Abstract
Maternal mRNA degradation is a critical event of the maternal‐to‐zygotic transition (MZT) that determines the developmental potential of early embryos. Nuclear Poly(A)‐binding proteins (PABPNs) are extensively involved in mRNA post‐transcriptional regulation, but their function in the MZT has not been investigated. In this study, we find that the maternally expressed PABPN1‐like (PABPN1L), rather than its ubiquitously expressed homolog PABPN1, acts as an mRNA‐binding adapter of the mammalian MZT licensing factor BTG4, which mediates maternal mRNA clearance. Female Pabpn1l null mice produce morphologically normal oocytes but are infertile owing to early developmental arrest of the resultant embryos at the 1‐ to 2‐cell stage. Deletion of Pabpn1l impairs the deadenylation and degradation of a subset of BTG4‐targeted maternal mRNAs during the MZT. In addition to recruiting BTG4 to the mRNA 3ʹ‐poly(A) tails, PABPN1L is also required for BTG4 protein accumulation in maturing oocytes by protecting BTG4 from SCF‐βTrCP1 E3 ubiquitin ligase‐mediated polyubiquitination and degradation. This study highlights a noncanonical cytoplasmic function of nuclear poly(A)‐binding protein in mRNA turnover, as well as its physiological importance during the MZT.
Keywords: early embryo development, female fertility, maternal‐effect gene, mRNA stability, RNA‐binding protein
Subject Categories: Development & Differentiation, RNA Biology
The maternal‐effect factor PABPN1‐like (PABPN1L) mediates maternal mRNA decay by acting as mRNA‐binding adapter of the mammalian MZT licensing factor BTG4 in the cytoplasm.
Introduction
An initial step of early embryonic development in all animals is the process called the “maternal‐to‐zygotic transition (MZT)”, by which developmental control passes from the maternal genome to the zygotic genome: The majority of maternal RNAs and proteins are eliminated, and the zygotic genome becomes transcriptionally active 1. The mechanisms that regulate the MZT have been extensively investigated in model organisms including Drosophila, zebrafish, and Xenopus, in which the embryos inherit large quantities of maternal materials due to the staggering volume of ooplasm 2, 3, 4, 5. In these species, the MZT is accomplished when thousands of blastomeres have formed, and along these lines is otherwise called the “mid‐blastula transition (MBT)” 6, 7. In mammals, however, oocytes are relatively small in size compared with those of other animal groups, and the MZT is “pre‐blastula” and occurs as early as the 1–4 cell stage after fertilization 8, 9, 10.
Since the maternal genome becomes transcriptionally silent when oocytes develop to their full size in antral ovarian follicles, the oocyte meiotic maturation, fertilization, and early embryo development, until zygotic gene activation (ZGA), are principally regulated by timely translational activation and degradation of specific maternally derived mRNAs stored in the ooplasm 5, 11, 12. Each of these events is tightly regulated and is interceded by a myriad of RNA‐binding proteins that coat the mRNA from its birth in the nucleus until its eventual degradation in the cytoplasm 3, 13, 14. While the vast majority of eukaryotic mRNAs in somatic cells are polyadenylated immediately after their transcription in the nucleus 15, a large number of maternal mRNAs are stored in the growing oocyte with a short poly(A) tail of 20 to 40 nucleotides and are translationally repressed 16. Upon oocyte maturation or after fertilization, the poly(A) tail of these dormant mRNAs is elongated to 80–250 residues and the mRNAs are translationally activated 17. In both cases, the elongated poly(A) tails are covered by poly(A)‐binding proteins (PABPs) 18, 19.
Two structurally distinct groups of PABPs have been identified in vertebrates. A nuclear PABP (PABPN1), present in all cells of organisms, contains a single RNA recognition motif (RRM) in the central region and a nuclear localization signal (NLS) at the C‐terminus 20. Conversely, a group of cytoplasmic PABPs (PABPCs) contains four RRMs at their N‐terminus and a unique C‐terminal PABP domain 21. According to textbook models, PABPN1 modulates polyadenylation, processing, and nuclear export of newly synthesized mRNAs, whereas PABPCs stabilize poly(A) RNA in the cytoplasm and also enhance translation 18. While these conventional roles are critically important, both PABP families expanded recently both in number and in function 22, 23.
In Xenopus and mouse, an oocyte‐specifically expressed cytoplasmic PABP, PABPC1L (also known as embryonic PAB, ePAB), stabilizes maternal mRNAs and promotes their translation 24, 25, 26. Knockout of the Pabpc1l gene in mice causes defects of oocyte meiotic maturation and ovulation 27, 28. Subsequently, a Pabpn1‐like (Pabpn1l) gene was identified in Xenopus, mouse, and the human genome, with high mRNA expression in oocyte and early embryos; however, its physiological function remained unknown 29, 30.
In addition, maternal mRNA deadenylation and degradation are the core event of the MZT and a prerequisite for ZGA. B‐cell translocation gene‐4 (BTG4), a meiotic cell cycle‐coupled MZT licensing factor in mouse, recruits the CNOT7 catalytic subunit of CCR4‐NOT deadenylase to maternal mRNAs and triggers their degradation 31, 32, 33. However, BTG4 per se does not contain an RNA‐binding domain 34. How BTG4 interacts with RNA and mediates mRNA decay during the MZT is still unknown.
In this study, we identified PABPN1L as a poly(A) adaptor of BTG4 and investigated the potential functional significance of these RNA‐binding proteins during the MZT. Pabpn1l null female mice were sterile. Genetic deletion of Pabpn1l impairs the deadenylation and degradation of a subset of maternal mRNAs during the MZT. As a result, embryos derived from Pabpn1l −/− females arrested at the 1‐ to 2‐cell stage after fertilization. The fact that Pabpn1l and Btg4 knockout mice phenotype each other provided in vivo evidence that PABPN1L mediated cytoplasmic mRNA decay during the MZT by recruiting BTG4 and CCR4‐NOT deadenylase to the 3ʹ‐poly(A) tail of maternal transcripts. Collectively, this study demonstrates new biochemical and physiological functions of poly(A)‐binding proteins during the MZT in mammals.
Results
Maternal PABPN1L is crucial for the MZT
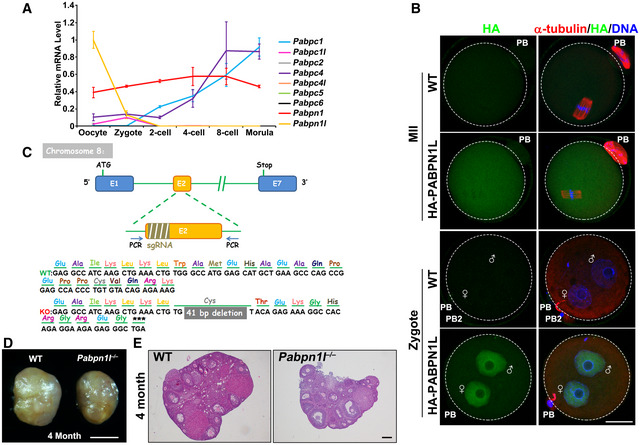
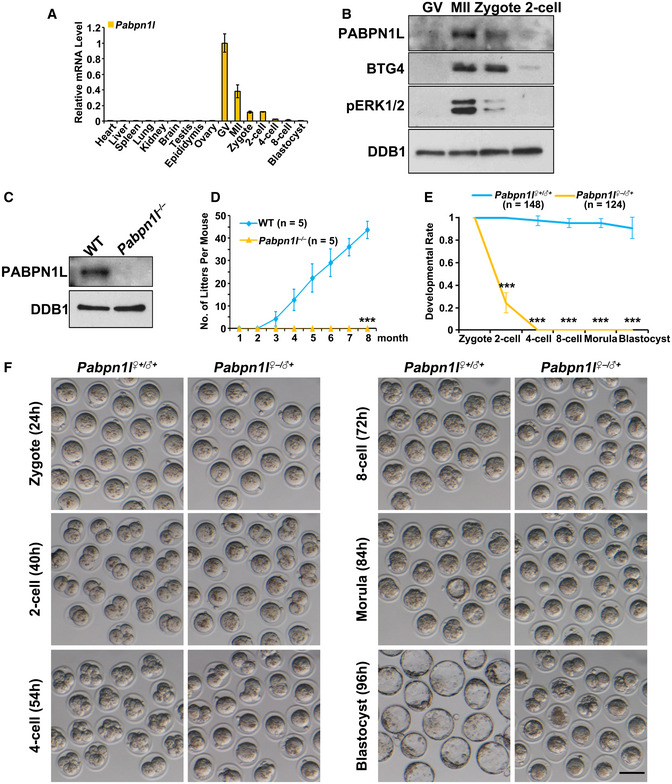
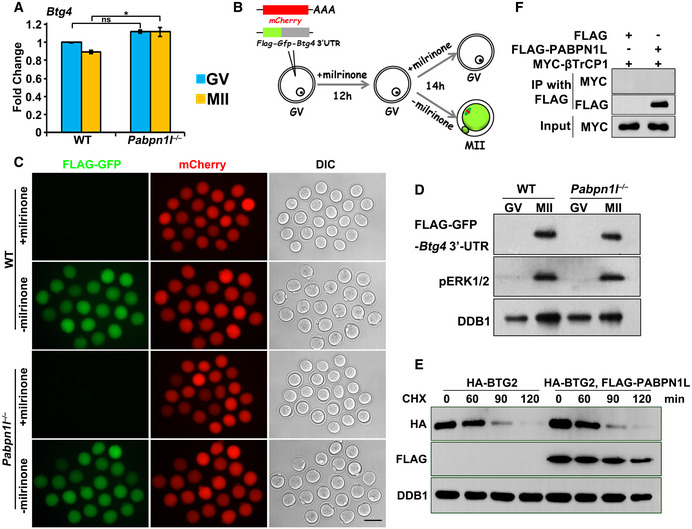
Recent oocytic transcriptome analyses 35 have demonstrated that the Pabpn1l transcript was abundantly expressed in mouse oocytes, and its expression level was the highest among all PABPs detected (Fig EV1A). The oocyte‐specific high‐level expression of Pabpn1l was confirmed by quantitative RT–PCR (RT–qPCR) (Fig 1A). In spite of the fact that Pabpn1l mRNA levels were high in germinal vesicle (GV) oocytes, the expression of the PABPN1L protein was detected only after meiotic resumption; this expression reached a maximal level at the MII stage and quickly decreased after fertilization (Fig 1B). The expression window of PABPN1L spatiotemporally overlapped with the expression pattern of BTG4 and the MZT process 31 (Figs 1B and EV1B). To study the in vivo function of Pabpn1l, we generated a Pabpn1l knockout mouse strain utilizing the CRISPR‐CAS9 system. This mutant line contained a 41‐nucleotide deletion in exon 2 (E2) of the Pabpn1l gene. The deletion caused a reading‐frame shift and created a premature stop codon (Fig EV1C). The absence of PABPN1L proteins in oocytes derived from the Pabpn1l −/− mice was confirmed by Western blot (Fig 1C).
