry
- PPI
Protein-Protein Interaction
- PRM
Parallel Reaction Monitoring
- TMT
Tandem Mass Tag
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author, GJ, upon reasonable request.
FUNDING
This research was funded by the “Education Department of Anhui Natural Science Research Project China” under Grant (KJ2017A245) and the Postgraduate innovation scholarship of Bengbu medical college (Byycx1844).
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport B.L., Demetriou G.S., Moodley S.D., Benn C.A. When and how do I use neoadjuvant chemotherapy for breast cancer? Curr. Treat. Options Oncol. 2014;15(1):86–98. doi: 10.1007/s11864-013-0266-0. [DOI] [PubMed] [Google Scholar]
- 3.Tang Y., Wang Y., Kiani M.F., Wang B. Classification, treatment strategy, and associated drug resistance in breast cancer. Clin. Breast Cancer. 2016;16(5):335–343. doi: 10.1016/j.clbc.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Jeselsohn R., Brown M. 2016.
- 5.Tamm I., Wang Y., Sausville E., Scudiero D.A., Vigna N., Oltersdorf T., Reed J.C. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58(23):5315–5320. [PubMed] [Google Scholar]
- 6.Salvesen G.S., Duckett C.S. IAP proteins: Blocking the road to death’s door. Nat. Rev. Mol. Cell Biol. 2002;3(6):401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 7.Vasilikos L., Spilgies L.M., Knop J., Wong W.W. Regulating the balance between necroptosis, apoptosis and inflammation by inhibitors of apoptosis proteins. Immunol. Cell Biol. 2017;95(2):160–165. doi: 10.1038/icb.2016.118. [DOI] [PubMed] [Google Scholar]
- 8.Lee E.F., Harris T.J., Tran S., Evangelista M., Arulananda S., John T., Ramnac C., Hobbs C., Zhu H., Gunasingh G., Segal D., Behren A., Cebon J., Dobrovic A., Mariadason J.M., Strasser A., Rohrbeck L., Haass N.K., Herold M.J., Fairlie W.D. BCL-XL and MCL-1 are the key BCL-2 family proteins in melanoma cell survival. Cell Death Dis. 2019;10(5):342. doi: 10.1038/s41419-019-1568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mescolini G., Lupini C., Felice V., Guerrini A., Silveira F., Cecchinato M., Catelli E. Molecular characterization of the meq gene of Marek’s disease viruses detected in unvaccinated backyard chickens reveals the circulation of low- and high-virulence strains. Poult. Sci. 2019;98(8):3130–3137. doi: 10.3382/ps/pez095. [DOI] [PubMed] [Google Scholar]
- 10.Philchenkov A., Miura K. The IAP protein family, SMAC mimetics and cancer treatment. Crit. Rev. Oncog. 2016;21(3-4):185–202. doi: 10.1615/CritRevOncog.2016017032. [DOI] [PubMed] [Google Scholar]
- 11.Chesi M., Mirza N.N., Garbitt V.M., Sharik M.E., Dueck A.C., Asmann Y.W., Akhmetzyanova I., Kosiorek H.E., Calcinotto A., Riggs D.L., Keane N., Ahmann G.J., Morrison K.M., Fonseca R., Lacy M.Q., Dingli D., Kumar S.K., Ailawadhi S., Dispenzieri A., Buadi F., Gertz M.A., Reeder C.B., Lin Y., Chanan-Khan A.A., Stewart A.K., Fooksman D., Bergsagel P.L. IAP antagonists induce anti-tumor immunity in multiple myeloma. Nat. Med. 2016;22(12):1411–1420. doi: 10.1038/nm.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren K., Chong D., Ma L., Zhang Z., Zhao S. Effects of LCL161, a SMAC mimetic on the proliferation and apoptosis in hepatocellular carcinoma cells. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2016;41(9):898–904. doi: 10.11817/j.issn.1672-7347.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Gerges S., Rohde K., Fulda S. Cotreatment with SMAC mimetics and demethylating agents induces both apoptotic and necroptotic cell death pathways in acute lymphoblastic leukemia cells. Cancer Lett. 2016;375(1):127–132. doi: 10.1016/j.canlet.2016.02.040. [DOI] [PubMed] [Google Scholar]
