“Mutator” tumor cells that cannot correct DNA replication errors exhibit an extremely high mutation rate that accelerates their evolution. But this gamble puts them at risk for extinction....
Keywords: DNA replication, spontaneous polyploidization, antimutator, mutation spectra
Abstract
Mutations affecting DNA polymerase exonuclease domains or mismatch repair (MMR) generate “mutator” phenotypes capable of driving tumorigenesis. Cancers with both defects exhibit an explosive increase in mutation burden that appears to reach a threshold, consistent with selection acting against further mutation accumulation. In Saccharomyces cerevisiae haploid yeast, simultaneous defects in polymerase proofreading and MMR select for “antimutator” mutants that suppress the mutator phenotype. We report here that spontaneous polyploids also escape this “error-induced extinction” and routinely outcompete antimutators in evolved haploid cultures. We performed similar experiments to explore how diploid yeast adapt to the mutator phenotype. We first evolved cells with homozygous mutations affecting polymerase δ proofreading and MMR, which we anticipated would favor tetraploid emergence. While tetraploids arose with a low frequency, in most cultures, a single antimutator clone rose to prominence carrying biallelic mutations affecting the polymerase mutator alleles. Variation in mutation rate between subclones from the same culture suggests that there exists continued selection pressure for additional antimutator alleles. We then evolved diploid yeast modeling MMR-deficient cancers with the most common heterozygous exonuclease domain mutation (POLE-P286R). Although these cells grew robustly, within 120 generations, all subclones carried truncating or nonsynonymous mutations in the POLE-P286R homologous allele (pol2-P301R) that suppressed the mutator phenotype as much as 100-fold. Independent adaptive events in the same culture were common. Our findings suggest that analogous tumor cell populations may adapt to the threat of extinction by polyclonal mutations that neutralize the POLE mutator allele and preserve intratumoral genetic diversity for future adaptation.
THE high fidelity of DNA replication and repair prevents mutations that might otherwise lead to cancer. As first proposed > 40 years ago, cells with defects in these pathways exhibit an elevated mutation rate (mutator phenotype) and an accelerated path to malignancy (Loeb et al. 1974; Loeb 2016). The mutator hypothesis of cancer first gained support from the discovery that Lynch syndrome was caused by defects in mismatch repair (MMR), which normally corrects DNA replication errors (Lynch et al. 2009). Mouse studies later demonstrated that loss of polymerase proofreading by the major leading- and lagging-strand DNA polymerases (Polε and Polδ, respectively) also promoted tumorigenesis (Goldsby et al. 2001; Albertson et al. 2009). More recently, cancer genome sequencing has revealed exonuclease-domain mutations in the catalytic subunit genes for Polε (POLE) and Polδ (POLD1) in a subset of highly mutated colorectal and endometrial human cancers (Yoshida et al. 2011; Cancer Genome Atlas Network 2012; Palles et al. 2012; Cancer Genome Atlas Research Network et al. 2013; Church et al. 2013). Heterozygous mutations in POLE predominate in these “ultrahypermutated” cancers, with the most common POLE variant encoding a P286R substitution (Barbari et al. 2018). POLE-P286R confers strong mutator and cancer phenotypes when engineered into mice in the heterozygous state (Li et al. 2018). The corresponding allele in budding yeast (pol2-P301R) was found to confer a strong dominant mutator phenotype that far exceeds that of an exonuclease-null allele (pol2-4) (Kane and Shcherbakova 2014). Recent biochemical and crystallography experiments indicate that the yeast Polε P301R variant retains residual exonuclease activity, but encodes a hyperactive polymerase that excels at mispair extension (Xing et al. 2019) due to occlusion of the DNA-binding site of the exonuclease domain by the arginine substitution (Parkash et al. 2019). Many of the other POLE exonuclease domain alleles, which confer weaker mutator phenotypes than pol2-P301R when modeled in yeast, may also promote mispair extension rather than inhibit proofreading (Barbari et al. 2018).
Tandem defects in proofreading and MMR pathways produce greater than additive increases in mutation rate in bacteria and yeast (Morrison et al. 1993; Schaaper 1993; Morrison and Sugino 1994), indicating that polymerase fidelity and MMR act redundantly to limit replication errors. Correspondingly, some human tumors with heterozygous POLE or POLD1 mutations also carry mutations affecting MMR (Cancer Genome Atlas Network 2012; Cancer Genome Atlas Research Network et al. 2013). Far from incidental, the MMR defects appear to synergize with the polymerase variants to accelerate mutagenesis. In so doing, they create mutational signatures consistent with the interaction between these two pathways (Campbell et al. 2017; Haradhvala et al. 2018; Hodel et al. 2018). Likewise, patients with inherited biallelic MMR deficiency frequently develop rapidly mutating tumors with heterozygous mutations affecting the fidelity of Polε or Polδ (Shlien et al. 2015). Thus, powerful mutator phenotypes based on the synergistic interactions between polymerase fidelity and MMR defects represent a reoccurring event in cancer evolution.
As an evolutionary strategy, cells that employ a high mutation rate run the risk of error-induced extinction (EEX), where every one of their descendants eventually acquires a lethal mutation (Morrison et al. 1993; Fijalkowska and Schaaper 1996; Herr et al. 2011a). As an extreme manifestation of EEX, double-mutant haploid yeast spores deficient in Polδ proofreading and MMR cease dividing within 10 cellular generations (Morrison et al. 1993). Strong mutator phenotypes can also drive EEX of diploid yeast, although the maximum tolerated mutation rate (error threshold) is higher than in haploids, consistent with genetic buffering by the diploid genome (Herr et al. 2014). Colony formation slows and becomes irregular once mutation rates are within an order of magnitude of the diploid error threshold. High mutation loads may inherently restrict cellular longevity. Recent life span experiments in yeast demonstrate that strong diploid mutators exhibit a form of genetic anticipation: the longer a lineage exists, the higher the mutation burden and the shorter the cellular life span (Lee et al. 2019). Evidence suggests that error thresholds also exist in mammals. Mouse embryos completely deficient in both proofreading and MMR initiate development but fail in the second week of gestation (Albertson et al. 2009). Moreover, tumors with combined deficiencies in polymerase fidelity and MMR appear to reach an upper limit of ∼250 mutations/Mb, which has been interpreted as evidence for a mutation threshold in cancer (Shlien et al. 2015).
It is unclear whether strong mutator-driven cancers adapt to the threat of extinction and, if so, how. In yeast, one-third of haploid “eex” mutants with combined defects in proofreading and MMR acquire intragenic antimutator mutations that suppress the mutator polymerase (Herr et al. 2011a; Williams et al. 2013). Polymerase antimutators also arise in bacteria and bacteriophages expressing strong mutator polymerases, suggesting that these adaptive mechanisms are highly conserved (Reha-Krantz 1988; Fijalkowska and Schaaper 1995). The yeast antimutator mutations encode amino acid substitutions in a wide variety of positions within the polymerase, including the polymerase active site, DNA-binding domains, polymerase structural elements, or even the exonuclease domain itself (Herr et al. 2011a,b; Williams et al. 2013; Dennis et al. 2017). Since Polδ and Polε both play essential roles in DNA replication, these antimutator alleles found in haploid cells likely produce functional polymerases. Some substitutions may increase nucleotide selectivity while others may promote polymerase dissociation from mispaired primer termini, allowing extrinsic proofreading (Herr et al. 2011b). Evidence for extrinsic proofreading in yeast between Polδ and Polε exists in the literature (Morrison and Sugino 1994; Flood et al. 2015; Bulock et al. 2020), and processing of mispaired primer termini by other DNA repair pathways may also be possible (Herr et al. 2011b).
