Abstract
Leucine-rich-repeat-containing G protein-coupled receptor 6 (LGR6) has been identified as the stem cell marker in multiple normal tissues and malignancies. Previous studies implicated paradoxical functions of LGR6 as a tumor-suppressor gene or oncogene given to the specific context. To explore the exact role of LGR6 in triple-negative breast cancer (TNBC) that never has been studied before, in this study, we assessed LGR6 expression levels by RT-PCR and immunohistochemistry. LGR6 stable expressing/silenced cells were established, and functional assays on tumor proliferation, as well as metastasis, were conducted both in vitro and in vivo. Here, we found that LGR6 was overexpressed in TNBC, which correlated with poor disease-free and overall survivals. Functional assays both in vitro and in vivo showed that LGR6 promotes tumor proliferation and metastasis. LGR6 also increased the ability of tumor spheroid formation. Underlying mechanism exploration further revealed that the oncogenic role of LGR6 might be associated with the Wnt/β-catenin pathway. In conclusion, our findings first proved that LGR6 acts as an oncogene in (TNBC), indicating that LGR6 might be a potential therapeutic target for TNBC treatment.
Graphical Abstract
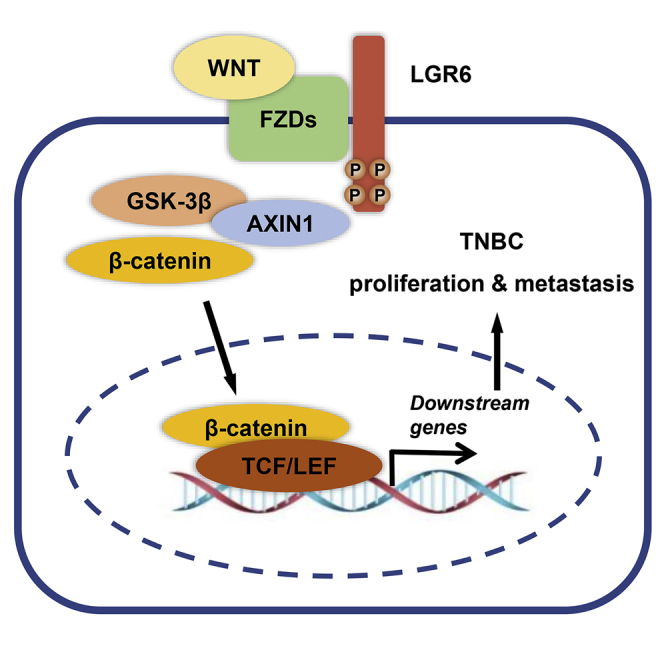
This study revealed the oncogenic role of LGR6 in triple-negative breast cancer (TNBC), which was highly expressed in TNBC and correlated with worse survivals. LGR6 promotes tumor proliferation and metastasis through Wnt/β-catenin signaling. These findings indicated that LGR6 might be a potential therapeutic target for TNBC treatment.
Introduction
Triple-negative breast cancer (TNBC) is one subtype of breast cancer that is defined as negative or a low expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor (EGF) receptor-2 (Her2). There is approximately 15% of breast cancer diagnosed as this subtype.1 Compared with luminal and Her2-enriched breast cancer, TNBC owns a higher ability of proliferation, invasion, and metastasis, leading to a more advanced stage and poor prognosis clinically.1,2 Due to lack of effective molecular targets, patients with TNBC have few choices on targeted therapies, making the way to cure TNBC tougher. Therefore, figuring out the underlying mechanisms of biological behavior of TNBC and finding novel molecular targets are under great need.
Leucine-rich-repeat-containing G protein-coupled receptor 6 (LGR6) is a member of the LGR family, which has been identified as a stem cell marker in many tissues, such as the skin,3,4 nail,5 lungs,6 ovary,7,8 breast,9,10 and taste bud.11 LGR4–6, as the homologous receptors, could activate Wnt/β-catenin signaling by binding to R-spondins (RSPOs),12,13 which have been proven important in tumor progression and metastasis.14 Previous studies implicated controversial functions of LGR6 as a tumor-suppressor gene or oncogene. Gong et al.13 reported that LGR6 plays the role of a tumor suppressor in colon and ovarian cancer, whereas some other studies found that LGR6 was overexpressed and associated with poor survivals in a variety of cancers. Krejs15 and Ke et al.16 revealed that LGR6 promotes the progression of gastric cancer through the Wnt/β-catenin and phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway. Ruan et al.7 reported that silencing LGR6 attenuates stemness and chemoresistance in ovarian cancer. Blaas et al.9 demonstrated that Lgr6+ progenitor cells own the ability to originate luminal mammary tumors. Genome-wide association studies identified LGR6 as an ER-negative and triple-negative-specific breast cancer germline susceptibility gene.17,18 However, the exact role of LGR6 in the development of TNBC and its underlying mechanisms are still unknown.
Here, we assessed the expression status of LGR6 in TNBC and the correlation with clinical survivals. LGR6-expressing or silenced cells were established, and a series of functional assays were performed in vitro and in vivo to evaluate the role of LGR6 in tumor proliferation and metastasis. In addition, the mechanisms were also explored by analyzing the association of LGR6 and the Wnt/β-catenin pathway, as well as TNBC-associated genes. Our results showed that LGR6 acts as an oncogene in TNBC and promotes tumor progression by the Wnt/β-catenin pathway, indicating that LGR6 may serve as a potential therapeutic target in TNBC.
