Abstract
Breast cancer is the most common type of cancer in women worldwide. Triple methylation of H4 lysine 20 (H4K20me3), a key component of epigenetic regulation of genomic integrity, is catalyzed by the methyltransferase, SUV420H2. Data on the expression status of SUV420H2 in breast cancer are limited. In the present study, the influence of SUV420H2 suppression on the proliferation of breast cancer cells was experimentally investigated. Subsequently, SUV420H2 expression was assessed in resectable breast cancer along with H4K20me3 status. SUV420H2 expression was knocked down in breast cells using small interfering RNA oligonucleotides. SUV420H2 expression was determined semi-quantitatively at the mRNA level. H4K20me3 was measured on extracted histone proteins using an approach similar to ELISA. Suppression of the SUV420H2 gene resulted in increased cell proliferation. Although the median SUV420H2 expression values were similar in tumor tissues and non-cancerous regions in the entire cohort (0.0022 and 0.0015, respectively; P=0.46), there was a notable difference in expression between tumor tissues and the adjacent non-cancerous region in the majority of patients. Increased SUV420H2 expression in tumors compared with healthy tissue was predominantly observed in patients with early-stage breast cancer, whereas reduced SUV420H2 expression was observed in tumors more frequently in patients with advanced stage diseases. There was no association between SUV420H2 expression and the tissue levels of H4K20me3. The results showed that SUV420H2 exhibited anti-proliferative activity in vitro, and exhibits a heterogeneous expression pattern in breast cancer tissues.
Keywords: breast cancer, epigenetics, histone methylation, triple methylation of H4 lysine 20, SUV420H2
Introduction
Breast cancer is the most common type of invasive malignancy in women worldwide, accounting for ~25% of all types of cancer, and ranking second after lung cancer in cancer-related deaths in women (1). Deaths from breast cancer are typically lower in developed countries, but the mortality is relatively higher in Turkey (2) and other developing countries (3). Despite substantial research efforts and improvements in personalized treatment, including targeted therapies, breast cancer remains a major health obstacle worldwide. This is partly associated with the heterogeneous nature of breast tumors, and hence a lack of appropriate and reliable biomarkers for early detection, prediction of therapy outcomes and disease recurrence (4).
Epigenetic changes are involved in the development of cancer. Alterations in post-translational histone modification pathways (PTHMs) are common in the development and progression of several types of cancer, including breast cancer (5). The methylation of histone proteins is vital for cells to perform their physiological functions; histone proteins serve several regulatory functions in chromatin formation, DNA damage repair, DNA replication and gene expression (6). Methylation of histones usually occurs on the lysine residues in the tails of histone H3 and H4(7). Methylation of H4 lysine 20 (H4K20) is evolutionarily conserved in mammalian cells, and occurs at three different levels as mono-, di- and triple-methylation. Triple-methylation of H4K20 (H4K20me3), catalyzed by the methyltransferase enzyme SUV420H2, is enriched in the gene-poor regions of the genome such as heterochromatin, telomeres, imprinted regions and repetitive elements, and is involved in transcriptional silencing of these regions (8,9). Loss of H4K20me3 in tumor tissues has been described as a hallmark of cancer (10-13). Therefore, the study of PTHMs has become an essential part of cancer research (14-18).
The loss of H4K20me3 in cultured breast cancer cells was found to be accompanied by reduced expression of histone methyltransferase SUV420H2(19). Invasive breast cancer cells (such as MDA-MB-231 or BT-474) were shown to express lower levels of SUV420H2 compared with less invasive breast cancer cells (20). Another study reported lower SUV420H2 expression in mesenchymal breast cancer cells compared with epithelial breast cancer cells (12). These data suggest that reduced SUV420H2 expression may serve as a marker of cancer progression. Accordingly, ectopic overexpression of SUV420H2 leads to reduced breast cancer cell invasion in vitro (20). Data on the expression status of SUV420H2 in breast cancer tissues are limited. Data retrieved from databases showed that breast cancer tissues tended to express lower levels of SUV420H2 compared with normal breast tissues (20).