The majority of eex mutants in the above haploid yeast genetic screens, which utilized plasmid shuffling, carried genetic changes that were extragenic to the plasmid-borne mutator polymerase gene and were never mapped (Herr et al. 2011a; Williams et al. 2013). Here, we performed new genetic screens that would allow us to more easily identify these loci using Mendelian segregation of mutator and antimutator phenotypes. In the process, we discovered that many eex mutants were, in fact, not antimutators, but spontaneous polyploids, which are buffered from mutation accumulation as described above. This finding led us to explore the competition between antimutators and spontaneous polyploids during the evolution of diploid mutators. These diploid mutator strains did not display an initial growth defect but became subject to selection during propagation. This scenario models a population of mutator tumor cells having to adapt to the threat of extinction during cancer progression.
Materials and Methods
Media and growth conditions
Yeast propagation and tetrad dissection followed standard procedures (Sherman 2002). Unless otherwise noted, reagents were purchased from Fisher Scientific (Pittsburgh, PA) or Sigma ([Sigma Chemical, St. Louis, MO). For rich liquid media, we used YPD (1% w/v yeast extract, 2% w/v peptone, and 2% w/v dextrose). For rich solid media we used YPD or synthetic complete (SC) [6.7 g Difco yeast nitrogen base without amino acids, 2% w/v dextrose, and 2 g/liter SC amino acid mix (SCM) (Bufferad)] supplemented with 2% w/v agar. For drop-out media, SCM powders lacking specified amino acids were either purchased from Bufferad or made to the same specifications (Kaiser et al. 1994). For haploid mutation rates, we used SC plates lacking arginine (SC-Arg) with 60 μg/ml canavanine (Can). For mutation rates with diploid strains we used SC_MSG-Arg [1.7 g Difco yeast nitrogen base without amino acids or ammonium sulfate, 1 g/liter monosodium glutamate (for a nitrogen source), 2% w/v dextrose, and 2 g/liter SCM-Arg)] with 60 μg/ml Can and 100 μg/ml nourseothricin (NTC) (Herr et al. 2014). We used similar plates lacking histidine and containing NTC (SC_MSG-His+NTC) to select for mating between CAN1::natMX and can1Δ::HIS3 cells (see below).
Yeast strains
The diploid parent strain used for these experiments, AH0401, is a BY4743 derivative engineered to be heterozygous at the CAN1 locus (Herr et al. 2014). One allele carries the natMX transgene (encoding resistance to NTC) inserted just downstream of the CAN1 coding sequence (CAN1::natMX), while the other CAN1 allele is deleted and replaced with HIS3 (can1Δ::HIS3). Selection for can1 mutants in the presence of Can and NTC allows us to score bona fide can1 mutants without the high background of can1Δ::HIS3/can1Δ::HIS3 mitotic recombinants (Herr et al. 2014). The four wild-type (WT) strains used for generating mapping strains (Supplemental Material, Figure S1) and for the mating-type switching experiments (Figures S3 and S4) were dissected from the same AH0401 tetrad: AH4002 (MATα CAN1::natMX), AH4003 (MATa CAN1::natMX), AH4010 (MATα can1Δ::HIS3), and AH4015 (MATa can1Δ::HIS3). AH12721 is a POL2/URA3::pol2-4 MSH2/msh2Δ::LEU2 strain isolated by mating freshly dissected spores from AH2801 (POL2/URA3::pol2-4 MSH6/msh6Δ::LEU2) (Kennedy et al. 2015) and AH5610 (MSH2/msh2Δ::LEU2). Zygotes were isolated by microdissection after 8 hr and allowed to form colonies, which were then genotyped. AH5610 was constructed for this study by transforming AH0401 with a LEU2 PCR product amplified from pRS416 with Msh2U and Msh2D as described (Williams et al. 2013). AH2601 is a previously described POL3/URA3::pol3-01 MSH6/msh6Δ::LEU2 strain (Lee et al. 2019) constructed in the AH0401 background. All strains derived from mating between pol3-01 msh6Δ spores used in the evolution experiment were designated AH164_NN, where “NN” refers to their coordinate in the 96-well plate. AH11304 is a URA3::pol2-P301R transformant of AH5610 in which a duplicated POL2 promoter flanks the URA3 transgene upstream of the POL2 coding sequence. We engineered the strain using a chimeric URA3::pol2P301R PCR product amplified in two fragments from pRS416-POL2 (Williams et al. 2013) with Phusion polymerase (New England Biolabs, Beverly, MA) and the following conditions: 98°, 1 min; 25 × (98°, 10 sec; 51°, 30 sec; 72°, 2 min); and 72°, 2 min. One fragment was amplified with pol2U (5′-ATGATGAAAGAGCACATTCTATCAAGATAACACTCTCAGGGGACAAGTATAGATTGTACTGAGAGTGCAC-3′) and POL2P301R-r (5′-CATTATTTGATCTACGGCGGAATCCcGGAATTTTAAAGGCGGCTTCGTGG-3′; pol2-P301R mutation, lower case). A second POL2 fragment was amplified with POL2P301R (5′-CCACGAAGCCGCCTTTAAAATTCCgGGATTCCGCCGTAGATCAAATAATG-3′) and pol2S7 (5′-ATGTGGATAACTTGGTCTGCG-3′). The two PCR fragments were gel purified, equimolar ratios were combined without primers in a second PCR reaction for 10 cycles (98°, 10 sec; 55°, 30 sec; and 72°, 5 min), outside primers pol2U and pol2S7 were added, and the reaction was continued for another 10 cycles. The entire pol2-P301R gene was initially confirmed by Sanger sequencing and then by whole-genome sequencing of the evolved clones.
Isolation of eex mutants
For pol3-01 msh6Δ eex mutants (Figure S1), we plated large numbers of random haploid spores onto media that selected for the URA3 and LEU2 transgenes tightly linked to each mutator allele. The pol3-01 msh6Δ cells do not form visible colonies unless they acquire an eex mutation. The isolation of pol2-4 msh2Δ eex mutants required a slightly different approach. During our initial tetrad analysis of the pol2-4/POL2 msh2Δ/MSH2 strain, we found that pol2-4 msh2Δ cells were not synthetically lethal, as they were in our earlier plasmid shuffling system, but grew slowly (Williams et al. 2013). To isolate eex mutants, we simply replated the slow-growing double-mutant spore clones for faster-growing colonies. Haploid yeast cells grow vegetatively as one of two mating types, MATa and MATα, which can mate to form MATa/MATα diploids. To isolate mapping strains, we mated each eex mutant in parallel with AH4002, AH4003, AH4010, and AH4015, and plated the mutants on SC_MSG-His+NTC. Candidates that only mated with one of the four strains were carried forward for genetic analyses.
Assay for mating-type switching or same-sex mating
We mixed ∼5 × 106 cells of two strains with the same mating type but opposite CAN1 alleles (can1Δ::HIS3 and CAN1::natMX) in a microtiter plate with YPD and incubated the cells at 30° for 8 hr without shaking. CAN1::natMX/can1Δ::HIS3 diploids were then selected on SC_MSG-His+NTC media. WT and pol2-4 msh6Δ mutator cells from the T1 transfer were used for Figure S3. WT cells picked from individual colonies were used for Figure S4. Mating type PCRs were performed as described (Harari et al. 2018).
Explaining the similar rates of mating-type switching and same-sex mating
Our finding that diploid colonies occur with similar frequencies indicates that a double-stranded DNA (dsDNA) break likely initiates both mating-type switching and same-sex mating. In MATa cells, the dsDNA break is repaired by gene conversion from the HML locus, converting the cell to a MATα cell, which then mates with an adjacent MATa cell to form a MATa/MATα diploid. In MATα cells, the dsDNA break likely disrupts expression of the MATα2 repressor, which normally suppresses the expression of genes required for the MATa program (Strathern et al. 1981). These phenotypic MATa cells then mate with adjacent MATα cells and repair the break through intra- or interchromosomal recombination.
Evolution experiments
To obtain URA3::pol2-4 msh2Δ::LEU2 double-mutant spores we dissected tetrads on SC-Ura-Leu. After 2 days of growth, we isolated single cells from the perimeter of each colony by microdissection and moved them below the colony to form new colonies (Figure S2A). We then suspended the main colonies in 100 μl of H2O in a 96-well format (Figure 1). Random empty wells were interspersed among the samples to monitor for contamination. We made glycerol stocks from part of the cell suspension. From the remaining cell suspensions, we diluted the cells 1:50 in YPD and grew them for 3 hr before removing aliquots for the first flow cytometry measurement, after which the cultures were grown to saturation and then subcloned as described below.
