Abstract
Cardiac metabolism plays a crucial role in producing sufficient energy to sustain cardiac function. However, the role of metabolism in different aspects of cardiomyocyte regeneration remains unclear. Working with the adult zebrafish heart regeneration model, we first find an increase in the levels of mRNAs encoding enzymes regulating glucose and pyruvate metabolism, including pyruvate kinase M1/2 (Pkm) and pyruvate dehydrogenase kinases (Pdks), especially in tissues bordering the damaged area. We further find that impaired glycolysis decreases the number of proliferating cardiomyocytes following injury. These observations are supported by analyses using loss‐of‐function models for the metabolic regulators Pkma2 and peroxisome proliferator‐activated receptor gamma coactivator 1 alpha. Cardiomyocyte‐specific loss‐ and gain‐of‐function manipulations of pyruvate metabolism using Pdk3 as well as a catalytic subunit of the pyruvate dehydrogenase complex (PDC) reveal its importance in cardiomyocyte dedifferentiation and proliferation after injury. Furthermore, we find that PDK activity can modulate cell cycle progression and protrusive activity in mammalian cardiomyocytes in culture. Our findings reveal new roles for cardiac metabolism and the PDK‐PDC axis in cardiomyocyte behavior following cardiac injury.
Keywords: cardiac regeneration, cardiomyocyte proliferation, glycolysis, metabolism, zebrafish
Subject Categories: Development & Differentiation, Metabolism
Cardiac metabolism regulates cardiomyocyte dedifferentiation and proliferation in injured zebrafish hearts as well as cell cycle progression in mammalian cardiomyocytes.

Introduction
Cardiac metabolism is essential to produce adenosine triphosphate (ATP), which is required for cardiac homeostasis and function (Neely & Morgan, 1974; Kolwicz et al, 2013). During development, embryonic cardiomyocytes (CMs) mainly utilize glucose metabolism for ATP production (Piquereau & Ventura‐Clapier, 2018). As the heart grows, CMs exhibit metabolic changes and rely mostly on fatty acid oxidation (Cho et al, 2006; Chung et al, 2007, 2010; Kreipke et al, 2016). It has also been shown that under conditions of cardiac disease, such as ischemia and heart failure, CMs revert to glucose as an energy source (Das et al, 1987; Doenst et al, 2013).
Unlike adult mammals, adult zebrafish regenerate their heart effectively after injury (Poss et al, 2002). During cardiac regeneration, CMs re‐express hand2 (Schindler et al, 2014), a transcription factor gene required for cardiac development, as well as an embryonic myosin (Sallin et al, 2015), and they also exhibit sarcomere disassembly (Jopling et al, 2010; Ben‐Yair et al, 2019; Beisaw et al, 2020). These and other studies (Kikuchi et al, 2010; Wu et al, 2016; Grajevskaja et al, 2018) suggest that in zebrafish, CMs undergo dedifferentiation to supply new CMs during cardiac regeneration. Thus, CMs may utilize similar regulatory mechanisms during development and regeneration, yet the role of cardiac metabolism in these processes remains unclear.
Pyruvate dehydrogenase kinase plays an essential role in pyruvate metabolism (Gudi et al, 1995; Takubo et al, 2013; Zhang et al, 2014). Glucose is initially converted to pyruvate through several glycolytic intermediates, and then, pyruvate is converted to lactate via anaerobic fermentation (Lehninger et al, 2005), or to acetyl‐coenzyme A (acetyl‐CoA) by the PDC (Behal et al, 1993). By inhibiting the conversion of pyruvate to acetyl‐CoA, PDK induces the conversion of pyruvate to lactate, via anaerobic fermentation (Korotchkina & Patel, 2001; Koukourakis et al, 2005). These studies indicate that PDK regulates the shift of pyruvate metabolism between glycolysis and OXPHOS (Korotchkina & Patel, 2001; Koukourakis et al, 2005; Takubo et al, 2013; Park et al, 2018). Higher expression of PDKs in cancer and stem cells, which rely mainly on glycolysis, has been associated with their enhanced proliferation and migration (Takubo et al, 2013; Zhang et al, 2014; Park et al, 2018).
Here, we first show that glycolytic enzymes are upregulated at the transcriptional level during cardiac regeneration. Pharmacological and genetic manipulations as well as CM‐specific modulation of glycolytic activity reveal its importance in CM proliferation after injury. We also found that modulation of pyruvate metabolism can affect mammalian CM cell cycle progression and cell behavior. Transcriptomic analyses show that PDK3 overexpression (OE) leads to higher levels of genes encoding cell cycle regulators in CMs. These data indicate that metabolism plays an important role in CM proliferation following cardiac injury.
Results and Discussion
Glycolysis and pyruvate metabolism modulate cardiomyocyte dedifferentiation and proliferation following cardiac injury
Adult mammalian CMs rely mainly on fatty acid oxidation as a source of energy (Cho et al, 2006; Chung et al, 2007, 2010; Kreipke et al, 2016). We started by performing proteomic analysis and found higher levels of fatty acid oxidation enzymes in adult zebrafish ventricles compared to those in juveniles (Fig 1A and B). These findings are consistent with mammalian models, in which adult CMs express higher levels of fatty acid oxidation enzymes (Puente et al, 2014; Fukushima et al, 2019).
Figure 1. Glycolysis and pyruvate metabolism play an important role following cardiac injury.

-
A30 and 150 days post fertilization (dpf) zebrafish ventricles were isolated and relative protein levels determined (n = 3 biological replicates).
-
BKEGG over‐representation analysis for selected categories of upregulated proteins in 150 dpf hearts compared to 30 dpf hearts.
-
CqPCR analysis of mRNA levels of glycolytic enzyme genes in 5 dpci remote and wound border cardiac tissue (n = 2–3 technical replicates using pooled cDNA from 10 ventricles for each condition (n = 2 for pkmb and pdk4, and n = 3 for the other genes)).
-
D, EImmunostaining of heart sections for PCNA and MEF2 (D) or N2.261 and MEF2 (E) in 5 dpci animals treated with PBS, 2‐DG, or DCA; magnified view of area in white boxes shown below; white dashed lines outline the wound area; arrowheads point to PCNA + (D) or N2.261+ (E) CMs; percentage of PCNA + or N2.261+ CMs in the border zone shown on the right (n = 4–5 ventricles).
It has been suggested that zebrafish CMs utilize similar regulatory mechanisms during cardiac development and regeneration (Poss et al, 2002; Jopling et al, 2010; Kikuchi et al, 2010; Wu et al, 2016). We have recently shown that glycolysis plays an important role during zebrafish cardiac development (Fukuda et al, 2019). Thus, we wanted to examine whether the expression levels of glycolytic enzyme genes were changed in zebrafish ventricles after cryoinjury. We found significant upregulation of many of them in the tissues bordering the damaged area when compared to remote areas (Fig 1C). This finding is supported by a single‐cell RNA‐sequencing analysis in regenerating zebrafish hearts (Honkoop et al, 2019). Notably, we found high upregulation of pdk2b, pdk3b, and pdk4 (Fig 1C), which encode key enzymes that regulate pyruvate metabolism and promote glycolysis in a number of cell types including CMs (Zhao et al, 2008) and cancer cells (Koukourakis et al, 2005; Leclerc et al, 2017; Peng et al, 2018). We also measured lactate levels, which are indicative of glycolytic activity, and found higher levels in regenerating cardiac tissues (Fig EV1A). These and other observations (Honkoop et al, 2019) suggest that adult zebrafish CMs switch from fatty acid oxidation to glycolytic metabolism in response to cardiac injury. In order to examine the role of glycolysis and Pdk function following cardiac injury, we treated zebrafish with 2‐deoxy‐D‐glucose (2‐DG) and dichloroacetate (DCA), which decrease glycolysis and Pdk function, respectively. Notably, these treatments led to a decrease in CM proliferation in the border zone following injury (Figs 1D and EV1B). We also found that the re‐expression of embryonic myosin, which has been reported to indicate CM dedifferentiation during cardiac regeneration (Sallin et al, 2015), was reduced when glycolysis or Pdk function was decreased (Fig 1E). These data suggest that glycolysis and pyruvate metabolism as regulated by Pdk play a role in border zone CMs following cardiac injury.