Figure EV1. Pabpn1l knockout in mouse and phenotype analyses.
-
AmRNA expression profiles of 9 PABPs detected in mouse oocytes and early embryos by RNA‐seq (GSE44183). The relative mRNA level of Pabpn1l in oocyte was set to 1.0, and fold changes of indicated mRNAs were normalized by Pabpn1l (in oocyte). n = 3 biological replicates. Error bars, SEM.
-
BImmunofluorescent staining of HA (green) and α‐tubulin (red) in WT MII oocytes and zygotes microinjected with mRNAs encoding HA‐PABPN1L. DNA was labeled by DAPI. Scale bar = 20 μm.
-
CA strategy for CRISPR/Cas9‐based mouse Pabpn1l knockout. Stop, stop codon; sgRNA, synthetic single‐guide RNA; E1, E2, E7, exon1, 2, 7.
-
DRepresentative images of ovaries from 4‐month‐old WT and Pabpn1l −/− mice. Scale bar = 500 μm.
-
EH&E staining results showing ovarian histology of 4‐month‐old WT and Pabpn1l −/− mice. Scale bar = 500 μm.
Figure 1. Expression and function of PABPN1L during MZT.
-
AQuantitative RT–PCR (RT–qPCR) results showing relative expression levels of mouse Pabpn1l in somatic tissues, oocytes, and preimplantation embryos. n = 3 biological replicates. Error bars indicate SEM.
-
BWestern blot results of PABPN1L, BTG4, and phosphorylated ERK1/2 (pERK1/2) in oocytes and embryos. Total protein from 400 oocytes or embryos is loaded in each lane. DNA damage‐binding protein 1 (DDB1) is blotted as a loading control.
-
CWestern blot results of PABPN1L in MII oocytes of wild‐type (WT) and Pabpn1l −/− females. DDB1 is blotted as a loading control.
-
DCumulative pup numbers of WT and Pabpn1l −/− female mice. n = 5 for each genotype. Error bars, SEM. ***P < 0.001 by two‐tailed Student's t‐test.
-
EQuantification of preimplantation embryos derived from WT and Pabpn1l −/− females when WT embryos reached the corresponding stages. The number of analyzed embryos is indicated (n). Error bars, SEM. ***P < 0.001 by two‐tailed Student's t‐test.
-
FRepresentative images of the embryos collected from the oviducts of mated WT and Pabpn1l −/− females at the indicated time points after hCG injection. Scale bar = 100 μm.
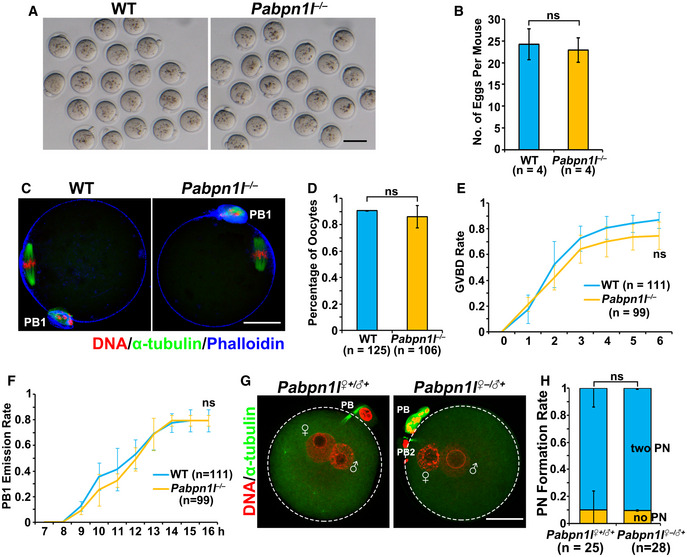
Pabpn1l −/− mice were viable and healthy. Males had normal fertility, but females were infertile (Fig 1D), although they had normal ovarian histology (Fig EV1D and E). After superovulation treatment, the Pabpn1l‐deleted females ovulated regular numbers of MII oocytes (Fig EV2A–D). Furthermore, the fully grown GV oocytes isolated from Pabpn1l null females underwent normal in vitro meiotic maturation (Fig EV2E and F). Zygotes derived from WT and Pabpn1l −/− oocytes (Pabpn1l ♀−/♂+) formed pronuclei with similar efficiency at 24 h after hCG injection (Fig EV2G and H). However, in contrast to the WT zygotes, Pabpn1l ♀−/♂+ zygotes arrested at the 1‐ to 2‐cell stage and failed to develop further (Fig 1E and F). These results indicated that Pabpn1l is a novel maternal‐effect gene and is crucial for the MZT.
Figure EV2. Phenotype analyses of Pabpn1l knockout oocytes.
-
ARepresentative images of MII oocytes collected from oviducts of WT and Pabpn1l −/− mice at 16 h after hCG injection. Scale bar = 100 μm.
-
BNumbers of MII oocytes being ovulated by WT and Pabpn1l −/− mice (n = 4 for each genotype) after superovulation. Error bars, SEM. ns: non‐significant, calculated by two‐tailed Student's t‐test.
-
CConfocal microscopy results showing spindle assembly and PB1 emission of oocytes collected from the oviducts of WT and Pabpn1l −/− mice. Scale bar = 20 μm.
-
DRate of normal spindle assembly in mature WT and Pabpn1l −/−oocytes.
-
E, FGVBD (E) and PB emission (F) rates in cultured oocytes derived from WT and Pabpn1l −/−females.
-
GImmunofluorescent staining for α‐tubulin (green) and DNA (red) in zygotes from WT and Pabpn1l −/− females 28 h after hCG injection. Scale bar = 20 μm.
-
HPercentage of zygotes with two pronuclei (PNs) from WT and Pabpn1l −/− females 28 h after hCG injection.
PABPN1L facilitates maternal mRNA clearance
To understand the role of PABPN1L during the MZT, we subjected WT and Pabpn1l −/− oocytes, as well as the derived embryos at the 1‐cell (zygote) and 2‐cell stages, to global RNA‐seq analyses. Gene expression levels were assessed as fragments per kilobase of transcript per million mapped reads (FPKM), and the relative mRNA copy number was evaluated using the External RNA Controls Consortium (ERCC) spike‐in. All samples were analyzed in duplicate and showed a high correlation (Rmin = 0.88; Raverage = 0.93; Table EV1). Compared to WT, only three and six transcripts were increased or decreased more than 3‐fold in Pabpn1l null oocytes at the GV stage, respectively (Fig 2A; Dataset EV1). In contrast, more transcripts were increased than decreased in Pabpn1l ♀−/♂+ zygotes (1,424 versus 5) (Fig 2A; Dataset EV1). Gene set enrichment analysis of the increased transcripts at the zygote stage revealed that 1,414 of the 1,424 increased transcripts were those being degraded in WT zygotes (Fig 2B). Furthermore, there was a substantial overlap between the transcripts that accumulated in Btg4 ♀−/♂+ and Pabpn1l ♀−/♂+ zygotes (Fig 2C), implying a collaboration between PABPN1L and BTG4 in maternal mRNA turnover. RT–qPCR results confirmed the RNA‐seq data and indicated that previously reported Btg4‐target transcripts in the MZT also accumulated after maternal Pabpn1l knockout (Fig 2D). To investigate the role of PABPN1L in mRNA turnover, a poly(A) tail (PAT) assay was performed, which recapitulates the poly(A) tail length changes in specific transcripts. The results showed the gradual shortening of poly(A) tails of the detected maternal mRNAs during the MZT in WT oocytes and embryos. However, the deadenylation of these transcripts was blocked or delayed after maternal Pabpn1l knockout (Fig 2E). These results indicated that PABPN1L is crucial for maternal mRNA clearance during the MZT.
Figure 2. Transcriptome analyses in Pabpn1l‐deleted oocyte and embryos during the MZT.
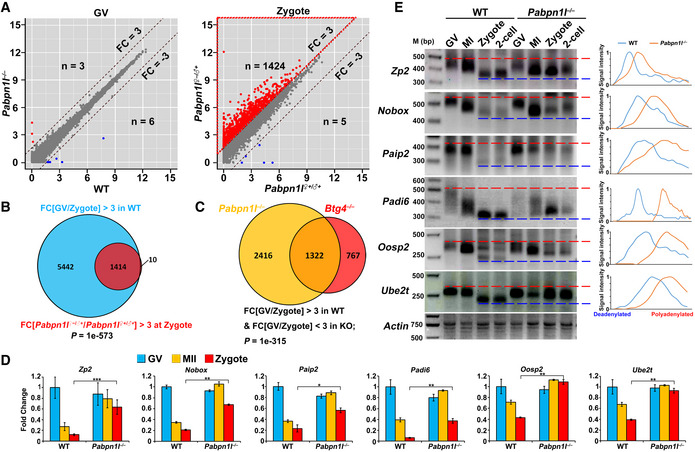
-
AScatter plot comparing the transcripts of GV oocytes and zygotes from WT and Pabpn1l −/− females. Transcripts that increased or decreased by more than 3‐fold in Pabpn1l‐deleted GV oocytes or zygotes are highlighted in red or blue, respectively.
-
BVenn diagrams showing the overlap in transcripts that are significantly degraded during the MZT (FPKM[GV/zygote] > 3) of WT oocytes and transcripts that are accumulated in this process after Pabpn1l knockout (FPKM[Pabpn1l ♀−/♂+/WT] in zygote) > 3).
-
CVenn diagrams showing the overlap in transcripts that are stabilized during the MZT in Pabpn1l ♀−/♂+ and Btg4 ♀−/♂+ zygotes (FPKM[GV/zygote] > 3 in WT, but < 3 in KO).
-
DRT–qPCR results for relative expression levels of the indicated maternal transcripts in oocytes and zygotes from WT and Pabpn1l −/− females. n = 3 biological replicates. Error bars, SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by two‐tailed Student's t‐test.
-
EPoly(A) tail assay results showing poly(A)‐tail length of indicated transcripts in WT and Pabpn1l‐deleted oocytes and embryos. The poly(A) tails of Actin are used as an internal control. Each sample was prepared from 100 oocytes or embryos. Plots show the averaged relative signal intensity (y‐axis) and length of the PCR products based on mobility (x‐axis).
Maternal PABPN1L is essential for ZGA
Because maternal Pabpn1l deletion causes zygotic developmental arrest, we further examined the potential effect of maternal PABPN1L on ZGA. Compared to WT, 269 and 718 transcripts were at abnormally high or low levels in Pabpn1l ♀−/♂+ 2‐cell embryos, respectively (Fig 3A; Dataset EV1). Gene set enrichment analysis revealed that there are 718 transcripts showed decreased levels in Pabpn1l ♀−/♂+ 2‐cell embryos when compared with WT (WT/Pabpn1l ♀−/♂+ ≥ 3). Among these transcripts, 419 belonged to genes that are activated during ZGA in the WT 2‐cell embryos (Fig 3B).