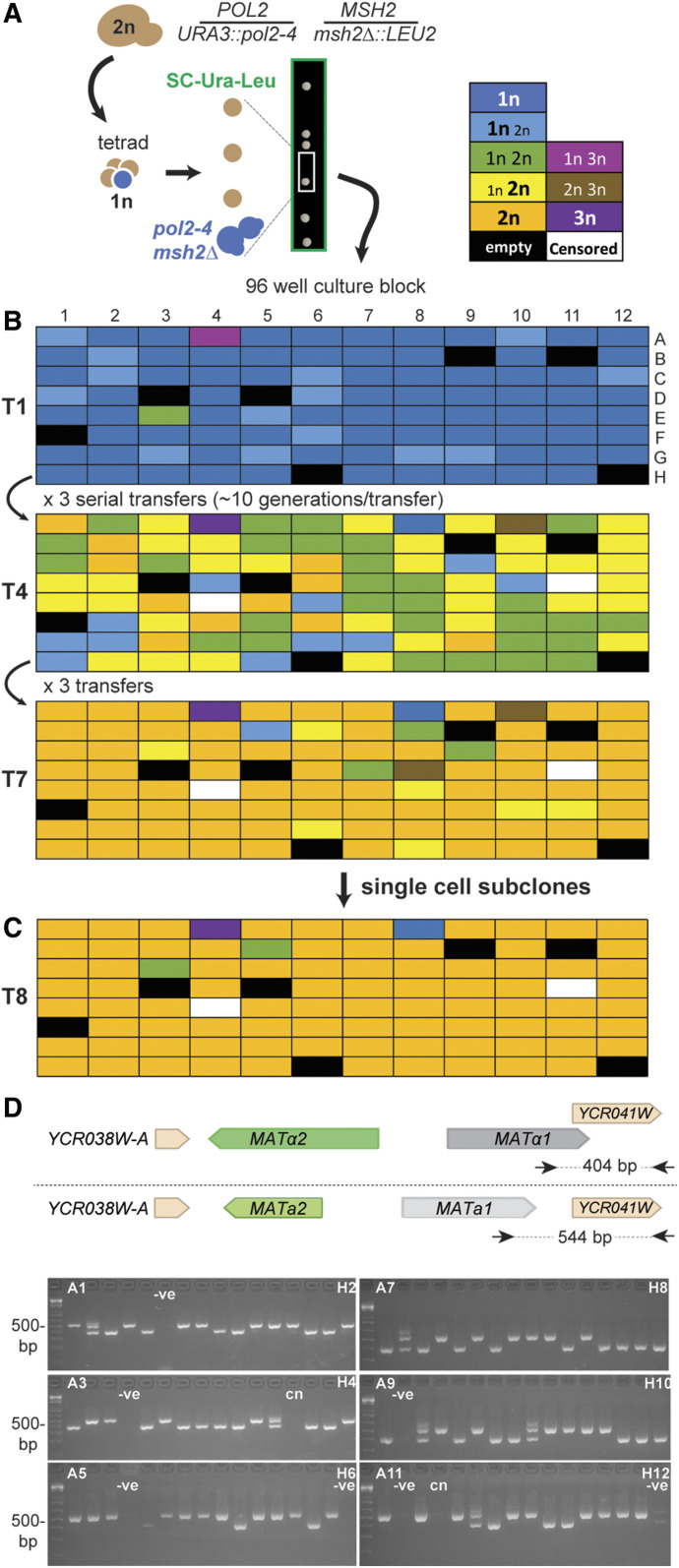
Figure 1.
Spontaneous polyploids routinely emerge during the evolution of pol2-4 msh2Δ haploids. (A) Isolation of pol2-4 msh2Δ haploids from sporulated parental diploid cells (2n). Diploid genotype indicated to the right. Arrows, from left to right, indicate manipulations of cells: sporulation, tetrad dissection, and inoculation into 96-well culture blocks. Following tetrad dissection, double-mutant pol2-4 msh2Δ spores (blue cells) selectively form colonies on media lacking uracil and leucine (green box). (B) Evolution of pol2-4 msh2Δ mutators. Independent isolates were cultured in a 96-well format by diluting 1:500 with each transfer (T). Ploidy was assessed by flow cytometry at T1, T4, and T7 (see Dataset S1 for histograms). Colors of boxes indicate the ploidy or mixture of ploidies of each culture: dark blue, 1n cells; light blue, more 1n than 2n; green, equal numbers of 1n and 2n; yellow, more 2n than 1n; orange, 2n; pink, mixture of 1n and 3n; brown, mixture of 2n and 3n; purple, 3n; black, blank; white, censored. (C) Ploidy of single-cell subclones isolated from T7. (D) Mating types of single-cell subclones. Top: schematic of MATa and MATα mating-type loci, PCR primers (arrows), and expected DNA sizes. Bottom: PCR products run on 2% agarose gels, labeled by their coordinates in the 96-well block; −ve, negative (no cells) and cn, controls; culture censored during evolution. The 500-bp fragment of the Invitrogen 1-kb plus ladder is indicated to the left.
To isolate mutator diploids for evolution experiments, AH2601 or AH11304 tetrads were dissected on SC-Ura-Leu plates and spores were separated from each other by ∼100 μm (the width of two diameters of the dissecting needle). After two-to-three divisions, dividing cells from different tetrads were placed next to each other to allow mating. One-half of the pairings produced zygotes after two to three additional divisions. Zygotes were moved to a defined coordinate on the plate and allowed to form a colony. In our initial experiment with AH2601, we noted that Ura− Leu+ spores germinated and divided once on this media and that we could isolate pol3-01/POL3 msh6Δ/msh6Δ heterozygotes by crossing them with more rapidly dividing Ura+ Leu+ cells. We used this approach to enrich for pol2-P301R/POL2 msh2Δ/msh2Δ strains in the AH11304 crosses. We also obtained numerous pol2-P301R/pol2-P301R msh2Δ/msh2Δ zygotes in the process, which allowed us to compare the relative colony-forming capacity of the two genotypes. Colonies were picked from the plate and suspended in 100 μl of H2O. We used 30 μl of the cell suspensions to inoculate 500 μl cultures, 40 μl for glycerol stocks, and the remaining cell suspension to measure mutation rates (see below). For AH11304-derived zygotes, we reserved 10 μl for pol2P301R genotyping. The pol2P301R allele creates a sequence (…AATTCCgGG…) that is 1 bp from a DraI site (CCCGGG). To genotype pol2-P301R, we amplified POL2 with pol2-P301R-dctF (5′-TGTGGTAATGGCATTTGATATAGAAACCACGAAGCCGCCTTTAAAATcCC-3′), which has a mismatch (lower case) that creates a SmaI site and pol2-P301R-dctF (5′-GGATACTCCGGTTTCGGTGTATACTCAAAGTCTTCAATATCC-TCAGAGA-3′). We digested the amplified products with SmaI in the NEB Cutsmart buffer and resolved the fragments on a 3% agarose gel. The 180-bp product is cleaved into fragments of 50 and 130 bp if the pol2-P301R allele is present.
In both evolution experiments, the strains were propagated at 30° with constant shaking in sterile Nunc 96-deep-well storage blocks (260252; Thermo Scientific), covered with an AeraSeal (Excel Scientific) gas-permeable disposable plate sealer. Each well had 500 μl of YPD and a single 3-mm glass bead to maintain cells in suspension. Subculturing steps were performed by transferring 1-μl volumes from saturated cultures to 500 μl of fresh YPD using an eight-channel pipettor. Frozen −80° glycerol stocks of the cultures were made every three transfers in 96-well format. To recover cultures from storage, the plates were first thawed at room temperature and then 5 μl were transferred into 250 μl of fresh media.