Figure EV1. Relative quantification of selected proteins from mass spectrometry data.

- Lactate levels were measured in 5 dpci wound border cardiac tissue after treatment with PBS, 2‐DG, or DCA compared to uninjured (n = 3 biological replicates, each consisting of tissue from 10 ventricles).
- Staining of heart sections for phospho histone‐3 (pHH3), MF20, and DNA (DAPI) in 5 dpci animals after treatment with PBS, 2‐DG, or DCA; magnified view of area in white boxes shown in the top right corner; arrowhead points to pHH3+ CMs; white dashed lines outline the wound area; percentage of pHH3+ CMs in the border zone shown on the right (n = 4–5 ventricles).
Genetic manipulation of metabolic enzymes affects cardiomyocyte dedifferentiation and proliferation following cardiac injury
Next, we assessed whether the loss of specific metabolic enzymes affected CM behavior after cardiac injury. We first examined the role of Pkm2. Mammalian Pkm gives rise to two splice isoforms, Pkm1 and Pkm2; PKM2 promotes glucose metabolism toward lactate, while PKM1 promotes glucose metabolism toward acetyl‐CoA (Lehninger et al, 2005; Christofk et al, 2008). Knockdown of PKM2 has been reported to lead to decreased glycolytic activity in human umbilical vein endothelial cells (Stone et al, 2018) and intestinal stem cells (Kim et al, 2019), as well as upregulation of PKM1 (Lunt et al, 2015; Zheng et al, 2016). Similarly, Pkm2 OE in mouse CMs leads to enhanced glycolysis (Magadum et al, 2020). The zebrafish genome contains two pkm genes (pkma and pkmb), with pkma encoding both an orthologue of mammalian Pkm1 (pkma1) and an orthologue of mammalian Pkm2 (pkma2), while pkmb encodes another orthologue of mammalian Pkm2 (Stone et al, 2018). Expression analysis in adult ventricles indicates that pkma is upregulated in tissues bordering the injured area, while pkmb is not (Fig 1C), suggesting a role for pkma following cardiac injury. In pkma2 mutant zebrafish, the pkma1 isoform, which promotes glucose metabolism toward acetyl‐CoA, remains intact, suggesting that the metabolism of pyruvate to lactate is impaired while pyruvate oxidation is increased. These data indicate that a zebrafish pkma2 mutant constitutes a potential model of impaired glycolysis, as pyruvate is expected to be converted preferentially to acetyl‐CoA and not lactate. Thus, we examined pkma2 −/−; pkmb +/− animals, which do not display obvious cardiac defects compared to WT (Fig EV2A–C), and found that they exhibited a reduction in the number of proliferating CMs after cardiac injury (Figs 2A and EV2D). Consistently, the expression of embryonic myosin was also decreased in pkma2 −/−; pkmb +/− animals (Fig 2B). We also examined pkma2 −/− animals and found that they exhibit a similar decrease in PCNA+ and N2.261+ CMs following cardiac injury (Figs EV2E and F). Together, these results suggest that reducing glycolysis by affecting Pkma2 function leads to an attenuation in CM dedifferentiation and proliferation during regeneration.
Figure EV2. Loss of pkma or ppargc1a does not lead to obvious alterations in heart size or embryonic myosin expression in uninjured hearts.

-
A, BStaining of heart sections for MF20, DNA (DAPI), and phalloidin (A) or MEF2, DNA (DAPI), and N2.261 (B) in uninjured WT, pkma2 −/−; pkmb +/−, and ppargc1a −/− animals; magnified view of area in white boxes shown on the right.
-
CPercentage of heart weight to body weight in adult WT, pkma2 −/−; pkmb +/−, and ppargc1a −/− animals (n = 7 animals of each genotype).
-
DStaining of heart sections for pHH3, MF20, and DNA (DAPI) in 5 dpci pkma2 +/−; pkmb +/− and pkma2 −/−; pkmb +/− animals; magnified view of area in white boxes shown in the top right corner; arrowheads point to pHH3+ CMs; white dashed lines outline the wound area; percentage of pHH3+ CMs in the border zone shown on the right (n = 4–5 ventricles of each genotype).
-
E, FImmunostaining of heart sections for PCNA and MEF2 (E) or N2.261 and MEF2 (F) in 5 dpci pkma2 +/− and pkma2 −/− animals; magnified view of area in white boxes shown in the top right corner; arrowheads point to PCNA+ (E) or N2.261+ (F) CMs; white dashed lines outline the wound area; percentage of PCNA+ or N2.261+ CMs in the border zone shown on the right (n = 3 ventricles of each genotype).
-
GStaining of heart sections for pHH3, MF20, and DNA (DAPI) in 5 dpci ppargc1a +/+, ppargc1a −/− animals; magnified view of area in white boxes shown in the top right corner; arrowheads point to pHH3+ CMs; white dashed lines outline the wound area; percentage of pHH3+ CMs in the border zone shown on the right (n = 4–5 ventricles of each genotype).
-
HqPCR analysis of ppargc1a, atp5pf, and ndufb5 mRNA levels in 5 dpci remote and wound border heart tissues (n = 3 technical replicates using pooled cDNA from 10 ventricles for each condition).
Figure 2. Loss of pkma2 and ppargc1a affects cardiomyocyte dedifferentiation and proliferation following cardiac injury.
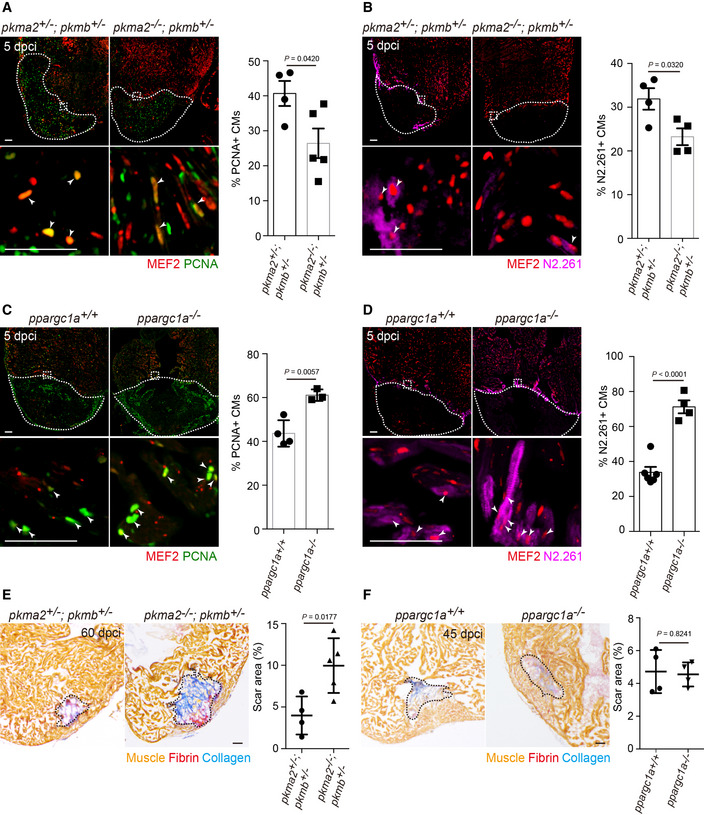
-
A–DImmunostaining of heart sections for PCNA and MEF2 (A, C) or N2.261 and MEF2 (B, D) in 5 dpci pkma2 +/−; pkmb +/− and pkma2 −/−; pkmb +/− (A, B) or ppargc1a +/+ and ppargc1a −/− (C, D) animals; magnified view of area in white boxes shown below; arrowheads point to PCNA + (A, C) or N2.261+ (B, D) CMs; white dashed lines outline the wound area; percentage of PCNA + (A, C) or N2.261+ (B, D) CMs in the border zone shown on the right (n = 4–6 ventricles of each genotype).
-
E, FAFOG staining of heart sections from 60 dpci pkma2 +/−; pkmb +/− and pkma2 −/−; pkmb +/− (E) or 45 dpci ppargc1a +/+ and ppargc1a −/− (F) animals; black dashed lines outline the scar area; scar area measured on the right (n = 3–5 ventricles of each genotype).
Conversely, in order to determine whether enhanced glycolytic activity can promote cardiac regeneration, we examined the role of peroxisome proliferator‐activated receptor gamma coactivator 1 alpha (ppargc1a), which encodes a key regulator of mitochondrial oxidative metabolism (Lin et al, 2005). Forced Ppargc1a expression in CMs enhances oxygen consumption and mitochondrial respiration in vitro and in vivo (Lehman et al, 2000), while Ppargc1a‐deficient mice exhibit decreased oxygen consumption in their left ventricular tissues (Lehman et al, 2008). Zebrafish ppargc1a mutant hearts exhibit reduced mitochondrial oxidative metabolism and increased glycolytic activity (Marin‐Juez et al, 2019) without obvious cardiac defects (Fig EV2A–C). Altogether, these data suggest that in ppargc1a mutant hearts, pyruvate oxidation is reduced while metabolism of pyruvate to lactate is increased, and thus, ppargc1a mutants constitute a potential model of increased glycolysis. First, we examined the mRNA expression levels of ppargc1a and its targets atp5pf and ndufb5 and found that these genes were down‐regulated in the cardiac tissues bordering the damaged area at 5 dpci (Fig EV2H). Notably, ppargc1a mutants exhibited an increase in proliferating CMs (Figs 2C and EV2G) as well as in the expression of embryonic myosin (Fig 2D) following cardiac injury, suggesting that promoting glycolysis in CMs can increase their dedifferentiation and proliferative potential. Furthermore, we examined pkma2 −/−; pkmb +/− animals at 60 dpci. We performed Acid Fuchsin Orange G (AFOG) staining to examine scar size. Notably, pkma2 −/−; pkmb +/− animals exhibited a significantly larger scar area compared to pkma2 +/−; pkmb +/− animals (Fig 2E). However, ppargc1a mutants did not exhibit a significant difference in scar area (Fig 2F). Together, these results provide further evidence that key regulators of pyruvate metabolism and glycolysis play an important role in CM dedifferentiation and proliferation in the early stages of cardiac regeneration.
Cardiomyocyte‐specific modulation of pyruvate metabolism regulates cardiomyocyte behavior following cardiac injury
Expression analysis showed that multiple pdk genes were highly upregulated in tissues bordering the damaged area following cardiac injury (Fig 1C). Pdk promotes the conversion of pyruvate to lactate by inactivating the PDC through phosphorylation of pyruvate dehydrogenase E1 alpha 1 subunit (Pdha1), a catalytic subunit of the PDC (Korotchkina & Patel, 2001; Rardin et al, 2009; Park et al, 2018). To impair glycolysis specifically in CMs, we used a form of Pdha1a (Pdha1aSTA), which cannot be phosphorylated by Pdk, leading to enhanced pyruvate conversion to acetyl‐CoA (Hitosugi et al, 2011; Fan et al, 2014; Fukuda et al, 2019). Using the HOTcre system (Hesselson et al, 2009), we generated a Tg(hsp70l:LOXP‐STOP‐LOXP‐pdha1aSTA‐T2A‐mCherry) line, which, when combined with the Tg(myl7:Cre‐ERT2) line and following tamoxifen and heat‐shock treatments, expresses pdha1aSTA specifically in CMs to impair glycolysis (Fig EV3A). Consistent with our previous results with DCA treatments, which inhibit Pdk function, the number of proliferating CMs following cardiac injury was decreased in Tg(myl7:Cre‐ERT2); Tg(hsp70l:LOXP‐STOP‐LOXP‐pdha1aSTA‐T2A‐mCherry) animals after tamoxifen‐induced recombination and heat‐shock treatments (Figs 3A and B, and EV3C). Conversely, in order to stimulate glycolysis in CMs, we generated a Tg(hsp70l:LOX‐STOP‐LOXP‐pdk3b‐T2A‐mCherry) line and found that when pdk3b was overexpressed in Tg(myl7:Cre‐ERT2); Tg(hsp70l:LOXP‐STOP‐LOXP‐pdk3b‐T2A‐mCherry) animals (Fig EV3B), the number of proliferating CMs following cardiac injury was increased (Figs 3C and EV3D). We also examined the effects of pdk3b OE in non‐injured hearts and saw no significant increase in the number of proliferating CMs (Fig EV3E). In line with their effects on CM proliferation, pdha1aSTA OE in CMs led to a decrease in the number of CMs exhibiting embryonic myosin expression, while pdk3b OE led to an increase (Fig 3D and E). Altogether, these results suggest that pyruvate metabolism as regulated by the PDK‐PDC axis plays an important role in CM dedifferentiation and proliferation following cardiac injury. We next examined whether metabolic modulation in CMs affects scar size during cardiac regeneration, and found that pdha1aSTA OE led to the retention of a larger scar area at 60 dpci (Fig 3F), while pdk3b OE did not affect scar area (Fig 3G). Together with the data using metabolic enzyme mutant models, these results suggest that in the early stages of cardiac regeneration, glycolysis promotes CM dedifferentiation and proliferation and that genetic manipulations to enhance glycolytic activity do not facilitate scar resolution at later stages.
Figure EV3. Validation of Cre‐ERT2‐mediated recombination in transgenic animals.

-
A, BStaining of heart sections for MF20, mCherry, and DNA (DAPI) in Tg(myl7:Cre‐ERT2); Tg(hsp70l:LOXP‐STOP‐LOXP‐pdha1aSTA‐T2A‐mCherry) (A) and Tg(myl7:Cre‐ERT2); Tg(hsp70l:LOXP‐STOP‐LOXP‐pdk3b‐T2A‐mCherry) (B) animals with or without tamoxifen and heat‐shock treatments. TAM, tamoxifen; HS, heat shock.
-
C, DStaining of heart sections for pHH3, MF20, and DNA (DAPI) in 5 dpci animals (Tg(hsp70l:LOXP‐STOP‐LOXP‐pdha1aSTA‐T2A‐mCherry) (C) and Tg(hsp70l:LOXP‐STOP‐LOXP‐pdk3b‐T2A‐mCherry) (D) alone (control) or in combination with Tg(myl7:Cre‐ERT2), all after tamoxifen and heat‐shock treatments); magnified view of area in white boxes shown in the top right corner; arrowheads point to pHH3+ CMs; white dashed lines outline the wound area; percentage of pHH3+ CMs in the border zone shown on the right (n = 4–5 ventricles of each genotype).
-
EStaining of heart sections for PCNA, MEF2, and DNA (DAPI) in animals (Tg(hsp70l:LOXP‐STOP‐LOXP‐pdk3b‐T2A‐mCherry) alone (control) or in combination with Tg(myl7:Cre‐ERT2), all after tamoxifen and heat‐shock treatments).
Figure 3. Cardiomyocyte‐specific modulation of pyruvate metabolism cell‐autonomously affects their dedifferentiation and proliferation following cardiac injury.
