Dear Editor,
Our previous study has demonstrated non‐small‐cell lung cancer (NSCLC) patients with high tissue‐based tumor mutation burden (TMB) derived encouraging benefits from immune checkpoint inhibitor (ICI). 1 The blood‐based TMB (bTMB) recently emerged as an encouraging non‐invasive approach to predict improvements in PFS of atezolizumab over docetaxel among previously treated NSCLC patients, but it failed to reproducibly stratify patients into groups with different OS. 2 The ctDNA maximum somatic allele frequency (MSAF) has been demonstrated affects the concordance between bTMB and tissue‐based TMB, 2 , 3 suggesting MSAF might potentially provide additional predictive value for the bTMB. Herein, we first performed individual patient data meta‐analysis using OAK and POPLAR randomized trial data 4 , 5 to assess the predictive value of adding MSAF to bTMB (bTMB‐MSAF algorithm) in identifying NSCLC patients who could benefit from atezolizumab over docetaxel. Moreover, we comprehensively investigated the blood‐based mutational landscape of NSCLC patients and developed nomogram considering oncogenic and clinicopathological variables to predict survival of ICI patients. Full methods are described in the Supporting Information.
In the whole intention‐to‐treat and EGFR wild‐type patients, compared with docetaxel, atezolizumab resulted in significantly longer OS (HR 0.72, 95% CI, 0.62‐0.83; P < .001; HR 0.67, 95% CI, 0.57‐0.78; P < .001; Figure 1A,B), but not PFS (Figures S2 and S3). EGFR mutant patients did not show significant difference in OS or PFS between two treatments (Figure S4). More clinicopathological subgroups are presented in Supporting Information Result S1 and Figures S5 and S6.
FIGURE 1.
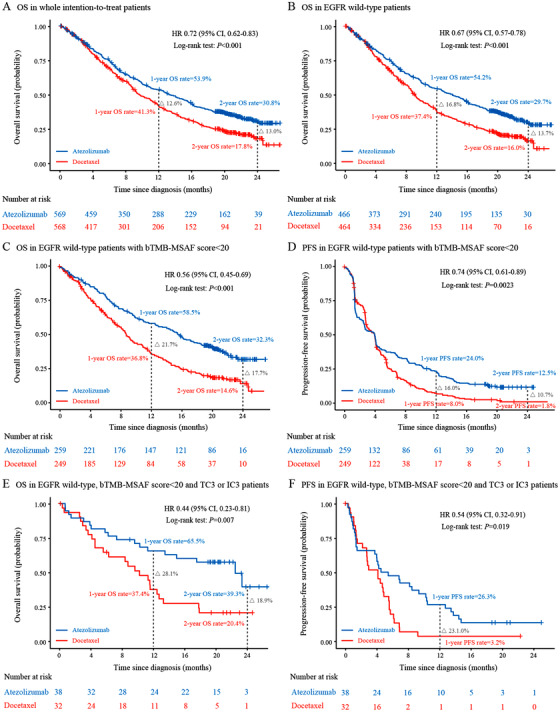
Survival analysis stratified by treatment. A, OS in the whole intention‐to‐treat patients. B, OS in EGFR wild‐type patients. C and D, OS and PFS in EGFR wild‐type patients with a bTMB‐MSAF score < 20, respectively. E and F, OS and PFS in EGFR wild‐type patients with a bTMB‐MSAF score < 20 and PD‐L1 expression of TC3 or IC3, respectively. HR, hazard ratio; CI, confidence interval; OS, overall survival; PFS, progression‐free survival; PD‐L1, programmed cell death ligand 1. TC3 or IC3 indicates that over 50% of tumor cells or over 10% of tumor infiltrating immune cells expressed PD‐L1
We found none of tested bTMB cut‐points could classify patients into groups with consistent differences in PFS and OS between atezolizumab and docetaxel in the EGFR wild‐type patients (Figure S7). Of note, a previously established bTMB threshold of 16 2 even produced paradoxical results; patients with bTMB < 16 had significant improvement in OS (HR 0.68, 95% CI, 0.52‐0.88) but not in PFS (HR 0.99, 95% CI, 0.82‐1.19); patients with bTMB ≥ 16 had significant improvement in PFS (HR 0.58, 95% CI, 0.36‐0.93) but not in OS (HR 0.48, 95% CI, 0.23‐1.02; Figure S7).
We next performed similar analysis at various MSAF cut‐points and found EGFR wild‐type patients with MSAF < 0.10 had improved OS (HR 0.62, 95% CI, 0.51‐0.75) and PFS (HR 0.83, 95% CI, 0.70‐0.98) when treated with atezolizumab compared with docetaxel, whereas patients with MSAF ≥ 0.10 showed no difference in OS or PFS (Figures S8‐S11). Additionally, a low MSAF significantly predict favorable OS and PFS among each of entire patients and atezolizumab‐treated patients (Figure S9). None of the tested bTMB or MSAF cut‐points could classify EGFR mutant patients into groups with different PFS or OS two treatments (Figures S10 and S11).
