Dear Editor,
Our previous results demonstrated that reducing lipid accumulation, called “Tumor Slimming,” can inhibit the growth and metastasis of tumor cells, 1 , 2 whereas we have hardly found drugs to directly affect the slimming of renal cancer cells. Here, we identified nine pivotal biomarkers and several small molecule drugs with complex bioinformatics methods, which may be helpful for clinical diagnosis and treatment for advanced renal cell carcinoma.
Renal clear cell carcinoma (ccRCC) is one of the most common cancers, with the highest fatality rate in renal cancer with obvious abnormal lipid accumulation, 3 although the mechanism and role of lipid accumulation are not fully known. Clarifications of pathological type and pathological grade are important for the cancer patient management. The treatment is adjusted according to the results of clinical diagnostic markers, with positive effects on the prognosis of patients. Due to the lack of specific tumor markers for renal cancer, cancers were metastasized when the disease was initially diagnosed with imaging. Therefore, the discover of effective biomarker is important for the treatment of renal cancer. There is still a high risk of local recurrence or distant metastasis after local or radical nephrectomies for early renal cancer. 4 Tyrosine kinase inhibitor (TKI) drugs are generally used for the treatment of advanced renal cancer, but still have poor effects on prognosis of thesurvival. 5 Cell cycle regulation and cell adhesion maintenance can promote the occurrence and development of tumor cells. 6 Epithelial‐mesenchymal transition (EMT) has an important clinical significance for cancer progression and drug resistance. 7 TKI can inhibit tumor cell cycle and EMT to affect tumor growth. 8 Unfortunately, TKI resistance developed and lead to poor progression in ccRCC. There is an urgent need to explore more effective prognostic biomarkers and understand the mechanism of the effects in order to determine new drug targets for new ccRCC treatment.
We performed a deep analysis of GSE150404 dataset for advanced ccRCC, which contained four grades with 15 ccRCC samples in each group. The differentially expressed genes (DEGs) between advanced ccRCC (grade III + IV) and early ccRCC (grade I + II) samples were identified by the GEO2R with |logFC| ≥ 2 or ≤ –2, and P‐value <.05. The 49 DEGs were detected, of which 2 were upregulated and 47 downregulated (Figures 1A and 1B). We performed GO analysis to explore the biological pathways and functions involved by DEGs with DAVID. Fatty acid and lipid metabolic processes and substance transport were the major functions among biological process. Moreover, we identified the genes from the DEGs using the protein‐protein interaction network (Figure S1A‐D).
FIGURE 1.
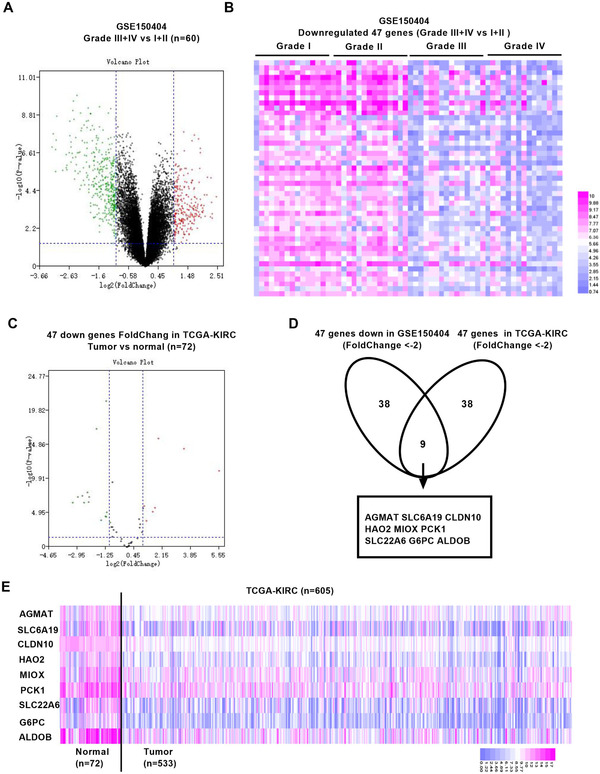
Analysis of pivotal biomarker expression in ccRCC. A, The volcano map of genes expression in ccRCC from GSE150404. B, The heat map of genes expression in ccRCC from GSE150404. Red dots represent the upregulated DEGs, green dots represent the downregulated DEGs, and black dots represent the genes with no difference in expression. C, The volcano map of genes expression in 72 matched normal and adjacent tissues of renal cancer in TCGA‐KIRC. D, Nine pivotal genes were significantly downregulated in kidney cancer tissues compared to adjacent tissues. E, The heat map of nine pivotal genes expression in total cases of TCGA‐KIRC
In order to further determine the most critical genes as pivotal biomarkers involved in the progression of renal cancer, we verified the expression of the selected genes in the Cancer Genome Atlas (TCGA‐KIRC) with 72 matched normal renal and ccRCC cancer tissues. We found that AGMAT, SLC6A19, CLDN10, HAO2, MIOX, PCK1, SLC22A6, G6PC, and ALDOB decreased significantly in ccRCC (Figures 1C, 1D, and S2A) and had obvious differences in expression from normal tissues in the entire database with 72 normal tissues and 533 tumor tissues (Figures 1E, S2B, S3A, and S3B). The Kaplan‐Meier analysis demonstrated that the low expression of selected genes prompted poor overall survival and undesirable disease‐free survival of ccRCC (Figures S4 and S5).