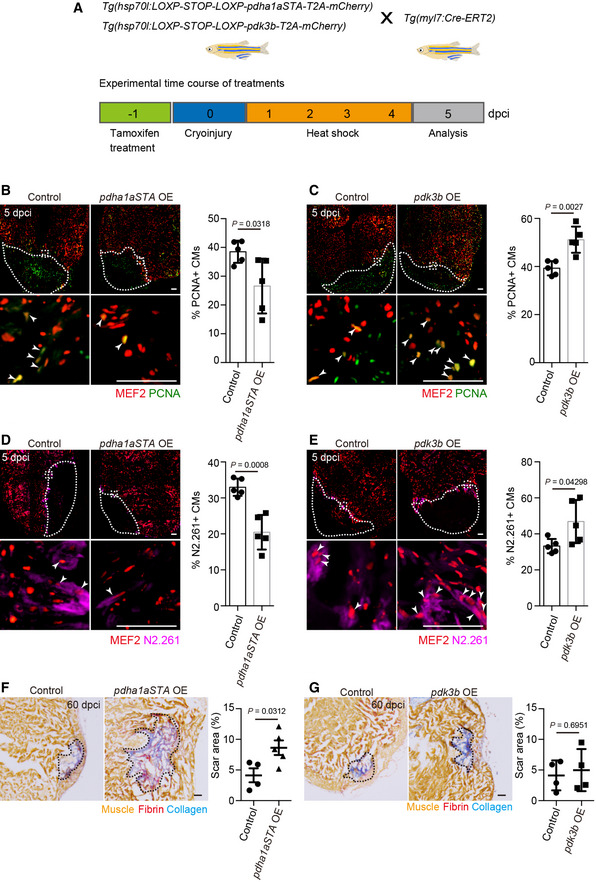
-
ATransgenic animals and experimental time course for data shown in (B–E).
-
B–EImmunostaining of heart sections for PCNA and MEF2 (B, C) or N2.261 and MEF2 (D, E) in 5 dpci animals (Tg(hsp70l:LOXP‐STOP‐LOXP‐pdha1aSTA‐T2A‐mCherry) and Tg(hsp70l:LOXP‐STOP‐LOXP‐pdk3b‐T2A‐mCherry) alone (control) or in combination with Tg(myl7:Cre‐ERT2), all after tamoxifen and heat‐shock treatments); magnified view of area in white boxes shown below; arrowheads point to PCNA + (B, C) or N2.261+ (D, E) CMs; white dashed lines outline the wound area; percentage of PCNA + (B, C) or N2.261+ (D, E) CMs in the border zone shown on the right (n = 4–5 ventricles of each genotype).
-
F, GAFOG staining of heart sections from 60 dpci animals (Tg(hsp70l:LOXP‐STOP‐LOXP‐pdha1aSTA‐T2A‐mCherry) (F) and Tg(hsp70l:LOXP‐STOP‐LOXP‐pdk3b‐T2A‐mCherry) (G) alone (control) or in combination with Tg(myl7:Cre‐ERT2), all after tamoxifen and heat‐shock treatments); black dashed lines outline the scar area; scar area measured on the right (n = 4–5 ventricles of each genotype).
Modulation of pyruvate metabolism regulates primary rat neonatal cardiomyocyte cell cycle progression and behavior
We also tested the role of pyruvate metabolism in primary rat neonatal cardiomyocytes (RNCMs). To examine the role of pyruvate metabolism modulation in CM cell cycle progression, Ki67 expression was analyzed in RNCMs overexpressing PDHA1STA or PDK3 in growth culture medium containing 10% bovine serum as well as in non‐growth culture medium. We found that PDHA1STA OE significantly decreased the number of Ki67+ RNCMs in growth conditions (Fig 4A), while PDK3 OE increased the number of Ki67+ RNCMs (Fig 4B) in non‐growth conditions. PDK3 OE also led to an increase in the number of pHH3+ RNCMs (Fig EV4C). We then determined cell numbers and found that PDHA1STA OE significantly decreased the number of RNCMs (Fig EV4D), while PDK3 OE did not lead to obvious changes (Fig EV4E). Next, we investigated the role of pyruvate metabolism in an in vitro scratch assay which is used to examine the ability of cells to fill a wound‐like gap in a confluent cell monolayer (Cormier et al, 2015). We first examined CM cell cycle progression in the scratch assay and found no significant differences in the number of Ki67+ CMs in the area close to the scratch border compared to the remote area (Fig EV4A and B). We then examined CM behavior and found that RNCMs close to the scratch border exhibited membrane protrusions (Fig 4C). Similar observations have been reported when analyzing zebrafish and mouse regeneration models, where CMs replenish to the injured area (Itou et al, 2012; Morikawa et al, 2015; Tahara et al, 2016; Marin‐Juez et al, 2019; preprint: Aharonov et al, 2020). We analyzed the effects of pyruvate metabolism modulation on CM behavior in the scratch assay and found that PDK3 OE led to an increase in the number of CM membrane protrusions (Figs 4C and EV4C). To gain insight into the signaling pathways and effectors that are regulated by PDK3 OE in RNCMs, we performed transcriptomic analysis. Notably, the resulting data indicate that PDK3 OE led to increased levels of genes encoding factors which promote DNA replication and cell cycle (Figs 4D–F and EV4F and G). Altogether, these data indicate that pyruvate metabolism regulated by PDK also promotes mammalian CM cell cycle progression and protrusive behavior.
Figure 4. Pyruvate metabolism modulation affects the behavior of rat neonatal cardiomyocytes in culture.
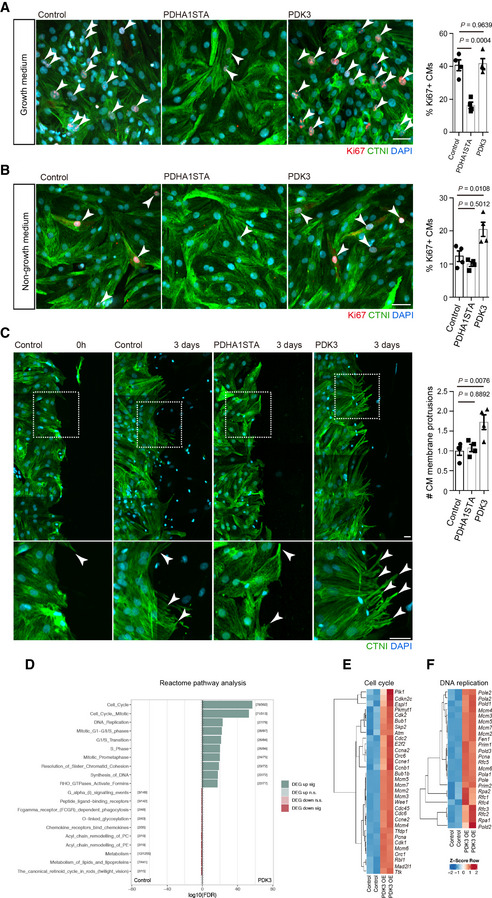
-
A, BStaining for Ki67, CTNI, and DNA (DAPI) in control, PDHA1STA OE, or PDK3 OE RNCMs cultured in growth (A) or non‐growth (B) medium; arrowheads point to Ki67+ RNCMs; percentage of Ki67+ RNCMs shown on the right (n = 4 biological replicates for each condition).
-
CStaining for CTNI and DNA (DAPI) in scratch assay using control, PDHA1STA OE, or PDK3 OE RNCMs immediately or at 3 days after generating the scratch; magnified view of area in white boxes shown below; arrowheads point to membrane protrusions of RNCMs; quantification of CM membrane protrusions shown on the right (n = 4 biological replicates for each condition).
-
DReactome over‐representation analysis of differentially regulated genes in PDK3 OE RNCMs compared to control.
-
E, FHeat map of gene expression for key regulators of cell cycle (E) and DNA replication (F) in PDK3 OE vs. control RNCMs.