In the present study, the influence of SUV420H2 suppression on the proliferation of breast cancer cells was first determined. Subsequently, SUV420H2 expression in breast cancer was assessed due to the limited availability of data on the expression of SUV420H2 histone methyltransferase in breast cancer tissues. The results showed that SUV420H2 expression in breast tumors relative to non-cancerous regions was heterogeneous and tended to decrease in more advanced tumors.
Materials and methods
Patients and cells
Patients with resectable breast cancer (n=102) with no secondary malignancies, were enrolled in the present study. Tissues were provided by Istanbul Training and Research Hospital from cases between March 2012 and November 2013. The present study was approved both by the Clinical Research Ethics Committee of Istanbul University (approval no. 2017/887) and the Ethics Committee of Istanbul Training and Research Hospital (approval no. 2012/1413-1201). Histological classification of breast tumors were performed in accordance with the World Health Organization guidelines (21). Tumor grading was performed according to the Nottingham modification of the Scarff-Bloom-Richardson grading system (22). Tumor staging was classified using the Tumor-Node-Metastsasis system adopted by American Joint Committee on Cancer (23). Tumor and adjacent non-cancerous tissue samples were stored at -80˚C. The mean age of the patients was 53 (range, 20-87) years. Table I depicts the clinical characteristics of the patients with breast cancer from whom the tumors were obtained.
Table I.
Clinicopathological characteristics of the breast cancer patients.
| Characteristics | N |
|---|---|
| Age, year | 52 |
| ≤53 | 50 |
| >53 | |
| Menopausal status | |
| Premenopausal | 36 |
| Postmenopausal | 66 |
| Tumor size | |
| T1 | 37 |
| T2 | 57 |
| T3-T4 | 8 |
| Nodal status | |
| N=0 | 43 |
| N≥1 | 59 |
| Stage | |
| I | 19 |
| II | 53 |
| III | 30 |
| Estrogen receptor | |
| Positive | 81 |
| Negative | 21 |
| Progesteron receptor | |
| Positive | 73 |
| Negative | 29 |
| HER2 expression | |
| Positive | 26 |
| Negative | 76 |
| Molecular classification | |
| Luminal A | 35 |
| Luminal B | 42 |
| Her2-like | 11 |
| Basal-like | 14 |
| Histologic Grade | |
| 1 | 8 |
| 2 | 67 |
| 3 | 27 |
| Nuclear Grade | |
| 1 | 2 |
| 2 | 65 |
| 3 | 35 |
For the in vitro studies, the hormone-sensitive breast cancer cell line, MCF-7, and the triple-negative MDA-MB-231 breast cancer cell line were used for silencing SUV420H2 expression. Cells with low passage numbers were grown in standard conditions (37˚C with 5% CO2 in a humidified incubator) in DMEM (Biochrom, Ltd.), containing 10% FBS (Biochrom, Ltd.) and penicillin and streptomycin.
SUV420H2 knockdown in breast cancer cells
The effect of SUV420H2 knockdown on the survival of breast cancer cells was assessed. For silencing experiments, commercially available small interfering (si)RNA oligonucleotides (SMARTpool siRNA; GE Healthcare Dharmacon, Inc.) targeting four different exons of the SUV420H2 gene (SUV420H2-siRNA) were used. Scramble siRNA was used as the negative control (nc-siRNA). The sequences of the siRNAs targeting SUV420H2 were: 5'-GUGAAGGUGCUCCGGGACA-3', 5'-GCGGUGAAGAGCUGUGACA-3', 5'-CGACAGAGUGACAGCACGA-3', and 5'-CUCAGCGCUGGAAACUUU-3'. The sequence of the scrambled siRNA was 5'-GCACGCUCCUACGAAUGCUAGUAAA-3'. A total of 40 h after seeding, ~2x105 cells were transfected using the cationic lipid-based commercial transfection agent Lipofectamine 2000® (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Cells were incubated under standard culture conditions for 48-72 h, after which, cells were harvested and stored at -80˚C until further use. SUV420H2 gene expression in transfected cells was analyzed as described below.