Figure 3. Maternal PABPN1L is essential for ZGA.
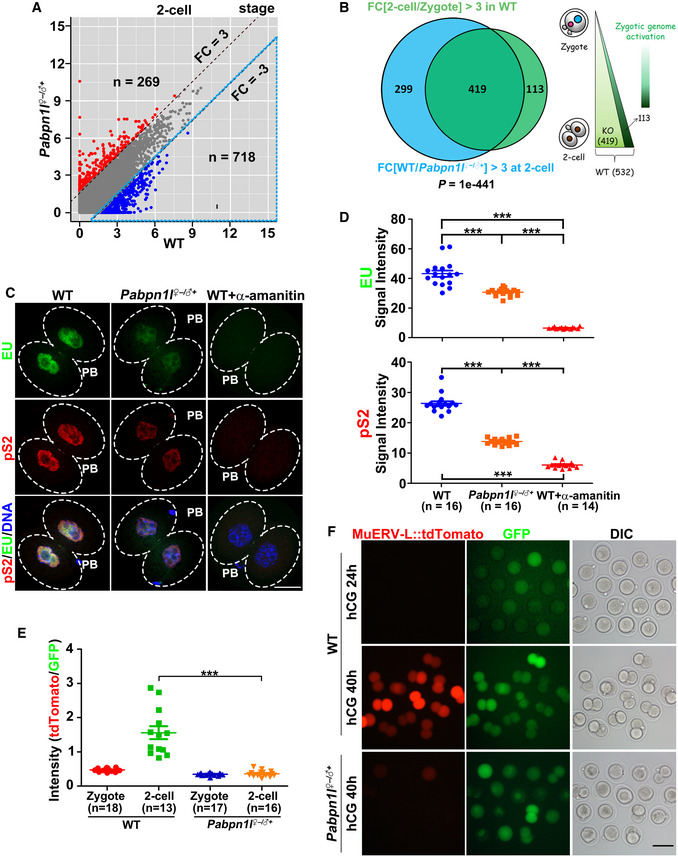
-
AScatter plot comparing the transcripts of 2‐cell embryos from WT and Pabpn1l −/− females. Transcripts that increased or decreased by more than 3‐fold in Pabpn1l‐deleted oocytes or embryos are highlighted in red or blue, respectively.
-
BVenn diagram and schematic showing the overlap in transcripts that are activated during zygote‐to‐2‐cell transition in WT (FPKM[2‐cell/zygote] > 3) and the transcripts that are decreased in Pabpn1l ♀−/♂+ compared to WT at the 2‐cell stage (FPKM[WT/Pabpn1l ♀−/♂+] > 3 in 2‐cell embryos).
-
C5‐Ethynyl uridine (EU) fluorescence (green) showing RNA transcription in WT and Pabpn1l ♀−/♂+ 2‐cell embryos. Some WT embryos were treated with α‐amanitin as early as the zygote stage and cultured to the 2‐cell stage. The phosphorylated RNA polymerase II CTD repeat YSPTSPS (pS2) (red) is co‐stained to label the RNA polymerase II activity. Nuclei are labeled by DAPI (blue). Scale bar = 20 μm.
-
DQuantification of EU and pS2 signals in (C). The numbers of analyzed embryos are indicated (n). Error bars, SEM. ***P < 0.001 by two‐tailed Student's t‐test.
-
ERelative intensity of tdTomato signals in (F). The numbers of analyzed embryos are indicated (n). Error bars, SEM. ***P < 0.001 by two‐tailed Student's t‐test.
-
FRepresentative images of MuERV‐L::tdTomato relative to GFP signal in the same embryo when WT embryos reached the corresponding stages. Zygotes were injected with the MuERV‐L::tdTomato reporter plasmid and polyadenylated Gfp mRNA (as a positive control of microinjection), then allowed to develop in vitro. Cultured embryos were imaged at 24 and 40 h after hCG injection. DIC, differential interference contrast. Scale bar = 100 μm.
To evaluate global transcription activity during ZGA, we performed 5′‐ethynyl uridine (EU) staining assay in which the EU labeled newly synthesized RNAs in the 2‐cell embryos. Phosphorylated RNA polymerase II CTD repeat YSPTSPS (pS2, also a marker of transcription activation) was co‐stained with the EU. As a negative control, some 2‐cell embryos were pre‐treated with α‐amanitin, an RNA polymerase II inhibitor, before ZGA (Fig 3C and D). EU and pS2 signals were detected in the nuclei of Pabpn1l ♀−/♂+ embryos but were weaker than those in WT embryos. These results indicated that maternal Pabpn1l deletion impairs the transcription activation in 2‐cell embryos. Results of RT–qPCR indicated that the expression pattern of representative zygotic genes was similar to those revealed by the RNA‐seq analysis (Appendix Fig S1A).
Previous studies showed that a large number of retrotransposons are expressed as a feature of 2‐cell embryos undergoing ZGA 36, 37. At the 2‐cell stage, the murine endogenous retrovirus with a leucine tRNA primer (MuERV‐L) element was transiently transcribed at the 2‐cell stage in WT embryos but was transcribed at a remarkably lower level after maternal Pabpn1l deletion (Appendix Fig S1B). Similarly, transcription of MuERV‐L target genes, including Guca1a, Tead4, Tdpoz1/4, and Zfp352, was also blocked after maternal Pabpn1l deletion (Appendix Fig S1B). We also microinjected zygotes (WT and Pabpn1l ♀−/♂+) with the MuERV‐L::tdTomato reporter plasmid (MuERV‐L 5′‐long terminal repeat (LTR) promoters upstream of the red fluorescent protein tdTomato) as previously described 36, 38 and monitored the expression of tdTomato during culture. TdTomato expression was observed in WT 2‐cell embryos but not in Pabpn1l ♀−/♂+ embryos, which arrested at the 1‐ to 2‐cell stage (Fig 3E and F). In contrast, Gfp mRNAs co‐injected with the MuERV‐L 5ʹ‐LTR reporter were equally expressed in all embryos (Fig 3F).
In summary, these results indicated that maternal PABPN1L‐mediated biochemical processes, most likely maternal mRNA turnover, are a prerequisite for ZGA in mouse early embryos.
PABPN1L binds both RNA and BTG4 and is involved in deadenylating maternal mRNAs
Based on these results, we hypothesized that PABPN1L may function as an RNA‐binding adapter of BTG4 during the MZT. Thus, BTG4‐RNA immunoprecipitation (RIP) assays were performed in the presence and absence of PABPN1L. Since it was technically challenging to perform extensive biochemical experiments in oocytes due to their low number, we co‐expressed PABPN1L and BTG4 in HeLa cells, which do not endogenously express these proteins, and in line with oocytes/zygotes, the PABPN1L possesses cytoplasm distributed characteristic in HeLa cells (Figs EV3A and EV1B). The results showed that representative transcripts, which were commonly targeted by BTG4 and PABPN1L according to RNA‐seq results, effectively interacted with BTG4 only in the presence of PABPN1L (Fig 4A). As a negative control, mRNAs were not enriched by the RRM‐deleted or Arg‐171 (an essential RNA‐binding residue in the RRM, discussed below)‐mutated PABPN1L in the RIP assay (Figs 4A and EV4B).
Figure EV3. PABPN1L interacts with BTG4.
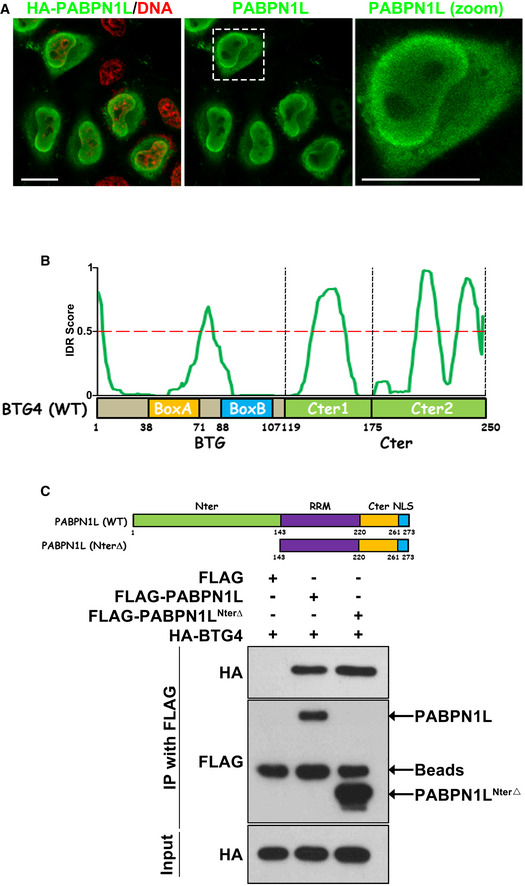
-
AImmunofluorescent staining of HA (green) and DNA (red) in HeLa cells transfected with a HA‐PABPN1L expressing plasmid. The white dotted box showing the PABPN1L signal was zoomed out on the right panel. Scale bar = 10 μm.
-
BA diagram showing of mouse BTG4 with regions of predicted disorder.
-
CDiagrams and co‐IP results showing that BTG4 binds with PABPN1L and its N‐terminal‐deleted form (NterΔ). Lysates from HeLa cells expressing HA‐BTG4 and FLAG‐PABPN1L were immunoprecipitated with an anti‐FLAG antibody. At least three independent experiments were done with consistent results.
Source data are available online for this figure.
Figure 4. PABPN1L is an RNA‐binding adapter of BTG4 involved in deadenylating maternal mRNAs.