Figure EV4. Ki67 immunostaining in scratch assay.

-
AStaining for Ki67, CTNI, and DNA (DAPI) in the scratch border and remote area; arrowheads point to Ki67+ RNCMs; and white dashed line marks the scratch border.
-
BPercentage of Ki67+ CMs in the 200 μm region adjacent to the scratch and in a remote area (n = 4 biological replicates for each condition).
-
CStaining for pHH3, CTNI, and DNA (DAPI) in control and PDK3 OE RNCMs; arrowhead points to pHH3+ RNCMs; percentage of pHH3+ RNCMs shown on the right (n = 4 biological replicates for each condition).
-
D, ERelative value of RNCM numbers (n = 3 biological replicates for each condition).
-
F, GGene set over‐representation analysis with KOBAS 2.0 using KEGG and Gene Ontology databases based on differentially regulated genes in PDK3 OE RNCMs compared to control.
Glycolysis plays an important role in the proliferation of cancer cells (Liberti & Locasale, 2016), endothelial cells (De Bock et al, 2013; Wilhelm et al, 2016), and neural progenitor cells (Zheng et al, 2016), as well as during cardiac (Wang et al, 2018; Honkoop et al, 2019; Magadum et al, 2020) and skeletal muscle (Wagner et al, 1976; Magadum & Engel, 2018) regeneration. Here, we showed that CM‐specific modulation of glycolysis also regulates CM dedifferentiation and proliferation during regeneration. In addition, our transcriptomic data show that PDK3 OE, which enhances glycolysis, leads to higher levels of cell cycle regulator gene expression in RNCMs. It will of course be important to investigate further how glycolysis regulates these processes.
Our data indicate that increased glycolysis by Pdk3 OE promotes CM dedifferentiation following cardiac injury. It was reported that in regenerating CMs, mitochondria exhibit an immature structure and that the levels of mitochondrial gene expression are reduced (Honkoop et al, 2019), indicating lower mitochondrial activity. In addition, mitochondrial function increases during CM maturation in the developing heart (Menendez‐Montes et al, 2016). Thus, reduced mitochondrial activity, and thereby reduced OXPHOS, may be important for CM dedifferentiation and proliferation during regeneration.
It has also been shown that the levels of glycolytic enzymes increase following cardiac ischemia and in failing hearts (Das et al, 1987; Doenst et al, 2013). However, adult mice do not exhibit robust cardiac regeneration. Recent data show that the upregulation of PKM2 levels following cardiac injury is limited and thus possibly insufficient to promote CM proliferation in an ischemic adult mouse model (Magadum et al, 2020), while Pkm2 OE induced CM proliferation after cardiac injury (Magadum et al, 2020). Together, these data suggest that the levels of glycolytic gene expression in mice following cardiac injury might be a limiting factor in terms of promoting CM proliferation. It will be interesting to further investigate the differences in metabolic changes between zebrafish and mouse cardiac injury models.
During cardiac regeneration, it is assumed that newly divided CMs have to undergo maturation to generate fully functional cardiac tissue. During development, the metabolic shift from glycolysis to OXPHOS plays an important role in CM differentiation and maturation (Cho et al, 2006; Chung et al, 2007). Our data in zebrafish indicate that while enhanced glycolysis by Pdk3 OE promotes CM proliferation during the early stages of cardiac regeneration, scar resolution at later stages does not appear to be significantly affected. However, it remains unclear whether, in conditions of Pdk3 OE, the newly formed CMs have undergone maturation and also whether CM maturation promotes scar resolution. Clearly, many questions remain to be addressed to fully understand the role of metabolism during cardiac regeneration.
Together, our data indicate that cardiac metabolism regulated by the PDK‐PDC axis promotes CM proliferation following cardiac injury in zebrafish as well as cell cycle progression of mammalian CMs in culture.
Materials and Methods
Zebrafish
All zebrafish husbandry was performed under standard conditions in accordance with FELASA guidelines (Aleström et al, 2019) and institutional (MPG) and national ethical and animal welfare guidelines. The following transgenic lines and mutants were used: Tg(cryaa:DsRed,‐5.1myl7:CreERT2) pd10 (Kikuchi et al, 2010) abbreviated Tg(myl7:CreERT2), pkma2s717 (Stone et al, 2018), pkmb s718 (Stone et al, 2018), and ppargc1a bns176 (Marin‐Juez et al, 2019). To generate Tg(cryaa:Cerulean,hsp70l:LOXP‐STOP‐LOXP‐pdha1aSTA‐T2A‐mCherry) bns343 abbreviated Tg(hsp70l:LOXP‐STOP‐LOXP‐pdha1aSTA‐T2A‐mCherry) and Tg(cryaa:Cerulean,hsp70l:LOXP‐STOP‐LOXP‐pdk3b‐T2A‐mCherry) bns344 abbreviated Tg(hsp70l:LOXP‐STOP‐LOXP‐pdk3b‐T2A‐mCherry), pdk3b (NM_001080688) and pdha1a (NM_213393) were isolated by RT–PCR and cloned into a vector containing a promoter of heat‐shock cognate 70‐kd protein, like (hsp70l), and two I‐SceI restriction sites. The following primers were used to amplify the cDNA: pdk3b (forward 5′‐TTCCACTAGTGCTCCACCGGTCCACCATGAAACTGTTTATCTG‐3′ and reverse 5′‐GCCCTCTCCACTGCCCTCGAGTCTGTTGACTTTGTATGTGGA‐3′); pdha1a (forward 5′‐TTCCACTAGTGCTCCACCGGTCCACCATGAGAAAGATGC‐3′ and reverse 5′‐GCCCTCTCCACTGCCCTCGAGGCTGATGGACTTGAGTTTG‐3′). Plasmids were then injected into one‐cell‐stage embryos with I‐SceI (NEB).
Cryoinjury and quantification
Cryoinjury was performed as previously described (González‐Rosa et al, 2011). Confocal images were processed using the ZEN software (ZEISS), and PCNA+, N2.261+, or pHH3+ CMs in the 50‐μm region adjacent to the injured area at 5 dpci were counted. The percentage of PCNA+, N2.261+, or pHH3+ CMs in the border zone was calculated by dividing the number of PCNA+, N2.261+, or pHH3+ CMs by the total number of CMs.
Pharmacological treatments
Adult zebrafish were treated with PBS, 2‐DG (1 nmol/mg body weight; Sigma‐Aldrich), or DCA (0.5 nmol/mg body weight; Sigma‐Aldrich) by intraperitoneal (IP) injections (Kinkel et al, 2010) from 1 to 4 dpci and analyzed at 5 dpci.
Tamoxifen and heat‐shock treatments
To induce Cre‐ERT2‐mediated recombination, animals were treated with 10 μM 4‐hydroxytamoxifen (4‐HT; Sigma‐Aldrich) by IP injection. Cryoinjury was performed 24 h after 4‐HT injection. Heat‐shock treatments were performed in a 37°C water bath for 1 h per day from 1 to 4 dpci, and animals were analyzed at 5 dpci. To examine 60 dpci hearts in transgenic animals (Fig 3F and G), heat‐shock treatments were performed five to seven times a week from 1 to 59 dpci.