Mutation rates
Mutation rates by fluctuation analysis were performed essentially as described (Herr et al. 2014). To determine the mutation rate of the zygote colonies, 10 μl of the original suspension (see above) were spot-plated directly on SC_MSG-Arg+CAN+NTC plates. Another 10 μl were used for a series of 10-fold serial dilutions. With each dilution, 10 μl were plated on the mutation rate plate and 10 μl on SC media to estimate the total number cells in each colony (Nt). Only one-half of the zygote colonies had the CAN1::natMX/can1Δ::HIS3 genotype suitable for mutation rate measurements. Thirty colonies with similar Nt values were used as replica colonies to estimate the average number of mutational events (m) by maximum likelihood using the newton.LD.plating function in the R package rSalvador (Zheng 2015). The confint.LD.plating function was used to calculate C.I.s. For mutation rates of all subclonal isolates (Figure 4A and Figure S2), replica colonies were obtained by plating serial dilutions onto SC media. For mutation rate calculations, we typically used mutation counts from eight replica colonies. In some assays of T1 subclones in Figure 4A, one or two replica colonies had very low mutation counts (0 or 1 mutants vs. 100’s). These clear outliers were censored under the assumption that they represented an antimutator or polyploid subclone. Cohorts with more than two such outliers were censored completely.
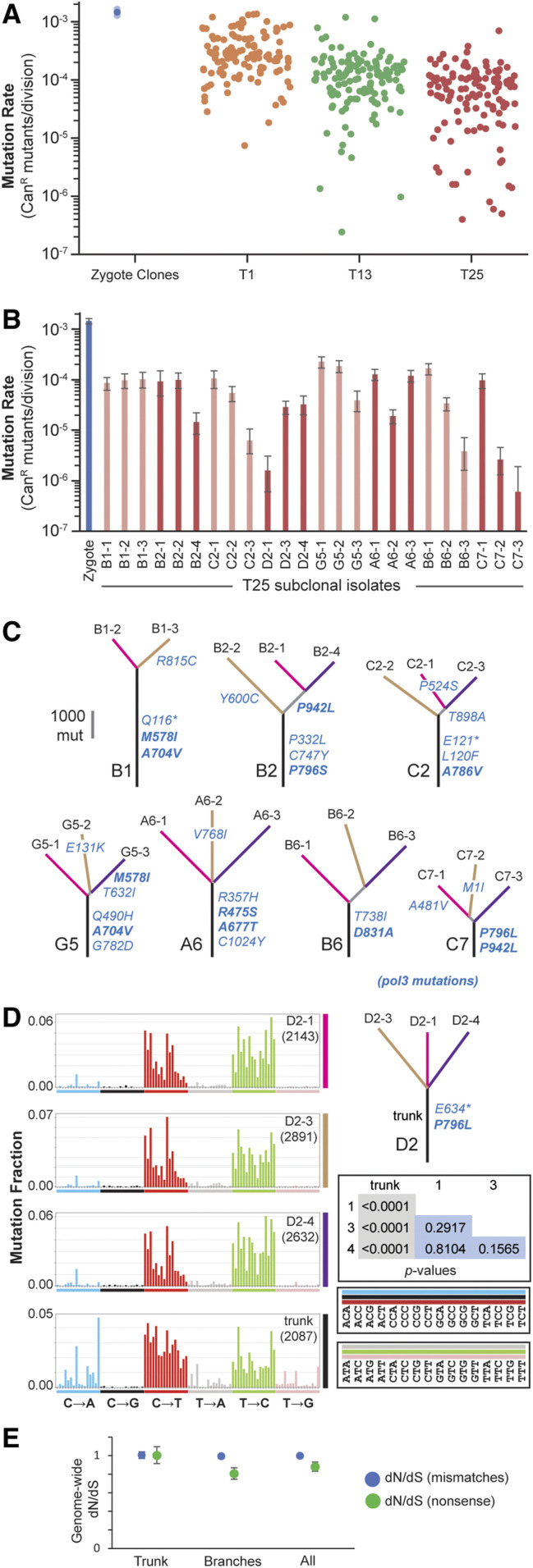
Figure 4.
Strong diploid mutators adapt through antimutator alleles. (A) Mutation rates of pol3-01/pol3-01 msh6Δ/msh6Δ subclones. Single-cell subclones were isolated after the first (T1), 13th (T13), and 25th (T25) transfers. Mutation rates were determined by a canavanine resistance (CanR) fluctuation assay (see Dataset S1 for mutation rates and 95% C.I.s). Solid circles, mutation rates; transparent circles, 95% C.I.s. (B) Variation in mutation rates depicted in (A) of T25 subclones from the same culture. Alternating shades of red indicate groups of subclones. (C) Phylogenetic trees of subclones depicted in (B) based on whole-genome sequencing. Observed pol3 mutations are in blue lettering using single-letter amino acid codes: *, nonsense mutation; bold type, known or likely antimutators. (D) Representative mutation spectra of shared (trunk) and unique mutations [see Figure S7 for mutation spectra of all trees in (C)]. Each mutation site (represented here by the pyrimidine base T or C) was categorized into one of the 96 mutation types created by subdividing the six general mutation types (C→A, C→G, C→T, T→A, T→C, T→G) into the 16 possible trinucleotide contexts (see boxes to the lower right for contexts; blue, black, and red bars correspond to mutation sites involving C; gray, green, and pink bars correspond to mutation sites involving T). Bar heights correspond to the fractions mutation types represent of the total mutations (given in the right-hand corner of each plot). Inset table: P-values of the hypergeometric test of similarity between mutation spectra. (E) Genome-wide (global) ratios of nonsynonymous and synonymous mutations (dN/dS) of trunk and branch mutations.
Flow cytometry
Overnight cultures were diluted 1:50 in fresh YPD and grown for 3–5 hr. Next, 100 μl of cells were recovered by centrifugation and washed twice in an equal volume of water by resuspending them and centrifuging again. We then resuspended the cells in 70% ethanol and incubated them for at least 1 hr at room temperature or overnight at 4°. We recovered the cells again by centrifugation, washed them once with water, and then resuspended them in 50 mM sodium citrate with RnaseA (10 μg/ml). The cells were heated to 95° for 15 min and then incubated for 3–4 hr at 37°. The cells were recovered by centrifugation and resuspended at a concentration of 106 cells/ml in 50 mM sodium citrate with 2 μM Sytox Green (Life Technologies). After 1 hr at room temperature, the cells were sonicated for 30 sec with 1-sec pulses in a Misonix cup horn sonicator and then analyzed on a BD Accuri c6 flow cytometer using the FL1 detector. Plots in Supplemental Material Dataset S1 are organized in the 96-well format used for each experiment. Histogram plots of the data were generated using the c6 software, which allows multiple plots to be overlaid. Plots used as controls are indicated at the top of each experiment.
Genome sequencing and analysis
We performed whole-genome sequencing of subclones from the AH164 evolved cultures as described (Herr et al. 2014; Lee et al. 2019). We used a custom sequencing analysis pipeline (eex_yeast_pileline.sh) that operates in a Unix command-line. It aligns the sequencing reads against a repeat-masked version of the R64-1-1 assembly of the S288C Saccharomyces cerevisiae genome using the Burrows–Wheeler Aligner (0.7.17). Discordant and split reads were removed using Samblaster (0.1.24). Picard tools (2.21.9) AddorReplaceReadGroups was used to add information to the header needed for later steps. After indexing the BAM files with Samtools (1.8), we sequentially processed them with the Genome Analysis Toolkit (GATK3) RealignerTargetCreator, IndelRealigner, LeftAlignIndels, BaseRecalibrator, and PrintReads to minimize false variant calls. We used SamTools to make a pileup file, and then VarScan (v2.3.9) mpileup2snp and mpileup2indel to call SNPs and insertion/deletions (indels), respectively. We used a variant frequency cutoff of 0.1 for SNPs to demonstrate that the sequenced genomes were indeed from 2n and not 4n cells (Dataset S2). We used a variant frequency cutoff of 0.3 for indels to minimize false positives at repetitive sequences due to frameshift errors during library preparation and filtered out SNPs and indels that were present in the parental strains. For AH11304 evolved cultures, we prepared DNA sequencing libraries with the NEBNext Ultra II FS DNA library kit, which utilizes enzymatic fragmentation instead of sonication. The same sequencing analysis pipeline was used except that, during the variant calls, we used a variant frequency cutoff of 0.22 for SNPs and 0.3 for indels.