We further determined a novel bTMB‐MSAF algorithm outperforming bTMB in predicting survival with atezolizumab versus docetaxel. Patients with lower versus higher bTMB‐MSAF scores had significantly better OS and PFS in entire patients and separate OAK and POPLAR cohorts (See details in Supporting Information Result 2 and Figures S12 and S13). The OS HR was more strongly correlated with the bTMB‐MSAF score (R 2 = 0.90) than with the bTMB (R 2 = 0.68) or with the MSAF (R 2 = 0.78) (Figure S14). The threshold of <20 gave the optimal clinical relevance, at which the lowest HRs for OS and PFS was identified (Figure S15).
EGFR wild‐type patients with a bTMB‐MSAF score < 20 had improved both OS (HR 0.56, 95% CI, 0.45‐0.69; P < .001) and PFS (HR 0.74, 95% CI, 0.61‐0.89; P = .0023) when treated with atezolizumab compared with docetaxel (Figure 1C,D). Clinicopathological variables were consistent between treatment arms below the bTMB‐MSAF score < 20 cut‐point (Table S4). Moreover, the EGFR wild‐type patients who concurrently had PD‐L1 expressing on over 50% tumor cells or over 10% of tumor‐infiltrating immune cells (TC3 or IC3) had the greatest benefits for OS (HR 0.44, 95% CI 0.23‐0.81; P = .007) and PFS (HR 0.54, 95% CI 0.32‐0.91; P = .019; Figure 1E,F; see details in Supporting Information Result 3, Figures S16 and S17, and Table S5). However, EGFR wild‐type patients with bTMB‐MSAF score ≥ 20 and EGFR mutant patients of any threshold showed no difference in OS or PFS two treatments (Figures S18 and S19).
We finally explored blood‐based mutational landscape associated with ICI efficacy in NSCLC (see details in Supporting Information Result 4, Figures S20‐22, and Table S6). The blood mutation status of TP53, KEAP1, and ATM were associated with OS following ICI and were incorporated with race, sex, histology, Eastern Cooperative Oncology Group Performance Status, sum of the longest diameter, number of metastasis sites, PD‐L1 expression, and treatment response information were used to develop clinicopathologic‐genomic nomogram, which was well predictive of atezolizumab efficacy (AUC = 0.855, 0.813, 0.860 for 1‐, 2‐, 3‐year OS; Figure 2A and Supporting Information Table S9 and had high clinical usefulness (Figure 2B). Full procedure of building and validating the nomogram was described in Supporting Information Result 5, Figures S23‐S36, and Tables S6‐S11.
FIGURE 2.
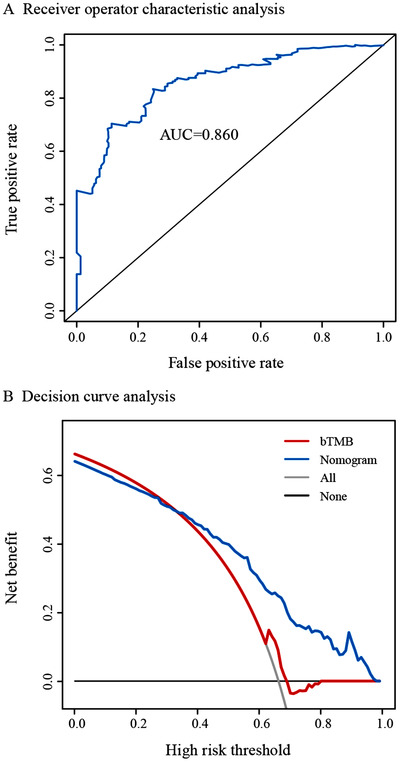
Clinicopathologic‐genomic nomogram to predict the survival of patients undergoing atezolizumab. A, Receiver operating characteristic curves correlating the nomogram with 3‐year overall survival of atezolizumab‐treated patients. B, Decision curve analysis comparing the clinically predictive usefulness between the nomogram and the bTMB
Based on the findings above, we recommend ICI as preferred treatment for EGFR wild‐type patients with a bTMB‐MSAF score < 20, especially those in the TC3 or IC3 subgroup. Our bTMB‐MSAF score markedly enhances the degree of correlation with OS compared with bTMB (R 2 = 0.90 vs 0.68), indicating 22% predictive information was additionally provided by adding MSAF.
This study is the first to provide evidence on the use of blood‐based genomic alterations as predictors of ICI efficacy, and we suggest KEAP1, TP53, and ATM be screened for mutations. The mechanisms for TP53 and KEAP1 mutations might be analogous with their functions in tissues 1 , 6 and the clinical relevance of ATM mutation might be explained by the its role in driving microsatellite instability. 7 More molecular signature such as epigenomics using lncRNAs has been identified as predictor of cancer ICI efficacy. 8 A main limitation of this study was that due to a lack of available data, we were unable to evaluate predictive ability of multi‐omics biomarkers.