Staining
Zebrafish heart sections and RNCMs were fixed in 4% paraformaldehyde. Anti‐MEF2 1:50 (sc‐313, Santa Cruz Biotechnology), anti‐PCNA 1:500 (PC10, Santa Cruz Biotechnology), anti‐N2.261 1:100 (N2.261, DSHB), anti‐DsRed 1:500 (632496, Takara), anti‐MHC 1:500 (MF20, DSHB), anti‐phospho‐histone H3 1:200 (06‐570, EMD Millipore), and anti‐cardiac troponin I (CTNI) 1:500 (ab56357, Abcam) were used. After washing with PBS, samples were stained with Alexa 568, Alexa 488, or Alexa 647 secondary antibodies 1:500 (Life Technologies), followed by 4′,6‐diamidine‐2′‐phenylindole dihydrochloride (DAPI) 1:2,000 (Merck) staining to visualize DNA. To count PCNA+, N2.261+, or pHH3+ CMs in regenerating ventricles, we analyzed more than three sections from each heart. Acid Fuchsin Orange G (AFOG) staining was performed using the A.F.O.G. Kit (BIOGNOST).
Cell culture
Rat neonatal (P1–P3) CMs were isolated as previously described (Fukuda et al, 2017), and CMs were further separated by density gradient centrifugation (Golden et al, 2012). Cells were plated onto 0.1% gelatin‐coated (Sigma) plates and cultured in DMEM/F12 (Gibco) supplemented with 5% horse serum, l‐glutamine, Na pyruvate, penicillin, and streptomycin at 37°C and 5% CO2. 10% FBS (HyClone) was added to the medium for the growth conditions. CMs were transfected with adenovirus vectors and cultured for 72 h in growth medium or non‐growth medium. Samples were then fixed with 4% PFA and stained with anti‐CTNI and DAPI. Five different 1,000 μm regions per sample were analyzed for CM numbers, and average values were used.
Lactate measurements
Lactate levels were measured using a PicoProbe Lactate Fluorometric Assay Kit (BioVision). For each condition, the cryoinjured area and adjacent tissue from 10 ventricles were collected in PBS and homogenized. Samples were centrifuged, and the supernatant was used to measure lactate levels according to manufacturer's protocol. Fluorescence was detected with a FLUOstar Omega instrument (BMG).
Adenovirus vectors
Adenovirus vectors for transfection into CMs were generated using the AdEasy system (Agilent Technologies). The adenovirus vector encoding Pdk3 was previously described (Fukuda et al, 2019). To amplify Pdha1 (NM_008810) from mouse tissues, the following primers were used: Pdha1 (forward 5′‐TAGAGATCTGGTACCGTCGACcACCATGAGGAAGATGCTTGCC‐3′ and reverse 5′‐GGATATCTTATCTAGAAGCTTAACTGACTGACTTAAACTTGATC). To generate Pdha1STA, the following primers were used: forward 5′‐TACCGCTACCATGGACACGCCATGAGTGACCCTGGAGTAAGCGCCCGCACTCGAGAAGAAATC‐3′ and reverse 5′‐GATTTCTTCTCGAGTGCGGGCGCTTACTCCAGGGTCACTCATGGCGTGTCCATGGTAGCGGTA‐3′
Scratch assay
CMs were cultured in growth medium until the plates were confluent and then transfected with adenovirus vectors encoding the gene of interest. After 24 h of culture, a sterilized plastic ruler was laid on the plate, and a sterile pipette tip was used to make a scratch in the CM layer as previously described (Cormier et al, 2015). After removing the dead cells, the plates were incubated for 3 days and then analyzed. To quantify CM membrane protrusions in the scratch assay, the number of CM membrane protrusions in an area of 1,000 μm height × 200 μm width was determined. The average value from two different regions per sample was used. Values were normalized to the average value of control.
Imaging
A Zeiss spinning disk confocal microscope (CSU‐X1, Yokogawa) and ORCA‐Flash4.0 sCMOS camera (Hamamatsu) were used to acquire images. Time‐lapse imaging for the scratch assay was performed using an SP8 microscope (Leica).
RT–qPCR
The miRNeasy Mini Kit (Qiagen) was used for total RNA extraction, and cDNA was synthesized using the SuperScript Second Strand Kit (Life Technologies). The CFX Connect Real‐Time System (Bio‐Rad) and DyNAmo ColorFlash SYBR Green qPCR Kit (Thermo Fisher Scientific) were used. Technical replicates were analyzed for each sample. Primer sequences are shown in Table EV1 and Ct values in Table EV2.
Statistical analysis
Benjamini–Hochberg‐corrected P values as well as standard errors for the fold changes represented by error bars were calculated using limma for proteomic analysis (Fig 1A and B). Comparative statistics between two sample groups was performed using two‐tailed unpaired t‐test and between more than two sample groups using ordinary one‐way ANOVA with Dunnett's multiple comparison test for parametric data. Benjamini–Hochberg correction was used for RNA‐seq data analysis.
Mass spectrometry
Zebrafish ventricles were homogenized with a grinder in SDS lysis buffer (4% SDS in 0.1 M Tris–HCl pH 7.6) and heated at 70°C for 10 min. For DNA shearing, samples were sonicated prior to sedimentation of cell debris by centrifugation at 16,000 g for 10 min. Cleared supernatants were subjected to estimation of protein concentration (DC protein assay, Bio‐Rad). Protein samples were separated according to their molecular weight by SDS–PAGE (NuPAGE 4–12% Bis‐Tris Gel, Thermo Scientific), stained by InstantBlue Coomassie staining (Expedeon), and subjected to in‐gel digestion (Shevchenko et al, 2006). Finally, samples were desalted and stored on STop And Go Extraction (STAGE) tips (Rappsilber et al, 2003) prior to analysis by liquid chromatography–tandem mass spectrometry (LC‐MS/MS). Capillary LC was performed in line to a Q Exactive HF mass spectrometer (Thermo Scientific) via an electrospray ionization (ESI) source using a nano‐UHPLC System (EASY‐nLC 1000, Thermo Scientific), as well as an in‐house packed 70 μm ID, 15‐cm reverse‐phase column emitter (ReproSil‐Pur 120 C18‐AQ, 1.9 μm, Dr. Maisch GmbH) with a buffer system comprising solvent A (5% acetonitrile and 1% formic acid) and solvent B (80% acetonitrile and 1% formic acid). Relevant instrumentation parameters were extracted using MARMoSET (Kiweler et al, 2019) and are included in the Appendix information. Raw data processing was done using the MaxQuant suite of algorithms (v. 1.6.8.0) (Cox & Mann, 2008) against the Danio rerio UniProtKB database (canonical and isoforms; downloaded on 2019/08/19; 62,099 entries) with parameters documented in the supplementary material and employing label‐free quantitation (Cox et al, 2014). Downstream bioinformatics analysis was performed using a limma‐based R pipeline (Ritchie et al, 2015) (https://github.com/bhagwataditya/autonomics), on logarithmized intensities provided by MaxQuant and including an over‐representation analysis by Fisher's exact test, testing for enrichment of KEGG annotation terms in protein groups significantly (P ≤ 0.05) upregulated in 150 dpf vs. 30 dpf hearts as compared to the detectome, the total of all detected protein groups.
RNA‐seq analysis
Total RNA was isolated from RNCMs using the miRNeasy Mini Kit (Qiagen). RNA and library preparation integrity were verified with LabChip GX Touch 24 (Perkin Elmer). 1 μg of total RNA was used as input for VAHTS Stranded mRNA‐seq Library Preparation following manufacturer's protocol (Vazyme). Sequencing was performed on a NextSeq 500 instrument (Illumina) using v2 chemistry, resulting in an average of 37 M reads per library with 1 × 75 bp single‐end setup. The resulting raw reads were assessed for quality, adapter content, and duplication rates with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Trimmomatic version 0.39 was employed to trim reads after a quality drop below a mean of Q20 in a window of 5 nucleotides (Bolger et al, 2014). Only reads between 30 and 150 nucleotides were cleared for further analyses. Trimmed and filtered reads were aligned vs. the Ensembl Rat genome version rn6 (version 94) using STAR 2.6.1d with the parameter “–outFilterMismatchNoverLmax 0.1” to increase the maximum ratio of mismatches to mapped length to 10% (STAR: ultrafast universal RNA‐seq aligner) (Dobin et al, 2013). The number of reads aligning to genes was counted with featureCounts 1.6.5 tool from the Subread package (Liao et al, 2014). Only reads mapping at least partially inside exons were admitted and aggregated per gene. Reads overlapping multiple genes or aligning to multiple regions were excluded. Differentially expressed genes were identified using DESeq2 version 1.26.0 (Love et al, 2014). Only genes with a minimum fold change of ± 2 (log2 = ± 1), a maximum Benjamini–Hochberg‐corrected P‐value of 0.05, and a minimum combined mean of 5 reads were deemed to be significantly differentially expressed. The Ensembl annotation was enriched with UniProt data (release 17.12.2018) based on Ensembl gene identifiers (Activities at the Universal Protein Resource (UniProt)). Gene Ontology analyses were performed using KEGG (https://www.genome.jp/kegg/kegg_ja.html), Reactome (https://reactome.org/), and KOBAS 2.0 (Xie et al, 2011). Two separate tests were performed per contrast using only either up‐ or down‐regulated genes for analysis. The results were combined keeping only gene sets that showed significant over‐representation at FDR < 0.2 in only one input list (i.e., that were clearly enriched for up‐ or down‐regulated genes, but not both). The Top10 gene sets considering enrichment FDR were selected per direction of regulation. The dashed line represents an FDR of 0.05, while the values in brackets denote the number of regulated genes vs. total genes in the respective pathway (Figs 4D and EV4F and G).
Author contributions
RF designed and conducted most experiments; RF and DYRS analyzed the data and wrote the paper; RM‐J and HES designed and performed the experiments; AB and RM‐J helped with the writing; RR helped with live imaging; CK and SG conducted transcriptome analysis; and AK, AMB, and JG conducted proteome analysis. All authors commented on the manuscript.
Conflict of interest
The authors declare no competing financial interests.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Movie EV1
Movie EV2
Movie EV3
Review Process File
Acknowledgements
We thank Beate Grohmann, Kosuke Mochizuki, Simon Perathoner, and Sylvia Jeratsch for help and support, and Jeroen Bakkers for communication. This work was supported by funds from the National Heart, Lung, and Blood Institute of the National Institutes of Health (award number 1F32HL143839) to A.B., the Max Planck Society and the Leducq Foundation to D.Y.R.S., and the DFG (project number 394046768) SFB1366/ project A4 to R.M.‐J. and D.Y.R.S.
EMBO Reports (2020) 21: e49752
Contributor Information
Ryuichi Fukuda, Email: ryuichi.fukuda@mpi-bn.mspg.de.
Didier YR Stainier, Email: didier.stainier@mpi-bn.mpg.de.
Data availability
The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez‐Riverol et al, 2019) partner repository with the dataset identifier PXD016235 (http://www.ebi.ac.uk/pride/archive/projects/PXD016235). The transcriptomics data have been deposited to the GEO database (accession number GSE147073; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147073).
References
- Aharonov A, Shakked A, Umansky KB, Savidor A, Kain D, Lendengolts D, Revach O‐Y, Morikawa Y, Dong J, Levin Y et al (2020) ERBB2 drives YAP activation and EMT‐like processes during cardiac regeneration. BioRxiv 10.1101/2020.01.07.897199 [PREPRINT] [DOI] [PubMed] [Google Scholar]
- Aleström P, D'Angelo L, Midtlyng PJ, Schorderet DF, Schulte‐Merker S, Sohm F, Warner S (2019) Zebrafish: housing and husbandry recommendations. Lab Anim 10.1177/0023677219869037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal RH, Buxton DB, Robertson JG, Olson MS (1993) Regulation of the pyruvate dehydrogenase multienzyme complex. Annu Rev Nutr 13: 497–529 [DOI] [PubMed] [Google Scholar]
- Beisaw A, Kuenne C, Guenther S, Dallman J, Wu CC, Bentsen M, Looso M, Stainier DYR (2020) AP‐1 contributes to chromatin accessibility to promote sarcomere disassembly and cardiomyocyte protrusion during zebrafish heart regeneration. Circ Res 10.1161/CIRCRESAHA.119.316167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Yair R, Butty VL, Busby M, Qiu Y, Levine SS, Goren A, Boyer LA, Burns CG, Burns CE (2019) H3K27me3‐mediated silencing of structural genes is required for zebrafish heart regeneration. Development 146: dev178632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park DJ, Park KS, Lee HK (2006) Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun 348: 1472–1478 [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452: 230–233 [DOI] [PubMed] [Google Scholar]
- Chung S, Dzeja PP, Faustino RS, Perez‐Terzic C, Behfar A, Terzic A (2007) Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med 4: S60–S67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP (2010) Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J Mol Cell Cardiol 48: 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier N, Yeo A, Fiorentino E, Paxson J (2015) Optimization of the wound scratch assay to detect changes in murine mesenchymal stromal cell migration after damage by soluble cigarette smoke extract. J Vis Exp 2015: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das DK, Engelman RM, Rousou JA, Breyer RH (1987) Aerobic vs anaerobic metabolism during ischemia in heart muscle. Ann Chir Gynaecol 76: 68–76 [PubMed] [Google Scholar]
- De Bock K, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, Quaegebeur A, Ghesquière B, Cauwenberghs S, Eelen G et al (2013) Role of PFKFB3‐driven glycolysis in vessel sprouting. Cell 154: 651–663 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doenst T, Nguyen TD, Abel ED (2013) Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res 113: 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Kang HB, Shan C, Elf S, Lin R, Xie J, Gu TL, Aguiar M, Lonning S, Chung TW et al (2014) Tyr‐301 phosphorylation inhibits pyruvate dehydrogenase by blocking substrate binding and promotes the Warburg effect. J Biol Chem 289: 26533–26541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Gunawan F, Beisaw A, Jimenez‐Amilburu V, Maischein HM, Kostin S, Kawakami K, Stainier DYR (2017) Proteolysis regulates cardiomyocyte maturation and tissue integration. Nat Commun 8: 14495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Aharonov A, Ong YT, Stone OA, El‐Brolosy M, Maischein HM, Potente M, Tzahor E, Stainier DYR (2019) Metabolic modulation regulates cardiac wall morphogenesis in zebrafish. eLife 8: e50161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A, Alrob OA, Zhang L, Wagg CS, Altamimi T, Rawat S, Rebeyka IM, Kantor PF, Lopaschuk GD (2019) Acetylation and succinylation contribute to maturational alterations in energy metabolism in the newborn heart. Am J Physiol Heart Circ Physiol 311: H347‐H363 [DOI] [PubMed] [Google Scholar]
- Golden HB, Gollapudi D, Gerilechaogetu F, Li J, Cristales RJ, Peng X, Dostal DE (2012) Isolation of cardiac myocytes and fibroblasts from neonatal rat pups. Methods Mol Biol 843: 205–214 [DOI] [PubMed] [Google Scholar]
- González‐Rosa JM, Martin V, Peralta M, Torres M, Mercader N (2011) Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 138: 1663–1674 [DOI] [PubMed] [Google Scholar]