Function pathway analysis showed the role of selected genes in KIRC, of which seven significantly inhibited EMT (Figure 2A). The potential drugs of small molecules that may affect normal cell gene expression were investigated in the CMap database. Nystatin, W‐13, and arachidonyltrifluoromethane had the highest negative enrichment scores (−0.935, –0.859, and −0.85, respectively) (Figure 2B). Small molecules are more likely to reverse the gene expression with higher negative enrichment scores. Interestingly, nystatin with highest negative enrichment scores (−0.935), used as an antifungal drug, can isolate cholesterol and increase the inhibitory effect on migration of endothelial cell, whereas little is known in ccRCC. Nystatin and endostatin can improve the antiangiogenesis and antitumor effects. 9 Previous studies have shown that the lipids accumulated in renal cancer cells are mainly cholesterol and triglycerides. 3 Lipid storage can maintain the integrity of the endoplasmic reticulum, maintaining the homeostasis of tumor cells and promoting tumor development. 10 Our previous results indicate that “Tumor Slimming” can inhibit the growth and metastasis of tumor cells, but direct effects of drugs on the slimming in renal cancer cells remain unclear. Nystatin can be a potential drug for renal cancer. Therefore, further cell and animal studies were required to reveal their potential role of the predicted drugs in renal cancer.
FIGURE 2.
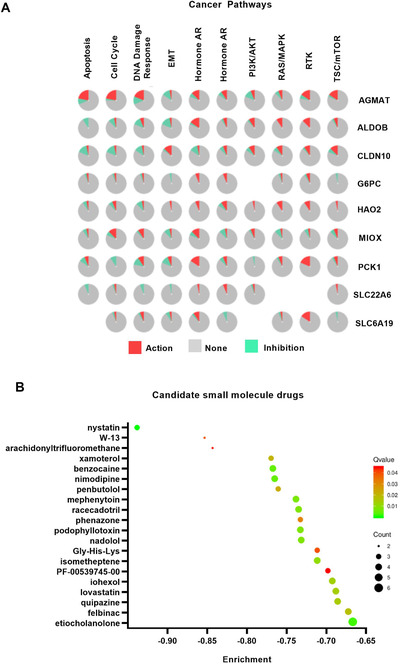
Pathways and drugs of pivotal biomarkers. A, Cancer pathways of pivotal biomarkers (activation or inhibition). B, Plot of top 20 drugs that could reverse the expression of pivotal biomarkers
In summary, we identified nine genes as potential biomarkers of ccRCC by comprehensive bioinformatics analysis. The low expression of these genes had the poor prognosis of advanced renal cancer. We also proposed several new small molecule compounds for renal cancer as they may reverse the expression of nine selected genes in renal cancer cells. This may be helpful to develop new effective biomarkers to diagnose and predict disease progression, and develop new drugs for advanced ccRCC.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
SUPPORTING INFORMATION
ACKNOWLEDGMENTS
We thank the National Natural Science Foundation of China (81902588, 81602218, and 81672528), the National Key Scientific Instrument Development Project of China (81927807), and National Key Research and Development Program of China (2017YFB1303100) for the support provided to this study.
REFERENCES
- 1. Xiong Z, Xiao W, Bao L, et al. Tumor cell “slimming” regulates tumor progression through PLCL1/UCP1‐mediated lipid browning. Adv Sci. 2019;6:1801862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xiao W, Xiong Z, Xiong W, et al. Melatonin/PGC1A/UCP1 promotes tumor slimming and represses tumor progression by initiating autophagy and lipid browning. J Pineal Res. 2019;67:e12607. [DOI] [PubMed] [Google Scholar]
- 3. Zhang Y, Udayakumar D, Cai L, et al. Addressing metabolic heterogeneity in clear cell renal cell carcinoma with quantitative Dixon MRI. JCI Insight. 2017;2:e94278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ravaud A, Motzer RJ, Pandha HS, et al. Adjuvant sunitinib in high‐risk renal‐cell carcinoma after nephrectomy. N Engl J Med. 2016;375:2246‐2254. [DOI] [PubMed] [Google Scholar]
- 5. Chen W, Hill H, Christie A, et al. Targeting renal cell carcinoma with a HIF‐2 antagonist. Nature. 2016;539:112‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang WH, Forde D, Lai AG. Dual prognostic role of 2‐oxoglutarate‐dependent oxygenases in ten cancer types: implications for cell cycle regulation and cell adhesion maintenance. Cancer Commun. 2019;39:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang TH, Wu ATH, Cheng TS, et al. In silico identification of thiostrepton as an inhibitor of cancer stem cell growth and an enhancer for chemotherapy in non‐small‐cell lung cancer. J Cell Mol Med. 2019;23:8184‐8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qiu H, Li J, Liu Q, et al. Apatinib, a novel tyrosine kinase inhibitor, suppresses tumor growth in cervical cancer and synergizes with paclitaxel. Cell Cycle. 2018;17:1235‐1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y, Wang S, Lu X, et al. Cholesterol sequestration by nystatin enhances the uptake and activity of endostatin in endothelium via regulating distinct endocytic pathways. Blood. 2011;117:6392‐6403. [DOI] [PubMed] [Google Scholar]
- 10. Qiu B, Ackerman D, Sanchez DJ, et al. HIF2alpha‐dependent lipid storage promotes endoplasmic reticulum homeostasis in clear‐cell renal cell carcinoma. Cancer Discov. 2015;5:652‐667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION