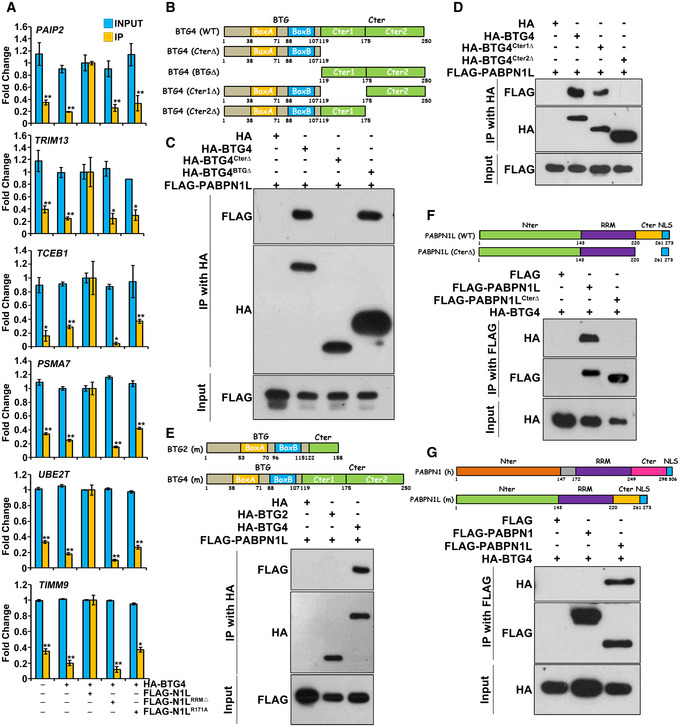
-
ARNA immunoprecipitation (RIP) results showing the interaction of BTG4 with indicated transcripts, in the presence or absence of PABPN1L (full length, RRM‐deleted, or R171A mutant). HeLa cells were co‐transfected with plasmids expressing FLAG‐tagged PABPN1L and HA‐tagged BTG4 for 48 h before immunoprecipitating with an anti‐HA antibody. RNAs recovered from the immunoprecipitants were subjected to RT–qPCR of the indicated transcripts. Fold change values of both input and IP samples were normalized by RIP results of HA‐BTG4 and FLAG‐PABPN1L co‐expression groups. n = 3 biological replicates. Error bars, SEM. The P‐value represents the two‐tailed Student's t‐test comparing the RIP results of BTG4 with the indicated transcripts in the presence of PABPN1L, *P < 0.05, **P < 0.01, and ***P < 0.001.
-
BDiagrams of mouse BTG4 constructs.
-
C, DCo‐IP and Western blot results showing interactions between PABPN1L and BTG4. Lysates from HeLa cells expressing HA‐BTG4 (WT or mutants shown in (B)) and FLAG‐PABPN1L were immunoprecipitated with an anti‐HA antibody. The immunoprecipitated proteins are detected by Western blot with the indicated antibodies.
-
EDiagrams of mouse BTG2 and BTG4 constructs, and co‐IP results showing that PABPN1L binds to BTG4 but not BTG2.
-
FDiagrams and co‐IP results showing BTG4 binding to PABPN1L. Lysates from HeLa cells expressing HA‐BTG4 and FLAG‐PABPN1L (full length and C‐terminal deleted) were immunoprecipitated with an anti‐FLAG antibody. The immunoprecipitated proteins are detected by Western blot with the indicated antibodies.
-
GDiagrams of PABPN1 and PABPN1L constructs, and co‐IP results showing that PABPN1L, but not PABPN1, binds to BTG4.
Figure EV4. Role of Arg‐171 of mouse PABPN1L in RNA binding.
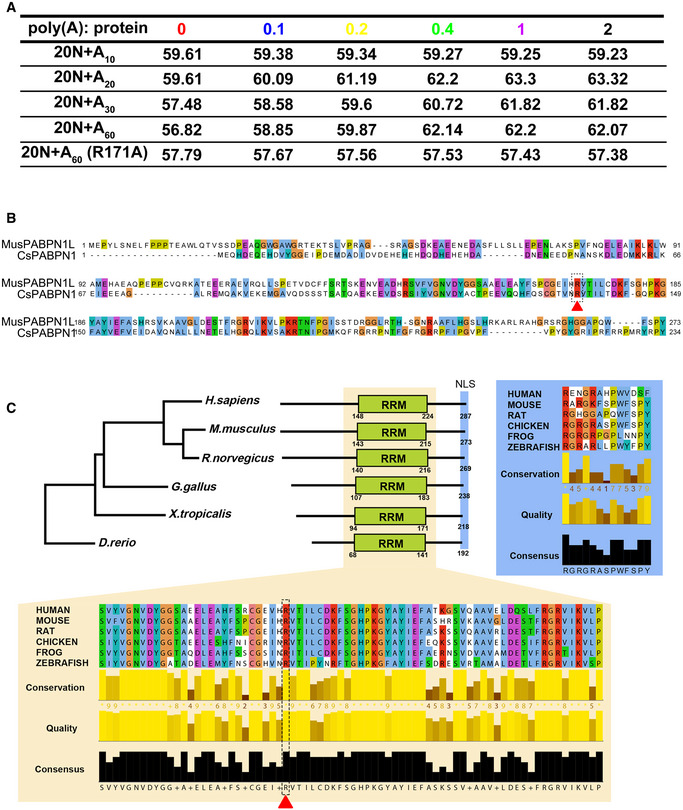
-
AA table showing the Tm in Fig 6A–C.
-
BSequence alignment of mouse PABPN1L (MusPABPN1L) and Citrus sinensis PABPN1(CsPABPN1). The broken circle and arrowhead indicate Arg‐171 of mouse PABPN1L.
-
CSequence alignment and a phylogenetic tree analysis of PABPN1L. PABPN1L aminol acid sequences from H. sapiens (A6NDY0), M. musculus (Q5XFR0), R. norvegicus (B0BNE4), G. gallus (F1NWW9), X. tropicalis (Q28GL9), and D. rerio (E9QGY6) were analyzed by Clustal Omega Multiple Sequence Alignment algorithm (https://www.ebi.ac.uk/Tools/msa/clustalo/). The phylogenetic tree was plotted based on results from sequence alignments using the neighbor‐joining tree algorithm without distance corrections. Sequence alignments and quality parameters of RNA recognition motif (RRM, highlighted in light yellow) and nucleus localization signal (NLS, highlighted in light blue) were generated by Jalview (http://www.jalview.org/). The broken circle and arrowhead indicate Arg‐171 of mouse PABPN1L.
PABPN1L co‐immunoprecipitated with full‐length BTG4 (Fig 4B and C). Deletion of the C‐terminal domain instead of the BTG domain abolished the BTG4‐PABPN1L interaction (Fig 4B and C). Based on intrinsically disordered region (IDR) prediction by IUPRED2 39, the C‐terminal region of BTG4 could be divided into two domains (termed Cter1 and Cter2) (Fig EV3B). Co‐IP results indicated that the BTG4‐PABPN1L interaction was mainly mediated by the Cter2 domain of BTG4 (Fig 4D). BTG2 is the best‐studied BTG family member in somatic cells and does not share homology with BTG4 in the C‐terminal domain 34, 40. It did not interact with PABPN1L in the Co‐IP experiment (Fig 4E). This result further indicated that the unique BTG4 Cter2 domain was specific for PABPN1L binding. Meanwhile, the PABPN1L domain mapping result indicated that PABPN1L bound to BTG4 through its C‐terminal domain (Fig 4F) instead of its N‐terminal domain (Fig EV3C). BTG4 did not bind to PABPN1, which showed homology to PABPN1L in the RRM but not in the C‐terminal domain (Fig 4G).
To test our hypothesis in vivo, we microinjected mRNAs encoding PABPN1L in Pabpn1l −/− oocytes and detected changes in maternal mRNA levels in meiotic maturation. RT–qPCR results indicated that exogenous expression of PABPN1L in Pabpn1l −/− oocytes reversed the maternal mRNA degradation defects (Fig 5A and B). In contrast, exogenous expression of BTG4 could not rescue maternal transcript clearance in Pabpn1l −/− oocytes, suggesting that the BTG4 function was PABPN1L‐dependent. To test if the RNA‐binding ability is essential to the function of PABPN1L, RRM‐deleted or Arg‐171 (an essential RNA‐binding residue in the RRM)‐mutated PABPN1L was expressed in Pabpn1l −/− oocytes (Fig 5C). Neither of them was able to mediate maternal mRNA clearance (Fig 5B). These results indicated that PABPN1L is an RNA‐binding adapter of BTG4‐CCR4‐NOT involved in mediating maternal mRNA deadenylation and degradation.
Figure 5. PABPN1L mediates BTG4 functions in vivo .
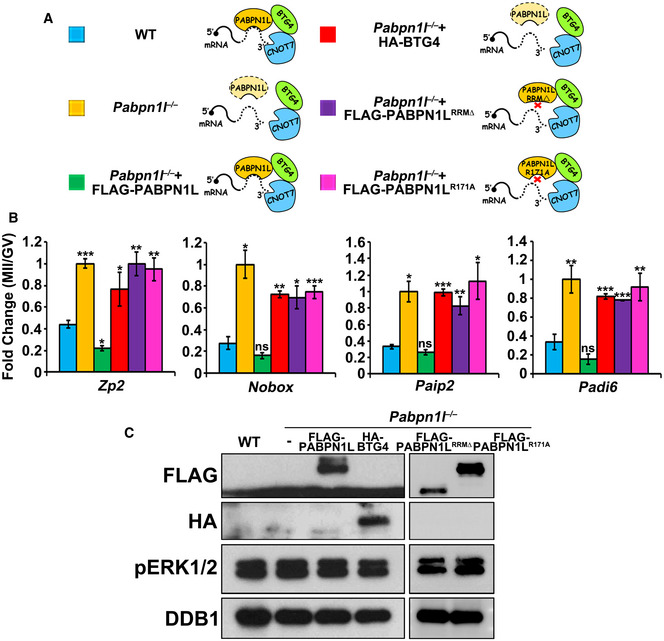
-
ADiagrams of the different treatments of (B).
-
BRT–qPCR results showing the relative expression level changes (MII/GV) of indicated transcripts. Fully grown GV oocytes of Pabpn1l −/− mice were microinjected with mRNAs encoding PABPN1L or BTG4 and were released from meiotic arrest at 12 h after microinjection. Total RNA was extracted from 10 oocytes in each sample. Error bars, SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by two‐tailed Student's t‐test compared to the first column. ns: non‐significant.
-
CWestern blot results showing the expression of exogenous PABPN1L (full length, RRM‐deleted, or R171A mutant) and BTG4 in Pabpn1l −/− oocytes at the MII stage. The total protein from 100 oocytes is loaded in each lane. pERK1/2 is blotted to indicate the developmental stages. DDB1 is blotted as a loading control. At least three independent experiments were done with consistent results.
Source data are available online for this figure.
Characterization of the poly(A)‐binding ability of PABPN1L
Although PABPN1L contains an evolutionarily conserved RRM, which is presumably to bind to poly(A), the poly(A)‐binding ability of this protein has not been experimentally determined. To further investigate the RNA‐binding properties of PABPN1L, a thermal shift assay (TSA) of PABPN1L was performed in the presence of the RNA substrate. Without the addition of RNA, PABPN1L (1 μM) had a melting temperature (Tm) of ~57°C (Figs 6A and EV4A). Oligonucleotides (20N) containing short poly(A) tails (20N+A10) did not alter the Tm of PABPN1L, suggesting that they did not interact with PABPN1L. In contrast, when PABPN1L was pre‐incubated with oligonucleotides containing poly(A) tails of 20, 30, and 60 bases (20N+A20‐60), the Tm increased in a concentration‐dependent manner (Figs 6A and B, and EV4A). As the concentration of 20N+A20‐30 increased from 0.1 to 1 μM, the Tm of PABPN1L increased. However, when the concentration of 20N+A20‐30 increased to 2 μM, the Tm of PABPN1L did not increase further, suggesting that all PABPN1L molecules were saturated by RNAs. In contrast, 20N+A60 saturated 1 μM PABPN1L at the concentration of 0.5 μM (Fig 6B). These results suggested that each PABPN1L molecule occupies 20‐ to 30‐adenosine bases in the poly(A) tail.