Real-time analysis of cell proliferation
Real-time analysis of cell proliferation was performed using an iCELLigence system (Roche Diagnostics GmbH). For analysis, ~2x104 cells were seeded into each well of the e-plate following baseline measurements using culture medium. siRNA transfection was performed as described above. The proliferation kinetics of the cells was monitored for up to 72 h after transfection.
Colony formation assays
The colony forming capacity of breast cancer cells was studied as a further measure of the effect of SUV420H2 knockdown. For this assay, 1x104 cells were seeded into each well of 6-well culture plates. Transfection was performed as described above, and cells were grown for 10 days under standard culture conditions. At the end of incubation period, the medium was removed, and cells were washed with PBS. Cells were fixed with methanol (100%) for 20 min at room temperature and stained with crystal violet for 30 min at room temperature. After washing, the plates were allowed to dry overnight. Subsequently, the number of colonies formed were counted under a light microscope (magnification, x40; Jenaval, Carl Zeiss AG).
Measurement of SUV420H2 expression in cultured cells and breast tissues
SUV420H2 expression was analyzed at the mRNA level. Total RNA was isolated from cells or tissues using TRIpure® RNA Isolation solution (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol. Extracted RNA was diluted in a final volume of 30 µl using RNase-free water and stored at -80˚C following assessment of integrity using agarose gel electrophoresis on a 2% gel and spectrophotometric purity analysis (Varioskan Flash; Thermo Fisher Scientific Inc.). cDNA synthesis was performed using the commercially available RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific Inc.) according to the manufacturer's protocol. cDNA samples were stored at -20˚C until further use.
SUV420H2 expression was determined semi-quantitatively using GAPDH as the reference gene. SYBR-Green (Thermo Fisher Scientific Inc.) was used as the fluorescent molecule for quantitative PCR. The sequences of the primers used were: SUV420H2 forward, 5'-GGCCCGCTACTTCCAGAG-3' and reverse, 5'-GCAGGATGGTAAAGCCACTT-3'; and GAPDH forward, 5'-GCTCTCTGCTCCTCCTGTTC-3' and reverse, 5'-ACGACCAAATCCGTTGACTC-3'. Measurements were performed twice in a LightCycler 480 instrument (Roche Diagnostics GmbH), and relative SUV420H2 expression was calculated using the 2-ΔΔCq method (24).
Quantification of H4K20me3 levels in tissue samples
For H4K20me3 measurement, histone proteins were extracted from the tissues using the commercially available EpiQuik Total Histone Extraction kit (Epigentek Group, Inc.). Briefly, 1 ml 1X lysis buffer (Epigentek Group, Inc.) was added to tissue lysates and centrifuged at 9,500 x g for 1 min at 4˚C. Supernatant was removed, and 200 µl lysis buffer was added to the pellet containing the cell nuclei and incubated for 30 min on ice. This was followed by a further centrifugation step (5 min, 12,000 rpm, 4˚C). The supernatant containing histone proteins was transferred to fresh tubes and 60 µl Balanced-DTT buffer (Epigentek Group, Inc.) was added. Quantities of histone proteins were spectrophotometrically measured using Varioskan (Thermo Fisher Scientific, Inc.) at 280 nm using FBS as a standard. The purity of the samples was evaluated by measuring the 260/280 nm ratio. Histones were aliquoted and stored at -80˚C for subsequent use.
Extracted histones were used to measure H4K20me3 levels. An ELISA-like measurement of H4K20me3 was performed using the EpiQuik Global Tri-Methylation Histone H4K20 Quantification kit (cat. no. P-3068-98; Epigentek Group, Inc.) according to the manufacturer's protocol. For measurement, 200 ng histone proteins was applied, and measurements were performed twice at 450 nm. H4K20me3 quantities were relatively calculated as the measured absorbance values, and were directly proportional to the amount of the modification.