- 14.Yang C., Wang H., Zhang B., Chen Y., Zhang Y., Sun X., Xiao G., Nan K., Ren H., Qin S. LCL161 increases paclitaxel-induced apoptosis by degrading cIAP1 and cIAP2 in NSCLC. J.Journal of experimental & clinical cancer research. CR (East Lansing Mich.) 2016;35(1):158. doi: 10.1186/s13046-016-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin G., Lan Y., Han F., Sun Y., Liu Z., Zhang M., Liu X., Zhang X., Hu J., Liu H., Wang B. SMAC mimetic-induced caspase-independent necroptosis requires RIP1 in breast cancer. Mol. Med. Rep. 2016;13(1):359–366. doi: 10.3892/mmr.2015.4542. [DOI] [PubMed] [Google Scholar]
- 16.Yousefi H. Long noncoding RNAs and exosomal lncRNAs: Classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2020;39(5):953–974. doi: 10.1038/s41388-019-1040-y. [DOI] [PubMed] [Google Scholar]
- 17.Weisberg E., Ray A., Barrett R., Nelson E., Christie A.L., Porter D., Straub C., Zawel L., Daley J.F., Lazo-Kallanian S., Stone R., Galinsky I., Frank D., Kung A.L., Griffin J.D. SMAC mimetics: Implications for enhancement of targeted therapies in leukemia. Leukemia. 2010;24(12):2100–2109. doi: 10.1038/leu.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao W., Lin Z. 2019.
- 19.Al-Alem L.F., Baker A.T., Pandya U.M., Eisenhauer E.L., Rueda B.R. Understanding and targeting apoptotic pathways in ovarian cancer. Cancers (Basel) 2019;11(11):E1631. doi: 10.3390/cancers11111631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanden Berghe T., Kaiser W.J., Bertrand M.J., Vandenabeele P. Molecular crosstalk between apoptosis, necroptosis, and survival signaling. Mol. Cell. Oncol. 2015;2(4):e975093. doi: 10.4161/23723556.2014.975093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramakrishnan V., Painuly U., Kimlinger T., Haug J., Rajkumar S.V., Kumar S. Inhibitor of apoptosis proteins as therapeutic targets in multiple myeloma. Leukemia. 2014;28(7):1519–1528. doi: 10.1038/leu.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C.W., Li Y., Hu S., Zhou W., Meng Y., Li Z., Zhang Y., Sun J., Bo Z., DePamphilis M.L., Yen Y., Han Z., Zhu W. DHS (trans-4,4′-dihydroxystilbene) suppresses DNA replication and tumor growth by inhibiting RRM2 (ribonucleotide reductase regulatory subunit M2). Oncogene. 2019;38(13):2364–2379. doi: 10.1038/s41388-018-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Zhi Q., Ye Q., Zhou C., Zhang L., Yan W., Wu Q., Zhang D., Li P., Huo K. SCYL1-BP1 affects cell cycle arrest in human hepatocellular carcinoma cells via Cyclin F and RRM2. Anticancer. Agents Med. Chem. 2016;16(4):440–446. doi: 10.2174/1871520615666150518093814. [DOI] [PubMed] [Google Scholar]
- 24.Lu A.G., Feng H., Wang P.X., Han D.P., Chen X.H., Zheng M.H. Emerging roles of the ribonucleotide reductase M2 in colorectal cancer and ultraviolet-induced DNA damage repair. World J. Gastroenterol. 2012;18(34):4704–4713. doi: 10.3748/wjg.v18.i34.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautam A., Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66(13):6497–6502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- 26.Fatkhutdinov N., Sproesser K., Krepler C., Liu Q., Brafford P.A., Herlyn M., Aird K.M., Zhang R. Targeting RRM2 and mutant BRAF is a novel combinatorial strategy for melanoma. Mol. Cancer Res. 2016;14(9):767–775. doi: 10.1158/1541-7786.MCR-16-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng S., Wang X., Weng Y.H., Jin X., Ji J.L., Guo L., Hu B., Liu N., Cheng Q., Zhang J., Bai H., Yang T., Xia X.H., Zhang H.Y., Gao S., Huang Y. siRNA knockdown of RRM2 effectively suppressed pancreatic tumor growth alone or synergistically with doxorubicin. J. Mol. Ther. Nucleic Acids. 