We wrote a python script to compare variants found in subclones from the same culture and create lists of shared and unique mutations found in each strain. The program only assesses positions in the genome with 18-fold depth coverage in all subclones (Dataset S2). We annotated the SNP lists using Annotate 0.1 (Pashkova et al. 2013) and the indel lists with a modified version of the program that reports the 3ʹ peptide produced by the frameshift (annotatefs2.py). We used the SNP lists to determine the 96 trinucleoide mutation spectra using snv-spectrum (now found as SnvSpectrum at https://github.com/clintval/nucleic). We wrote a python script that creates a 2 × 96 table of the output of snv-spectrum for two different variant lists, and then estimates P-values of the hypergeometric test (Adams and Skopek 1987) and the cosine similarity test, under the null hypothesis that they came from the same population (HypergeometicCosinSimTestPub.py).
Cosine similarity, commonly used in mutation signature analyses, is a measure of the angle between two vectors projected into an n-dimensional space, where n is the number of categories used for the comparison. In this case, the categories are the 96 trinucleotide contexts. The number of mutations at each context together define the direction of the vector. The closer two spectra, the smaller the angle, and the closer the cosine similarity is to 1. The statistical significance of such comparisons is typically not reported. However, the hypergeometric test, which is an expansion of Fisher’s exact test, has previously been used to test the null hypothesis that two spectra are the same. Formally, the hypergeometric probability of each possible contingency table is calculated and the number of tables with a higher probability than the observed table provides a measure of significance. With a 2 × 96 contingency table, an exact calculation of significance becomes computationally impractical but can be estimated using a Monte Carlo approach. Implementation of the test involves construction of 10,000 random 2 × 96 tables that maintain the same row and column totals as the original table (Agresti et al. 1979), and then calculation of the hypergeometric probability or cosine similarity of each table (Dataset S2). The fraction of random tables with a hypergeometric probability higher than the original table is an estimate for the probability that two spectra are the same. Likewise, the cosine similarity P-value represents the number of random tables whose cosine similarity was higher than the observed cosine similarity. We used the conservative Bonferroni multiple testing correction to estimate the P-value cutoff for significance for each set of comparisons (Dataset S2). Finally, we used the SNP lists described above to determine the ratio of nonsynonymous and synonymous mutations (dN/dS) of individual genes and of the entire genome using the R-package dNdscv (Martincorena et al. 2017) (https://github.com/im3sanger/dndscv).
Data availability
Strains and plasmids are available upon request. All whole-genome sequencing data are available from the Sequence Read Archive (BioProject identifier: PRJNA629499). Supplemental Dataset S1 contains flow cytometry results, mutation rates, and modeling of diploid mutator evolution. Supplemental Dataset S2 describes whole-genome sequencing results of diploid mutator strains. Supplemental Dataset S1 and S2, along with all computer scripts used in this work, can also be found at GitHub (https://github.com/mutatorUW/MutatorEvolution). Supplemental material available at figshare: https://doi.org/10.25386/genetics.12276461.
Results
Discovery of spontaneous polyploid eex mutants
The genetic screens that led to the discovery of spontaneous polyploid eex mutants began with diploid parent strains that were heterozygous for deletion of MSH6 (msh6Δ) and pol3-01 (Polδ-proofreading defective), or msh2Δ and pol2-4 (Polε-proofreading defective) (Figure S1, A and B). After inducing the diploids to undergo sporulation (meiosis), we isolated eex mutants that suppressed the EEX phenotypes of freshly isolated double-mutant haploid cells. Both parental strains were heterozygous for two distinct transgene insertions at the CAN1 locus: each eex mutant carried either CAN1::natMX, which confers sensitivity to Can and resistance to NTC, or can1Δ::HIS3, which confers Can resistance and histidine prototrophy. We crossed all eex mutants to WT haploids with the opposite CAN1 allele and mating type, and then isolated diploid mapping strains by plating for His+ and NTC-resistant colonies. We sporulated the resulting mapping strains and although they routinely formed tetrads, the majority of the mapping strains from both genetic screens failed to produce any viable spores (Figure S1B). Surprisingly, several mapping strains with intermediate spore viability produced progeny that carried both CAN1 alleles. Flow cytometry revealed that these unusual mapping strains were in fact tetraploids (4n) (Figure S1B), while the mapping strains with no viable spores were triploids (3n). We inferred that the original eex mutants for the 3n mapping strains were MATa/MATa or MATα/MATα diploids, and that the 4n strains were derived from MATa/MATa/MATa or MATα/MATα/MATα triploid eex mutants. Since polyploid eex mutants arose in both genetic screens, this escape mechanism is a general one, likely related to the ability of polyploids to better withstand mutation accumulation.
Evolution of haploid mutator cells
A tendency to pick larger candidate eex colonies could explain why polyploids were more common than antimutators in our genetic screens. To determine the relative success of these two adaptations in an unbiased manner, we isolated 89 independent pol2-4 msh2Δ spore clones by dissecting tetrads from POL2/pol2-4 MSH2/msh2Δ diploids on growth media that selected for the transgenes linked to the mutator alleles (Figure 1A). Freshly dissected pol2-4 msh2Δ haploids form colonies with an uneven perimeter, consistent with the onset of EEX and the emergence of eex mutant subclones with improved survival. After 2 days, we isolated four cells from the perimeter of each colony and moved them elsewhere on the plate to assess their colony-forming capacity, mutation rate, and ploidy (Figure S2A). Two-thirds of the cells isolated from the perimeters of the initial colonies failed to form a second colony, a manifestation of EEX. The 120 colonies from viable cells contained 12 that were uniformly polyploid [including one triploid (A4.1)] (Figure S2B and Dataset S1). To test for antimutators, we measured mutation rates of the haploid subclones that carried the CAN1::natMX allele. This involved subcloning these cells a second time to obtain replica colonies, which we assessed for ploidy. A high frequency of polyploid replica colonies prevented mutation rate measurements for 12 subclones. Thirteen subclones exhibited the expected mutation rate for pol2-4 msh2Δ cells, while 12 subclones displayed an antimutator phenotype (Figure S2C). These results indicate that antimutators and polyploids emerge with similar frequencies during the initial growth of pol2-4 msh2Δ haploids, and that polyploids continue to arise.
To investigate which eex strategy prevails during long-term growth, after removing individual cells for the above analysis, we suspended the original colonies in liquid growth media and evolved them in a 96-well format with interspersed empty wells to serve as controls for contamination. We repeatedly subcultured the cells by diluting them 1:500 (∼105 cells/inoculum) and growing them to saturation (∼108 cells), yielding an estimated 10–12 generations per transfer. We assessed ploidy at the initial time point (T1), and then after the fourth (T4) and the seventh transfer (T7). The ploidies of the initial colony suspensions were overwhelmingly haploid, although 15 out of 89 cultures at this early stage showed evidence of a minor diploid subclone (Figure 1B and Dataset S1). The colony suspension from the above triploid subclone (A4) contained both haploid and triploid cells in equal proportions. Prolonged growth of the original cultures revealed profound fitness differences between haploid antimutators and spontaneous polyploids (Figure 1B). At T7, only two cultures remained haploid: 10 were mixtures of 1n and 2n cells, and 73 cultures were uniformly 2n or 3n. Thus, polyploids routinely win out over antimutators during the adaptation of haploid cells to the mutator phenotype.