Statistical analysis
The numerical data from the in vitro tests were compared using a Student's t-test, and the data are presented as the mean ± standard deviation. SUV420H2 and H4K20me3 levels in tumor tissues and non-cancerous regions were compared using a paired samples t-test, and the data are presented as box plots. Statistical analysis of SUV420H2 and H4K20me3 in the patient subgroups was performed using a Kruskal-Wallis test and a χ2 test. Kaplan-Meier analysis and log rank tests were used to assess the survival curves and prognostic value of SUV420H2 expression in breast cancer. P<0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using SPSS version 18 (SPSS, Inc.).
Results
SUV420H2 knockdown in breast cancer cells results in stimulation of cell proliferation
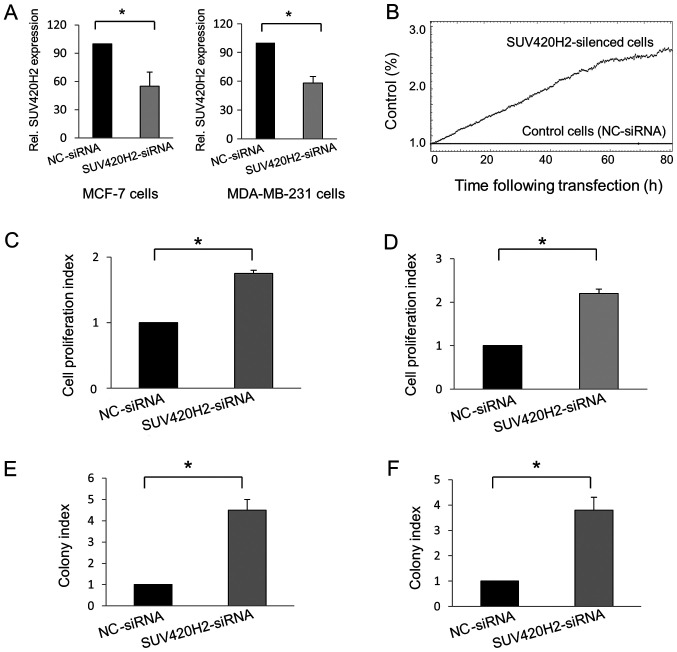
Knockdown experiments in MCF-7 and MDA-MB-231 cells were used to reduce the gene expression levels of SUV420H2. Compared with cells transfected with nc-siRNA, SUV420H2 expression was downregulated 45% in mean in SUV420H2-silenced MCF-7 cells (P=0.048) and 42% in MDA-MB-231 cells (P=0.01; Fig. 1A). The effect of SUV420H2 knockdown on cell proliferation relative to the control cells was assessed in real-time using the iCELLigence instrument (Fig. 1B). Knockdown of the SUV420H2 gene in both cell types resulted in increased cell proliferation compared with nc-siRNA-transfected MCF-7 (Fig. 1C) and MDA-MB-231 cells (Fig. 1D). The colony formation assay also showed that SUV420H2 knockdown resulted in cell proliferation; the number of colonies were increased in the SUV420H2-kncokdown cells compared with the nc-siRNA-transfected cells (Fig. 1E and F).
Figure 1.
SUV420H2 silencing in breast cancer cells and its effect on cell proliferation. (A) SUV420H2 expression was successfully knocked down in MCF-7 and MDA-MB-231 cells. (B) Real-time analysis of cell proliferation in the iCELLigence instrument in MDA-MB-231 cells following transfection with SUV420H2-siRNA relative to nc-siRNA transfected cells. Mean proliferation index following SUV420H2 knockdown in (C) MCF-7 and (D) MDA-MB-231 cells. Mean colony forming capacity following SUV420H2 knockdown in (E) MCF-7 and (F) MDA-MB-231 cells. Data are presented as the mean ± standard deviation. *P<0.05. nc, negative control; siRNA, small interfering RNA.
SUV420H2 gene expression status in breast tumor relative to the non-cancerous tissues does differ significantly
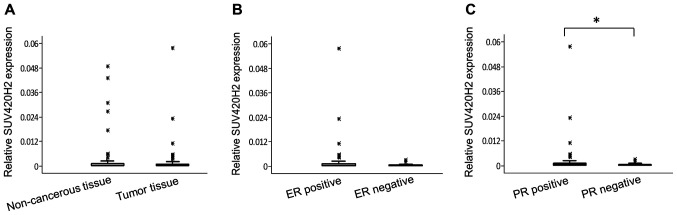
Compared with the reference gene, the expression levels of SUV420H2 were relatively low in breast tissue (both non-cancerous tissue and tumor tissue) (Fig. 2A). The relative median values of SUV420H2 expression in non-cancerous regions and tumor tissues were similar (0.0015 and 0.0022, respectively; P=0.46). There were no association between SUV420H2 expression and clinical parameters (data not shown) except for hormone receptor status; SUV420H2 expression was higher in hormone receptor-positive tumors. The median SUV420H2 expression was 2.4-fold higher in ER+ tumors (Fig. 2B; P=0.12) and 2.8-fold higher in PR+ tumors (Fig. 2C; P=0.047) compared with hormone receptor-negative tumors.
Figure 2.
Levels of SUV420H2 expression in breast tissues. (A) Distribution of relative SUV420H2 expression in breast tumors and the matching non-cancerous regions. (B) Distribution of relative SUV420H2 expression in ER-positive and negative tumors. (C) Distribution of relative SUV420H2 expression in PR-positive and negative tumors. The box plots show the median values, the interquartile range (25 and 75%) and the maximum and minimum values. *P<0.05. PR, progesterone receptor; ER, estrogen receptor.
Although the median SUV420H2 expression was similar in tumor tissues and non-cancer regions in the entire cohort, there were notable differences between tumor tissue and adjacent non-cancerous regions in the majority of individual patients. SUV420H2 expression in tumor tissue significantly increased (at least 2.5-fold) in 31 patients out of 102 (30%) compared with non-cancerous regions, whereas a notable decrease of SUV420H2 expression (by at least 2.5-fold) in tumor tissue was observed in 30 patients (29%). Interestingly, the patients in whom SUV420H2 expression was higher in the tumor compared with the non-cancerous tissue tended to have early-stage breast cancer, whereas in the group with a decrease of SUV420H2 expression in tumors was observed, the patients frequently presented with more advanced staged cancer, with significant differences for both nodal status and disease stage (Table II).
Table II.
Status of the SUV420H2 expression in the breast tumors.
| Nodal status | Stage | |||||
|---|---|---|---|---|---|---|
| Change in SUV420H2 expressiona | N0 | N≥1 | P-value | I-II | III | P-value |
| Increased, n=31 | 20 | 11 | 0.001c | 27 | 4 | 0.02b |
| Decreased (n=30) | 7 | 23 | 18 | 12 | ||
aRelative to non-cancerous region with at least a 2.5-fold change.
bP<0.05,
cP<0.01.
SUV420H2 gene expression is not associated with H4K20me3 levels
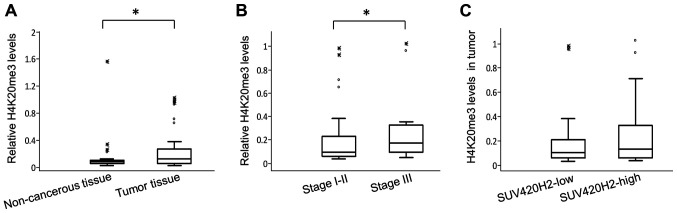
H4K20me3 levels in breast tumor tissues and adjacent non-cancerous regions were also assessed. The relative quantities of H4K20me3 in tumor tissues were significantly higher compared with the adjacent non-cancerous regions (median values 0.1124 and 0.087, respectively; P=0.001; Fig. 3B). Patients with stage III breast cancer had 2-fold higher H4K20me3 levels in their tumors compared with patients with stage I-II (relative median levels 0.17 vs. 0.88, respectively; P=0.04; Fig. 3B).
Figure 3.
H4K20me3 levels in breast tissues. (A) Relative H4K20me3 levels in breast tumors and accompanying non-cancerous tissues. (B) Relative H4K20me3 levels in patients with stage III disease vs. stage I-II disease. (C) Relative H4K20me3 levels in tumors with low and high SUV420H2 expression. The box plots show the median values, the interquartile range (25 and 75%) and the maximum and minimum values. *P<0.05. H4k20me3, triple methylation of H4 lysine 20.
There was no association between SUV420H2 expression and H4K20me3 levels in tumors. Accordingly, H4K20me3 levels were similar (0.1 vs. 0.13, respectively; P=0.68) in patients with lower SUV420H2 expression (below median) and in those with high expression (above median) in tumors (Fig. 3C). Similarly, patients in whom SUV420H2 expression was increased in tumors compared with adjacent non-cancerous regions had similar H4K20me3 levels in their tumor tissues to those in whom SUV420H2 expression was decreased in tumors.
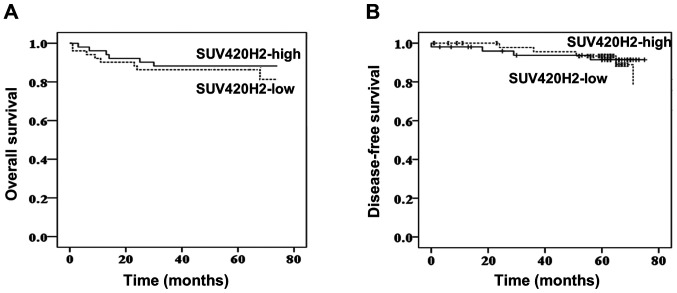
SUV420H2 gene expression does not exhibit prognostic value in patients with breast cancer
The patients were followed up for 75 months. The overall survival (OS) and disease-free survival (DFS) were compared between the low SUV420H2-expressing group and high SUV420H2-expressing group (stratified by the median value). Median OS times were 63 and 65 months for low and high expression groups, respectively (P=0.734; Fig. 4A). Similarly, DFS was very similar for both expression groups (P=0.86; Fig. 4B) indicating that the SUV420H2 expression in tumor tissues had no prognostic value.
Figure 4.
Survival analysis in patients with breast cancer. Kaplan-Meier analysis of overall and disease-free survival in patients with low and high SUV420H2 expression.
Discussion
The experimental evidence in the present study and previously published data suggest that SUV420H2 suppresses the proliferation, migration and invasiveness of breast cancer cells (20). However, SUV420H2 was found to be an epigenetic regulator of epithelial-to-mesenchymal transition in pancreatic cancer cells (25), suggesting that the role of SUV420H2 in tumor progression may be cell-type specific.
Data on the expression status of SUV420H2 in cancer in general and in breast cancer are limited (11,20,25), despite the well-defined tumor suppressive role of SUV420H2 in breast cancer cells in vitro (11,20). In the present study, SUV420H2 expression in breast cancer was examined and found similar levels of SUV420H2 expression in the entire cohort in paired samples comparison between tumor tissues and adjacent non-cancerous tissues. In individual patients however, a heterogeneous pattern of SUV420H2 expression status in tumors relative to non-cancerous regions was observed. In patients with early-stage breast tumors, SUV420H2 expression in tumor relative to adjacent non-cancerous regions was frequently higher. In contrast, in patients with larger tumors and/or lymphatic metastasis, SUV420H2 expression in tumor tissues relative to non-cancerous tissues tended to decrease. These results suggest that SUV420H2 expression decreases as breast cancer progresses. Data retrieved from databases such as The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), and Methylation and Expression database of Normal and Tumor tissues (MENT) revealed that SUV420H2 expression is lower in breast cancer tissues compared with normal tissues (20). In contrast to the present study, the previous study was not a paired samples comparison of tumors with adjacent non-cancerous tissues in individual patients. These data may thus be summarized by stating that in a substantial portion of patients with breast cancer, SUV420H2 expression is decreased in breast tumors compared with normal tissue, and the loss of SUV420H2 expression may be an indicator of breast cancer progression. A limitation of the present study was that the protein expression levels of SUV420H2 were not assessed, which may have provided additional information on its role in tumor development and progression. Based on TCGA data, SUV420H2 expression was found to be increased in tumor tissues in pancreatic cancer compared with equivalent normal tissues, and high levels of SUV420H2 were correlated with a loss of epithelial characteristics in progressively invasive cancer (25). This contradictory state compared with breast cancer implies a heterogeneous role of SUV420H2 in cancer development and progression.
