Abstract
Globally, chickpea production is severely affected by salinity stress. Understanding the genetic basis for salinity tolerance is important to develop salinity tolerant chickpeas. A recombinant inbred line (RIL) population developed using parental lines ICCV 10 (salt-tolerant) and DCP 92-3 (salt-sensitive) was screened under field conditions to collect information on agronomy, yield components, and stress tolerance indices. Genotyping data generated using Axiom®CicerSNP array was used to construct a linkage map comprising 1856 SNP markers spanning a distance of 1106.3 cM across eight chickpea chromosomes. Extensive analysis of the phenotyping and genotyping data identified 28 quantitative trait loci (QTLs) explaining up to 28.40% of the phenotypic variance in the population. We identified QTL clusters on CaLG03 and CaLG06, each harboring major QTLs for yield and yield component traits under salinity stress. The main-effect QTLs identified in these two clusters were associated with key genes such as calcium-dependent protein kinases, histidine kinases, cation proton antiporter, and WRKY and MYB transcription factors, which are known to impart salinity stress tolerance in crop plants. Molecular markers/genes associated with these major QTLs, after validation, will be useful to undertake marker-assisted breeding for developing better varieties with salinity tolerance.
Keywords: chickpea, salinity, quantitative trait loci, stress susceptibility index (SSI), stress tolerance index (STI), candidate genes
1. Introduction
Chickpea (Cicer arietinum L.) is one of the most important dietary grain legumes, and has a small genome size of ~740 Mb [1]. This crop is highly valued for its intrinsic potential for symbiotic nitrogen fixation. It is the primary source of human dietary proteins, vitamins, and essential minerals, and is valuable for food security in the developing world [2]. Due to its high nutrient content, chickpea is ideal for feeding the global population to combat issues related to nutritional food security. The crop is grown on over 17.8 million hectares (ha), producing 17.2 million metric tons (t). India is the world’s leading producer with 11.4 million metric tons, or about 66% of total world’s production [3]. Average chickpea productivity is less than 1 t/ha, far below its global yield potential of 3–4 t/ha in optimum growing conditions. Chickpea productivity is severely affected by several biotic (including Fusarium wilt, Ascochyta blight, Botrytis grey mould, dry root rot, and pod borer) and abiotic (including salinity, drought, and heat) stresses, causing production losses of up to 70% [4,5,6].
Among different abiotic stresses, salinity stress is the second major abiotic stress after drought that limits chickpea productivity and reduces total global production by approximately 8–10% [7,8]. Notably, almost 80 million ha of the world’s arable land is prone to salinity stress [7]. Globally, 20% (45 million ha) of irrigated land and 2% (32 million ha) of dryland are constrained by salinity [9]. In the past few decades, salinity has become one of the major threats to crop productivity, as it influences plant growth at different developmental stages. Salt stress impacts plants by affecting germination, growth, reproduction, and the ability to biologically fix nitrogen [10]. Salinity affects vital physiological functions, hormonal regulation and nutritional balance [11,12], reduce carbon fixation [13], causes flower abortion, reduces flower numbers and pod setting, and eventually limits crop yield [14].
Chickpea is intrinsically salt-sensitive, unlike cereals [7], and salinity causes osmotic imbalance to tissues through specific ion toxicity, nutritional imbalance, and disturbed hormonal interactions, which affects overall growth and development [15,16,17,18]. Increased salt concentration alters grain composition and yield [19,20]. Salinity also increases leaf necrosis and chlorosis, which leads to leaf senescence and reduced photosynthesis in grain legumes [14,21]. While chickpea is sensitive to salt stress, there is a range of variation available for salinity tolerance in germplasm collection [22,23,24].
The development of genetically tolerant cultivars is a preferred strategy for managing salinity stress as plants preserve the quality of the final product. In this direction, mapping of genomic regions/quantitative trait loci (QTLs) responsible for salinity tolerance is a prerequisite for using molecular breeding for developing new cultivars. Several attempts have been made to understand the molecular basis of salt tolerance in many plant species. For instance, QTLs for traits associated with salinity tolerance have been mapped in legumes such as soybean [25], Medicago truncatula [26], and cereals such as barley [27] and bread wheat [28]. Advances in next-generation sequencing (NGS) technology are paving the way towards development of high-density SNP-based platforms for genotyping [29]. The development of high-throughput genotyping platforms such as the Axiom®CicerSNP array has facilitated genetics research in chickpea [30]. Although several mapping studies have identified QTLs for biotic tolerance [31] and drought tolerance [32,33] in chickpea, few studies have identified QTLs for salinity tolerance [34,35]. Furthermore, a very few QTLs have been identified for yield components governing salinity tolerance [36].
Stress tolerance indices, which indicate the comparative performance of genotypes under normal and stress conditions, are the key salinity stress tolerance component traits. Although these traits have been used to map salinity and heat stress tolerance in rice [37,38,39,40], no such report is available in chickpea. Evaluating the crop performance under both control and stress scenarios will serve as a better strategy to identify QTLs and develop crop varieties for enhanced stress tolerance [41]. Such relative performance has practical relevance since genotypes with low yield potential under control conditions often show higher tolerance to stress than high-yielding genotypes. Therefore, it is important to identify QTLs governing salinity stress tolerance for yield and yield components that can be used effectively in marker-assisted breeding in chickpea.
With an objective to dissect the genetic basis of salt tolerance in chickpea, the present study has been undertaken with following objectives: (a) develop a dense genetic map for ICCV 10 (salt stress-tolerant) × DCP 92-3 (salt stress-sensitive) population, (b) identify QTLs for salinity stress tolerance in chickpea, and (c) identify genes underlying major QTLs associated with salinity tolerance component traits in chickpea.
2. Results
2.1. Phenotypic Variation for Salinity Tolerance Component Traits
RILs, along with parents, were analyzed for their phenotypic performance in control and salt-stressed environments. Salt stress significantly reduced yield in the salinity-sensitive parent (DCP 92-3), being seven-fold more than the salinity-tolerant parent (ICCV 10) across seasons (Table 1). During the 2015–16 and 2016–17 crop seasons, a normal frequency distribution was observed for all traits analyzed, except for stress tolerance index (STI) and stress susceptibility index (SSI) for yield per plant (Supplementary Figures S1 and S2).
Table 1.
Phenotypic variation for the parameters evaluated in the control and salinity treatments in the ICCV 10 × DCP 92-3 RIL population.
| S. No. | RILs | ICCV 10 | DCP 92-3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Traits | Treatment | Minimum | Maximum | Range | Mean | SD * | CV ** | |||
| Karnal 2015–16 | ||||||||||
| 1 | PH (cm) | Control | 29 | 41 | 12 | 37.1 | 6.2 | 0.2 | 46.3 | 51.0 |
| 2 | Saline | 27.5 | 29.5 | 2 | 27.3 | 4.5 | 0.2 | 31.0 | 22.0 | |
| 3 | NB | Control | 8 | 9 | 1 | 9.2 | 2.7 | 0.3 | 8.6 | 6.5 |
| 4 | Saline | 4 | 5.5 | 1.5 | 4.1 | 1.3 | 0.3 | 5.0 | 3.0 | |
| 5 | PPP | Control | 61 | 87 | 26 | 65.1 | 21.9 | 0.3 | 84.0 | 32.0 |
| 6 | Saline | 12 | 35 | 23 | 22.7 | 12.3 | 0.5 | 39.0 | 10.0 | |
| 7 | 100SW (g) | Control | 12 | 12.8 | 0.8 | 13.7 | 1.7 | 0.1 | 11.4 | 12.9 |
| 8 | Saline | 6.3 | 10 | 3.7 | 8.7 | 2.2 | 0.3 | 12.2 | 10.2 | |
| 9 | YPP (g) | Control | 2.0 | 27.2 | 25.2 | 11.1 | 4.8 | 0.4 | 17.2 | 18.8 |
| 10 | Saline | 0.29 | 11.8 | 11.5 | 2.8 | 2.2 | 0.8 | 7.2 | 1.0 | |
| 11 | SSI_YP | NA | 1 | 1 | 0 | 1.0 | 0.1 | 0.1 | 1.0 | 1.0 |
| 12 | STI_YP | NA | 0 | 0.1 | 0.1 | 0.1 | 0.1 | 1.1 | 0.2 | 0.0 |
| 13 | SSI_100SW | NA | 0.3 | 1 | 0.7 | 0.7 | 0.3 | 0.4 | −0.1 | 0.4 |
| 14 | STI_100SW | NA | 0.4 | 0.6 | 0.2 | 0.6 | 0.2 | 0.3 | 0.7 | 0.7 |
| Karnal 2016–17 | ||||||||||
| 1 | PH (cm) | Control | 48.3 | 54.7 | 6.4 | 46.5 | 8.7 | 0.2 | 58.67 | 53.33 |
| 2 | Saline | 27.5 | 39.3 | 11.8 | 31.6 | 10.7 | 0.3 | 29.00 | 22.00 | |
| 3 | NB | Control | 8.1 | 9 | 0.9 | 9.8 | 3.3 | 0.3 | 7.33 | 7.00 |
| 4 | Saline | 2.3 | 4.3 | 2 | 2.6 | 7.3 | 2.9 | 2.33 | 2.00 | |
| 5 | PPP | Control | 14 | 164 | 150 | 50.9 | 21.8 | 0.4 | 56.00 | 45.00 |
| 6 | Saline | 19.7 | 30 | 10.3 | 20.9 | 11.1 | 0.5 | 18.33 | 27.33 | |
| 7 | 100SW (g) | Control | 11.6 | 13.7 | 2.1 | 13.7 | 1.7 | 0.1 | 11.35 | 12.95 |
| 8 | Saline | 4.4 | 8.8 | 4.4 | 5.9 | 7.7 | 1.3 | 13.10 | 7.15 | |
| 9 | YPP (g) | Control | 2.6 | 26.6 | 24 | 11.6 | 4.9 | 0.4 | 10.20 | 11.47 |
| 10 | Saline | 0.18 | 7.8 | 7.6 | 1.3 | 0.9 | 9.1 | 1.09 | 1.19 | |
| 11 | SSI_YP | NA | 0.998 | 1.011 | 0.013 | 1.0 | 0.0 | 0.0 | 1.65 | 1.57 |
| 12 | STI_YP | NA | 0.013 | 0.028 | 0.015 | 0.0 | 0.0 | 0.7 | 0.20 | 0.18 |
| 13 | SSI_100SW | NA | 0.7 | 1.2 | 0.5 | 1.0 | 0.3 | 0.3 | 0.21 | 0.61 |
| 14 | STI_100SW | NA | 0.3 | 0.7 | 0.4 | 0.5 | 0.3 | 0.2 | 1.05 | 1.05 |
* Standard deviation. ** Coefficient of variation (PH: plant height; NB: branch number per plant; PPP: pods number per plant; 100SW: 100 seed weight; YPP: yield per plant; SSI_YP: SSI for yield per plant; STI_YP: STI for yield per plant; SSI_100SW: SSI for 100 seed weight; STI_100SW: STI for 100 seed weight; NA: not applicable).