- Grajevskaja V, Camerota D, Bellipanni G, Balciuniene J, Balciunas D (2018) Analysis of a conditional gene trap reveals that tbx5a is required for heart regeneration in zebrafish. PLoS ONE 13: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi R, Bowker‐Kinley MM, Kedishvili NY, Zhao Y, Popov KM (1995) Diversity of the pyruvate dehydrogenase kinase gene family in humans. J Biol Chem 270: 28989–28994 [DOI] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Beinat M, Stainier DYR (2009) Distinct populations of quiescent and proliferative pancreatic β‐cells identified by HOTcre mediated labeling. Proc Natl Acad Sci USA 106: 14896–14901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL et al (2011) Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell 44: 864–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkoop H, de Bakker DE, Aharonov A, Kruse F, Shakked A, Nguyen PD, de Heus C, Garric L, Muraro MJ, Shoffner A et al (2019) Single‐cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. eLife 8: e50163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, Kawakami Y (2012) Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 139: 4133–4142 [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC (2010) Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464: 606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DYR, Poss KD (2010) Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lee YS, Kang SW, Kim S, Kim TY, Lee SH, Hwang SW, Kim J, Kim EN, Ju JS et al (2019) Loss of PKM2 in Lgr5+ intestinal stem cells promotes colitis‐associated colorectal cancer. Sci Rep 9: 6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel MD, Eames SC, Philipson LH, Prince VE (2010) Intraperitoneal injection into adult zebrafish. J Vis Exp 42: 2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiweler M, Looso M, Graumann J (2019) MARMoSET – extracting publication‐ready mass spectrometry metadata from RAW files. Mol Cell Proteomics 18: 1700–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolwicz SC Jr, Purohit S, Tian R (2013) Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res 113: 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotchkina LG, Patel MS (2001) Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J Biol Chem 276: 37223–37229 [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Harris AL (2005) Pyruvate dehydrogenase and pyruvate dehydrogenase kinase expression in non small cell lung cancer and tumor‐associated stroma. Neoplasia 7: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreipke R, Wang Y, Miklas JW, Mathieu J, Ruohola‐Baker H (2016) Metabolic remodeling in early development and cardiomyocyte maturation. Semin Cell Dev Biol 52: 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc D, Pham DN, Levesque N, Truongcao M, Foulkes WD, Sapienza C, Rozen R (2017) Oncogenic role of PDK4 in human colon cancer cells. Br J Cancer 116: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP (2000) Peroxisome proliferator‐activated receptor gamma coactivator‐1 promotes cardiac mitochondrial biogenesis. J Clin Invest 106: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JJ, Boudina S, Banke NH, Sambandam N, Han X, Young DM, Leone TC, Gross RW, Lewandowski ED, Abel ED et al (2008) The transcriptional coactivator PGC‐1alpha is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol 295: H185–H196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM (2005) Principles of biochemistry, 4th edn New York, NY: Worth; [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liberti MV, Locasale JW (2016) The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 41: 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM (2005) Metabolic control through the PGC‐1 family of transcription coactivators. Cell Metab 1: 361–370 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo PNM et al (2015) Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell 57: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadum A, Engel FB (2018) PPARβ/δ: linking metabolism to regeneration. Int J Mol Sci 19: 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadum A, Singh N, Kurian AA, Munir I, Mehmood T, Brown K, Sharkar M, Chepurko E, Sassi Y, Oh JG et al (2020) Pkm2 regulates cardiomyocyte cell cycle and promotes cardiac regeneration. Circulation 141: 1249–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin‐Juez R, El‐Sammak H, Helker CSM, Kamezaki A, Mullapuli ST, Bibli S‐I, Foglia MJ, Fleming I, Poss KD, Stainier DYR (2019) Coronary revascularization is regulated by epicardial and endocardial cues and forms a scaffold for cardiomyocyte repopulation. Dev Cell 51: 503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez‐Montes I, Escobar B, Palacios B, Gómez MJ, Izquierdo‐Garcia JL, Flores L, Jiménez‐Borreguero LJ, Aragones J, Ruiz‐Cabello J, Torres M et al (2016) Myocardial VHL‐HIF signaling controls an embryonic metabolic switch essential for cardiac maturation. Dev Cell 39: 724–739 [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Zhang M, Heallen T, Leach J, Tao G, Xiao Y, Bai Y, Li W, Willerson JT, Martin JF (2015) Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo‐deficient mice. Sci Signal 8: ra41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely JR, Morgan HE (1974) Relationship between carbohydrate and lipid metabolism and the energy balance of heart muscle. Annu Rev Physiol 36: 413–459 [DOI] [PubMed] [Google Scholar]
- Park S, Jeon JH, Min BK, Ha CM, Thoudam T, Park BY, Lee IK (2018) Role of the pyruvate dehydrogenase complex in metabolic remodeling: differential pyruvate dehydrogenase complex functions in metabolism. Diabetes Metab J 42: 270–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, Cui B, Wang HF, Zhao Y, An F et al (2018) Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 37: 1062–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Riverol Y, Csordas A, Bai J, Bernal‐Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M et al (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquereau J, Ventura‐Clapier R (2018) Maturation of cardiac energy metabolism during perinatal development. Front Physiol 9: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT (2002) Heart regeneration in zebrafish. Science 298: 2188–2190 [DOI] [PubMed] [Google Scholar]
- Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, Grinsfelder D, Rothermel BA, Chen R, Garcia JA et al (2014) The oxygen‐rich postnatal environment induces cardiomyocyte cell‐cycle arrest through DNA damage response. Cell 157: 565–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Rardin MJ, Wiley SE, Naviaux RK, Murphy AN, Dixon JE (2009) Monitoring phosphorylation of the pyruvate dehydrogenase complex. Anal Biochem 389: 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res 43: e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallin P, de Preux Charles AS, Duruz V, Pfefferli C, Jazwinska A (2015) A dual epimorphic and compensatory mode of heart regeneration in zebrafish. Dev Biol 399: 27–40 [DOI] [PubMed] [Google Scholar]
- Schindler YL, Garske KM, Wang J, Firulli BA, Firulli AB, Poss KD, Yelon D (2014) Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 141: 3112–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Stone OA, El‐Brolosy M, Wilhelm K, Liu X, Romao AM, Grillo E, Lai JKH, Gunther S, Jeratsch S, Kuenne C et al (2018) Loss of pyruvate kinase M2 limits growth and triggers innate immune signaling in endothelial cells. Nat Commun 9: 4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara N, Brush M, Kawakami Y (2016) Cell migration during heart regeneration in zebrafish. Dev Dyn 245: 774–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K, Nagamatsu G, Kobayashi CI, Nakamura‐Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, Soga T et al (2013) Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KR, Max SR, Grollman EM, Koski CL (1976) Glycolysis in skeletal muscle regeneration. Exp Neurol 52: 40–48 [DOI] [PubMed] [Google Scholar]
- Wang X, Ha T, Liu L, Hu Y, Kao R, Kalbfleisch J, Williams D, Li C (2018) TLR3 mediates repair and regeneration of damaged neonatal heart through glycolysis dependent YAP1 regulated miR‐152 expression. Cell Death Differ 25: 966–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm K, Happel K, Eelen G, Schoors S, Oellerich MF, Lim R, Zimmermann B, Aspalter IM, Franco CA, Boettger T et al (2016) FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature 529: 216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, Noel ES, Grun D, Berezikov E, Engel FB et al (2016) Spatially resolved genome‐wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev Cell 36: 36–49 [DOI] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39: W316–W322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Hulver MW, McMillan RP, Cline MA, Gilbert ER (2014) The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr Metab 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Jeoung NH, Burgess SC, Rosaaen‐Stowe KA, Inagaki T, Latif S, Shelton JM, McAnally J, Bassel‐Duby R, Harris RA et al (2008) Overexpression of pyruvate dehydrogenase kinase 4 in heart perturbs metabolism and exacerbates calcineurin‐induced cardiomyopathy. Am J Physiol Heart Circ Physiol 294: H936–H943 [DOI] [PubMed] [Google Scholar]
- Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L, Ma L, Hamm M, Gage FH, Hunter T (2016) Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife 5: e13374 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Movie EV1
Movie EV2
Movie EV3
Review Process File
Data Availability Statement
The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez‐Riverol et al, 2019) partner repository with the dataset identifier PXD016235 (http://www.ebi.ac.uk/pride/archive/projects/PXD016235). The transcriptomics data have been deposited to the GEO database (accession number GSE147073; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147073).