There was no significant association between expression of SUV420H2 and the age, menopausal status, tumor stage, lymphatic metastasis, histologic grade or nuclear grade of the patients. However, there was an association between SUV420H2 expression and the hormone receptor status. SUV420H2 expression was higher in patients with hormone receptor-positive tumors compared with hormone receptor-negative tumors. The functional basis of this association and its relationship with prognosis should be investigated in future studies.
Based on the in vitro data and tissue analysis, the loss of H4K20me3 in tumor tissues has been described as a hallmark of cancer (10-13). Similar to the heterogeneity in SUV420H2 expression in breast cancer, the H4K20me3 expression pattern in breast tumors seems to be heterogeneous. Elsheikh et al (26) reported in their immunohistochemical study, which consisted of a large series of breast tumors, that H4K20me3 expression was increased in breast tumors in 70% of patients with invasive breast carcinoma compared with normal tissue, whereas it was decreased in the remaining 30%. Yokoyama et al (11) also described a heterogeneous pattern of H4K20me3 levels in breast tumors using immunohistochemistry, where the reduction of H4K20me3 in tumors compared with non-cancerous regions was more frequently observed. In the present study, the status of H4K20me3 in tumor tissue relative to non-cancerous tissue among the patients was also heterogeneous, although an increase was more frequently observed. It is possible that the existing heterogeneity in H4K20me3 expression in tumors may be amplified by variations in methodologies. In the present study, H4K20me3 levels were measured on extracted histone proteins, whereas previous studies used immunohistochemistry. The assessment of H4K20me3 by immunohistochemistry may be a more informative method of assessing distribution morphologically in tumor tissue.
There was no prognostic predictive role for SUV420H2 expression in breast cancer identified in the present study, based on a 75 month follow-up period. However, longer follow-up times are required to more accurately evaluate the prognostic role of SUV420H2 expression in breast cancer.
In conclusion, the results of the present study indicate that the methyltransferase SUV420H2, which exhibits anti-tumor activity in vitro, is heterogeneously expressed in breast tumors relative to matched non-cancerous regions of the breast. The reduction of SUV420H2 in tumors relative to non-cancerous regions was apparently more frequent in patients with more advanced disease, which suggested that tumor cells which downregulated SUV420H2 expression during progression were more likely to survive or proliferate. These data may provide a basis for further analysis of SUV420H2 in future studies examining the potential SUV420H2 as a target for therapeutic interventions in patients with breast cancer.
Acknowledgements
The present study forms part of a Ph.D. thesis of Ms Hüsniye Isin (Istanbul University).
Funding
The present study was supported by the Scientific Research Projects Coordination Unit of Istanbul University (grant no. TDK-2017-27336).
Availability of data and materials
The datasets used and/or analyzed during the present study and patient consent are available from the corresponding author on reasonable request.
Authors' contributions
HI and UG were responsible for the conception and design of the study. CKT, DCT and DK were involved in the provision of study materials and clinical data, and critically revised the manuscript. HI and EÖ analyzed and interpreted the data as well as drafted the manuscript. All authors read and approved the final version of the manuscript for publication.