2018;12:805–816. doi: 10.1016/j.omtn.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton T.R., Kashour T., Wright J.A., Amara F.M. Cellular signaling pathways affect the function of ribonucleotide reductase mRNA binding proteins: mRNA stabilization, drug resistance, and malignancy. Int. J. Oncol. 2003;22(1):21–31. doi: 10.3892/ijo.22.1.21. [Review]. [DOI] [PubMed] [Google Scholar]
- 29.Wang A., Zhao C., Liu X., Su W., Duan G., Xie Z., Chu S., Gao Y. Knockdown of TBRG4 affects tumorigenesis in human H1299 lung cancer cells by regulating DDIT3, CAV1 and RRM2. Oncol. Lett. 2018;15(1):121–128. doi: 10.3892/ol.2017.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrandina G., Mey V., Nannizzi S., Ricciardi S., Petrillo M., Ferlini C., Danesi R., Scambia G., Del Tacca M. Expression of nucleoside transporters, deoxycitidine kinase, ribonucleotide reductase regulatory subunits, and gemcitabine catabolic enzymes in primary ovarian cancer. Cancer Chemother. Pharmacol. 2010;65(4):679–686. doi: 10.1007/s00280-009-1073-y. [DOI] [PubMed] [Google Scholar]
- 31.Satow R., Shitashige M., Kanai Y., Takeshita F., Ojima H., Jigami T., Honda K., Kosuge T., Ochiya T., Hirohashi S., Yamada T. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin. Cancer Res. 2010;16(9):2518–2528. doi: 10.1158/1078-0432.CCR-09-2214. [DOI] [PubMed] [Google Scholar]
- 32.Zuckerman J.E., Hsueh T., Koya R.C., Davis M.E., Ribas A. siRNA knockdown of ribonucleotide reductase inhibits melanoma cell line proliferation alone or synergistically with temozolomide. J. Invest. Dermatol. 2011;131(2):453–460. doi: 10.1038/jid.2010.310. [DOI] [PubMed] [Google Scholar]
- 33.Grade M., Hummon A.B., Camps J., Emons G., Spitzner M., Gaedcke J., Hoermann P., Ebner R., Becker H., Difilippantonio M.J., Ghadimi B.M., Beissbarth T., Caplen N.J., Ried T. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int. J. Cancer. 2011;128(5):1069–1079. doi: 10.1002/ijc.25453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kittler R., Putz G., Pelletier L., Poser I., Heninger A.K., Drechsel D., Fischer S., Konstantinova I., Habermann B., Grabner H., Yaspo M.L., Himmelbauer H., Korn B., Neugebauer K., Pisabarro M.T., Buchholz F. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432(7020):1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 35.Liu X., Zhou B., Xue L., Yen F., Chu P., Un F., Yen Y. Ribonucleotide reductase subunits M2 and p53R2 are potential biomarkers for metastasis of colon cancer. J. Clin Colorectal Cancer. 2007;6(5):374–381. doi: 10.3816/CCC.2007.n.007. [DOI] [PubMed] [Google Scholar]
- 36.Liang W.H., Li N., Yuan Z.Q., Qian X.L., Wang Z.H. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol. Carcinog. 2019;58(4):461–473. doi: 10.1002/mc.22941. [DOI] [PubMed] [Google Scholar]