Results
Increased Expression of LGR6 in TNBC Cell Lines and Tissues
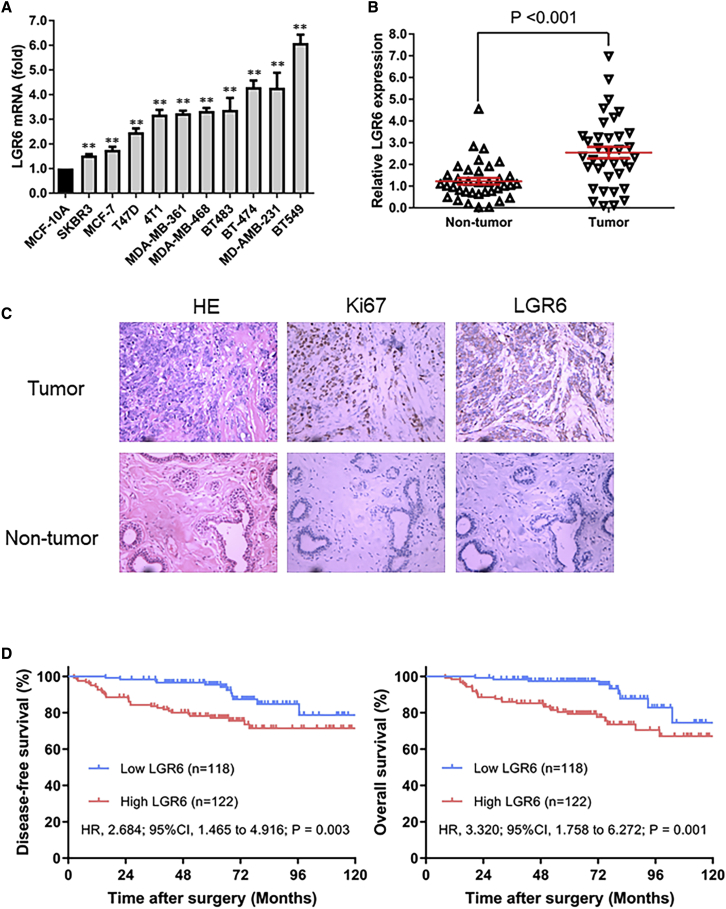
We measured the expression levels of LGR6 in both breast cancer cell lines and tissues. Quantitative real-time PCR was performed in a variety of mammary cell lines, and LGR6 was found to be overexpressed in breast cancer cell lines, compared with human mammary epithelial cell line MCF-10A. Among all breast cancer cell lines, the highest expression level was observed in the TNBC cell line BT549 (Figure 1A). To further confirm LGR6 expression in tissues, we extended our findings to 36 pairs of TNBC tumor samples and adjacent nontumor samples. LGR6 overexpression (defined as a more than 2-fold increase) was detected in 20 (55.5%) patients. The mean fold change of LGR6 expression in TNBC tumor tissues was significantly higher than that in the paired nontumor tissues (p < 0.001, paired Student’s t test; Figure 1B).
Figure 1.
Increased Expression of LGR6 in TNBC
(A) LGR6 expression in normal mammary cell lines, non-TNBC cell lines, and TNBC cell lines was tested by quantitative real-time PCR. ∗p < 0.05, p < 0.01. (B) The relative expression levels of LGR6 were tested by quantitative real-time PCR in 36 pairs of TNBC specimens. (C) IHC staining of LGR6 and Ki67 expression in two pairs of matched TNBC tumor and adjacent nontumor samples. Original magnification, ×200. (D) Kaplan-Meier analysis showed high expression of LGR6 correlated with poor disease-free survival and overall survival.
High Expression of LGR6 Correlated with Poor Survivals in TNBC
To explore the correlation between LGR6 expression and survivals in patients with TNBC, we analyzed tumor tissues from 240 patients with TNBC using immunohistochemistry (IHC) staining. The basics of clinic-pathological characteristics were listed in Table 1. LGR6 expression was classified into a high and low group, the staining of which was shown in Figure 1C. High expression of LGR6 was detected in 122 patients, accounting for 53.0% of all. Survival analysis showed that TNBC patients with high expression of LGR6 have a worse disease-free survival (hazard ratio, 2.684; 95% confidence interval [CI], 1.465 to 4.916; p = 0.003) and overall survival (hazard ratio, 3.320; 95%CI, 1.758 to 6.272; p = 0.001) than those with low expressions (Figure 1D). Multivariate Cox regression showed that LGR6 expression is an independent prognostic factor in TNBC (Tables 2 and 3). These results suggested that LGR6 is a potential prognostic factor and may act as an oncogene involved in the progression of TNBC.
Table 1.
Correlation between Clinic-Pathologic Characteristics and LGR6 Expression
| Characteristics | Low LGR6 (n = 118) | High LGR6 (n = 122) | p Value |
|---|---|---|---|
| Age | |||
| ≤35 years | 9 (7.6) | 18 (14.8) | 0.081 |
| >35 years | 109 (92.4) | 104 (85.2) | |
| Menopausal Status | |||
| Pre | 79 (66.9) | 65 (53.3) | 0.031 |
| Post | 39 (33.1) | 57 (46.7) | |
| Tumor Size | |||
| ≤2 cm | 35 (29.7) | 31 (25.4) | 0.461 |
| >2 cm | 83 (70.3) | 91 (74.6) | |
| Lymph Nodes Invasion | |||
| Yes | 63 (53.4) | 54 (44.3) | 0.157 |
| No | 55 (46.6) | 68 (55.7) | |
| TNM staging | |||
| I–II | 92 (78.0) | 96 (78.7) | 0.892 |
| III–IV | 26 (22.0) | 26 (21.3) | |
| Histologic Grade | |||
| I–II | 71 (60.2) | 73 (59.8) | 0.958 |
| III–IV | 47 (39.8) | 49 (40.2) | |
Data were numbers of patients with percentage in parentheses.