Mechanism of polyploidization in haploid mutator cultures
In the haploid evolution experiment, cells could have become polyploid by either mating or failed cytokinesis (Ganem et al. 2007). Although deletion of Homing Endonuclease (HO) stabilizes mating type in our studies, mating-type switching may still occur through random dsDNA breaks at the MAT locus, which are then repaired using the transcriptionally silent HML (MATα) or HMR (MATa) loci (Haber 2012). Conceivably, the rates of these events could be elevated in mutator cells. To determine the relative contributions of mating and spontaneous polyploidization, we isolated a single colony from each evolved culture and performed flow cytometry. Of the 87 isolates, there were 83 diploids, 1 haploid, 1 triploid, and 2 mixed cultures with both 1n and 2n cells (Figure 1C). Mating-type PCR assays revealed 41 MATa/MATa, 36 MATα/MATα and 6 MATa/MATα diploids (Figure 1D). Three of the MATa/MATα isolates were heterozygous at the CAN1 locus (CAN1::natMX/can1Δ::HIS3) (Dataset S2), providing evidence for rare mating events between cells from neighboring wells. The three remaining MATa/MATα cells were homozygous for CAN1::natMX or can1Δ::HIS3, and could have arisen through contamination or mating-type switching. We specifically investigated the frequency of mating-type switching with both MATa and MATα cells. Diploids formed in both same-sex pairings with similar frequencies, but while MATa × MATa crosses nearly always gave rise to MATa/MATα diploids, MATα × MATα crosses produced MATα/MATα diploids (Figures S3 and S4). Same-sex mating has previously been observed (Strathern et al. 1981). Given the similarities in frequencies, a dsDNA break at the MAT locus likely initiates both mating-type switching of MATa cells and MATα same-sex mating (see Materials and Methods). The three MATa/MATα clones that were homozygous for the CAN1::natMX or can1Δ::HIS3 allele, which arose by mating-type switching, suggests that only a small percentage of the 36 MATα/MATα cultures in the evolution experiment arose by same-sex mating. Together, these observations argue that the majority of MATa/MATa or MATα/MATα isolates in our evolving populations arose by spontaneous polyploidization due to failed cytokinesis, which recent studies indicate occurs with a frequency of 7 × 10−5 in haploid yeast (Harari et al. 2018). Since tetraploids have been observed in various cancers, we reasoned that spontaneous polyploidization could conceivably represent a conserved mechanism of escape from EEX for mutator-driven tumors.
Tetraploidization further protects spontaneous diploids
As a first test of whether tetraploidy confers an advantage in a diploid mutator population, we utilized the above spontaneous pol2-4/pol2-4 msh2Δ/msh2Δ diploids. We isolated MATa/MATa and MATα/MATα cells from different evolved cultures by microdissection, and mated their daughter cells. We moved the parental diploid cells as well as the resulting tetraploid zygotes to distinct locations on the agar plates to form colonies (Figure 2A). We then mixed each parent diploid strain separately with their tetraploid progeny at different ratios (100:1, 10:1, 1:1, 1:10, and 1:100) and grew them in competition. The tetraploids overtook the parental mutator diploids in seven out of eight competitions. These experiments suggest that tetraploid mutators would potentially have a sizeable fitness advantage over the diploid mutator population from which they arose. However, an important caveat to this experiment is that both diploid parents likely carry homozygous mutations that first arose in the original haploid mutators. These genetic liabilities would become heterozygous in their tetraploid offspring. In diploid cells that spontaneously acquire a mutator phenotype, mutations would accumulate from the beginning in a buffered, heterozygous state.
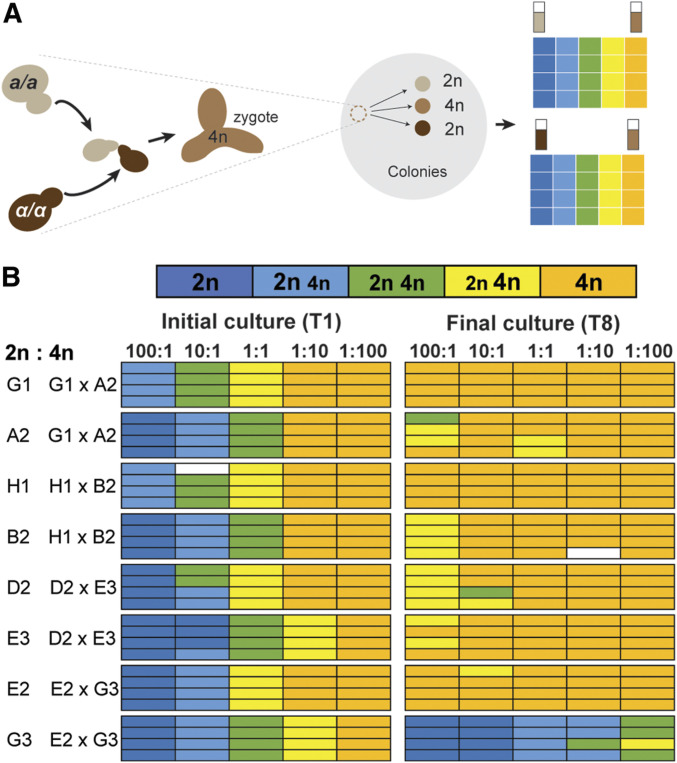
Figure 2.
Competition between diploid mutator parents and tetraploid offspring. (A) Experimental design. MATα/MATα and MATa/MATa subclones from the pol2-4 msh2Δ evolved cultures (see Figure 1B) were mated. Cell suspensions of the resulting tetraploid and parental diploid colonies were counted, mixed, and then competed through eight serial transfers. (B) Ploidy measurements after the first (T1) and eighth (T8) serial transfers (see Dataset S1 for histograms). Letters and numbers to the left indicate locations in the 96-well culture block described in Figure 1. Colors of boxes indicate the ploidy or mixture of ploidies of each culture: dark blue, 2n cells; light blue, more 2n than 4n; green, equal numbers of 2n and 4n; yellow, more 4n than 2n; orange, 4n; white, censored.
Adaptation of diploid yeast to a strong sublethal mutator phenotype
To explore how diploid cells with a near-WT mutation burden adapt to a strong mutator phenotype, we began with pol3-01/pol3-01 msh6Δ/msh6Δ cells, which form robust colonies despite having a mutation rate (1 × 10−3 CanR mutants/division) just below the diploid error threshold (Figure 3A) (Herr et al. 2014). Using a mathematical model in which the decrease in fitness was driven solely by lethal homozygous inactivation of essential genes (Herr et al. 2014), we estimated that a tetraploid or antimutator eex mutant would overtake a pol3-01/pol3-01 msh6Δ/msh6Δ diploid culture in 200 generations (Figure S5 and Dataset 1). With two sets of homozygous mutator alleles driving the mutator phenotype, we anticipated that tetraploids would be at their greatest advantage relative to antimutators.