Figure 6. Characterization of the PABPN1L poly(A) binding ability.
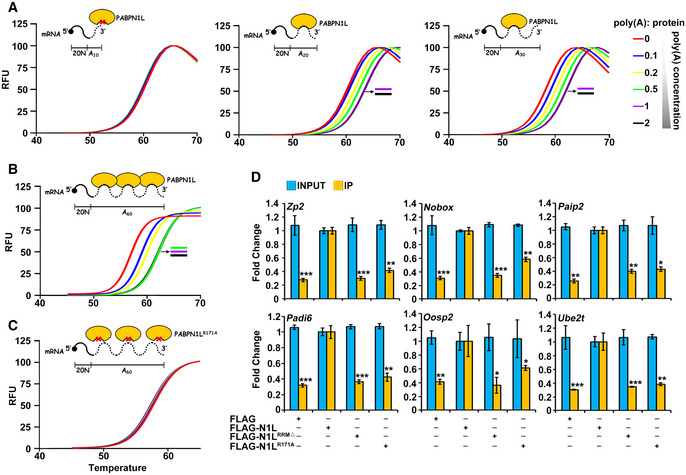
-
A–CDiagrams and averaged fluorescence traces of the thermal stability assays of PABPN1L in the presence and absence of poly(A) variants (n = 6). The RNA substrate comprised 20 non‐poly(A) nucleotides followed by a 10/20/30/60‐adenosine poly(A) tail. PABPN1L‐bound substrates were prepared with 2/1/0.5/0.2/0.1 RNA molecules per PABPN1L. The midpoint of the unfolding curve was determined as the melting temperature (Tm).
-
DRNA immunoprecipitation results showing the interaction of PABPN1L with indicated transcripts. Fully grown GV oocytes of WT mice were microinjected with mRNAs encoding PABPN1L (WT, RRM‐deleted, or R171A mutant) and were released from meiotic arrest at 12 h after microinjection. Total RNAs were extracted from 350 MI stage oocytes in each sample. RNAs recovered from the immunoprecipitants were subjected to RT–qPCR of the indicated transcripts. Fold change values of both Input and IP samples were normalized by RIP results of the FLAG‐PABPN1L microinjection group. Error bars, SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by two‐tailed Student's t‐test comparing with RNA levels pulled down by PABPN1L WT in the second column.
Based on previous nuclear magnetic resonance (NMR) chemical shift analysis upon Poly(A) binding experiments of citrus PABPN1 41, Arg‐136 of citrus PABPN1 is indispensable for the poly(A) interaction. By sequence analysis, we found that PABPN1L has the conserved R171 site (Fig EV4B), and this residue is conserved in the RRM of vertebrate PABPN1L (Fig EV4C). Thus, we performed experiments using the R171 mutated PABPN1L (PABPN1LR171A). The Tm of PABPN1LR171A remained unchanged in the absence or presence of 20N+A60 (Figs 6C and EV4A), indicating that the Arg‐171 mutation abolished the RNA‐binding ability of PABPN1L. The results of the RNA immunoprecipitation assay indicated that both the RRM deletion and Arg‐171 mutation decreased the RNA‐binding ability of PABPN1L in maturing oocytes (Fig 6D). Furthermore, expression of PABPN1LR171A in Pabpn1l −/− oocytes by mRNA microinjection failed to induce maternal mRNA decay as the PABPN1LWT did (Fig 5A–C), indicating that Arg‐171 is functionally essential for PABPN1L in vivo.
PABPN1L stabilizes BTG4 in mature oocytes
To identify other potential mechanisms that may cause MZT arrest in Pabpn1l ♀−/♂+ zygotes, we detected the expression of key maternal factors after Pabpn1l depletion. Activation of the meiosis kinases ERK1/2 is a triggering signal of translational activation of maternal mRNAs during the MZT 12, 42. ERK1/2‐mediated phosphorylation and degradation of the cytoplasmic polyadenylation element‐binding protein‐1 (CPEB1) are a prerequisite for releasing the maternal mRNAs from translational dormancy 43. However, ERK1/2 phosphorylation and CPEB1 degradation occurred normally in Pabpn1l‐deleted oocytes (Fig 7A), indicating that these early steps of the MZT were PABPN1L‐independent.
Figure 7. The role of PABPN1L in stabilizing BTG4 during oocyte maturation.
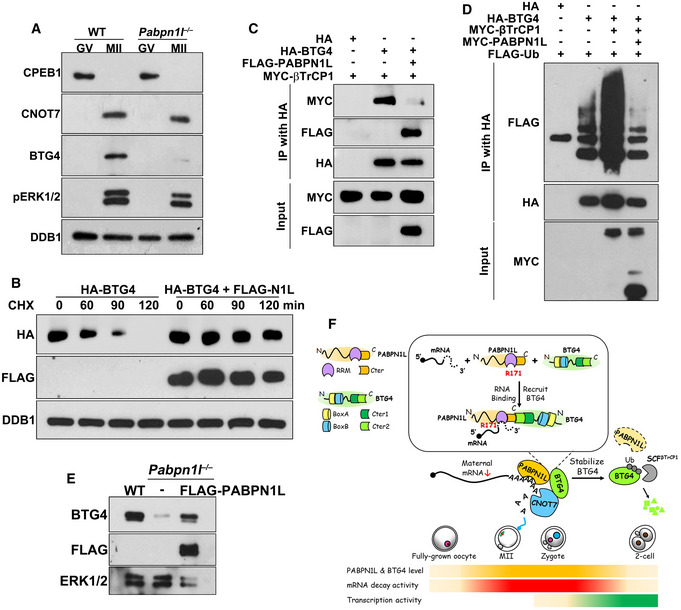
-
AWestern blot results showing the levels of the indicated proteins in WT and Pabpn1l −/− oocytes. Total protein from 100 oocytes is loaded in each lane. DDB1 is blotted as a loading control.
-
BHeLa cells were transfected with plasmids expressing HA‐BTG4 and FLAG‐PABPN1L for 12 h and then treated by cycloheximide (CHX, 10 μM). Cells were harvested at the indicated time points for Western blots. DDB1 is a loading control.
-
CCo‐IP and Western blot results showing the interaction of βTrCP1 with BTG4 in the presence or absence of PABPN1L.
-
DCo‐IP and Western blot results showing BTG4 polyubiquitination. HeLa cells transfected with plasmids encoding the indicated proteins were lysed and subjected to IP with an anti‐HA affinity gel. Input cell lysates and precipitates are immunoblotted with the indicated antibodies.
-
EWestern blot results showing endogenous BTG4 levels in WT and Pabpn1l −/− oocytes at the MII stage. FLAG‐PABPN1L was expressed in Pabpn1l −/− oocytes by mRNA microinjected at the GV stage. Total proteins from 100 oocytes are loaded in each lane. ERK1/2 is blotted as a loading control.
-
FA schematic model for PABPN1L Function during the MZT. During oocyte maturation and fertilization, PABPN1L recruits BTG4 and CCR4‐NOT deadenylase to the poly(A) tails of maternal transcripts and mediates cytoplasmic mRNA decay. At the molecular level, PABPN1L‐Cter and BTG4‐Cter2 mediate the BTG4‐PABPN1L interaction, and arginine‐171 determines the poly(A)‐binding ability of PABPN1L. Furthermore, PABPN1L protects BTG4 from being polyubiquitinated by SCFβTrCP1 and degraded to create an expression window of BTG4 to mediate maternal mRNA decay. These events are prerequisites for ZGA. The gradient ribbons represent the indicated protein level and activity. Deep color represents a high level and activity. The functional domains of PABPN1L and BTG4 proteins are illustrated.
BTG4 and CNOT7 (a catalytic subunit of CCR4‐NOT deadenylase that binds to BTG4) are ERK1/2‐ and CPEB1‐downstream MZT regulators 31, 42. While CNOT7 was normally expressed in mature Pabpn1l −/− oocytes, BTG4 failed to accumulate in MII Pabpn1l −/− oocytes (Fig 7A). The Btg4 mRNA level in oocytes was not affected by Pabpn1l deletion (Fig EV5A). Furthermore, the result of a Flag‐GFP‐Btg4 3′‐UTR reporter assay indicated that the translational activation that is regulated by the Btg4 3′‐UTR was intact in the maturing Pabpn1l −/− oocytes (Fig EV5B–D). Thus, the BTG4 protein turnover was monitored in HeLa cells after treatment with cycloheximide (CHX), a protein synthesis inhibitor 44. The BTG4 protein was mostly degraded within 2 h after CHX treatment but was stabilized in cells expressing PABPN1L (Fig 7B). In contrast, the time‐dependent BTG2 degradation was not prevented by PABPN1L overexpression (Fig EV5E). This result indicated that PABPN1L specifically stabilizes BTG4.
Figure EV5. Translational activity of Btg4 3′‐UTR reporter in WT and Pabpn1l −/− oocytes.
-
ART–qPCR results showing the relative levels of Btg4 transcripts in WT and Pabpn1l −/− oocytes. n = 3 biological replicates. Error bars, SEM. *P < 0.05 by two‐tailed Student's t‐test. ns: non‐significant.
-
BA diagram of the 3′‐UTR reporter assay.
-
CExpression of FLAG‐GFP fused with Btg4 3′‐UTR in WT and Pabpn1l −/− oocytes. An in vitro‐transcribed and polyadenylated mCherry mRNA was co‐microinjected as a positive control. Scale bar = 100 μm.
-
DWestern blot result showing the expression of FLAG‐GFP fused with Btg4 3′‐UTR. Total proteins from 100 oocytes were loaded in each lane. Phosphorylated ERK1/2 was blotted to indicate the developmental stages. DDB1 was blotted a loading control.
-
EHeLa cells were transfected with plasmids expressing HA‐BTG2 and FLAG‐PABPN1L for 12 h and then treated by CHX (10 μM). Cells were harvested at the indicated time points for Western blot analysis.
-
FCo‐IP and Western blot results showing that βTrCP1 did not interact with PABPN1L. Lysates from HeLa cells expressing MYC‐βTrCP1 and FLAG‐PABPN1L were immunoprecipitated with an anti‐FLAG antibody.