Table 2.
Multivariate Analysis of Risk Factors for Disease-Free Survival
| Characteristics | Hazard Ratio | 95% Confidence Interval | p Value |
| LGR6 Status | |||
| Low expression | 1 | 0.002 | |
| High expression | 2.886 | 1.454 to 5.729 | |
| Age | |||
| ≤35 years | 1 | 0.034 | |
| >35 years | 0.426 | 0.194 to 0.936 | |
| Menopausal Status | |||
| Pre | 1 | 0.310 | |
| Post | 0.690 | 0.337 to 1.413 | |
| Tumor Size | |||
| ≤2 cm | 1 | 0.330 | |
| >2 cm | 1.633 | 0.609 to 4.378 | |
| Lymph Nodes Invasion | |||
| No | 1 | 0.118 | |
| Yes | 1.960 | 0.844 to 4.553 | |
| TNM staging | |||
| I–II | 1 | 0.012 | |
| III–IV | 2.598 | 1.230 to 5.484 | |
| Histologic Grade | |||
| I–II | 1 | 0.177 | |
| III–IV | 1.547 | 0.821 to 2.916 | |
Table 3.
Multivariate Analysis of Risk Factors for Overall Survival
| Characteristics | Hazard Ratio | 95% Confidence Interval | p Value |
| LGR6 Status | |||
| Low expression | 1 | 0.001 | |
| High expression | 3.674 | 1.701 to 7.937 | |
| Age | |||
| ≤35 years | 1 | 0.099 | |
| >35 years | 0.501 | 0.221 to 1.139 | |
| Menopausal Status | |||
| Pre | 1 | 0.197 | |
| Post | 0.603 | 0.280 to 1.300 | |
| Tumor Size | |||
| ≤2 cm | 1 | 0.184 | |
| >2 cm | 2.082 | 0.706 to 6.140 | |
| Lymph Nodes Invasion | |||
| No | 1 | 0.102 | |
| Yes | 2.048 | 0.866 to 4.842 | |
| TNM staging | |||
| I–II | 1 | 0.081 | |
| III–IV | 2.019 | 0.918 to 4.442 | |
| Histologic Grade | |||
| I–II | 1 | 0.217 | |
| III–IV | 1.525 | 0.780 to 2.980 | |
LGR6 Promotes Cell Proliferation and Tumorigenesis in TNBC
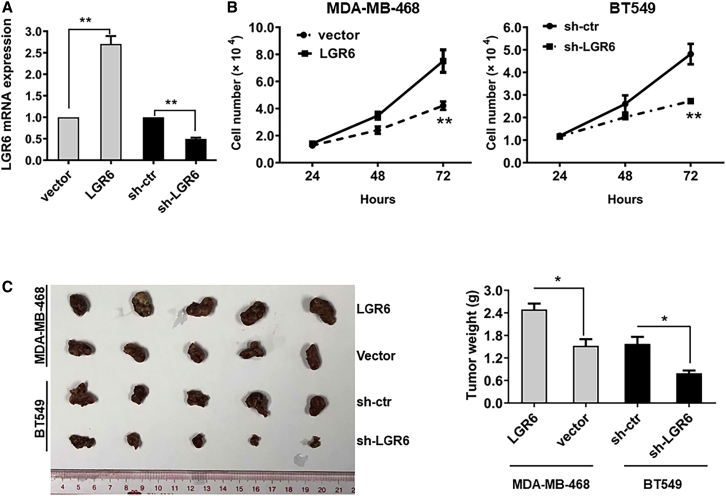
To further testify the role of LGR6 in the development of TNBC, stable LGR6 overexpressing cells (LGR6-MDA-MB-468) and stable LGR6 silenced cells (shLGR6-BT549) were established by plasmid construction and short hairpin RNA (shRNA) transfection. Empty vector-transfected cells (vector-MDA-MB-468) and scramble shRNA-transfected cells (shcrl-BT549) were used as controls. LGR6 levels were significantly overexpressed in MB-468-LGR6 cells and downregulated in shcrl-BT549 cells compared to the controls (p < 0.01; Figure 2A). Cell growth assays were performed, and the cell growth rate in LGR6-MDA-MB-468-transfected cells was significantly higher than that in controls, whereas silenced LGR6 in BT549 cells led to decreased cell growth rates (p < 0.01; Figure 2B). Animal experiments were performed, and LGR6-modulated breast cancer cells were injected into the flanks of nude mice. After 4 weeks, the mice were euthanized, and the tumors were harvested. The volume and weight of tumors induced by LGR6-MDA-MB-468 cells and shLRG6-BT549 were significantly larger and heavier (p < 0.05 for LGR6-MDA-MB-468 cells; p < 0.05 for shLRG6-BT549 cells) than those induced by controls (Figure 2C left and right). Our findings indicated that LGR6 could promote cell proliferation and tumorigenesis in TNBC.
Figure 2.
LGR6 Promotes Tumorigenesis in TNBC
(A) LGR6 expression was confirmed by quantitative real-time PCR in LGR6 stable expressing/knockdown cells, and β-actin was used as a loading control. (B) Cell proliferation assays were conducted to compare the cell growth rates between LGR6-MDA-MB-468 and control cells and between shLGR6-BT549 and control cells. (C) Representative images of xenografts and a summary of tumor weight in nude mice. The weights of xenograft tumors are summarized in the right panel. All results are expressed as the mean ± standard deviation of three independent experiments (∗p < 0.05).