- 37.Fang Z., Gong C., Liu H., Zhang X., Mei L., Song M., Qiu L., Luo S., Zhu Z., Zhang R., Gu H., Chen X. E2F1 promote the aggressiveness of human colorectal cancer by activating the ribonucleotide reductase small subunit M2. Biochem. Biophys. Res. Commun. 2015;464(2):407–415. doi: 10.1016/j.bbrc.2015.06.103. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., Liu X., Warden C.D., Huang Y., Loera S., Xue L., Zhang S., Chu P., Zheng S., Yen Y. Prognostic and therapeutic significance of ribonucleotide reductase small subunit M2 in estrogen-negative breast cancers. BMC Cancer. 2014;14:664. doi: 10.1186/1471-2407-14-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee B., Ha S.Y., Song D.H., Lee H.W., Cho S.Y., Park C.K. High expression of ribonucleotide reductase subunit M2 correlates with poor prognosis of hepatocellular carcinoma. Gut Liver. 2014;8(6):662–668. doi: 10.5009/gnl13392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mah V., Alavi M., Márquez-Garbán D.C., Maresh E.L., Kim S.R., Horvath S., Bagryanova L., Huerta-Yepez S., Chia D., Pietras R., Goodglick L. Ribonucleotide reductase subunit M2 predicts survival in subgroups of patients with non-small cell lung carcinoma: effects of gender and smoking status. PLoS One. 2015;10(5):e0127600. doi: 10.1371/journal.pone.0127600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osako Y., Yoshino H., Sakaguchi T., Sugita S., Yonemori M., Nakagawa M., Enokida H. Potential tumor-suppressive role of microRNA-99a-3p in sunitinib-resistant renal cell carcinoma cells through the regulation of RRM2. Int. J. Oncol. 2019;54(5):1759–1770. doi: 10.3892/ijo.2019.4736. [DOI] [PubMed] [Google Scholar]
- 42.Lu H., Lu S., Yang D., Zhang L., Ye J., Li M., Hu W. MiR-20a-5p regulates gemcitabine chemosensitivity by targeting RRM2 in pancreatic cancer cells and serves as a predictor for gemcitabine based chemotherapy. J. Biosci. Rep. 2019;39(5) doi: 10.1042/BSR20181374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun H., Yang B., Zhang H., Song J., Zhang Y., Xing J., Yang Z., Wei C., Xu T., Yu Z., Xu Z., Hou M., Ji M., Zhang Y. RRM2 is a potential prognostic biomarker with functional significance in glioma. Int. J. Biol. Sci. 2019;15(3):533–543. doi: 10.7150/ijbs.30114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu; Jiamin, P.; Yayun, Z.; Bei, X.; Jianchao, W. Silencing RRM2 inhibits multiple myeloma by targeting the Wnt/β catenin signaling pathway. J. Mol. Medic. Rep. 2019;20(3):2159–2166. doi: 10.3892/mmr.2019.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M., Mangion J., Jacotot E., Costantini P., Loeffler M., Larochette N., Goodlett D.R., Aebersold R., Siderovski D.P., Penninger J.M., Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397(6718):441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 46.Miramar M.D., Costantini P., Ravagnan L., Saraiva L.M., Haouzi D., Brothers G., Penninger J.M., Peleato M.L., Kroemer G., Susin S.A. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 2001;276(19):16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 47.Li L.Y., Luo X., Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412(6842):95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 48.Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391(6662):43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 49.Ingham M., Schwartz G.K. Cell-cycle therapeutics come of age. J. Clin. Oncol. 2017;35(25):2949–2959. doi: 10.1200/JCO.2016.69.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Q., Geng Y., Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411(6841):1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 51.Yu Q., Sicinska E., Geng Y., Ahnström M., Zagozdzon A., Kong Y., Gardner H., Kiyokawa H., Harris L.N., Stål O., Sicinski P. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9(1):23–32. doi: 10.1016/j.ccr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Zhu S., Mott R.T., Fry E.A., Taneja P., Kulik G., Sui G., Inoue K. Cooperation between Dmp1 loss and cyclin D1 overexpression in breast cancer. Am. J. Pathol. 