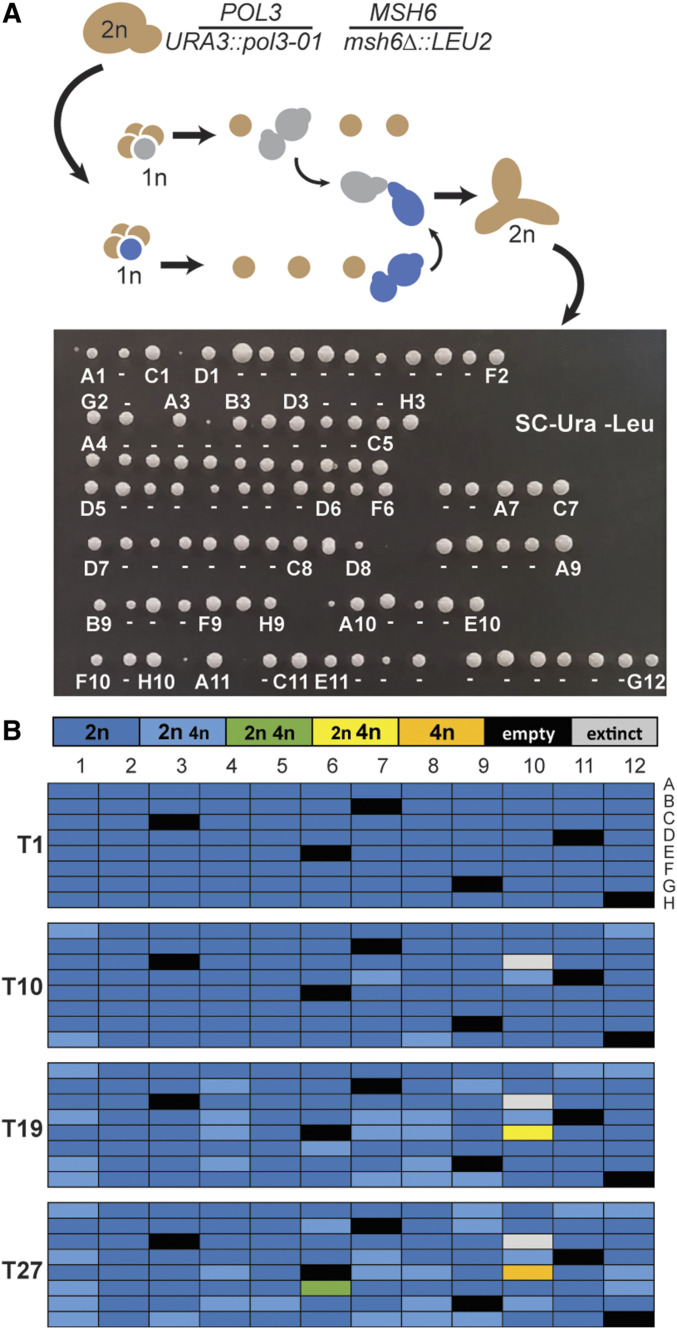
Figure 3.
Strong diploid mutators maintain diploidy during evolution. (A) Isolation of pol3-01/pol3-01 msh6Δ/msh6Δ zygotes. Top: double-mutant pol3-01 msh6Δ haploids, identified by dissecting tetrads on selective media (SC-Ura-Leu), were mated to form zygotes. Zygotes were then moved to new locations on the plate to form colonies. Bottom: growth phenotypes of the initial zygote colonies. Colonies were picked from left to right. Those used for evolution are indicated by either their 96-well coordinate or a dash (−). Colonies without a label or dash were not evolved. (B) Ploidies of evolved cultures after one (T1), 10 (T10), 19 (T19), and 27 (T27) serial transfers (see Dataset S1 for histograms). Colors of boxes indicate the ploidy or mixture of ploidies of each culture: dark blue, 2n cells; light blue, more 2n than 4n; green, equal numbers of 2n and 4n; yellow, more 4n than 2n; orange, 4n; black, blank; gray, censored.
To set up the evolution experiment, we first isolated numerous independent pol3-01/pol3-01 msh6Δ/msh6Δ zygotes by mating pol3-01 msh6Δ haploids that were freshly dissected from the pol3-01/POL3 msh6Δ/MSH6 parental strain (Figure 3A). We evolved 89 zygote clones through ∼300 generations, monitoring ploidy at the T1, T10, T19, and T27 transfers (Figure 3B). A single pure tetraploid culture emerged at the end of the experiment. The remaining cultures were overwhelmingly diploid, suggesting that they adapted through antimutator mutations. To assay for antimutator phenotypes, we isolated independent subclones from T25 CAN1::natMX/can1Δ::HIS3 cultures, assessed their ploidy, and measured mutation rates. Almost all subclones were diploids (Dataset S1) and exhibited a clear antimutator phenotype (Figure 4A). Antimutators also dominated the T13 cultures and were evident as early as T1 (Figure 4A). T1 cells from every culture formed variably sized colonies, which were notably smaller than T13 colonies, suggesting that strong selection for eex mutants exists by ∼30 cellular generations (20 generations/colony and 10 generations for T1) (Figure S6).
Independent T25 isolates from the same pol3-01/pol3-01 msh6Δ/msh6Δ culture sometimes displayed different mutation rates (Figure 4B). We wondered whether they represented independent adaptations, and therefore sequenced the genomes of eight sets of subclones. In each set, the subclones could be arranged into a phylogenetic tree with ∼1300–4500 shared single-nucleotide variants (SNVs) in the trunk and ∼1400 to 3000 unique SNVs in the branches (Figure 4, C and D). This indicates that a single adapted clone rose to prominence in each culture and then diverged. Thus, differences in mutation rates between subclones likely reflect the presence of additional antimutator mutations in some isolates but not others.
The mutations that delineate the phylogenetic trees bear the imprint of the mutator and antimutator phenotypes involved in the evolution of these cultures. We classified the shared and unique SNVs according to the 96 trinucleotide contexts defined by the six different mutation subtypes (C:G→A:T, C:G→G:C, C:G→T:A, T:A→A:T, T:A→C:G, and T:A→G:C), flanked by all possible 5ʹ and 3ʹ nucleotides (Figure 4D). We then compared mutation spectra using the hypergeometric and cosine similarity tests. By both tests, the spectra of the trunk mutations were largely the same between different cultures, consistent with each culture beginning with a common mutator phenotype (Figure 4D, Figure S7, and Dataset S2). The spectrum of unique mutations in each branch of a given tree differed substantially from that of the trunk mutations (P < 0.0001), although generally not from each other, consistent with the antimutator phenotype arising prior to their divergence (Figure 4D and Figure S7). The antimutator alleles caused particularly sharp decreases in the relative abundance of C:G→A:T mutations in the branches of all trees. Despite these similarities, the spectra of unique mutations from unrelated evolved cultures often differed statistically from each other, suggesting that the antimutator alleles altered replication fidelity in distinct ways (Figure 4D and Dataset S2). Thus, acquisition of an antimutator phenotype that fundamentally changes the mutation spectrum coincides with the key adaptive event in each culture.
To understand how the cells attenuated the mutator phenotype, we annotated the mutations (Dataset S2) and compared the affected genes. All eight strain sets acquired multiple pol3 mutations prior to their divergence (Figure 4, C and D, see trunks). Polδ plays an essential role in DNA replication. Both pol3-01 alleles contribute to the onset of EEX, but only one allele needs to remain functional for cells to survive. Thus, the premature nonsense codons observed in three trees (B1, C2, and D2) contribute to mutator suppression by inactivating one of the two pol3-01 alleles, which allows the other pol3-01 allele with an antimutator mutation to serve as the sole source for Polδ. Many missense mutations affect the same amino acid residue as (or are identical to) known antimutator mutations [A704V(2x), A786V, A677T, D831A, R475S, and R815C] (Herr et al. 2011a, 2014; Dennis et al. 2017). The other missense mutations affecting pol3-01 may inactivate polymerase function or may be bona fide antimutator alleles that have not previously been identified. Candidate antimutator mutations include P942L and M578I, which were each observed in two independent lineages, as well as mutations affecting P796, which were isolated three independent times [P796L(2x) and P796S], once in combination with a stop codon. In some lineages, additional pol3 alleles were observed in the branches. In two cases (G5-3 and A6-2), these may account for the variation in mutation rates between subclones from the same culture (Figure 4B), but in other cases they clearly do not (B2-4, C2-3, D2-1, B6-2, B6-3, and C7-3), implicating other loci in the attenuation of mutation rates.
We looked for genes under positive selection by comparing the dN/dS across the genome using dNdscv, which normalizes the observed dN/dS ratio by the 192-trinucleotide mutation spectra of each gene to assess whether the observed ratio departs from neutrality (dN/dS = 1) (Martincorena et al. 2017). We analyzed the mutations from the trunks of the eight lineages and found that only pol3 mutations are under positive selection (q = 0.004, corrected P-value using Benjamini and Hochberg’s false discovery rate) (Dataset S2). Mutations in other genes, within the resolution of this data set, accumulate in a neutral fashion. Repeating this analysis for the 23 unique sets of mutations from the branches failed to reveal any other genes under strong selection despite evidence for their existence, suggesting that more observations are needed for sufficient statistical power, especially if there are multiple antimutator genes. Interestingly, when branch and trunk mutations were analyzed together, pol3 mutations did not emerge from the multiple testing correction as significant, underscoring the importance of using phylogenetic information for the identification of candidate antimutators by dNdScv.