The salinity tolerance component traits were subjected to Pearson’s correlation analysis to identify relationships between them. For simplicity, we classified significant correlation values into three different degrees, viz. high, moderate, and low. For instance, a high degree of correlation represented correlation coefficient values between ±0.50 and ±1.0 and was considered to be a strong correlation. A moderate degree of correlation represented coefficient values between ±0.30 and ±0.49, while coefficient values below ±0.29 were classified as low degree of correlation. In this context, we evaluated the correlations between traits that were measured during the 2015–16 season (Table 2, Supplementary Figure S3a). A strong positive correlation was observed between SSI for yield per plant and yield per plant evaluated under control conditions, and between STI for yield per plant and yield per plant under stress conditions. In contrast, SSI for yield per plant and yield per plant measured under salinity stress showed a strong negative correlation. Furthermore, STI for yield per plant displayed a medium positive correlation with pods per plant, 100-seed weight and plant height, all evaluated under salinity stress conditions. STI for 100-seed weight showed a small positive correlation with STI for yield per plant and yield per plant assessed under stress conditions.
Table 2.
Pearson correlation analysis for salinity tolerance component traits evaluated during the 2015–16 and 2016–17 seasons.
| PH_C | NB_C | PPP_C | 100SW_C | YPP_C | PH_S | NB_S | PPP_S | 100SW_S | YPP_S | SSI_YP | STI_YP | SSI_100SW | STI_100SW | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2015–16 | ||||||||||||||
| PH_C | 1 | |||||||||||||
| NB_C | 0.11 ns | 1 | ||||||||||||
| PPP_C | 0.07 ns | −0.05 ns | 1 | |||||||||||
| 100SW_C | −0.12 ns | −0.04 ns | −0.10 ns | 1 | ||||||||||
| YPP_C | 0.27 *** | −0.23 ** | −0.03 ns | −0.03 ns | 1 | |||||||||
| PH_S | −0.01 ns | 0.00 ns | −0.05 ns | −0.10 ns | −0.07 ns | 1 | ||||||||
| NB_S | 0.01 ns | −0.07 ns | 0.03 ns | −0.09 ns | −0.07 ns | 0.24 ** | 1 | |||||||
| PPP_S | 0.00 ns | −0.17 * | 0.13 ns | −0.07 ns | 0.08 ns | 0.31 *** | 0.37 *** | 1 | ||||||
| 100SW_S | 0.18 * | −0.20 * | −0.04 ns | 0.22 ** | 0.06 ns | −0.02 ns | 0.03 ns | 0.17 * | 1 | |||||
| YPP_S | 0.01 ns | −0.09 ns | 0.04 ns | −0.02 ns | −0.14 ns | 0.44 *** | 0.15 ns | 0.43 *** | 0.33 *** | 1 | ||||
| SSI_YP | 0.20 ** | −0.13 ns | 0.00 ns | −0.07 ns | 0.52 *** | −0.20 * | −0.02 ns | −0.14 ns | −0.19 * | −0.55 *** | 1 | |||
| STI_YP | 0.16 * | −0.24 ** | 0.06 ns | −0.02 ns | 0.42 *** | 0.32 *** | 0.11 ns | 0.43 *** | 0.33 *** | 0.68 *** | 0.02 ns | 1 | ||
| SSI_100SW | −0.22 ** | 0.19 * | 0.00 ns | 0.25 ** | −0.08 ns | −0.03 ns | −0.08 ns | −0.19 * | −0.88 *** | −0.34 *** | 0.15 * | −0.32 *** | 1 | |
| STI_100SW | 0.12 ns | −0.17 * | −0.08 ns | 0.57 *** | 0.07 ns | −0.06 ns | −0.02 ns | 0.10 ns | 0.91 *** | 0.24 ** | −0.15 * | 0.27 *** | −0.62 *** | 1 |
| 2016–17 | ||||||||||||||
| PH_C | 1 | |||||||||||||
| NB_C | −0.10 ns | 1 | ||||||||||||
| PPP_C | 0.67 *** | −0.19 ** | 1 | |||||||||||
| 100SW_C | −0.10 ns | −0.03 ns | −0.06 ns | 1 | ||||||||||
| YPP_C | 0.67 *** | −0.19 ** | 0.67 *** | −0.06 ns | 1 | |||||||||
| PH_S | 0.07 ns | 0.10 ns | 0.01 ns | −0.03 ns | 0.09 ns | 1 | ||||||||
| NB_S | 0.05 ns | −0.18 * | 0.07 ns | 0.19 ** | 0.18 * | 0.33 *** | 1 | |||||||
| PPP_S | 0.12 ns | −0.13 ns | 0.14 * | 0.00 ns | 0.16 * | 0.47 *** | 0.48 *** | 1 | ||||||
| 100SW_S | −0.06 ns | −0.15 * | 0.05 ns | 0.06 ns | 0.01 ns | 0.11 ns | 0.08 ns | 0.07 ns | 1 | |||||
| YPP_S | −0.15 * | −0.13 ns | 0.01 ns | 0.00 ns | −0.06 ns | 0.09 ns | 0.00 ns | 0.10 ns | 0.28 *** | 1 | ||||
| SSI_YP | 0.45 *** | 0.00 ns | 0.31 *** | −0.13 ns | 0.52 *** | −0.06 ns | 0.09 ns | 0.02 ns | −0.17 * | −0.78 *** | 1 | |||
| STI_YP | 0.25 *** | −0.22 ** | 0.38 *** | −0.02 ns | 0.50 *** | 0.13 ns | 0.11 ns | 0.17 * | 0.29 *** | 0.78 *** | −0.29 *** | 1 | ||
| SSI_100SW | 0.01 ns | 0.13 ns | −0.07 ns | 0.31 *** | −0.05 ns | −0.11 ns | −0.03 ns | −0.07 ns | −0.92 *** | −0.26 *** | 0.10 ns | −0.29 *** | 1 | |
| STI_100SW | −0.09 ns | −0.15 * | 0.02 ns | 0.45 *** | −0.02 ns | 0.09 ns | 0.13 ns | 0.07 ns | 0.91 *** | 0.25 *** | −0.22 ** | 0.25 *** | −0.69 *** | 1 |
*** p < 0.001; ** p < 0.01; * p < 0.05; ns, non-significant. (PH: plant height; NB: branch number per plant; PPP: pods number per plant; 100SW: 100 seed weight; YPP: yield per plant; SSI_YP: SSI for yield per plant; STI_YP: STI for yield per plant; SSI_100SW: SSI for 100 seed weight; STI_100SW: STI for 100 seed weight; S: under saline condition; C: under control condition).
We also assessed the correlation between traits evaluated during the 2016–17 crop season (Table 2, Supplementary Figure S3b). Here, STI for yield per plant displayed a strong positive correlation with yield per plant measured under salinity stress. Although SSI for yield per plant was strongly and positively related to yield per plant measured under control conditions, it showed a strong negative correlation with yield per plant measured under stress conditions.
Furthermore, STI for yield per plant showed a positive and moderate correlation with pods per plant and yield per plant, both measured under control scenarios. The SSI for yield per plant was positively and moderately correlated with pods per plant and plant height under control conditions. Furthermore, a small and positive correlation was observed between STI for yield per plant and plant height measured under control, while SSI for yield per plant showed a small and negative correlation with 100-seed weight measured under stress conditions.
2.2. High-Density Linkage Map
Of the 50,591 SNP probes on the Axiom®CicerSNP array, 5123 SNPs were polymorphic between both parents and displayed segregation within the population. In the constructed linkage map, 1856 markers were assigned to eight linkage groups. The integrated map had a total length of 1106.3 cM with an average distance of 0.59 cM between adjacent markers. The number of loci onto eight linkage groups ranged from 56 (CaLG04) to 487 (CaLG07) and the length ranged from 45.6 cM (CaLG08) to 270.7 cM (CaLG06) with a mean value of 138.2 cM (Table 3). The marker density of the linkage groups varied significantly, with the highest marker density on CaLG01 with 327 markers distributed over a distance of 147.7 cM, while CaLG04 had the lowest density with 56 markers distributed over a distance of 87 cM (Table 3).
Table 3.
Distribution of markers on the eight linkage groups (LGs) of the chickpea genetic map for the ICCV 10 × DCP 92-3 RIL population.
| S. No. | LGs | Genetic Distance (cM) | Number of Markers Mapped | Inter Marker Distance (cM) |
|---|---|---|---|---|
| 1 | CaLG1 | 147.69 | 327 | 0.5 |
| 2 | CaLG2 | 120.4 | 192 | 0.6 |
| 3 | CaLG3 | 98.9 | 158 | 0.6 |
| 4 | CaLG4 | 87 | 56 | 1.6 |
| 5 | CaLG5 | 74 | 86 | 0.9 |
| 6 | CaLG6 | 270.75 | 476 | 0.6 |
| 7 | CaLG7 | 262 | 487 | 0.5 |
| 8 | CaLG8 | 45.6 | 74 | 0.6 |
| Total | 1106.34 | 1856 | 0.6 |
2.3. QTLs for Salinity Tolerance Component Traits
The phenotypic and genotypic data were analyzed for identification of QTLs to understand the genetic basis of salinity tolerance in the RIL mapping population derived from ICCV 10 × DCP 92-3. The QTL analysis was performed for seven yield and yield-related traits, namely, pod number per plant (PPP), 100 seed weight (100SW), yield per plant (YPP), SSI for yield per plant (SSI_YP), SSI for 100 seed weight (SSI_100SW), STI for yield per plant (STI_YP), and STI for 100 seed weight (STI_100SW), and two agronomic traits, i.e., plant height (PH) and branch number per plant (NB). The QTLs that contributed >10% of the phenotypic variation explained (PVE) were considered as major QTLs. Furthermore, if QTL for a given trait in a particular treatment appeared in both years, it was considered as a consistent QTL [32].
2.3.1. QTLs for Yield and Yield-Related Traits
A total of nine major and 12 minor QTLs were detected for seven yield and yield-related traits in the two seasons (2015–16 and 2016–17), as follow:
Stress susceptible index (SSI) and stress tolerance index (STI): SSI and STI are important measures for estimating the effect of salinity tolerance on yield; therefore, QTLs for these traits can be potential targets to improve the salinity response in chickpea. In the case of SSI, four major and two minor QTLs were identified. Among the major QTLs, one major consistent QTL for SSI_YP was identified on CaLG06 in 2015–16 (PVE 12.2%) and 2016–17 (PVE 28.3%), with flanking markers AX-123640392 and AX-123640389. Two other major QTLs for SSI_YP (qSSIYP3.1; PVE 10.0%) and SSI_100SW (qSSI100SW3.1; PVE 10.1%) on CaLG03 were identified (Table 4). Two minor QTLs for SSI_YP (qSSIYP6.2; PVE 8.3%) and SSI_100SW (qSSI100SW2.1; PVE 8.9%) on CaLG06 and CaLG02, respectively, were also identified (Table 4).
Table 4.
Summary of the major and minor QTLs for various salinity tolerance component traits.
| Trait Name | QTL Name | Year | Treatment | LG | Position (cM) | Marker Interval | LOD Value |
PVE (%) | Additive Effect | Allele-Contributing Parent |
|---|---|---|---|---|---|---|---|---|---|---|
| Yield and Yield-Related Traits | ||||||||||
| SSI_YP | qSSIYP6.1 | 2016–17 | – | CaLG06 | 226.1 | AX-123640392–AX-123640389 | 5.7 | 28.4 | 0.1 | DCP92-3 |
| qSSIYP6.1 | 2015–16 | – | CaLG06 | 226.1 | AX-123640392–AX-123640389 | 4.8 | 12.2 | 0.1 | DCP92-3 | |
| qSSIYP3.1 | 2016–17 | – | CaLG03 | 50.79 | AX-123659975–AX-123622699 | 5.3 | 10.0 | 0 | DCP92-3 | |
| qSSIYP6.2 | 2016–17 | – | CaLG06 | 260.98 | AX-123635094–AX-123635091 | 3.8 | 8.3 | 0 | DCP92-3 | |
| SSI_100SW | qSSI100SW3.1 | 2016–17 | – | CaLG03 | 50.79 | AX-123659975–AX-123622699 | 4.6 | 10.1 | 0.1 | DCP92-3 |
| qSSI100SW2.1 | 2015–16 | – | CaLG02 | 62.91 | AX-123659415–AX-123620733 | 3.7 | 8.9 | 0.1 | DCP92-3 | |
| STI_YP | qSTIYP5.1 | 2016–17 | – | CaLG05 | 20.31 | AX-123653399–AX-123653409 | 4.8 | 8.6 | 0 | DCP92-3 |
| qSTIYP6.1 | 2016–17 | – | CaLG06 | 268.65 | AX-123640437–AX-123655585 | 4.3 | 8.1 | 0 | DCP92-3 | |
| STI_100SW | qSTI100SW3.1 | 2016–17 | – | CaLG03 | 50.79 | AX-123659975–AX-123622699 | 8.7 | 17.1 | −0.1 | ICCV10 |
| 100SW | q100SWS3.1 | 2016–17 | Saline | CaLG03 | 50.79 | AX-123659975–AX-123622699 | 6.9 | 13.9 | −0.8 | ICCV10 |
| q100SWS3.1 | 2015–16 | CaLG03 | 56.21 | AX-123641496–AX-123622502 | 3.1 | 8.8 | −0.7 | ICCV10 | ||
| q100SWS2.1 | 2015–16 | CaLG02 | 39.16 | AX-123620430–AX-123659350 | 3.4 | 7.7 | −0.6 | ICCV10 | ||
| q100SWC5.1 | 2015–16 | Control | CaLG05 | 58.7 | AX-123631523–AX-123631537 | 3.2 | 10.1 | −0.9 | ICCV10 | |
| q100SWC7.1 | 2015–16 | CaLG07 | 92.67 | AX-123636120–AX-123636108 | 3 | 7.8 | 0.8 | DCP92-3 | ||
| YPP | qYPPS6.1 | 2016–17 | Saline | CaLG06 | 270.65 | AX-123635072–AX-123640437 | 3.5 | 7.1 | 0.2 | DCP92-3 |
| qYPPC6.1 | 2015–16 | Control | CaLG06 | 236.66 | AX-123655575–AX-123663343 | 6.5 | 13.8 | −0.8 | ICCV10 | |
| qYPPC5.1 | 2015–16 | CaLG05 | 20.31 | AX-123653409–AX-123653399 | 4.7 | 10.2 | 7.8 | DCP92-3 | ||
| qYPPC5.1 | 2016–17 | CaLG05 | 20.31 | AX-123653399–AX-123653409 | 4.9 | 8.7 | 7.4 | DCP92-3 | ||
| qYPPC7.1 | 2015–16 | CaLG02 | 120.16 | AX-123620346–AX-123620222 | 3.2 | 6.5 | 6.7 | DCP92-3 | ||
| qYPPC4.1 | 2016–17 | CaLG04 | 87.63 | AX-123630936–AX-123652553 | 3.7 | 6.5 | 6.5 | DCP92-3 | ||
| PPP | qPPP8.1 | 2015–16 | Saline | CaLG08 | 36.06 | AX-123638292–AX-123664233 | 3.3 | 8.1 | 3.6 | DCP92-3 |
| Agronomic traits | ||||||||||
| PH | qPHC5.2 | 2016–17 | Saline | CaLG05 | 60.33 | AX-123653281–AX-123631517 | 4.1 | 10.0 | 3.6 | DCP92-3 |
| qPHC5.1 | 2016–17 | Control | CaLG05 | 16.55 | AX-123662454–AX-123631761 | 6.1 | 11.8 | 3.9 | DCP92-3 | |
| qPHC7.1 | 2015–16 | CaLG07 | 230.14 | AX-123635844–AX-123655878 | 3.8 | 9.2 | 8.9 | DCP92-3 | ||
| qPHC6.1 | 2015–16 | CaLG06 | 44.23 | AX-123654072–AX-123633446 | 3.4 | 8.1 | −4.9 | ICCV10 | ||
| NB | qNBC8.1 | 2015–16 | Control | CaLG08 | 44.91 | AX-123638389–AX-123638445 | 5.5 | 12.7 | −1.7 | ICCV10 |
| qNBC8.2 | 2015–16 | CaLG08 | 10.22 | AX-123657789–AX-123638459 | 4 | 8.9 | 1.4 | DCP92-3 | ||
| qNBC8.3 | 2016–17 | CaLG08 | 44.91 | AX-123638389–AX-123638445 | 3.5 | 6.1 | −1.4 | ICCV10 | ||
(SSI_YP: SSI for yield per plant; SSI_100SW: SSI for 100 seed weight; STI_YP: STI for yield per plant; STI_100SW: STI for 100 seed weight; 100SW: 100 seed weight; YPP: yield per plant; PPP: pods number per plant; PH: plant height; NB: number of branches per plant).
Similarly, in the case of STI, one major QTL for STI_100SW (qSTI100SW3.1; PVE 17.1%) on CaLG03 with flanking markers AX-123659975 and AX-123622699 and two minor QTLs for STI_YP (qSTIYP5.1; PVE 8.6% and qSTIYP6.1; PVE 8.1%) were identified in the 2016–17 season (Table 4).
100 seed weight (100SW): A total of five QTLs for 100SW (three for saline and two for control treatment) were identified across seasons. One major QTL (q100SWS3.1; PVE 13.9%) on CaLG03 with flanking markers AX-123659975 and AX-123622699 in 2016–17, and two minor QTLs (q100SWS3.1; PVE 8.8% and q100SWS2.1; PVE 7.7%) in 2015–16 for 100SW under saline treatment were identified. Similarly, under control treatment, one major QTL (q100SWC5.1; PVE 10.1%) and one minor QTL (q100SWC7.1; PVE 7.8%) were identified (Table 4).
Yield per plant (YPP): In the case of YPP, six QTLs, including two major and four minor, were identified under saline and control treatment. Under saline treatment, we could get only one minor QTL (qYPPS6.1; PVE 7.1%) on CaLG06 in 2016–17 season (Table 4). However, in 2015–16 under control treatment, two major QTLs namely, qYPPC6.1 (PVE 13.8%) and qYPPC5.1 (PVE 10.2%) on CaLG06 and CaLG05, respectively, were identified, of which qYPPC5.1 was found to be consistent as it appears in 2015–16 as well as in 2016–17 (Table 4). Similarly, three minor QTLs (qYPPC5.1; PVE 8.7%, qYPPC7.1; PVE 6.5% and qYPPC4.1; PVE 6.5%) were detected on CaLG05, CaLG02, and CaLG04, respectively (Table 4).
Pod number per plant (PPP): In the case of PPP, although no major QTL was identified, one minor QTL (qPPP8.1; PVE 8.1%) was identified on CaLG08 in the salinity treatment in 2015–16, and one minor QTL with 9.1% PVE was identified on CaLG07 in the control (Table 4).
2.3.2. QTLs for Agronomic Traits
A total of seven QTLs including three major and four minor QTLs were detected for two agronomic traits in two seasons (2015–16 and 2016–17) as follows:
Plant height (PH): In the case of PH, two major QTLs in the 2016–17 season, one each in saline and control treatment, were identified. Under saline treatment, one major QTL (qPHC5.2; PVE 10.0%) on CaLG05 flanked by AX-123653281 and AX-123631517 SNP markers was identified. Similarly, under control treatment, one major QTL (qPHC5.1; PVE 11.8%) in 2016–17 on CaLG05 flanked by AX-123662454 and AX-123631761 markers and two minor QTLs (qPHC7.1; PVE 9.2% and qPHC6.1; PVE 8.1%) in 2015–16 were identified (Table 4).
Number of branches per plant (NB): One major QTL (qNBC8.1; PVE 12.7) was identified for NB on CaLG08 flanked by AX-123638389 and AX-123638445 markers in 2015–16 under control treatment. This QTL was considered as a consistent QTL as it also appeared in 2016–17 season (Table 4). Two minor QTLs (qNBC8.2; PVE 8.9% and qNBC8.3; PVE 6.1%) were also identified for NB under control condition on CaLG08 in 2015–16 and 2016–17, respectively. No QTL was identified for NB under the salinity treatment across the seasons (Table 4).
2.4. Candidate Genes for Salinity Tolerances
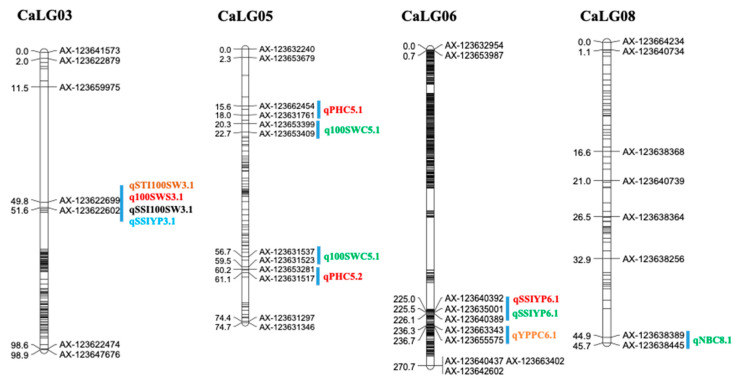
The genes located in the genomic regions for the identified QTLs were extracted. A total of 1121 genes were present in all QTL regions, based on the reference chickpea genome [1]. Of the 1121 genes, 136 putative candidate genes (88 on CaLG03, 25 on CaLG06, 14 on CaLG02, six on CaLG07, and one each on CaLG04 and CaLG08) belong to several gene families, including kinases, genes encoding transcription factors, and ion channels encoding proteins in response to salinity stress, among others. These genomic regions on CaLG03 and CaLG06 are of great interest as they hold QTLs for traits related to yield under salinity (Supplementary Table S1, Figure 1, and Supplementary Figure S4). The major QTL intervals AX-123622602 and AX-123622699 on CaLG03 and AX-123640392 and AX-123655575 on CaLG06 harbored 113 key genes that are reportedly involved in salinity and other abiotic stress tolerances in chickpea and other crops (Supplementary Table S2).
Figure 1.
Major quantitative trait loci (QTLs) for various salinity tolerance component traits identified in the DCP 92-3 × ICCV 10 RIL population.
A detailed analysis of QTLs from CaLG03 showed that QTLs for four traits (STI_100SW, SSI_100SW, 100SW, SSI_YP) were flanked by AX-123622602 and AX-123622699 markers. Here, the ~3.3 Mb region between these two markers was selected to identify candidate genes. The ~3.3 Mb region contained 763 predicted genes. Functional annotation of the candidate genes revealed their roles in various biotic and abiotic stress responses. For instance, a MYB transcription factor, which is mainly involved in the regulation of plant growth and development under abiotic stress, such as salinity tolerance, was identified in this region [42,43,44]. Similarly, a C3HC4-type RING zinc finger protein, which reportedly improve and enhances salt tolerance in various crops, was identified [45,46]. An abscisic acid insensitive (ABI) 5, a typical subfamily of proteins belonging to the basic domain/leucine zipper (bZIP) transcription factors, was recently reported to contribute to salt stress tolerance in Arabidopsis thaliana via abscisic acid signaling [47]. Notably, the ABI5 gene was also found in the predicted QTL region. The stress-responsive NAC transcription factor family of proteins has been studied extensively for its role in salinity stress tolerance in transgenic plants [48,49]. We identified the presence of the NAC-domain-containing protein in the QTL region. In addition to the above genes, some trait-specific candidate genes, such as E3-ubiquitin-protein ligase, WRKY transcription factor, and zinc finger proteins, were also identified. The gene encoding E3 ubiquitin ligase activity has been shown to regulate grain width and weight in rice [50]. Furthermore, while the differential expression of the WRKY transcription factor gene alters seed size in soybean [51], a zinc finger CCHC-type protein is involved in regulating seed size in Medicago truncatula [52]. However, further fine mapping and validation of these genes would be needed to pinpoint the candidate genes regulating traits of interest.
Analysis of the QTLs from the CaLG06 genomic region revealed that QTLs for three traits, namely, STI_YP, SSI_YP, and YPP, were flanked by AX-123640392 and AX-123655575 markers spanning a ~0.1 Mb region. The predicted ~0.1 Mb region encompassed 155 genes. Analysis of the genes underlying the QTL interval identified several potential candidates with a predicted role in salinity stress tolerance. For example, overexpression of cation/proton antiporter genes encoding cellular Na+/H+ exchanger proteins can upregulate plant performance under salinity stress [53]. A cation/H+ antiporter 4-like gene was identified in this region. Further, a plant calmodulin-binding family protein, predicted to be involved in salinity stress responses in plants during early germination, was also detected [54]. Membrane proteins containing a transmembrane domain have been demonstrated to play important roles in conferring salinity tolerance in Chenopodium quinoa [55]. In this region, the presence of transmembrane domain proteins is anticipated to be involved in conferring salinity tolerance in the genotypes. Notably, histidine kinases act as sensory molecules and are involved in the transduction of environmental signals in plants, fungi, and prokaryotes. A previous study identified the involvement of histidine kinases in perception or transduction of salt stress signals in Synechocystis sp. [56]. The presence of a histidine kinase gene underlying the QTL region is thought to be involved in the perception of salt stress signals and the regulation of salt-inducible genes. We anticipate that further fine mapping and cloning of the candidate genes underlying the major QTL regions will unravel the salinity tolerance mechanism in chickpea.
3. Discussion
Recent technological advances have led to the evolution of breeding methodologies that have the potential to accelerate the breeding process [57]. Plant response to abiotic stresses, such as salinity, is a complex trait that is governed by many genes; therefore, breeding approaches for such complex stress tolerance and crop stability have been challenging. The use of marker-assisted selection/marker-assisted breeding in this regard has helped to simplify things to a certain extent. Marker-assisted breeding has successfully produced crops with tolerance to biotic stresses and has increased yield in many crops [57,58,59].
Salinity stress appears to be a polygenic and quantitative trait that is regulated by several genes under diverse environments [60]. To understand the genetic basis of salinity stress, significant efforts have been made to identify QTLs associated with salinity tolerance, with several QTLs mapped on different chromosomes of chickpea by multiple research groups in the last decade. In a study by Vadez et al. [36], QTLs for seed weight, pod number, and harvest index in the salinity treatment were obtained on CaLG07 using a RIL population derived from a cross between salt-tolerant JG62 and salt-sensitive ICCV 2. Similarly, a minor QTL for yield that explained 8% PVE on CaLG07 was identified in chickpea [34]. In another study, two key genomic regions on CaL05 and CaL07, harboring QTLs for yield and other salinity-associated traits, were reported in a RIL population derived from the cross ICCV 2 × JG 11, using SSR and SNP markers [35].
Recent advances in genome-based approaches have endorsed the development of high-throughput approaches for genotyping, enabling the identification of and access to desirable alleles, with different QTLs having the potential to affect desired responses. In the current study, using a reasonably large RIL population, high-density genetic map, and phenotyping under controlled conditions, we identified 28 QTLs for nine traits across seasons and treatments in chickpea. Importantly, two genomic regions on CaLG03 and CaLG06, harboring 10 QTLs for salinity stress indices and yield-related traits, were detected.
Identification of QTLs for relative performance of genotypes under stress and controlled conditions has significant over identification of QTLs based on phenotypic performance in the stress environment alone [41]. QTLs regulating salinity stress tolerance by utilizing stress tolerance indices, which differentiate the performance of crops both under control and stress conditions, have been reported in several crops, but not in chickpea. In rice, genomic regions governing heat stress tolerance [40] and salinity stress tolerance have been successfully mapped using stress indices [37,38,39,61]. In the present study, major QTLs for salinity stress indices SSI_YP, SSI_100SW, STI_100SW, and yield component (100SW) under salinity have been identified. These QTLs, after validation, may play a key role in marker-assisted breeding programs to produce saline-tolerant lines in chickpea. Salt-tolerant genotypes will have lower SSI values, indicating the smaller difference in yield between control and stress treatment, with the reverse being true for susceptible genotypes [37,62].
Identifying the genes involved in the salinity stress response is the first step towards gaining the necessary knowledge for genetics, physiology, and breeding. The application of genomics-based knowledge with breeding platforms is expected to provide useful insights into the molecular responses of plants to salinity. Plants alleviate the salinity stress effects of increased Na+ in cells by excluding and sequestering Na+. Increased cytosolic Na+ concentration is detected and, in turn, the response pathways to stress ion channels, calcium-dependent kinases, histidine kinases, and membrane receptors, etc., activate to regulate gene expression, resulting in the synthesis of compatible solutes to abate the destructive effects of increased Na+ ions in the cytoplasm [63]. Plant hormones also play a vital role in regulating plant responses to salinity. Plants induce ABA synthesis to mediate the plant responses through the ABA-dependent signal transduction pathway. To maintain cellular ionic and osmotic homeostasis, ABA induces osmolyte synthesis and stomatal closure in the guard cells, and other related mechanisms leading to the maintenance of cellular ionic and osmotic homeostasis [64,65,66].
The putative candidate genes found in the QTL regions in this study have been experimentally validated for their role in the salinity stress response in several earlier studies in different plant species. For example, genes identified on CaLG03 and CaLG06 that encode several kinases, calcium-dependent protein kinases (CDPKs), mitogen-activated protein kinases (MAPKs), histidine kinases (HKs), sucrose non-fermenting related kinases (SnRK1), transcription factors such as WRKY, basic leucine zipper (bZIP), MYB/MYC, and cation calcium exchanger, were reported to be playing a vital role in the salinity stress response [67,68,69,70]. Similarly, candidate genes coding for proteins related to cation calcium exchanger, cation antiporter 4 (regulates plasma membrane antiporter activity), and potassium channel AKT1 (involved in regulating K+/Na+ ratio) led to salinity stress tolerance in Arabidopsis [71,72]. The transmembrane protein-encoding genes found in the QTL region on CaLG03 are candidate genes for seed weight in chickpea [73].
Among the 113 putative candidate genes found in the QTL regions on CaLG03 and CaLG06, most were involved in osmoregulation, which help plants to cope with salinity and other biotic stresses (Table S3). The identification of probable candidate genes for salinity tolerance on small genomic regions on CaLG03 and CaLG06 make these regions promising for future use in genomics-assisted breeding (GAB) to improve salt and other abiotic stress tolerance in chickpea.
4. Materials and Methods
4.1. Development of RIL Population
Two diverse chickpea parental lines DCP 92-3 and ICCV 10 that differ in salinity tolerance were used to develop a chickpea mapping population comprising of 201 RILs (F8) at the ICAR—Indian Institute of Pulses Research, Kanpur. ICCV 10 is a desi-type genotype and is highly tolerant to salinity stress, while DCP 92-3 is susceptible to salinity. The population was advanced using the single seed descent method.
4.2. Phenotyping for Salinity Stress
The RIL population was screened for two consecutive years from November to April (2015–16 and 2016–17) at Karnal, Haryana, India located between 29°43′ N longitude and 76°58′ E latitude. Karnal has an average elevation of 240 m from mean average sea level (m.a.s.l) and received total rainfall of 85.8 mm during January, 7.8 mm in March, and 3.4 mm in April, during the crop season 2015–16. The average temperature during cropping season ranged between 11.92 °C and 26.85 °C with minimum temperature of 6.8 °C in December 2015. The initial humidity during seedling stage was 91.87% (max) and 46.06% (min). During 2016–17 season, the average temperature was between 11.82 °C and 27.36 °C, with a minimum temperature of 6.78 °C in the month of January 2017. The total rainfall received during January was 85.8 mm, 7.8 mm in March, and 3.4 mm in April, 2017. The average humidity of 86% (max) and 11% (min) was observed. Overall, the two cropping seasons had cool environmental conditions suitable for crop growth and expression. The experiment was done under control conditions; hence, it did not affect the level of salinity stress in micro-plot. The mapping population was screened using the micro-plot screening method described below.
A total of 201 RILs, along with the parents and Karnal Chana-1 as a positive check, were sown in micro-plots (2 × 2 m2) top covered with 2 mm polycarbonate sheet to protect it from unseasonal rains. The experiment was organized in an augmented design to evaluate seed yield and other yield contributing traits in control and saline (ECiw 6 dS/m) conditions at ICAR-Central Soil Salinity Research Institute (CSSRI), Karnal. Initial soil salinity ranged from ECe 0.92–0.97 dS/m. Saline irrigation of ECiw 6 dS/m was applied at 30, 60, and 90 day intervals after sowing. Soil samples were taken from time to time to measure the salinity build-up of ECiw 6 dS/m in the soil.
The genotype response to salt stress was expressed as the stress susceptibility index (SSI; [61]) and stress tolerance index (STI; [37]) using the formulae:
SSI = (1 − Ys/Yp)/SI;SI = 1 − Ȳs/Ȳp [61] and stress tolerance index
STI = Yp × Ys/Ȳp2
where Ys and Yp are the yield of genotypes evaluated under saline (stress) and non-saline (nonstress) condition, and Ȳs and Ȳp are the mean yield of all genotypes evaluated under stress and non-stress conditions.