BTG1 and BTG2 are polyubiquitinated by the SKP1‐Cullin 1‐F‐box protein (SCF) ubiquitin E3 ligase and then degraded. β‐transducin repeat‐containing protein 1 (βTrCP1) is the substrate adaptor of SCF in mediating BTG2 polyubiquitination 45, 46. In Co‐IP experiments, βTrCP1 interacted with BTG4 (Fig 7C) but not with PABPN1L (Fig EV5F). Overexpression of βTrCP1 increased the polyubiquitination of BTG4 (Fig 7D). However, overexpression of PABPN1L abolished the βTrCP1‐BTG4 interaction (Fig 7C) as well as βTrCP1‐mediated BTG4 polyubiquitination (Fig 7D). Furthermore, ectopic expression of PABPN1L in Pabpn1l −/− oocytes restored endogenous BTG4 accumulation after meiotic maturation (Fig 7E). These observations suggested that PABPN1L not only recruits BTG4 to mRNA 3′‐UTRs but also stabilizes the BTG4 protein by preventing it from being polyubiquitinated by SCFβTrCP1.
Discussion
A poly(A) tail plays versatile roles in mRNA post‐transcriptional regulation 47. Throughout its generation and removal, poly(A) tails associate with PABPs 48. Two structurally different PABP groups have been identified: nuclear PABPs (PABPNs) and cytoplasmic PABPs (PABPCs) 21, 48. As indicated by their names, the two groups of PABPs presumably interact with poly(A)s in the nuclear and cytoplasmic compartments, respectively 18, 21, 49. Based on amino acid similarity, PABPN1L was homologous to the ubiquitously expressed PABPN1, but neither its biochemical function nor its physiological importance had been investigated before this study. The best known molecular functions of PABPN1 include the following: (i) directing nuclear mRNA poly(A) tail elongation immediately after pre‐mRNA transcription; (ii) determining alternative 3′‐UTR selection during pre‐mRNA processing; and (iii) mediating nuclear export of mature mRNAs 50, 51. PABPN1 is thought to be replaced by PABPCs on poly(A) tails once the mRNAs are transported to the cytoplasm. It was unclear if PABPNs were also involved in cytoplasmic mRNA regulation, particularly in circumstances where the nuclear membrane does not exist, such as dividing oocytes.
Results of both in vitro and in vivo approaches in this study suggested that PABPN1L had at least two novel cytoplasmic functions that were indispensable for the MZT: (i) mediating the binding between mRNA poly(A) tails and the BTG4‐CCR4‐NOT complex; (ii) stabilizing the BTG4 protein in mature oocytes. Although Pabpn1l mRNA was highly expressed in fully grown GV oocytes, the PABPN1L protein level only accumulated after germinal vesicle breakdown, when the nuclear membrane no longer existed, and the rate of maternal transcript turnover increased. PABPN1L then bound to poly(A) tails of cytoplasmic mRNAs through its RRM and facilitated their degradation during the MZT. These new observations raise the question of why the cytoplasmic functions of PABPN1L cannot be substituted by PABPCs.
Traditionally, it is believed that the tails of cytoplasmic mRNAs are exclusively coated by PABPCs 23. The major biochemical functions of PABPCs include the following: (i) facilitating the assembly of the translation initiating complex; (ii) protecting the poly(A) tail from being digested by RNA deadenylases; (iii) anchoring mRNA in certain cytoplasmic compartments; and (iv) mediating cytoplasmic mRNA polyadenylation together with other 3′‐UTR‐binding proteins 18. The last function is particularly important in germ cells and neurons because mRNAs are frequently deadenylated and then re‐adenylated in these cell types 52. However, transcripts of most PABPCs (Pabpc1‐6) are detected in mouse oocytes and early embryos at very low levels, except Pabpc1‐like (Pabpc1l, also known as embryonic poly(A)‐binding protein (ePAB)) 35, 53.
PABPC1L‐dependent mRNA cytoplasmic polyadenylation and translation are required for Xenopus oocyte maturation 27. Pabpc1l knockout mice are viable and healthy, but females are infertile due to multiple abnormalities related to oocyte maturation, folliculogenesis, cumulus expansion, and ovulation. Pabpc1l null oocytes are smaller in size, contain peripheral germinal vesicles, and are loosely associated with cumulus cells 28. Chromatin reorganization into the SN configuration and transcriptional silencing does not occur in Pabpc1l‐deleted oocytes 54. Collectively, these phenotypes are very different from those observed in Pabpn1l −/− mice: The main functions of PABPC1L are to protect stored mRNAs from undergoing premature deadenylation and to regulate mRNA translation during oogenesis; in contrast, PABPN1L was dispensable for oogenesis but was crucial for the MZT by functioning together with BTG4.
This study revealed that PABPN1L was likely the previously undiscovered poly(A)‐binding adapter of BTG4 in the MZT. BTG/TOB proteins interact with CNOT7/8 with their conserved BTG domain to facilitate mRNA deadenylation 55. TOB1/2 have conserved PAM2 domains in the C‐terminal region, which bind poly(A)‐binding proteins and recruit CNOT7/8 to poly(A) tails 56. However, BTG4 lacked PAM2 domains 34. Instead, BTG4 interacted with PABPN1L and CNOT7 through its C2‐terminal region and N‐terminal BTG domain, respectively. Further evidence included the following: (i) PABPN1L interacted with BTG4 and poly(A) using distinct domains, and increased the BTG4‐mRNA binding efficiency in RIP assays; (ii) maternal Pabpn1l and Btg4 deletion caused the abnormal accumulation of similar transcripts during the MZT; (iii) the observation that Pabpn1l and Btg4 null mice phenocopy each other provides in vivo evidence that PABPN1L is required for BTG4‐mediated cytoplasmic mRNA decay during the MZT 31; and (iv) female mice expressing the C‐terminal deleted BTG4 (Btg4 ∆C/∆C) are infertile with embryos arresting at the 1‐ or 2‐cell stage 31, indicating that the PABPN1L‐binding ability was essential for BTG4 function in vivo.
BTG family proteins are short‐lived in somatic cells and are promptly degraded by the ubiquitin‐proteasome system 45, 46. However, the mechanisms regulating the stability of BTG4 during the MZT have not yet been clarified. BTG4, in contrast to other BTG/TOB proteins, is relatively stable in oocytes. This study revealed that BTG4 stability was maintained by PABPN1L, which protected BTG4 from polyubiquitination by SCFβTrCP1, and created an expression window for BTG4 to mediate the MZT‐coupled maternal mRNA decay.
In summary, poly(A)‐binding protein PABPN1L functioned in the cytoplasm, by acting as a placeholder which tethered BTG4 and CCR4‐NOT deadenylase to the poly(A) tails of their target mRNAs (Fig 7F). This study provides new mechanistic insight into the MZT and highlights PABPN1L as a maternal factor that potentially affects human fertility.
Materials and Methods
Animals
All mouse strains had a C57B6 background. Pabpn1l −/− mice were generated using the CRISPR‐CAS9 system as illustrated in Fig EV1C. Founder mice were identified by genotyping PCR and crossed with wild‐type C57B6 partners to ensure germline transmission and to avoid any potential off‐targeting effects. A Pabpn1l mutant stain contained a 41‐nucleotide deletion, and a stop codon (sequence: AGAGAGGGCTGA) in exon 2 was used in all experiments. Mice were bred under SPF conditions in a controlled environment of 20–22°C, with a 12/12 h light/dark cycle, 50–70% humidity, and food and water provided ad libitum. Animals were treated with respect based on the Animal Research Committee guidelines of Zhejiang University. The experiments were randomized and were performed with blinding to the conditions of the experiments. No statistical method was used to predetermine sample size.
Superovulation and fertilization
Female mice (21–23 days old) were intraperitoneally injected with 5 IU of PMSG. After 44 h, mice were then injected with 5 IU of hCG. After an additional 16 h, oocyte/cumulus masses were surgically removed from the oviducts. To obtain early embryos, female mice were mated with 10‐ to 12‐week‐old WT males. Successful mating was confirmed by the presence of vaginal plugs. Embryos were harvested from oviducts at the indicated times post‐hCG injection.
Oocyte collection and in vitro culture
Mice at 21–23 days of age were injected with 5 IU of PMSG and humanely euthanized 44 h later. Oocytes at the GV stage were harvested in M2 medium (Sigma‐Aldrich; M7167) and cultured in mini‐drops of M16 medium (Sigma‐Aldrich; M7292) covered with mineral oil at 37°C in a 5% CO2 atmosphere.
In vitro transcription and mRNA microinjection
To prepare mRNAs for microinjection, expression vectors were linearized and in vitro transcribed using the SP6 message mMACHINE Kit (Invitrogen; AM1340) for 4 h at 37°C. Transcribed mRNAs were polyadenylated using the mMACHINE Kit (Invitrogen; AM1350). For microinjection, fully grown GV oocytes were harvested in M2 medium with 2 μM milrinone to inhibit spontaneous GVBD. All microinjections were performed using an Eppendorf TransferMan NK2 micromanipulator. Denuded oocytes were injected with 5–10 pl samples per oocyte. After injection, oocytes were washed and cultured in the M16 medium at 37°C with 5% CO2.
Western blot analysis
Oocytes were lysed with SDS sample buffer and heated for 5 min at 95°C. Total oocyte proteins were separated by SDS–PAGE and electrophoretically transferred to PVDF membranes, followed by blocking in TBST containing 5% defatted milk for 30 min. After probing with primary antibodies and incubated with an HRP‐linked secondary antibody, bound antibodies were detected using the Super Signal West Femto maximum sensitivity substrate. The primary antibodies and dilution factors used are listed in Table EV2.
Cell culture, plasmid transfection, and immunoprecipitation
HeLa cells were obtained from ATCC and were recently authenticated and tested for contamination. Cells were cultured in DMEM plus 10% fetal bovine serum and 1% penicillin–streptomycin solution at 37°C in a humidified 5% CO2 incubator. Mouse Pabpn1l, Pabpn1, Btg4, Btg2, and β‐Trcp1 cDNAs were PCR amplified from a mouse ovarian cDNA pool and ligated into pcDNA‐based eukaryote expression vectors. Transient plasmid transfection was accomplished using Lipofectamine 2000 (Invitrogen). After 48 h of transfection, cells were harvested in a lysis buffer containing 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10% glycerol, and 0.5% NP‐40. After centrifugation, the supernatant was subjected to immunoprecipitation with different affinity gels (Sigma). After incubation at 4°C for 4 h, beads were washed with lysis buffer. The bead‐bound proteins were eluted using SDS sample buffer for Western blot analysis.