LGR6 Increased Stemness and Promotes Tumor Metastasis in TNBC
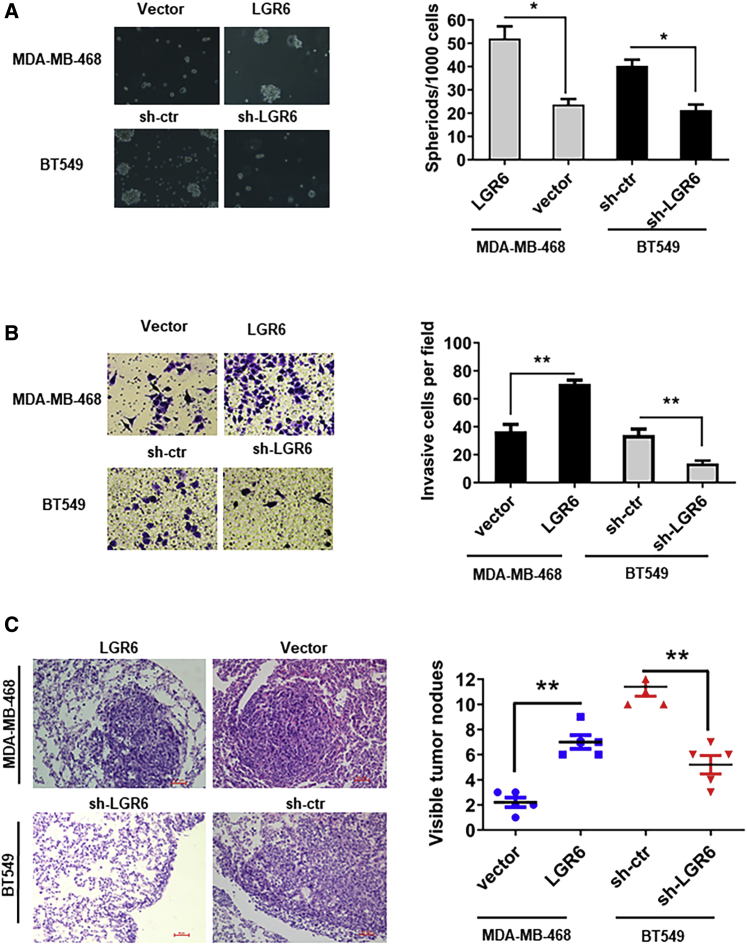
Previous studies reported that LGR6 marks stem cells of a variety of tissues, including mammary gland cells, and we explored whether LGR6 could increase the stemness of TNBC cells. It was found that compared with shcrl-BT549 cells, the ability of sphere forming in shLGR6-BT549 cells was significantly decreased. Furthermore, spheroids of shLGR6-BT549 cells were observed to be surrounded by a cluster of loosely associated cells, which are susceptible to disruption, whereas shcrl-BT549 cells form compact spheroids that are not easily mechanically disrupted. More and larger spheroids are formed by LGR6-MDA-MB-468 cells than by the control cells (p < 0.05; Figure 3A). To further assess the ability of migration promoted by LGR6, a Matrigel invasion assay was conducted in vitro, and it was found that LGR6 expressing cells own higher invasiveness than controls (p < 0.01 for LGR6-MDA-MB-468 cells; p < 0.01 for shLGR6-BT549 cells; Figure 3B). To testify effect of LGR6 on tumor metastasis in vivo, we injected LGR6-modulated MB-468/BT549 cells into nude mice (six mice per group) through the tail vein. After 8 weeks, the mice were sacrificed, and metastatic nodules of lungs were dissected. Hematoxylin and eosin staining was used to confirm the nodules on the lungs of the mice as metastatic tumors (Figure 3C, left). LGR6-MDA-MB-468 cells presented a stronger ability of metastasis than controls, and LGR6 silencing significantly reduced the efficiency of lung metastasis originating from BT549 cells (p < 0.01, Student’s t test; Figure 3C, right). All of these results indicated that LGR6 could promote tumor invasion and metastasis, both in vitro and in vivo.
Figure 3.
LGR6 Increases the Stemness and Promotes Metastasis in TNBC
(A) LGR6 increased the sphere-forming ability in LGR6-MDA-MB-468 cells and decreased it in LGR6 silenced BT549 cells (∗p < 0.05, ∗∗p < 0.01). (B) Transwell invasion assay demonstrating that LGR6 promotes cell invasion. Representative images of invaded cells are shown in the left panel, and the results are summarized in the right panel. The results are expressed as the mean ± standard deviation of three independent experiments (∗p < 0.05, ∗∗p < 0.01). (C) Representative images of H&E-stained sections derived from metastatic nodules at the lung surface. Original magnification, ×100. The micrometastases in the H&E-stained sections of individual mice were calculated. Each group had 5 mice. Bars, 50 μm. All data are shown as mean ± standard deviation (∗∗p < 0.01).