The SSI is a measure of reduction in yield caused by a stress, relative to a favorable environment. Salt-tolerant genotypes would have a lower SSI value, indicating a smaller yield difference with the control than the susceptible genotypes with a larger yield difference. The STI identifies genotypes that produce higher yields in the control and under stress conditions (higher STI value), which is more desirable than only under stress conditions.
4.3. High Density Genotyping
Genomic DNA was collected from pooled young leaves of RILs and both parents using the CTAB method [74]. DNA quality was tested in 1% agarose gel and quantified using NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific Inc. Waltham, MA, USA). DNA concentration was adjusted to a minimum of 50 ng/μL. RILs were genotyped using an Axiom®CicerSNP array with 50,590 probes distributed on all eight linkage groups as described in Roorkiwal et al. [30].
4.4. Genetic Map Construction and QTL Analysis
A total of 5123 polymorphic SNP markers were used to construct a genetic linkage map using Join Map v. 4.1 (https://www.kyazma.nl/index.php/JoinMap) [75] as described in [31]. To find QTLs responsible for salinity tolerance, we used multi-seasonal field phenotyping data for yield and agronomical traits associated with salinity tolerance of parental accessions and 201 RILs from the mapping population derived from DCP 92-3 × ICCV 10. SNP genotyping data, obtained from Axiom®CicerSNP array-based SNP genotyping, was correlated with the aforementioned phenotyping data using Windows QTL Cartographer ver. 2.5 [76] (composite interval mapping at significant LOD (logarithm of odds) threshold >3.0 at a p < 0.05) as per earlier studies [77,78].
4.5. Mining of Candidate Genes
To identify candidate genes, present within the QTL region, flanking markers were subjected to BLAST against chickpea reference genome assembly, and candidate genes in the QTL regions were retrieved.
4.6. Statistical Analysis
Pearson’s correlation analysis was conducted using the “Hmisc” package in R. Frequency distributions of the component traits within the RIL population were analyzed and plotted with “UsingR” package within the R environment.
5. Conclusions
The present study, using a DCP 92-3 × ICCV 10 RIL mapping population and 50K SNP genotyping array, identified two genomic regions that harbor QTLs for salinity tolerance in a ~3.3 Mb region on CaLG03 and ~0.1 Mb region on CaLG06 with major QTLs for yield and salinity tolerance. The genes present in the identified QTL regions in this study reportedly play a key role in salinity tolerance. Further sequencing and functional validation are required to identify the candidate genes in these QTL regions responsible for salt tolerance. Nonetheless, the high-density linkage map, major QTLs, and genomic regions identified in this study can be used in GAB, after suitable markers are developed and validated, to breed high-yielding salinity-tolerant chickpea varieties.
Acknowledgments
M.R. and R.K.V. thank the Department of Science and Technology, Government of India for providing funding support through the INSPIRE Faculty Scheme and Early Career Research Award—SERB and the JC Bose Fellowship, respectively. R.B. acknowledges funding support from the Council of Scientific and Industrial Research (CSIR), India for the award of research fellowship. The work reported in this article was undertaken as part of the CGIAR Research Program on Grain Legumes and Dryland Cereals. ICRISAT is a member of the CGIAR Consortium.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/14/5058/s1. Table S1: Summary of major and minor QTLs identified on CaLG03 and CaLG06. Table S2: List of genes present in identified QTL regions. Table S3: List of putative candidate genes associated with salinity stress response on CaLG03 and CaLG06. Figure S1: Frequency distribution of salinity tolerance component traits evaluated in 2015–16. PH: plant height; NB: number of branches per plant; PPP: pod number per plant; 100SW: 100 seed weight; YPP: yield per plant; SSI-YP: SSI for yield per plant; STI-YP: STI for yield per plant; SSI-100SW: SSI for 100 seed weight; STI-100SW: STI for 100 seed weight. Figure S2: Frequency distribution of salinity tolerance component traits evaluated in 2016–17. PH: plant height; NB: branch number per plant; PPP: pod number per plant; 100SW: 100 seed weight; YPP: yield per plant; SSI-YP: SSI for yield per plant; STI-YP: STI for yield per plant; SSI-100SW: SSI for 100 seed weight; STI-100SW: STI for 100 seed weight. Figure S3: Correlation matrices for salinity tolerance component traits evaluated across two seasons. Correlation matrix for (a) 2015–16 season, and (b) 2016–17 season. PH: plant height; NB: branch number per plant; PPP: pod number per plant; 100SW: 100 seed weight; YPP: yield per plant; SSI-YP: SSI for yield per plant; STI-YP: STI for yield per plant; SSI-100SW: SSI for 100 seed weight; STI-100SW: STI for 100 seed weight. Figure S4: Major QTLs for salinity tolerance component traits identified in DCP 92-3 × ICCV 10 RIL population. (a) QTLs on CaLG06 in 2015–16, (b) QTLs on CaLG03 in 2015–16, (c) QTLs on CaLG06 in 2016–17, and (d) QTLs on CaLG03 in 2016–17.
Author Contributions
M.R. and R.K.V. conceived the idea and provided critical inputs to the concept. K.R.S. planed the experiment. K.R.S. and P.M. generated the genotyping data. N.K., M.S.S., C.B., P.C.S., S.K.S., S.S., J.S., M.P., S.P.P.R. and A.M. generated phenotyping data. P.M., M.R., R.B., A.B., S.R.S., S.P. and K.R.S. conducted phenotyping, genotyping data & QTL mapping analysis including the statistical analysis. P.M., M.R., R.B., K.R.S., N.P.S., K.H.M.S. and R.K.V. contributed to analyse, interpret data and wrote the manuscript. All authors contributed to the final reading and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
“This research was funded by Indian Council of Agricultural Research (ICAR) through Incentivizing Research in Agriculture; Department of Agriculture and Cooperation & Farmers Welfare, Ministry of Agriculture and Farmers Welfare, Government of India; Department of Biotechnology (DBT), Government of India through Indo-Australian Biotechnology Fund (IABF) and Bill and Melinda Gates Foundation (BMGF)” and “The APC was funded by Bill and Melinda Gates Foundation”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Varshney R.K., Song C., Saxena R.K., Azam S., Yu S., Sharpe A.G., Cannon S., Baek J., Rosen B.D., Tar’an B., et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013;31:240–246. doi: 10.1038/nbt.2491. [DOI] [PubMed] [Google Scholar]
- 2.Jukanti A.K., Gaur P.M., Gowda C.L., Chibbar R.N. Nutritional quality and health benefits of chickpea (Cicer arietinum L.): A review. Br. J. Nutr. 2012;108:S11–S26. doi: 10.1017/S0007114512000797. [DOI] [PubMed] [Google Scholar]
- 3.FAOSTAT 2018. [(accessed on 30 May 2020)]; Available online: https://www.fao.org/faostat/en/#data.