In conclusion, a novel bTMB‐MSAF algorithm was established by adding MSAF to bTMB, which could precisely identify NSCLC subsets deriving OS and PFS benefits from atezolizumab over docetaxel, and could synergize with PD‐L1 expression. We suggest that considering blood oncogenic alterations and clinicopathological to establish the blood‐based genomic nomogram could aid in selecting candidates to receive ICI.
AUTHOR CONTRIBUTIONS
Y.Y.Y., Y.J.C., A.L.L., Q.Y.O., X.D.T., Z.H.L., H.H., and H.R.Y. jointly designed the study. Y.Y.Y., Y.J.C., A.L.L., and Q.Y.O. drafted of the manuscript. X.D.T., Z.H.L., H.H., and H.R.Y. provided supervision. All authors contributed to the data collection, data analysis and interpretation, manuscript revision, and approval of the final version.
FUNDING INFORMATION
This study was supported by grants from the National Science and Technology Major Project (2020ZX09201021), the Medical artificial intelligence project of Sun Yat‐Sen Memorial Hospital (YXRGZN201902), the National Natural Science Foundation of China (81572596, 81972471, U1601223), the Natural Science Foundation of Guangdong Province (2017A030313828), the Guangzhou Science and Technology Major Program (201704020131), the Guangdong Science and Technology Department (2017B030314026), the Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (pdjh2019a0212), National Students’ Innovation and Entrepreneurship training program (201910571001), and Guangdong Medical University College Students’ Innovation Experiment Project (ZZZF001).
AVAILABILITY OF DATA AND MATERIALS
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request (yaoherui@mail.sysu.edu.cn).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was approved by the ethics committee of the Sun Yat‐sen Memorial Hospital of Sun Yat‐sen University. The requirement for informed consent of study participants was waived because the human data were obtained from publicly available datasets.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
MEETING PRESENTATION
Preliminary results of this study were presented in part as a mini–oral presentation at the ESMO Asia 2019 Congress; November 22–24, 2019; Singapore.
Supporting information
Supporting information
ACKNOWLEDGMENTS
We sincerely thank Profs David R. Gandara and David S. Shames for kindly providing us the individual‐patient level data of the OAK and POPLAR trials. In addition, we acknowledge all of the investigators and patients who participated in the OAK and POPLAR trials for their contributions.
Contributor Information
Zhihua Li, Email: lzhdoct@163.com.
Hai Hu, Email: huhai@mail.sysu.edu.cn.
Herui Yao, Email: yaoherui@mail.sysu.edu.cn.
REFERENCES
- 1. Yu Y, Zeng D, Ou Q et al. Association of survival and immune‐related biomarkers with immunotherapy in patients with non‐small cell lung cancer: a meta‐analysis and individual patient‐level analysis. JAMA Netw Open. 2019;2(7):e196879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gandara DR, Paul SM, Kowanetz M et al. Blood‐based tumor mutational burden as a predictor of clinical benefit in non‐small‐cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441‐1448. [DOI] [PubMed] [Google Scholar]
- 3. Zhou C, Yuan Z, Ma W et al. Clinical utility of tumor genomic profiling in patients with high plasma circulating tumor DNA burden or metabolically active tumors. J Hematol Oncol. 2018;11(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fehrenbacher L, Spira A, Ballinger M et al. Atezolizumab versus docetaxel for patients with previously treated non‐small‐cell lung cancer (POPLAR): a multicentre, open‐label, phase 2 randomised controlled trial. Lancet. 2016;387(10030):1837‐46. [DOI] [PubMed] [Google Scholar]
- 5. Rittmeyer A, Barlesi F, Waterkamp D et al. Atezolizumab versus docetaxel in patients with previously treated non‐small‐cell lung cancer (OAK): a phase 3, open‐label, multicentre randomised controlled trial. Lancet. 2017;389(10066):255‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong ZY, Zhong WZ, Zhang XC et al. Potential predictive value of TP53 and KRAS mutation status for response to PD‐1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012‐3024. [DOI] [PubMed] [Google Scholar]
- 7. Kim JW, Im SA, Kim MA, et al. Ataxiatelangiectasia mutated protein expression with microsatellite instability in gastric cancer as prognostic marker. Int J Cancer. 2014;134:72‐80. [DOI] [PubMed] [Google Scholar]
- 8. Yu Y, Zhang W, Li A, et al. Association of Long Noncoding RNA Biomarkers With Clinical Immune Subtype and Prediction of Immunotherapy Response in Patients With Cancer. JAMA Netw Open. 2020;3(4):e202149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request (yaoherui@mail.sysu.edu.cn).