LGR6 Regulates the Wnt/β-Catenin Pathway in TNBC
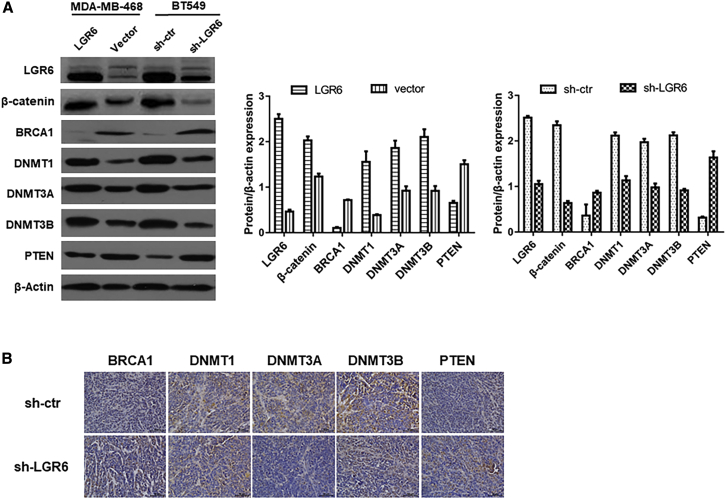
To evaluate the correlation between LGR6 and the Wnt/β-catenin pathway, we performed western blot analysis on LGR6-regulated and control MBA-MB-468/BT549 cells. It was observed that inhibition of LGR6 suppressed the expression of β-catenin, DNA methyltransferase 1 (Dnmt1), Dnmt3a, and Dnmt3b. We also examined the expression of two suppressive oncogenes and found that silenced LGR6 increased the expression of BRCA1 and PTEN (Figure 4A). To further confirm the correlation of these markers, IHC was performed on tumors from a xenograft model (Figure 4B). We found that the expression of Dnmt1, Dnmt3a, and Dnmt3b was downregulated, whereas BRCA1 and PTEN were upregulated in tumors, which originated from shLGR6-BT549 cells. These findings indicated that the oncogenic role of LGR6 may be associated with the Wnt/β-catenin pathway and DNA methylation in TNBC.
Figure 4.
LGR6 Regulates the Wnt/β-Catenin Pathway in TNBC
(A) The protein expression of LGR6, β-catenin, BRCA1, Dnmt1, Dnmt3a, Dnmt3b, and PTEN in LGR6-MDA-MB-468 and control cells and between shLGR6-BT549 and control cells detected by western blot. β-actin was used as the loading control. The band intensities are quantified and normalized to β-actin intensities (∗p < 0.05, ∗∗p < 0.01). (B) Immunohistochemistry assay of BRCA1, Dnmt1, Dnmt3a, Dnmt3b, and PTEN expressions in shLGR6-BT549 and control cell xenografts.
Discussion
This study explored the expression status and the exact role of LGR6 in TNBC. We found that LGR6 was upregulated in TNBC cell lines and tumor tissues. Overexpression of LGR6 correlated with poor disease-free survival and overall survivals in patients with TNBC. LGR6 promotes tumor growth and metastasis both in vitro and in vivo, which might be associated with the Wnt/β-catenin pathway. Therefore, our findings reveal that LGR6 acts as an oncogene in TNBC and may serve as a novel therapeutic target for TNBC.
LGR proteins, including LGR4–6, are Wnt signaling mediators that have been explored a lot in various malignancies in recent years.12,13,19,20 LGR5 was the first and most concerned LGR member since Barker et al.21 identified the stem cells in small intestine and colon stem/progenitor cells by LGR5 via in vivo lineage tracing. Numerous studies have revealed the significant role of LGR5 in stem cell identification and stemness maintenance in both normal tissues and cancers.22, 23, 24 However, subsequent research found that although homologs, LGR6 and LGR5 may have distinctly different distribution and function. Huang et al.4 demonstrated that Lgr6, but not Lgr5, acts as an epithelial stem cell marker in squamous cell carcinomas (SCCs) in vivo. Lee et al.25 found that Lgr6+ cells comprise smooth muscle cells and promote airway differentiation of epithelial progenitors via Wnt-Fgf10 cooperation, whereas Lgr5+ cells are located in alveolar compartments and promote alveolar differentiation of epithelial progenitors through Wnt activation. A study of ovarian cancer showed that LGR6 was dramatically upregulated, but LGR5 was slightly upregulated compared with normal controls.8 For breast cancer, Yang et al.26 have reported that LGR5 promotes breast cancer progression and maintains stem-like cells through activation of Wnt/β-catenin signaling; however, the status and function of LGR6 in breast cancer are less concerning. In our study, we assessed the expression levels of LGR6 in cell lines and found that LGR6 was significantly upregulated in TNBC cell lines compared with others. Tissue arrays and survival analysis testified that the overexpression of LGR6 in TNBC and the association with poor survivals are consistent with the survival analysis from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) database, whereas contradicted the outcomes from The Cancer Genome Atlas (TCGA) database. The discrepancy among ours, METABRIC’s, and TCGA’s may be attributed to the fact that there are multiple subtypes within TNBC, as it is a heterogeneous disease. According to the findings revealed by Lehmann et al. in 201127, cluster analysis identified 6 TNBC subtypes displaying unique gene expression and ontologies, including 2 basal-like (BL1 and BL2), an immunomodulatory (IM), a mesenchymal (M), a mesenchymal stem-like (MSL), and a luminal androgen receptor (LAR) subtype. MDA-MD-468 belongs to the BL1 subtype, and BT549 belongs to the M subtype, which share the similar genetic background. Therefore, our findings may be limited to some subtypes of TNBC. Further validation is still needed, especially in a Chinese TNBC population database and other subtypes that were not represented in this study.28 To further explore the exact role of LGR6, LGR6 stable expressing or silenced cells were established, and experiments in vitro and in vivo showed that LGR6 could promote tumor proliferation and metastasis. Our findings proved the oncogenic role of LGR6 in TNBC through the Wnt/β-catenin pathway, which is consistent with genome-wide association study (GWAS) screening results that LGR6 is a susceptible gene for ER-negative breast cancer or TNBC.17,18