2013;183(4):1339–1350. doi: 10.1016/j.ajpath.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen F., Liu C., Zhang J., Xu W., Zhang Y. Progress of CDK4/6 inhibitor palbociclib in the treatment of cancer. Anticancer. Agents Med. Chem. 2018;18(9):1241–1251. doi: 10.2174/1871521409666170412123500. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz G.K., Shah M.A. Targeting the cell cycle: A new approach to cancer therapy. J. Clin. Oncol. 2005;23(36):9408–9421. doi: 10.1200/JCO.2005.01.5594. [DOI] [PubMed] [Google Scholar]
- 55.Massagué J.G. 1 cell-cycle control and cancer. Nature. 2004;432(7015):298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 56.Brookes S., Gagrica S., Sanij E., Rowe J., Gregory F.J., Hara E., Peters G. Evidence for a CDK4-dependent checkpoint in a conditional model of cellular senescence. Cell Cycle. 2015;14(8):1164–1173. doi: 10.1080/15384101.2015.1010866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finn R.S., Dering J., Conklin D., Kalous O., Cohen D.J., Desai A.J., Ginther C., Atefi M., Chen I., Fowst C., Los G., Slamon D.J. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wardell S.E., Ellis M.J., Alley H.M., Eisele K., VanArsdale T., Dann S.G., Arndt K.T., Primeau T., Griffin E., Shao J., Crowder R., Lai J.P., Norris J.D., McDonnell D.P., Li S. Efficacy of SERD/SERM hybrid-CDK4/6 inhibitor combinations in models of endocrine therapy-resistant breast cancer. Clin. Cancer Res. 2015;21(22):5121–5130. doi: 10.1158/1078-0432.CCR-15-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malorni L., Curigliano G., Minisini A.M., Cinieri S., Tondini C.A., D’Hollander K., Arpino G., Bernardo A., Martignetti A., Criscitiello C., Puglisi F., Pestrin M., Sanna G., Moretti E., Risi E., Biagioni C., McCartney A., Boni L., Buyse M., Migliaccio I., Biganzoli L., Di Leo A. Palbociclib as single agent or in combination with the endocrine therapy received before disease progression for estrogen receptor-positive, HER2-negative metastatic breast cancer: Trend trial. Ann. Oncol. 2018;29(8):1748–1754. doi: 10.1093/annonc/mdy214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haraguchi N., Inoue H., Tanaka F., Mimori K., Utsunomiya T., Sasaki A., Mori M. Cancer stem cells in human gastrointestinal cancers. Hum. Cell. 2006;19(1):24–29. doi: 10.1111/j.1749-0774.2005.00004.x. [DOI] [PubMed] [Google Scholar]
- 61.Corbeil D., Röper K., Hellwig A., Tavian M., Miraglia S., Watt S.M., Simmons P.J., Peault B., Buck D.W., Huttner W.B. The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J. Biol. Chem. 2000;275(8):5512–5520. doi: 10.1074/jbc.275.8.5512. [DOI] [PubMed] [Google Scholar]
- 62.Corbeil D., Röper K., Fargeas C.A., Joester A., Huttner W.B. Prominin: A story of cholesterol, plasma membrane protrusions and human pathology. Traffic. 2001;2(2):82–91. doi: 10.1034/j.1600-0854.2001.020202.x. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y.K., Zhu Y.L., Qiu F.M., Zhang T., Chen Z.G., Zheng S., Huang J. Activation of Akt and MAPK pathways enhances the tumorigenicity of CD133+ primary colon cancer cells. Carcinogenesis. 2010;31(8):1376–1380. doi: 10.1093/carcin/bgq120. [DOI] [PubMed] [Google Scholar]