Ethics approval and consent to participate
The present study was approved both by the Clinical Research Ethics Committee of Istanbul University (approval no. 2017/887) and the Ethics Committee of Istanbul Training and Research Hospital (approval no. 2012/1413-1201), (Istanbul, Turkey). All patients provided informed written consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F. Breast cancer. Nat Rev Dis Primers. 2019;5(66) doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 2.Dogan N, Toprak D. Female breast cancer mortality rates in Turkey. Asian Pac J Cancer Prev. 2014;15:7569–7573. doi: 10.7314/apjcp.2014.15.18.7569. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 4.Sauter ER. Reliable biomarkers to identify new and recurrent cancer. Eur J Breast Health. 2017;13:162–167. doi: 10.5152/ejbh.2017.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Z, Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol. 2019;20(245) doi: 10.1186/s13059-019-1870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller JL, Grant PA. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell Biochem. 2013;61:289–317. doi: 10.1007/978-94-007-4525-4_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Jia S. Degrees make all the difference: The multifunctionality of histone H4 lysine 20 methylation. Epigenetics. 2009;4:273–276. doi: 10.4161/epi.4.5.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svobodová Kovaříková A, Legartová S, Krejčí J, Bártová E. H3K9me3 and H4K20me3 represent the epigenetic landscape for 53BP1 binding to DNA lesions. Aging (Albany NY) 2018;10:2585–2605. doi: 10.18632/aging.101572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jørgensen S, Schotta G, Sørensen CS. Histone H4 lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tryndyak VP, Kovalchuk O, Pogribny IP. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther. 2006;5:65–70. doi: 10.4161/cbt.5.1.2288. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama Y, Matsumoto A, Hieda M, Shinchi Y, Ogihara E, Hamada M, Nishioka Y, Kimura H, Yoshidome K, Tsujimoto M, Matsuura N. Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res. 2014;16(R66) doi: 10.1186/bcr3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Nuland R, Gozani O. Histone H4 lysine 20 (H4K20) methylation, expanding the signaling potential of the proteome one methyl moiety at a time. Mol Cell Proteomics. 2016;15:755–764. doi: 10.1074/mcp.R115.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 14.Füllgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 15.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Investig. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanmugam MK, Arfuso F, Arumugam S, Chinnathambi A, Jinsong B, Warrier S, Wang LZ, Kumar AP, Ahn KS, Sethi G, Lakshmanan M. Role of novel histone modifications in cancer. Oncotarget. 2017;19:11414–11426. doi: 10.18632/oncotarget.23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noberini R, Osti D, Miccolo C, Richichi C, Lupia M, Corleone G, Hong SP, Colombo P, Pollo B, Fornasari L, et al. Extensive and systematic rewiring of histone post-translational modifications in cancer model systems. Nucleic Acids Res. 2018;46:3817–3832. doi: 10.1093/nar/gky224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson NE, Tryndyak VP, Beland FA, Pogribny IP. An in vitro investigation of metabolically sensitive biomarkers in breast cancer progression. Breast Cancer Res Treat. 2012;133:959–968. doi: 10.1007/s10549-011-1871-x. [DOI] [PubMed] [Google Scholar]
- 20.Shinchi Y, Hieda M, Nishioka Y, Matsumoto A, Yokoyama Y, Kimura H, Matsuura S, Matsuura N. SUV420H2 suppresses breast cancer cell invasion through down regulation of the SH2 domain-containing focal adhesion protein tensin-3. Exp Cell Res. 2015;334:90–99. doi: 10.1016/j.yexcr.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ (eds) WHO classification of tumours of the breast. In: WHO Classification of Tumours. Vol 4. 4th edition. IARC, Lyon. 2012 [Google Scholar]
- 22.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 23.Singletary SE, Allred C, Ashley P, Bassett LW, Berry D, Bland KI, Borgen PI, Clark G, Edge SB, Hayes DF, et al. Revision of the American Joint Committee on Cancer staging system for breast cancer. J Clin Oncol. 2002;20:3628–3636. doi: 10.1200/JCO.2002.02.026. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Viotti M, Wilson C, McCleland M, Koeppen H, Haley B, Jhunjhunwala S, Klijn C, Modrusan Z, Arnott D, Classon M, et al. SUV420H2 is an epigenetic regulator of epithelial/mesenchymal states in pancreatic cancer. J Cell Biol. 2018;217:763–777. doi: 10.1083/jcb.201705031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study and patient consent are available from the corresponding author on reasonable request.