Dnmt1, Dnmt3a, and Dnmt3b are members of the mammalian Dnmt family, which is responsible for genomic DNA methylation and plays important roles in normal development and genosome stability.29 Dnmt dysregulation perturbs the homeostasis of various tissue types, such as lung, intestine, hematopoietic cells, neural cells, and the epidermis.30,31 Somatic mutations of Dnmts may lead to tumorigenesis and cancer progression.32 Poomipark’s team33 reported that Dnmt1 was involved in acquired resistance to chemotherapy in lung cancer, whereas Dnmt3a and Dnmt3b associated with cervical cancer progression.34 In this study, we found that the expression of Dnmts was positively correlated with LGR6, indicating that DNA methylation might be involved in the oncogenic function of LGR6. Mounting previous studies have demonstrated that conjunction of LGR4–6 and RPSO1–4 could activate the Wnt/β-catenin signaling pathway, which is pivotal in inducing and maintaining cancer stem cell features in a variety of malignancies. Similarly, Dnmts were also found to be important for the regulation of stem cells in several studies. Trowbridge et al.35 reported that Dnmt1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Rinaldi et al.36 revealed that Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. All of these findings, including ours, indicated that there might be some underlying mechanisms between LGR6 and DNA methylation in TNBC that are not yet explicit and need further investigation. In addition, two common tumor-suppressor genes, BRCA1 and PTEN, were analyzed in TNBC. We found that the expression of BRCA1 and PTEN was negatively correlated with LGR6, confirming that LGR6 functions as an oncogene to promote tumorigenesis in TNBC.
In conclusion, our study first proved that LGR6 functions as an oncogene in TNBC. LGR6 promotes tumor progression, and its oncogenic role may associate with the Wnt/β-catenin pathway. All of these findings provided some new insights into the role of LGR6 in the development of TNBC, which may help to better understand the biological behavior of TNBC and find out a potential approach to treat TNBC.
Materials and Methods
Tissue Samples and Cell Lines
A total of 36 paired fresh samples (tumor and adjacent nontumor tissues) were obtained from patients with TNBC who received surgery at Sun Yat-sen University Cancer Center (Guangzhou, Guangdong, China). All resected tissues were instantaneously infiltrated in RNAlater (Ambion, Texas). This study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center, and all patients signed the informed consent before participation. All cell lines (normal: MCF-10A; non-TNBC: SKBR-3, MCF-7, T47D, MDA-MB-361, BT483, BT474; TNBC: 4T1, MDA-MB-468, MDA-MB-231, BT549) used in this study were obtained from the American Type Culture Collection and reauthenticated by short tandem repeat (STR) profile analysis every 6 months after resuscitation (Beijing Microread Genetics, China).
Tissue Microarray (TMA) and IHC
TMA analyses were performed on tumor samples from 240 patients with negative breast cancer at Sun Yat-sen University Cancer Center (Guangzhou, Guangdong, China). The procedure of the TMA and IHC staining has been described previously.37 According to the percentage of positive cells and staining intensity,38 LGR6 expression was classified into high and low expression groups. Scores 0–4 were used to evaluate the staining intensity: 0–1 (no staining), 1–2 (weak staining), 2–3 (medium staining), and 3–4 (strong staining). The percentage of positive cells was divided into four rankings by 25%, 50%, and 75%. The intensity and quantity determined the final score, from 0 to 4. High expression was defined as scores 3–4, and low expression was defined as scores 0–2. All samples were accessed by at least two blinded pathologists.
RNA Extraction and Quantitative Real-Time PCR
RNA was isolated using the TRIzol reagent (Invitrogen). Quantitative real-time PCR was performed with SYBR Premix Ex Taq (Takara), as previously described.39 β-actin was used as an internal control. The primers of LGR6 and β-actin were purchased from Invitrogen (Shanghai, China). The primer sequences were as follows: LGR6 forward: 5′-TGACGGCTTACCTGGACCTCA-3′, reverse: 5′-AGAGAATGCTTGTCCTGGGATG-3′; β-actin forward: 5′-GGACTTCGAGCAAGAGATGG-3′, reverse: 5′-ATCTGCTGGAAGGTGGACAG-3′.
Cell Proliferation and Invasion Assay
For cell proliferation assay, cells were plated on 96-well plates at the concentration of 1 × 103/well. Cell numbers were counted after 1, 2, and 3 days of incubation by a Coulter Counter (Beckman Coulter, USA), and the cell proliferation rate was calculated using a Cell Counting Kit 8 (CCK-8) (Dojindo, Japan). Cell invasion assay was evaluated by Transwell, according to the manufacturer’s instructions. Briefly, cells with serum-free medium were added to the upper chamber of the Transwell (8 μm pore size; BD Biosciences, USA), and 10% fetal bovine serum was added to the lower chamber. After incubation for 48 h, cells adhering to the lower membrane of the inserts were stained with crystal violet and counted. All experiments were conducted in triplicate independently.
Plasmid Construction and Transfection
The full-length LGR6 cDNA and shRNA against LGR6 (GeneCopoeia) were subcloned into the eukaryotic expression vector pcDNA3.1 (Invitrogen), as previously described.40 The most effective silenced shLGR6 was chosen from 3 candidates for the following experiments (Figure S1). Plasmid transfection was performed using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions. Cells transfected with empty vector were used as controls. Stable LGR6 expressing/silenced clones were selected by Geneticin (Roche Diagnostic). Puromycin was used to select stable cells.