- 4.Turner N.C., Abbo S., Berger J.D., Chaturvedi S.K., French R.J., Ludwig C., Mannur D.M., Singh S.J., Yadava H.S. Osmotic adjustment in chickpea (Cicer arietinum L.) results in no yield benefit under terminal drought. J. Exp. Bot. 2007;58:187–194. doi: 10.1093/jxb/erl192. [DOI] [PubMed] [Google Scholar]
- 5.Samineni S., Siddique K.H.M., Gaur P.M., Colmer T.D. Salt sensitivity of the vegetative and reproductive stages in chickpea (Cicer arietinum L.): Podding is a particularly sensitive stage. Environ. Exp. Bot. 2011;71:260–268. doi: 10.1016/j.envexpbot.2010.12.014. [DOI] [Google Scholar]
- 6.Muehlbauer F.J., Sarker A. Economic importance of chickpea: Production, value, and world trade. In: Varshney R.K., Thudi M., Muehlbauer F., editors. The Chickpea Genome. Springer; Cham, Germany: 2017. pp. 5–12. [DOI] [Google Scholar]
- 7.Flowers T.J., Gaur P.M., Gowda C.L., Krishnamurthy L., Samineni S., Siddique K.H., Turner N.C., Vadez V., Varshney R.K., Colmer T.D. Salt sensitivity in chickpea. Plant Cell Environ. 2010;33:490–509. doi: 10.1111/j.1365-3040.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 8.Atieno J., Li Y., Langridge P., Dowling K., Brien C., Berger B., Varshney R.K., Sutton T. Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci. Rep. 2017;1:1–11. doi: 10.1038/s41598-017-01211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 10.van Hoorn J.W., Katerji N., Hamdy A., Mastrorilli M. Effect of salinity on yield and nitrogen uptake of four grain legumes and on biological nitrogen contribution from the soil. Agric. Water Manag. 2001;51:87–98. doi: 10.1016/S0378-3774(01)00114-7. [DOI] [Google Scholar]
- 11.Ashraf M., Akram N.A., Arteca R.N., Foolad M.R. The physiological, biochemical and molecular roles of brassinosteroids and salicylic acid in plant processes and salt tolerance. Crit. Rev. Plant Sci. 2010;29:162–190. doi: 10.1080/07352689.2010.483580. [DOI] [Google Scholar]
- 12.Liang W., Ma X., Wan P., Liu L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018;495:286–291. doi: 10.1016/j.bbrc.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 13.Garg N., Singla P. Stimulation of nitrogen fixation and trehalose biosynthesis by naringenin (Nar) and arbuscular mycorrhiza (AM) in chickpea under salinity stress. Plant. Growth Regul. 2016;80:5–22. doi: 10.1007/s10725-016-0146-2. [DOI] [Google Scholar]
- 14.Khan H.A., Siddique K.H.M., Colmer T.D. Vegetative and reproductive growth of salt-stressed chickpea are carbon-limited: Sucrose infusion at the reproductive stage improves salt tolerance. J. Exp. Bot. 2017;68:2001–2011. doi: 10.1093/jxb/erw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flexas J., Bota J., Loreto F., Cornic G., Sharkey T.D. Diffusive and metabolic limitations to photosynthesis under drought and salinity in C3 plants. Plant Biol. 2004;6:269–279. doi: 10.1055/s-2004-820867. [DOI] [PubMed] [Google Scholar]
- 16.Murillo-Amador B., Yamada S.T., Yamaguchi E., Rueda-Puente E., Avila-Serrano N., Garcia-Hernandez J.L., Lopez-Aguilar R., Troyo-Dieguez E., Nieto-Garibay A. Influence of calcium silicate on growth, physiological parameters and mineral nutrition in two legume species under salt stress. J. Agron. Crop Sci. 2007;193:413–421. doi: 10.1111/j.1439-037X.2007.00273.x. [DOI] [Google Scholar]
- 17.He Y., Fu J., Yu C., Wang X., Jiang Q., Hong J., Lu K., Xue G., Yan C., James A., et al. Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. J. Exp. Bot. 2015;66:6877–6889. doi: 10.1093/jxb/erv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patil G., Do T., Vuong T.D., Valliyodan B., Lee J.D., Chaudhary J., Shannon J.G., Nguyen H.T. Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 2016;6:19199. doi: 10.1038/srep19199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lõpez-Aguilar R., Orduňo-Cryz A., Lucero-Arce A., Murillo-Amador B., Troyo-Diéguez E. Response to salinity of three grain legumes for potential cultivation in arid areas. Soil Sci. Plant Nutr. 2003;49:329–336. doi: 10.1080/00380768.2003.10410017. [DOI] [Google Scholar]
- 20.Manchanda G., Garg N. Salinity and its effects on the functional biology of legumes. Acta Physiol. Plant. 2008;30:595–618. doi: 10.1007/s11738-008-0173-3. [DOI] [Google Scholar]
- 21.Sehrawat N., Bhat K.V., Sairam R.K., Jaiwal P.K. Identification of salt resistant wild relatives of mungbean (Vigna radiata L. Wilczek) Asian J. Plant Sci. Res. 2013;3:41–49. [Google Scholar]
- 22.Vadez V., Krishnamurthy L., Serraj R., Gaur P.M., Upadhyaya H.D., Hoisington D.A., Varshney R.K., Turner N.C., Siddique K.H.M. Large variation in salinity tolerance in chickpea is explained by differences in sensitivity at the reproductive stage. Field Crops Res. 2007;104:123–129. doi: 10.1016/j.fcr.2007.05.014. [DOI] [Google Scholar]
- 23.Krishnamurthy L., Turner N.C., Gaur P.M., Upadhyaya H.D., Varshney R.K., Siddique K.H.M., Vadez V. Consistent variation across years in salinity resistance in a diverse range of chickpea (Cicer arietinum L.) genotypes. J. Agron. Crop Sci. 2011;197:214–227. doi: 10.1111/j.1439-037X.2010.00456.x. [DOI] [Google Scholar]
- 24.Turner N.C., Colmer T.D., Quealy J., Pushpavalli R., Krishnamurthy L., Kaur J., Singh G., Siddique K.H.M., Vadez V. Salinity tolerance and ion accumulation in chickpea (Cicer arietinum L.) subjected to salt stress. Plant Soil. 2013;365:347–361. doi: 10.1007/s11104-012-1387-0. [DOI] [Google Scholar]
- 25.Hamwieh A., Tuyen D., Cong H., Benite E., Takahashi R., Xu D. Identification and validation of a major QTL for salt tolerance in soybean. Euphytica. 2011;179:451–459. doi: 10.1007/s10681-011-0347-8. [DOI] [Google Scholar]
- 26.Arraouadi S., Badri M., Abdelly C., Huguet T., Aouani M.E. QTL mapping of physiological traits associated with salt tolerance in Medicago truncatula recombinant inbred lines. Genomics. 2012;99:118–125. doi: 10.1016/j.ygeno.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen V., Ribot S., Dolstra O., Niks R., Visser R.F., Linden C.G. Identification of quantitative trait loci for ion homeostasis and salt tolerance in barley (Hordeum vulgare L.) Mol. Breed. 2013;31:137–152. doi: 10.1007/s11032-012-9777-9. [DOI] [Google Scholar]
- 28.Genc Y., Oldach K., Gogel B., Wallwork H., McDonald G.K., Smith A.B. Quantitative trait loci for agronomic and physiological traits for a bread wheat population grown in environments with a range of salinity levels. Mol. Breed. 2013;32:39–59. doi: 10.1007/s11032-013-9851-y. [DOI] [Google Scholar]
- 29.Varshney R.K., Nayak S.N., May G.D., Jackson S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009;27:522–530. doi: 10.1016/j.tibtech.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Roorkiwal M., Jain A., Kale S.M., Doddamani D., Chitikineni A., Thudi M., Varshney R.K. Development and evaluation of high-density Axiom®CicerSNP Array for high-resolution genetic mapping and breeding applications in chickpea. Plant Biotechnol. J. 2018;16:890–901. doi: 10.1111/pbi.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabbavarapu M.M., Sharma M., Chamarthi S.K., Swapna N., Rathore A., Thudi M., Gaur P.M., Pande S., Singh S., Kaur L., et al. Molecular mapping of QTLs for resistance to Fusarium wilt (race 1) and Ascochyta blight in chickpea (Cicer arietinum L.) Euphytica. 2013;193:121–133. doi: 10.1007/s10681-013-0959-2. [DOI] [Google Scholar]
- 32.Varshney R.K., Thudi M., Nayak S.N., Gaur P.M., Kashiwagi J., Krishnamurthy L., Jaganathan D., Koppolu J., Bohra A., Tripathi S., et al. Genetic dissection of drought tolerance in chickpea (Cicerarietinum, L.) Theor. Appl. Genet. 2014;127:445–462. doi: 10.1007/s00122-013-2230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kale S.M., Jaganathan D., Ruperao P., Chen C., Punna R., Kudapa H., Thudi M., Roorkiwal M., Katta M.A., Doddamani D., et al. Prioritization of candidate genes in “QTL-hotspot” region for drought tolerance in chickpea (Cicer arietinum L.) Sci. Rep. 2015;19:15296. doi: 10.1038/srep15296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samineni S. Master’s Thesis. The University of Western Australia; Perth, Australia: 2010. Physiology, Genetics and Molecular Mapping of Salt Tolerance in Chickpea (Cicer arietinum L.) [Google Scholar]
- 35.Pushpavalli R., Krishnamurthy L., Thudi M., Gaur P.M., Rao M.V., Siddique K.H., Colmer T.D., Turner N.C., Varshney R.K., Vadez V. Two key genomic regions harbour QTLs for salinity tolerance in ICCV 2 × JG 11 derived chickpea (Cicer arietinum L.) recombinant inbred lines. BMC Plant Biol. 2015;15:124. doi: 10.1186/s12870-015-0491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vadez V., Krishnamurthy L., Thudi M., Anuradha C., Colmer T.D., Turner N.C., Siddique K.H.M., Gaur P.M., Varshney R.K. Assessment of ICCV 2 × JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTL for seed yield and yield components. Mol. Breed. 2012;30:9–21. doi: 10.1007/s11032-011-9594-6. [DOI] [Google Scholar]
- 37.Fernandez G.C.J. Adaptation of Food Crops to Temperature and Water Stress. Volume 410. AVRDC; Taipei, Taiwan: 1992. Effective Selection Criteria for Assessing Plant Stress Tolerance; pp. 257–270. [Google Scholar]
