Dear Editor,
Lower grade glioma (LGG) is an infiltrative neoplasm that would progress to glioblastoma (GBM). So far, the molecular mechanism of LGG has not been fully elucidated indicating that more biomarkers are needed. Recently, cancer immunotherapy appears to be an effective way in cancer treatment. However, the discoveries of immunotherapy in gliomas mainly involved in GBM, few investigations appeared in LGG. 1 , 2 Capping actin protein, gelsolin like (CAPG) is one of the members of gelsolin protein superfamily that has been reported overexpressed in gliomas. 3 However, less is known about the role of CAPG in LGG, especially its role in regulation of tumor environment.
We found that CAPG expression was notably higher in LGG tumors compared with normal tissues (Figure S1A‐B). Further, we discovered that CAPG expressions were significantly associated with age, tumor grade, and histological subtypes in LGG (Figure S1C). Next, we analyzed the correlation between expression and methylation of CAPG in LGG tumors and found that the methylation levels of five methylation sites (cg04881903, cg26476820, cg27139457, cg19627006, and cg07215695) were strongly negatively correlated with CAPG expression, whereas one (cg27090078) showed weak negative association (Figure S2A). Survival analysis results suggested that high levels of methylation in these six sites were associated with better survival in LGG patients (Figure S2B). Further validation results identified that patients of LGG with higher expression of CAPG had a short survival time (Figure S2C). To exam the role of CAPG as a biomarker in LGG, we analyzed gene alterations including mutation, putative copy number variation, and mRNA expression of CAPG, IDH1, IDH2, and TERT. The results suggested that gene alteration of CAPG and IDH1 was mutually exclusive (q < .001 in TCGA PanCancer Atlas) (Figure 1). Collectively, these results indicated that CAPG is a prognostic factor and might be used as a new subtype biomarker in LGG.
FIGURE 1.
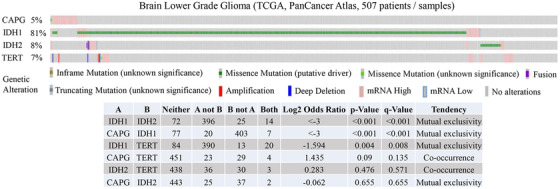
Gene alteration of CAPG and IDH1 is mutually exclusive in LGG. Gene alterations including mutation, putative copy number variation, and mRNA expression of CAPG, IDH1, IDH2, and TERT in LGG samples from TCGA, PanCancer Atlas
Subsequently, we investigated biological process of CAPG involved in LGG via gene set enrichment analysis (GSEA). Results displayed that overexpression of CAPG was significantly correlated with immune regulation, including T cell proliferation and differentiation (Figure S3). These results indicated that CAPG might be involved in the regulation of tumor microenvironment. We next investigated the relationship between CAPG expression and immune infiltration level in LGG. Results revealed that higher expression of CAPG correlated with more immune infiltration of immune cells (Figure 2A). Furthermore, we examined the associations between CAPG expressions and markers of various immune cells in LGG. Results displayed that markers of M1 macrophages showed no correlations with CAPG expression (NOS2 and PTGS2), whereas markers of M2 macrophage, CD163, VSIG4, and MS4A4A showed positive correlations (Figures 2D and 2E; Table S1). These results exhibit a potential regulating effect of CAPG in polarization of tumor associated macrophages (TAMs). Moreover, we observed that high CAPG expression was related to high infiltration levels of dendritic cells (DCs). Markers of DC exhibited notable correlations with CAPG expressions. These results further indicated a strong relationship between CAPG and DCs infiltration. Additionally, significant correlations between CAPG and markers of T cell exhaustion was determined (Table S1). TIM‐3, which is a key gene regulating T cell exhaustion, 4 is significantly and positively correlated with CAPG expression, indicating that high expression of CAPG might play a vital role in T cell exhaustion mediated by TIM‐3. Consequently, our results suggested that CAPG might play an important role in immune escape in LGG cancer microenvironment.
FIGURE 2.

Correlations of CAPG expression with immune cell infiltration in LGG. A, The expression of CAPG is significantly negatively correlated with tumor purity and positively related to infiltrating levels of B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell. B‐E, Plots of correlations between CAPG expression and markers of monocytes (B), TAMs (C), M1 (D), and M2 macrophages (E)
In conclusion, we discovered that CAPG is overexpressed in LGG tumor samples and associated with poor survival. Six methylation sites were identified to be negatively correlated with CAPG expression and patient's survival. Interestingly, we found that gene alterations of CAPG and IDH1 were mutually exclusive in LGG, which indicates that CAPG might be used as a new marker for subtype classification of LGG. Additionally, increased expression of CAPG correlates with enhanced immune infiltration of immune cells in LGG. CAPG potentially participates in the regulation of TAMs, DCs, and T cell exhaustion. Accordingly, CAPG is a potential prognosis biomarker and probably involved in the regulation of tumor environment in LGG.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supporting information
ACKNOWLEDGMENTS
This work was supported by the Guangxi Natural Science Foundation (2017GXNSFBA198240 and 2018GXNSFAA050055), the Project of Open Laboratory of Key Laboratory of Electromagnetic Radiation Protection, Ministry of Education (2017DCKF001), and Middle‐Aged and Young Teachers in Colleges and Universities in Guangxi Basic Ability Promotion Project (2017KY0295 and 2018KY0049).
AVAILABILITY OF DATA AND MATERIALS
All the data and materials are available.
CONSENT FOR PUBLICATION
All the authors consent for publication.
AUTHOR CONTRIBUTIONS
LW designed the study and wrote the paper. JW and LF analyzed the data. All authors approved the final manuscript.
REFERENCES
- 1. Sippel TR, White J, Nag K, et al. Neutrophil degranulation and immunosuppression in patients with GBM: restoration of cellular immune function by targeting arginase I. Clin Cancer Res. 2011;17(22):6992‐7002. [DOI] [PubMed] [Google Scholar]
- 2. Perng P, Lim M. Immunosuppressive mechanisms of malignant gliomas: parallels at non‐CNS sites. Front Oncol. 2015;5:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yun DP, Wang YQ, Meng DL, et al. Actin‐capping protein CapG is associated with prognosis, proliferation and metastasis in human glioma. Oncol Rep. 2018;39(3):1011‐1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang YH, Zhu C, Kondo Y, et al. CEACAM1 regulates TIM‐3‐mediated tolerance and exhaustion. Nature 2015;517(7534):386‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