Western Blotting
Protein was extracted using radioimmunoprecipitation assay (RIPA) lysis buffer with a proteinase inhibitor. Proteins in the lysates were separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidenedifluoride (PVDF) membrane (Millipore, USA). To block the nonspecific binding, membranes were incubated at room temperature for 1 h with 5% skim milk powder. Subsequently, the membranes were incubated for 12 h at 4°C with antibodies against LGR6, β-catenin, Dnmt1, Dnmt3a, Dnmt3b, BRCA1, and PTEN, purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The target proteins were detected by chemiluminescence. β-actin served as a protein-loading control.
Spheroid Culture and Formation
Cells at a density of 1 × 103 cells/mL were seeded into 6-well ultra-low cluster plates (Corning) and cultured in DMEM/F-12 medium with 1% B27 (Invitrogen, Carlsbad, CA, USA), EGF (20 ng/mL; Invitrogen), basic FGF (bFGF) (10 ng/mL; Invitrogen), and insulin (10 mg/mL; Sigma), as previously described.26 After 5 days, the number of the spheres (spherical, tight, nonadherent masses >50 mm in diameter) was counted, and spheroid formation efficiency (colonies/input cells 100%) was calculated under an inverse microscope.
Mouse Xenograft Model
LGR6 stable expressing/silenced cells were collected and suspended in PBS at a concentration of 1 × 106 cells/mL. For tumorigenesis experiments, the cells were inoculated subcutaneously into the dorsal flanks of each nude mouse with 100 μL of cancer cell suspension (5 in each group). Tumor size was measured every 4 days. After 28 days, the mice were sacrificed, and the tumors were dissected and weighed. For metastasis, 1 × 105 of cells were injected into the tail vein of the nude mouse (5 weeks old, 5 in each group). 8 weeks later, all mice were euthanized, and the pulmonary metastatic nodules were carefully examined and counted. All procedures on animals were performed strictly according to the institutional guidelines and approved by the Institute Research Ethics Committee of Sun Yat-Sen University Cancer Center.
Statistics
Comparisons between two groups were analyzed by Student’s t test, one-way ANOVA, and chi-square test. Data are shown as the mean ± standard deviation of three independent experiments unless specifically indicated. The survivals were analyzed Kaplan-Meier curves and log-rank tests. All differences were statistically significant at the p <0.05 level. Statistical analysis was processed using SPSS 16.0 software.
Author Contributions
Conception and Design, Collection and/or Assembly of Data, Data Analysis and Interpretation, Manuscript Writing, and Final Approval of the Manuscript, Y.K., X.O., X.L., Y.Z., and G.G.; Conception and Design, Data Analysis and Interpretation, Financial Support, and Final Approval of the Manuscript, N.L. and P.L.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
This work was supported by funds from the National Natural Science Foundation of China (81672598) and Science and Technology Planning Project of Guangzhou (201704020188).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.06.020.
Contributor Information
Ning Lyu, Email: lvning@sysucc.org.cn.
Peng Liu, Email: liupeng@sysucc.org.cn.
Supplemental Information
References
- 1.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Bauer K.R., Brown M., Cress R.D., Parise C.A., Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 3.Snippert H.J., Haegebarth A., Kasper M., Jaks V., van Es J.H., Barker N., van de Wetering M., van den Born M., Begthel H., Vries R.G. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 4.Huang P.Y., Kandyba E., Jabouille A., Sjolund J., Kumar A., Halliwill K., McCreery M., DelRosario R., Kang H.C., Wong C.E. Lgr6 is a stem cell marker in mouse skin squamous cell carcinoma. Nat. Genet. 2017;49:1624–1632. doi: 10.1038/ng.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehoczky J.A., Tabin C.J. Lgr6 marks nail stem cells and is required for digit tip regeneration. Proc. Natl. Acad. Sci. USA. 2015;112:13249–13254. doi: 10.1073/pnas.1518874112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guinot A., Oeztuerk-Winder F., Ventura J.J. miR-17-92/p38α Dysregulation Enhances Wnt Signaling and Selects Lgr6+ Cancer Stem-like Cells during Lung Adenocarcinoma Progression. Cancer Res. 2016;76:4012–4022. doi: 10.1158/0008-5472.CAN-15-3302. [DOI] [PubMed] [Google Scholar]
- 7.Ruan X., Liu A., Zhong M., Wei J., Zhang W., Rong Y., Liu W., Li M., Qing X., Chen G. Silencing LGR6 Attenuates Stemness and Chemoresistance via Inhibiting Wnt/β-Catenin Signaling in Ovarian Cancer. Mol. Ther. Oncolytics. 2019;14:94–106. doi: 10.1016/j.omto.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schindler A.J., Watanabe A., Howell S.B. LGR5 and LGR6 in stem cell biology and ovarian cancer. Oncotarget. 2017;9:1346–1355. doi: 10.18632/oncotarget.20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaas L., Pucci F., Messal H.A., Andersson A.B., Josue Ruiz E., Gerling M., Douagi I., Spencer-Dene B., Musch A., Mitter R. Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat. Cell Biol. 2016;18:1346–1356. doi: 10.1038/ncb3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim E., Wu D., Pal B., Bouras T., Asselin-Labat M.L., Vaillant F., Yagita H., Lindeman G.J., Smyth G.K., Visvader J.E. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren W., Lewandowski B.C., Watson J., Aihara E., Iwatsuki K., Bachmanov A.A., Margolskee R.F., Jiang P. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. USA. 2014;111:16401–16406. doi: 10.1073/pnas.1409064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 13.Gong X., Carmon K.S., Lin Q., Thomas A., Yi J., Liu Q. LGR6 is a high affinity receptor of R-spondins and potentially functions as a tumor suppressor. PLoS ONE. 2012;7:e37137. doi: 10.1371/journal.pone.0037137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelstein B., Kinzler K.W. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 15.Krejs G.J. Gastric cancer: epidemiology and risk factors. Dig. Dis. 2010;28:600–603. doi: 10.1159/000320277. [DOI] [PubMed] [Google Scholar]