- 38.Pandit A., Rai V., Bal S., Sinha S., Kumar V., Chauhan M., Gautam R.K., Singh R., Sharma P.C., Singh A.K., et al. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryza sativa L.) Mol. Genet. Genom. 2010;284:121–136. doi: 10.1007/s00438-010-0551-6. [DOI] [PubMed] [Google Scholar]
- 39.Tiwari S., Sl K., Kumar V., Singh B., Rao A.R., Mithra Sv A., Rai V., Singh A.K., Singh N.K. Mapping QTLs for salt tolerance in rice (Oryza sativa L.) by bulked segregant analysis of recombinant inbred lines using 50K SNP chip. PLoS ONE. 2016;11:e0153610. doi: 10.1371/journal.pone.0153610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shanmugavadivel P.S., Sv A.M., Prakash C., Ramkumar M.K., Tiwari R., Mohapatra T., Singh N.K. High resolution mapping of QTLs for heat tolerance in rice using a 5K SNP array. Rice. 2017;10:28. doi: 10.1186/s12284-017-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raman A., Verulkar S., Mandal N., Variar M., Shukla V., Dwivedi J., Singh B., Singh O., Swain P., Mall A., et al. Drought yield index to select high yielding rice lines under different drought stress severities. Rice. 2012;5:31. doi: 10.1186/1939-8433-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dossa K., Mmadi M.A., Zhou R., Liu A., Yang Y., Diouf D., You J., Zhang X. Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis. AoB Plants. 2020;12:plz081. doi: 10.1093/aobpla/plz081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y., Bao X., Zhi Y., Wu Q., Guo Y., Yin X., Zeng L., Li J., Zhang J., He W., et al. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 2019;10:168. doi: 10.3389/fpls.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai X., Xu Y., Ma Q., Xu W., Wang T., Xue Y., Chong K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007;143:1739–1751. doi: 10.1104/pp.106.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agarwal P., Khurana P. TaZnF, a C3HC4 type RING zinc finger protein from Triticum aestivum is involved in dehydration and salinity stress. J. Plant Biochem. Biotechnol. 2020 doi: 10.1007/s13562-019-00546-8. [DOI] [Google Scholar]
- 46.Wu W., Cheng Z., Liu M., Yang X., Qiu D. C3HC4-type RING finger protein NbZFP1 is involved in growth and fruit development in Nicotiana benthamiana. PLoS ONE. 2014;9:e99352. doi: 10.1371/journal.pone.0099352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang H.C., Tsai M.C., Wu S.S., Chang I.F. Regulation of ABI5 expression by ABF3 during salt stress responses in Arabidopsis thaliana. Bot. Stud. 2019;60:16. doi: 10.1186/s40529-019-0264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He Z., Li Z., Lu H., Huo L., Wang Z., Wang Y., Ji X. The NAC protein from Tamarix hispida, ThNAC7, confers salt and osmotic stress tolerance by increasing reactive oxygen species scavenging capability. Plants. 2019;8:221. doi: 10.3390/plants8070221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.An J.P., Yao J.F., Xu R.R., You C.X., Wang X.F., Hao Y.J. An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol. Plant. 2018;164:279–289. doi: 10.1111/ppl.12724. [DOI] [PubMed] [Google Scholar]
- 50.Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 51.Gu Y., Li W., Jiang H., Wang Y., Gao H., Liu M., Chen Q., Lai Y., He C. Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size. J. Exp. Bot. 2017;68:2717–2729. doi: 10.1093/jxb/erx147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radkova M., Revalska M., Kertikova D., Iantcheva A. Zinc finger CCHC-type protein related with seed size in model legume species Medicago truncatula. Biotechnol. Biotechnol. Equip. 2019;33:278–285. doi: 10.1080/13102818.2019.1568914. [DOI] [Google Scholar]
- 53.Ma Y.C., Augé R.M., Dong C., Cheng Z.M. Increased salt tolerance with overexpression of cation/proton antiporter 1 genes: A meta-analysis. Plant Biotechnol. J. 2017;15:162–173. doi: 10.1111/pbi.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shkolnik D., Finkler A., Pasmanik-Chor M., Fromm H. Calmodulin-binding transcription activator 6: A key regulator of Na+ homeostasis during germination. Plant Physiol. 2019;180:1101–1118. doi: 10.1104/pp.19.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmöckel S.M., Lightfoot D.J., Razali R., Tester M., Jarvis D.E. Identification of putative transmembrane proteins involved in salinity tolerance in Chenopodium quinoa by integrating physiological data, RNAseq, and SNP analyses. Front. Plant Sci. 2017;8:1023. doi: 10.3389/fpls.2017.01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marin K., Suzuki I., Yamaguchi K., Ribbeck K., Yamamoto H., Kanesaki Y., Hagemann M., Murata N. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA. 2003;100:9061–9066. doi: 10.1073/pnas.1532302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varshney R.K., Singh V.K., Hickey J.M., Xun X., Marshall D.F., Wang J., Edwards D., Ribaut J.M. Analytical and decision support tools for genomics-assisted breeding. Trends Plant Sci. 2016;21:354–363. doi: 10.1016/j.tplants.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 58.Pandey M.K., Roorkiwal M., Singh V.K., Ramalingam A., Kudapa H., Thudi M., Chitikineni A., Rathore A., Varshney R.K. Emerging Genomic Tools for Legume Breeding: Current Status and Future Prospects. Front. Plant Sci. 2016;7:455. doi: 10.3389/fpls.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roorkiwal M., Bharadwaj C., Barmukh R., Dixit G.P., Thudi M., Gaur P.M., Chaturvedi S.K., Fikre A., Hamwieh A., Kumar S., et al. Integrating genomics for chickpea improvement: Achievements and opportunities. Theor. Appl. Genet. 2020;133:1703–1720. doi: 10.1007/s00122-020-03584-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foolad M.R. Recent advances in genetics of salt tolerance in tomato. Plant Cell Tissue Org. 2004;76:101–119. doi: 10.1023/B:TICU.0000007308.47608.88. [DOI] [Google Scholar]
- 61.Kumar V., Singh A., Mithra S.V., Krishnamurthy S.L., Parida S.K., Jain S., Tiwari K.K., Kumar P., Rao A.R., Sharma S.K., et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa) DNA Res. 2015;22:133–145. doi: 10.1093/dnares/dsu046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer R.A., Maurer R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust. J. Agric. Res. 1978;29:892–912. doi: 10.1071/AR9780897. [DOI] [Google Scholar]
- 63.Mahajan S., Pandey G.K., Tuteja N. Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 2008;471:146–158. doi: 10.1016/j.abb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 64.O’Brien J.A., Benková E. Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 2013;4:451. doi: 10.3389/fpls.2013.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015;20:219–229. doi: 10.1016/j.tplants.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 66.Lan Z., Krosse S., Achard P., van Dam N.M., Bede J. DELLA proteins modulate Arabidopsis defences induced in response to caterpillar herbivory. J. Exp. Bot. 2014;65:571–583. doi: 10.1093/jxb/ert420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urao T., Yakubov B., Satoh R., Yamaguchi-Shinozaki K., Seki M., Hirayama T., Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tran L.S., Urao T., Qin F., Maruyama K., Kakimoto T., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schulz P., Herde M., Romeis T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013;163:523–530. doi: 10.1104/pp.113.222539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 71.Jaspers P., Blomster T., Brosché M., Salojärvi J., Ahlfors R., Vainonen J.P., Reddy R.A., Immink R., Angenent G., Turck F., et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009;60:268–279. doi: 10.1111/j.1365-313X.2009.03951.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhou H., Zhao J., Yang Y., Chen C., Liu Y., Jin X., Chen L., Li X., Deng X.W., Schumaker K.S., et al. Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell. 2012;24:5106. doi: 10.1105/tpc.112.106393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh V.K., Khan A.W., Jaganathan D., Thudi M., Roorkiwal M., Takagi H., Garg V., Kumar V., Chitikineni A., Gaur P.M., et al. QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol. J. 2016;14:2110–2119. doi: 10.1111/pbi.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jain A., Roorkiwal M., Kale S., Garg V., Yadala R., Varshney R.K. InDel markers: An extended marker resource for molecular breeding in chickpea. PLoS ONE. 2019;14:3. doi: 10.1371/journal.pone.0213999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Ooijen J.W. JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma BV; Wageningen, The Netherlands: 2006. [Google Scholar]
- 76.Wang S., Basten C.J., Zeng Z.B. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University; Raleigh, NC, USA: 2012. [Google Scholar]
- 77.Bajaj D., Upadhyaya H.D., Khan Y., Das S., Badoni S., Shree T., Kumar V., Tripathi S., Gowda C.L., Singh S., et al. A combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea. Sci. Rep. 2015;5:9264. doi: 10.1038/srep09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saxena M.S., Bajaj D., Das S., Kujur A., Kumar V., Singh M., Bansal K.C., Tyagi A.K., Parida S.K. An integrated genomic approach for rapid delineation of candidate genes regulating agro-morphological traits in chickpea. DNA Res. 2014;21:695. doi: 10.1093/dnares/dsu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.