Protein expression and purification
The cDNA sequence of PABPN1L was inserted into a modified pRSF‐Duet vector with 6XHis‐SUMO‐tag and a ubiquitin‐like protease (ULP1) cleavage site at the N‐terminal. The recombinant proteins were overexpressed in BL21 (DE3) strain in Lysogeny broth medium. Cells were cultured at 37°C until OD600 = 0.8 and then added with 0.2 mM isopropyl‐β‐D‐thiogalactopyranoside (IPTG) to induce the protein expression for an additional 12 h at 18°C. Cell pellets were suspended in buffer A (25 mM Tris–HCl [pH 8.5], 1.5 M NaCl, 5 mM 2‐hydroxy‐1‐ethanethiol) and lysed by French Press (JNBIO; cat No. JN‐3000PLUS). After high‐speed centrifugation, the lysate supernatant was collected and loaded on Ni2+ Column. The protein with the N‐terminal His6‐SUMO tag was eluted by buffer B (25 mM Tris–HCl [pH 8.5], 0.5 M NaCl, 5 mM 2‐hydroxy‐1‐ethanethiol, 0.5 M imidazole). After digestion with ULP1, the elutes were loaded on HiTrap Q HP column (GE Healthcare; cat No. 17115401) to remove 6XHis‐SUMO tag, followed by gel filtration on HiLoad 16/600 Superdex 75 pg column (GE Healthcare; cat No. GE28‐9893‐33) pre‐equilibrated with buffer C (25 mM HEPES [pH 7.8], 150 mM NaCl, 2 mM DTT, 1 mM MgCl2).
Thermal shift assay
Samples containing WT or R171A mutant PABPN1L (1 μM) in elution buffer (25 mM HEPES [pH 7.8], 150 mM NaCl, 2 mM DTT in PBS) with 25×SYPRO Orange (Invitrogen; S6651) were pipetted in quadruplicate into a 96‐well plate and subjected to heat denaturation using a Bio‐Rad CFX384 Touch Real‐Time PCR Detection System. The temperature was increased from 25°C to 100°C in 0.3°C increments, and at each increment, fluorescent intensities were acquired using HEX detector (excitation 515–535 nm, emission 560–580 nm). PABPN1L/PABPN1LR171A proteins were analyzed alone and in the presence of the poly(A) (20N+A10/20/30/60: CAGCUCCGCAUCCCUUUCCCA10/20/30/60). The fluorescence intensities for the six replicates were averaged, normalized to the maximum fluorescence intensity, and plotted as a function of temperature to obtain melting curves, which were fitted with a sigmoidal function in GraphPad Prism to determine the midpoint of transition or the apparent Tm.
RNA‐seq analysis
Transcripts in oocytes/embryos were amplified using the Smart‐seq2 protocol 57. Briefly, each sample included 10 oocytes/embryos were lysed in 2 μl lysis buffer (0.2% Triton X‐100 and 2 IU/μl RNase inhibitor) followed by reverse transcription with the SuperScript III reverse transcriptase and amplification by PCR for 10 cycles. RNA samples were sequenced on the Illumina HiSeq platform as paired‐end 150‐base reads. Raw reads were trimmed with Trimmomatic‐0.36 to 150 bp and mapped to the mouse genome (mm9) with TopHat (v2.0.11). The mapped reads were subsequently assembled into transcripts guided by reference annotation (University of California at Santa Cruz [UCSC] gene models) with Cufflinks version 2.2.1. The expression level of each gene was quantified with normalized FPKM based on the FPKM of exogenous ERCC. Genes with FPKM < 1 in all samples were excluded, and for the remaining genes, all FPKM values smaller than 1 were set to 1 in subsequent analyses.
Statistical analyses were implemented with R (http://www.rproject.org). The Spearman correlation coefficient (rs) was calculated using the cor function, and the complete linkage hierarchical algorithm was used to cluster the genes. Quality controls of RNA‐seq results were provided as Tables EV1 and EV3, respectively. The FPKMs of the RNA‐seq results are listed in Dataset EV2.
RNA isolation and RT–qPCR analysis
Total RNA was extracted using the RNeasy Mini kit (Qiagen; cat. No. 74106) according to the manufacturer's instructions. cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen; cat. No. 18080200). Quantitative PCR was performed using a Power SYBR Green PCR Master Mix and an Applied Biosystems 7500 Real‐Time PCR System. Relative mRNA levels were calculated by normalizing to the levels of endogenous Gapdh mRNA. The relative transcript levels of samples were compared with the control, and the fold changes are demonstrated. For each experiment, qPCR reactions were carried out in triplicate. Primer sequences are listed in Table EV4.
Ribonucleoprotein immunoprecipitation (RIP) assay
The RIP assay procedure was modified from a previously described method 58. Briefly, 350 oocytes for each sample or HeLa cells were collected and lysed in polysome lysis buffer (50 mM Tris–HCl [pH 7.4], 1% Triton X‐100, 150 mM NaCl, 5 mM EDTA, protease inhibitor cocktail, and RNase inhibitor). A total of 10% of the cell lysate supernatant was used as the “input”, while 90% was subjected to immunoprecipitation with agarose beads conjugated with IgG or FLAG antibodies. After incubation at 4°C for 4 h, bead‐bound RNAs were extracted using an RNeasy Mini Kit (Qiagen, 74106) and reverse‐transcribed using M‐MLV Reverse Transcriptase (Invitrogen). Relative cDNA abundance was analyzed by qPCR.
Poly(A) tail assay
Total RNA was isolated from 100 oocytes using the RNeasy Mini kit (Qiagen; cat. No. 74106). Oligo(dT)12‐18 allowed to saturate the poly(A) tails of the mRNAs. The Oligo(dT)12‐18 was ligated together at 42°C by T4 DNA ligase (NEB; cat. No. M0202). Then, oligo(dT)‐anchored P1 (5′‐GCGAGCTCCGCGGCCGCGT12‐3′) was added at a fivefold molar excess with respect to Oligo(dT)12‐18, and the temperature was lowered to 12°C to favor the hybridization and ligation of the oligo(dT)‐anchor to the extreme 3′ end of the poly(A) tail. Reverse transcription was performed using the SuperScript III (Invitrogen; cat. No. 18080200) with Oligo(dT)‐anchored P1. The products were used in PCRs with gene‐specific primers (Table EV4) and Oligo(dT)‐anchor P1 (5′‐GCGAGCTCCGCGGCCGCGT12‐3′). The PCR conditions were as follows: 30 s at 94°C, 20 s at 58°C, and 40 s at 72°C for 35 cycles. PCR products were analyzed on a 2% agarose gel, and images were captured during exposure to ultraviolet light. Signals were quantified using the “Plot profiles” function of ImageJ software, normalized using the maximum signal intensity in each lane, and the averaged values of three biological replicates were plotted.
Immunofluorescence
Oocytes and embryos were fixed in 4% paraformaldehyde in PBS for 30 min and permeabilized in PBS containing 0.2% Triton X‐100 for 20 min. After being blocked with 1% bovine serum albumin in PBS, the oocytes were incubated with primary antibodies for 1 h and sequentially labeled with Alexa Fluor 594‐ or 488‐conjugated secondary antibodies and 4′,6‐diamidino‐2‐phenylindole (DAPI) for 30 min. Oocytes were imaged using a Zeiss LSM710 confocal microscope. The antibodies used are listed in Table EV2.
Detection of transcription in oocytes
To detect the transcriptional activity, oocytes were cultured in M16 medium containing 1 mM 5‐ethynyl uridine (EU) for 1 h. EU staining was performed using a Click‐iT® RNA Alexa Fluor® 488 Imaging Kit (Life Technologies) according to the manufacturer's instructions. The mean signals were measured across the middle of each oocyte and quantified using ImageJ software.
MuERV‐L 5′‐LTR::td Tomato reporter assay
The plasmid containing MuERV‐L 5′‐LTR::td Tomato reporter was previously reported and gifted by the authors 36. Zygotes were collected from oviducts of mated female mice at 24 h after hCG treatment and microinjected with the plasmids containing MuERV‐L 5′‐LTR::td Tomato reporter. Embryos were allowed to develop in vitro for another 24 h before imaging. In vitro‐transcribed and polyadenylated mRNAs encoding GFP were co‐injected as a positive control reporter.
Statistical analysis
Results are given as means SEM. Each experiment included at least three independent samples and was repeated at least three times. Results for two experimental groups were compared by two‐tailed unpaired Student's t‐tests. Statistically significant values of P < 0.05, P < 0.01, and P < 0.001 are indicated by asterisks (*), (**), and (***), respectively. All tests and P values are provided in the corresponding legends and/or figures.
Author contributions
H‐YF conceived the project. H‐YF and L‐WZ designed and analyzed experiments. L‐WZ, HC, Y‐ZZ, Y‐WW, S‐BP, and LC performed experiments. LS and Y‐ZZ performed RNA‐seq analysis. L‐WZ and H‐YF wrote the paper. L‐WZ, HC, and Y‐ZZ contributed equally to this work.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Dataset EV1
Dataset EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 7
Acknowledgements
We thank Dr. Samuel L. Pfaff for sharing the MuERV‐L 5ʹ‐LTR::tdTomato reporter plasmid. This study was funded by the National Natural Science Foundation of China (31930031, 31890781, 31671558), the National Key Research and Development Program of China (2017YFC1001500, 2016YFC1000600), and The Key Research and Development Program of Zhejiang Province (2017C03022).