- 16.Ke J., Ma P., Chen J., Qin J., Qian H. LGR6 promotes the progression of gastric cancer through PI3K/AKT/mTOR pathway. OncoTargets Ther. 2018;11:3025–3033. doi: 10.2147/OTT.S149303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Closas M., Couch F.J., Lindstrom S., Michailidou K., Schmidt M.K., Brook M.N., Orr N., Rhie S.K., Riboli E., Feigelson H.S. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 2013;45:392–398e3982. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purrington K.S., Slager S., Eccles D., Yannoukakos D., Fasching P.A., Miron P., Carpenter J., Chang-Claude J., Martin N.G., Montgomery G.W., GENICA Network Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis. 2014;35:1012–1019. doi: 10.1093/carcin/bgt404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmon K.S., Gong X., Lin Q., Thomas A., Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu S.Y., Kudo M., Chen T., Nakabayashi K., Bhalla A., van der Spek P.J., van Duin M., Hsueh A.J. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol. Endocrinol. 2000;14:1257–1271. doi: 10.1210/mend.14.8.0510. [DOI] [PubMed] [Google Scholar]
- 21.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 22.Barker N., Rookmaaker M.B., Kujala P., Ng A., Leushacke M., Snippert H., van de Wetering M., Tan S., Van Es J.H., Huch M. Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep. 2012;2:540–552. doi: 10.1016/j.celrep.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Jaks V., Barker N., Kasper M., van Es J.H., Snippert H.J., Clevers H., Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 24.Plaks V., Brenot A., Lawson D.A., Linnemann J.R., Van Kappel E.C., Wong K.C., de Sauvage F., Klein O.D., Werb Z. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3:70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J.-H., Tammela T., Hofree M., Choi J., Marjanovic N.D., Han S., Canner D., Wu K., Paschini M., Bhang D.H. Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell. 2017;170:1149–1163.e12. doi: 10.1016/j.cell.2017.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L., Tang H., Kong Y., Xie X., Chen J., Song C., Liu X., Ye F., Li N., Wang N., Xie X. LGR5 Promotes Breast Cancer Progression and Maintains Stem-Like Cells Through Activation of Wnt/β-Catenin Signaling. Stem Cells. 2015;33:2913–2924. doi: 10.1002/stem.2083. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y.-Z., Ma D., Suo C., Shi J., Xue M., Hu X., Xiao Y., Yu K.-D., Liu Y.-R., Yu Y. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell. 2019;35:428–440.e5. doi: 10.1016/j.ccell.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 30.Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., Bock C., Vasanthakumar A., Gu H., Xi Y. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Challen G.A., Sun D., Mayle A., Jeong M., Luo M., Rodriguez B., Mallaney C., Celik H., Yang L., Xia Z. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin B., Robertson K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poomipark N., Flatley J.E., Hill M.H., Mangnall B., Azar E., Grabowski P., Powers H.J. Methyl Donor Status Influences DNMT Expression and Global DNA Methylation in Cervical Cancer Cells. Asian Pac. J. Cancer Prev. 2016;17:3213–3222. [PubMed] [Google Scholar]
- 34.El-Awady R.A., Hersi F., Al-Tunaiji H., Saleh E.M., Abdel-Wahab A.H., Al Homssi A., Suhail M., El-Serafi A., Al-Tel T. Epigenetics and miRNA as predictive markers and targets for lung cancer chemotherapy. Cancer Biol. Ther. 2015;16:1056–1070. doi: 10.1080/15384047.2015.1046023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trowbridge J.J., Snow J.W., Kim J., Orkin S.H. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinaldi L., Datta D., Serrat J., Morey L., Solanas G., Avgustinova A., Blanco E., Pons J.I., Matallanas D., Von Kriegsheim A. Dnmt3a and Dnmt3b Associate with Enhancers to Regulate Human Epidermal Stem Cell Homeostasis. Cell Stem Cell. 2016;19:491–501. doi: 10.1016/j.stem.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 37.Way T.D., Huang J.T., Chou C.H., Huang C.H., Yang M.H., Ho C.T. Emodin represses TWIST1-induced epithelial-mesenchymal transitions in head and neck squamous cell carcinoma cells by inhibiting the β-catenin and Akt pathways. Eur. J. Cancer. 2014;50:366–378. doi: 10.1016/j.ejca.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 38.Tang H., Liu P., Yang L., Xie X., Ye F., Wu M., Liu X., Chen B., Zhang L., Xie X. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol. Cancer Ther. 2014;13:3185–3197. doi: 10.1158/1535-7163.MCT-14-0243. [DOI] [PubMed] [Google Scholar]
- 39.Tang H., Huang X., Wang J., Yang L., Kong Y., Gao G., Zhang L., Chen Z.S., Xie X. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol. Cancer. 2019;18:23. doi: 10.1186/s12943-019-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong Y., Yang L., Wei W., Lyu N., Zou Y., Gao G., Ou X., Xie X., Tang H. CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics. 2019;11:1163–1176. doi: 10.2217/epi-2019-0093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.