- 64.Shmelkov S.V., Butler J.M., Hooper A.T., Hormigo A., Kushner J., St Milde T., Clair R., Baljevic M., White I., Jin D.K., Chadburn A., Murphy A.J., Valenzuela D.M., Gale N.W., Thurston G., Yancopoulos G.D., D’Angelica M., Kemeny N., Lyden D., Rafii S. CDI33 expression is not restricted to stem cells, and both CDI33+ and CDI33-metastatic colon cancer cells initiate tumors. J. Clin. Invest. 2008;118(6):2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bidlingmaier S., Zhu X., Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J. Mol. Med. (Berl.) 2008;86(9):1025–1032. doi: 10.1007/s00109-008-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song J., Zhang J., Wang J., Cao Z., Wang J., Guo X., Dong W. β1 integrin modulates tumor growth and apoptosis of human colorectal cancer. Oncol. Rep. 2014;32(1):302–308. doi: 10.3892/or.2014.3168. [DOI] [PubMed] [Google Scholar]
- 67.Song J., Zhang J., Wang J., Wang J., Guo X., Dong W. β1 integrin mediates colorectal cancer cell proliferation and migration through regulation of the Hedgehog pathway. Tumour Biol. 2015;36(3):2013–2021. doi: 10.1007/s13277-014-2808-x. [DOI] [PubMed] [Google Scholar]
- 68.Kim J.Y., Beart R.W., Shibata D. Stability of colon stem cell methylation after neo-adjuvant therapy in a patient with attenuated familial adenomatous polyposis. BMC Gastroenterol. 2005;5(1):19–19. doi: 10.1186/1471-230X-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Z., Zhou X., Liu Y., Gong C., Wei X., Zhang T., Ma D., Gao Q. Activation of integrin β1 mediates the increased malignant potential of ovarian cancer cells exerted by inflammatory cytokines. Anticancer. Agents Med. Chem. 2014;14(7):955–962. doi: 10.2174/1871520614666140613123108. [DOI] [PubMed] [Google Scholar]
- 70.Alfieri C., Chang L., Zhang Z., Yang J., Maslen S., Skehel M., Barford D. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature. 2016;536(7617):431–436. doi: 10.1038/nature19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gayyed M.F., El-Maqsoud N.M., Tawfiek E.R., El Gelany S.A., Rahman M.F. A comprehensive analysis of CDC20 overexpression in common malignant tumors from multiple organs: Its correlation with tumor grade and stage. Tumour Biol. 2016;37(1):749–762. doi: 10.1007/s13277-015-3808-1. [DOI] [PubMed] [Google Scholar]
- 72.Hou M.F., Luo C.W., Chang T.M., Hung W.C., Chen T.Y., Tsai Y.L., Chai C.Y., Pan M.R. The NuRD complex-mediated p21 suppression facilitates chemoresistance in BRCA-proficient breast cancer. Exp. Cell Res. 2017;359(2):458–465. doi: 10.1016/j.yexcr.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 73.O’Connor L., Strasser A., O’Reilly L.A., Hausmann G., Adams J.M., Cory S., Huang D.C. Bim: A novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17(2):384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wan L., Tan M., Yang J., Inuzuka H., Dai X., Wu T., Liu J., Shaik S., Chen G., Deng J., Malumbres M., Letai A., Kirschner M.W., Sun Y., Wei W. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev. Cell. 2014;29(4):377–391. doi: 10.1016/j.devcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng Y., Lv X., Wang X., Wang B., Shao X., Huang Y., Shi L., Chen Z., Huang J., Huang P. MiR-181b promotes chemoresistance in breast cancer by regulating Bim expression. Oncol. Rep. 2016;35(2):683–690. doi: 10.3892/or.2015.4417. [DOI] [PubMed] [Google Scholar]
- 76.Woo S.M., Min K.J., Kwon T.K. Melatonin-mediated Bim up-regulation and cyclooxygenase-2 (COX-2) down-regulation enhances tunicamycin-induced apoptosis in MDA-MB-231 cells. J. Pineal Res. 2015;58(3):310–320. doi: 10.1111/jpi.12217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, GJ, upon reasonable request.