EMBO Reports (2020) 21: e49956
Data availability
RNA‐seq data have been deposited in the NCBI Gene Expression Omnibus database. GEO accession number: GSE139072 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139072)
References
- 1. Tadros W, Lipshitz HD (2009) The maternal‐to‐zygotic transition: a play in two acts. Development 136: 3033–3042 [DOI] [PubMed] [Google Scholar]
- 2. Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE (2009) An essential role for the RNA‐binding protein Smaug during the Drosophila maternal‐to‐zygotic transition. Development 136: 923–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Despic V, Dejung M, Gu M, Krishnan J, Zhang J, Herzel L, Straube K, Gerstein MB, Butter F, Neugebauer KM (2017) Dynamic RNA‐protein interactions underlie the zebrafish maternal‐to‐zygotic transition. Genome Res 27: 1184–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rosa A, Brivanlou AH (2017) Role of MicroRNAs in zygotic genome activation: modulation of mRNA during embryogenesis. Methods Mol Biol 1605: 31–43 [DOI] [PubMed] [Google Scholar]
- 5. Svoboda P, Franke V, Schultz RM (2015) Sculpting the transcriptome during the oocyte‐to‐embryo transition in mouse. Curr Top Dev Biol 113: 305–349 [DOI] [PubMed] [Google Scholar]
- 6. Blythe SA, Wieschaus EF (2015) Coordinating cell cycle remodeling with transcriptional activation at the Drosophila MBT. Curr Top Dev Biol 113: 113–148 [DOI] [PubMed] [Google Scholar]
- 7. Iwao Y, Uchida Y, Ueno S, Yoshizaki N, Masui Y (2005) Midblastula transition (MBT) of the cell cycles in the yolk and pigment granule‐free translucent blastomeres obtained from centrifuged Xenopus embryos. Dev Growth Differ 47: 283–294 [DOI] [PubMed] [Google Scholar]
- 8. Jukam D, Shariati SAM, Skotheim JM (2017) Zygotic genome activation in vertebrates. Dev Cell 42: 316–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J et al (2013) Single‐cell RNA‐Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20: 1131–1139 [DOI] [PubMed] [Google Scholar]
- 10. Li L, Lu X, Dean J (2013) The maternal to zygotic transition in mammals. Mol Aspects Med 34: 919–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sha QQ, Zhang J, Fan HY (2019) A story of birth and death: mRNA translation and clearance at the onset of Maternal‐toZygotic transition in mammals. Biol Reprod 101: 579–590 [DOI] [PubMed] [Google Scholar]
- 12. Sha QQ, Dai XX, Dang Y, Tang F, Liu J, Zhang YL, Fan HY (2017) A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocytes. Development 144: 452–463 [DOI] [PubMed] [Google Scholar]
- 13. Sysoev VO, Fischer B, Frese CK, Gupta I, Krijgsveld J, Hentze MW, Castello A, Ephrussi A (2016) Global changes of the RNA‐bound proteome during the maternal‐to‐zygotic transition in Drosophila . Nat Commun 7: 12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wessels HH, Imami K, Baltz AG, Kolinski M, Beldovskaya A, Selbach M, Small S, Ohler U, Landthaler M (2016) The mRNA‐bound proteome of the early fly embryo. Genome Res 26: 1000–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang X, Virtanen A, Kleiman FE (2010) To polyadenylate or to deadenylate: that is the question. Cell Cycle 9: 4437–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conti M, Franciosi F (2018) Acquisition of oocyte competence to develop as an embryo: integrated nuclear and cytoplasmic events. Hum Reprod Update 24: 245–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lim J, Lee M, Son A, Chang H, Kim VN (2016) mTAIL‐seq reveals dynamic poly(A) tail regulation in oocyte‐to‐embryo development. Genes Dev 30: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wigington CP, Williams KR, Meers MP, Bassell GJ, Corbett AH (2014) Poly(A) RNA‐binding proteins and polyadenosine RNA: new members and novel functions. Wiley Interdiscip Rev RNA 5: 601–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Charlesworth A, Meijer HA, de Moor CH (2013) Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA 4: 437–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banerjee A, Apponi LH, Pavlath GK, Corbett AH (2013) PABPN1: molecular function and muscle disease. FEBS J 280: 4230–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goss DJ, Kleiman FE (2013) Poly(A) binding proteins: are they all created equal? Wiley Interdiscip Rev RNA 4: 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicholson AL, Pasquinelli AE (2019) Tales of detailed Poly(A) tails. Trends Cell Biol 29: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ozturk S, Uysal F (2017) Poly(A)‐binding proteins are required for translational regulation in vertebrate oocytes and early embryos. Reprod Fertil Dev 29: 1890–1901 [DOI] [PubMed] [Google Scholar]
- 24. Friend K, Brook M, Bezirci FB, Sheets MD, Gray NK, Seli E (2012) Embryonic poly(A)‐binding protein (ePAB) phosphorylation is required for Xenopus oocyte maturation. Biochem J 445: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Esencan E, Kallen A, Zhang M, Seli E (2019) Translational activation of maternally derived mRNAs in oocytes and early embryos and the role of embryonic poly(A) binding protein (EPAB). Biol Reprod 100: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uysal F, Ozturk S (2019) Embryonic poly(A)‐binding protein is differently expressed and interacts with the messenger RNAs in the mouse oocytes and early embryos. J Cell Biochem 120: 4694–4709 [DOI] [PubMed] [Google Scholar]
- 27. Guzeloglu‐Kayisli O, Lalioti MD, Aydiner F, Sasson I, Ilbay O, Sakkas D, Lowther KM, Mehlmann LM, Seli E (2012) Embryonic poly(A)‐binding protein (EPAB) is required for oocyte maturation and female fertility in mice. Biochem J 446: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lowther KM, Favero F, Yang CR, Taylor HS, Seli E (2017) Embryonic poly(A)‐binding protein is required at the preantral stage of mouse folliculogenesis for oocyte‐somatic communication. Biol Reprod 96: 341–351 [DOI] [PubMed] [Google Scholar]
- 29. Cosson B, Braun F, Paillard L, Blackshear P, Beverley Osborne H (2004) Identification of a novel Xenopus laevis poly (A) binding protein. Biol Cell 96: 519–527 [DOI] [PubMed] [Google Scholar]
- 30. Sakugawa N, Miyamoto T, Sato H, Ishikawa M, Horikawa M, Hayashi H, Ishikawa M, Sengoku K (2008) Isolation of the human ePAB and ePABP2 cDNAs and analysis of the expression patterns. J Assist Reprod Genet 25: 215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu C, Ji SY, Sha QQ, Dang Y, Zhou JJ, Zhang YL, Liu Y, Wang ZW, Hu B, Sun QY et al (2016) BTG4 is a meiotic cell cycle‐coupled maternal‐zygotic‐transition licensing factor in oocytes. Nat Struct Mol Biol 23: 387–394 [DOI] [PubMed] [Google Scholar]
- 32. Liu Y, Lu X, Shi J, Yu X, Zhang X, Zhu K, Yi Z, Duan E, Li L (2016) BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis. J Mol Cell Biol 8: 366–368 [DOI] [PubMed] [Google Scholar]
- 33. Pasternak M, Pfender S, Santhanam B, Schuh M (2016) The BTG4 and CAF1 complex prevents the spontaneous activation of eggs by deadenylating maternal mRNAs. Open Biol 6:160184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winkler GS (2010) The mammalian anti‐proliferative BTG/Tob protein family. J Cell Physiol 222: 66–72 [DOI] [PubMed] [Google Scholar]
- 35. Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE et al (2013) Genetic programs in human and mouse early embryos revealed by single‐cell RNA sequencing. Nature 500: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB (2004) Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606 [DOI] [PubMed] [Google Scholar]
- 38. Rong Y, Ji SY, Zhu YZ, Wu YW, Shen L, Fan HY (2019) ZAR1 and ZAR2 are required for oocyte meiotic maturation by regulating the maternal transcriptome and mRNA translational activation. Nucleic Acids Res 47: 11387–11402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meszaros B, Erdos G, Dosztanyi Z (2018) IUPred2A: context‐dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res 46: W329–W337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tirone F (2001) The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol 187: 155–165 [DOI] [PubMed] [Google Scholar]
- 41. Domingues MN, Sforca ML, Soprano AS, Lee J, de Souza T, Cassago A, Portugal RV, de Mattos Zeri AC, Murakami MT, Sadanandom A et al (2015) Structure and mechanism of dimer‐monomer transition of a plant Poly(A)‐binding protein upon RNA interaction: insights into its poly(A) tail assembly. J Mol Biol 427: 2491–2506 [DOI] [PubMed] [Google Scholar]
- 42. Dai XX, Jiang JC, Sha QQ, Jiang Y, Ou XH, Fan HY (2018) A combinatorial code for mRNA 3′‐UTR‐mediated translational control in the mouse oocyte. Nucleic Acids Res 47: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keady BT, Kuo P, Martinez SE, Yuan L, Hake LE (2007) MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J Cell Sci 120: 1093–1103 [DOI] [PubMed] [Google Scholar]
- 44. Sha QQ, Dai XX, Jiang JC, Yu C, Jiang Y, Liu J, Ou XH, Zhang SY, Fan HY (2018) CFP1 coordinates histone H3 lysine‐4 trimethylation and meiotic cell cycle progression in mouse oocytes. Nat Commun 9: 3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sasajima H, Nakagawa K, Kashiwayanagi M, Yokosawa H (2012) Polyubiquitination of the B‐cell translocation gene 1 and 2 proteins is promoted by the SCF ubiquitin ligase complex containing betaTrCP. Biol Pharm Bull 35: 1539–1545 [DOI] [PubMed] [Google Scholar]
- 46. Sasajima H, Nakagawa K, Yokosawa H (2002) Antiproliferative proteins of the BTG/Tob family are degraded by the ubiquitin‐proteasome system. Eur J Biochem 269: 3596–3604 [DOI] [PubMed] [Google Scholar]
- 47. Reyes JM, Ross PJ (2016) Cytoplasmic polyadenylation in mammalian oocyte maturation. Wiley Interdiscip Rev RNA 7: 71–89 [DOI] [PubMed] [Google Scholar]
- 48. Eliseeva IA, Lyabin DN, Ovchinnikov LP (2013) Poly(A)‐binding proteins: structure, domain organization, and activity regulation. Biochemistry 78: 1377–1391 [DOI] [PubMed] [Google Scholar]
- 49. Mangus DA, Evans MC, Jacobson A (2003) Poly(A)‐binding proteins: multifunctional scaffolds for the post‐transcriptional control of gene expression. Genome Biol 4: 223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Klerk E, Venema A, Anvar SY, Goeman JJ, Hu O, Trollet C, Dickson G, den Dunnen JT, van der Maarel SM, Raz V et al (2012) Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res 40: 9089–9101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jenal M, Elkon R, Loayza‐Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K et al (2012) The poly(A)‐binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149: 538–553 [DOI] [PubMed] [Google Scholar]
- 52. Brook M, Smith JW, Gray NK (2009) The DAZL and PABP families: RNA‐binding proteins with interrelated roles in translational control in oocytes. Reproduction 137: 595–617 [DOI] [PubMed] [Google Scholar]
- 53. Sha QQ, Yu JL, Guo JX, Dai XX, Jiang JC, Zhang YL, Yu C, Ji SY, Jiang Y, Zhang SY et al (2018) CNOT6L couples the selective degradation of maternal transcripts to meiotic cell cycle progression in mouse oocyte. EMBO J 37: e99333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lowther KM, Mehlmann LM (2015) Embryonic Poly(A)‐binding protein is required during early stages of mouse oocyte development for chromatin organization, transcriptional silencing, and meiotic competence. Biol Reprod 93: 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doidge R, Mittal S, Aslam A, Winkler GS (2012) Deadenylation of cytoplasmic mRNA by the mammalian Ccr4‐Not complex. Biochem Soc Trans 40: 896–901 [DOI] [PubMed] [Google Scholar]
- 56. Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB (2007) Human TOB, an antiproliferative transcription factor, is a poly(A)‐binding protein‐dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol 27: 7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R (2014) Full‐length RNA‐seq from single cells using Smart‐seq2. Nat Protoc 9: 171–181 [DOI] [PubMed] [Google Scholar]
- 58. Zhang J, Zhang YL, Zhao LW, Guo JX, Yu JL, Ji SY, Cao LR, Zhang SY, Shen L, Ou XH et al (2018) Mammalian nucleolar protein DCAF13 is essential for ovarian follicle maintenance and oocyte growth by mediating rRNA processing. Cell Death Differ 26: 1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Dataset EV1
Dataset EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 7
Data Availability Statement
RNA‐seq data have been deposited in the NCBI Gene Expression Omnibus database. GEO accession number: GSE139072 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139072)