Being able to classify mutations according to the phylogenetic tree also provided evidence for negative selection acting against the mutator phenotype. For subclasses of trunk mutations involving missense or nonsense mutations, which arose early during mutator evolution, the genome-wide (or global) dN/dS ratio was 1.0, indicative of neutrality (Figure 4E) (Dataset S2). However, the genome-wide dN/dS ratio in the branches for nonsense mutations was 0.8, indicating that negative selection begins to act against the most deleterious class of mutations during extended propagation of these lines.
Modeling adaptation to the POLE-P286R mutator phenotype in cancer
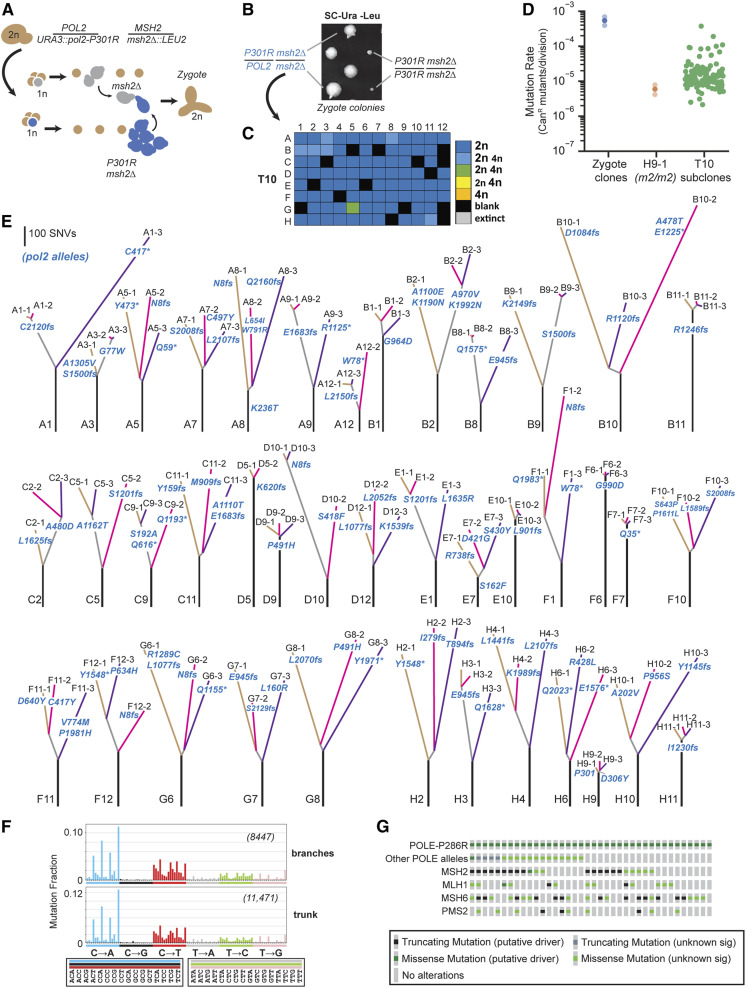
Having established that diploid mutators rapidly evolve to avoid extinction in long-term culture, we sought to understand how cells would adapt to the most common POLE mutator allele in cancer, P286R. Most POLE-P286R tumors appear microsatellite-stable and bear mutational signatures that suggest that they are MMR proficient (Haradhvala et al. 2018). However, one POLE-P286R patient curated on cBioPortal (Cerami et al. 2012) carried two distinct null alleles of MSH6 (TCGA-IB-7651) (Dataset S1) and a clear mutation signature consistent with synergy between POLE-P286R and MMR deficiency (Haradhvala et al. 2018). To model this extreme scenario, we took a similar approach as before and isolated numerous independent pol2-P301R/POL2 msh2Δ/msh2Δ zygotes by crossing freshly dissected pol2-P301R msh2Δ and POL2 msh2Δ haploid spores (Figure 5A). Whereas concurrently isolated double homozygous mutants formed small colonies of ∼1 × 105 cells, the pol2-P301R/POL2 msh2Δ/msh2Δ zygotes grew robustly (Figure 5B and Figure S8) and we evolved 81 independent clones through 10 transfers (∼120 generations). By T10, one culture had evolved a prominent tetraploid subclone (Figure 5C). To test for antimutator phenotypes in the others, we isolated three subclones from 44 CAN1::natMX/can1Δ::HIS3 cultures. Genotyping revealed 1 out of the 132 isolates had lost the heterozygous pol2-P301R allele through mitotic recombination (H9-1). Most of the other isolates had mutation rates > 10-fold lower than the original zygote clones, and many as low as the mitotic recombinant, which serves as an internal msh2Δ/msh2Δ control (Figure 5D) (Dataset S1). Thus, as with pol3-01/pol3-01 msh6Δ/msh6Δ diploids, there is strong selection pressure for pol2-P301R/POL2 msh2Δ/msh2Δ cells to suppress their mutator phenotype, even though they do not show an obvious initial growth defect.
Figure 5.
Modeling adaptation of POLE-P286R mismatch repair-deficient cancers to EEX. (A) Isolation of pol2-P301R/POL2 msh2Δ/msh2Δ zygotes. POL2 msh2Δ spores slowly divide once on SC-Ura-Leu media, allowing mating with more rapidly dividing pol2-P301R msh2Δ cells. (B) Representative growth phenotypes of the initial zygote colonies (see Figure S8 for all colonies). (C) Ploidy of evolved cultures after 10 (T10) serial transfers. Colors of boxes indicate the ploidy or mixture of ploidies of each culture: dark blue, 2n cells; light blue, more 2n than 4n; green, equal numbers of 2n and 4n; yellow, more 4n than 2n; orange, 4n; black, blank; gray, censored. (see Dataset S1 for histograms). (D) Mutation rates as determined by a canavanine resistance (CanR) fluctuation assay (see Dataset S1 for mutation rates and 95% C.I.s). Solid circles, mutation rates; transparent circles, 95% C.I.s (for zygote clones only). (E) Phylogenetic trees based on whole-genome sequencing of the subclones. Line length, the number of shared or unique SNVs; lettering at the bottom of each tree, culture position in 96-well plate; lettering at the end of each branch, subclone identifier number; pol2 mutations are in blue lettering: fs, frameshift; *, nonsense mutation. (F) Spectra of grouped mutations from trunks and branches. Format is the same as in Figure 4. Trunk spectrum determined from all trunk mutations combined. Branch spectrum determined from combined mutations from branches with truncating mutations at E945 or before (A1-3, A5-1, A5-2, A5-3, A8-1, A12-2, B3-3, C11-1, F1-2, F1-3, F12-2, G6-2, G7-1, H2-2, H2-3, and H3-1/H3-2). (G) Oncoprint of POLE-P286R tumors on cBioPortal showing presence of additional POLE alleles and mutations affecting mismatch repair components (see Dataset S1).
To understand the basis for the antimutator phenotypes, we sequenced the genomes of 119 subclones from 40 evolved cultures. The subclones averaged 665 ± 178 (SD) SNVs and 225 ± 35 (SD) indels, corresponding to 60 SNVs/Mb and 20 indels/Mb (Dataset S2). We annotated all variants (Dataset S2) and constructed phylogenetic trees using the SNVs (Figure 5E). Secondary mutations in pol2 were found in all subclones, suggesting that they are adaptive (Figure 5E and Dataset S2). In 6 out of 40 evolved cultures, the pol2 mutation preceded the divergence of the subclones (B1, B11, D9, F6, F7, and H11). As they diverged, the subclones accumulated few additional mutations, providing strong evidence for a direct role of the pol2 mutations in mutator suppression. Altogether, we observed 88 candidate adaptations in pol2, including 45 frameshifts, 23 nonsense mutations, and 36 missense (or combinations of missense) mutations. Two mutant alleles, I279fs (H2-2) and D306Y (H9-2,3), were close enough to P301R to confirm that they reside on the same chromosome. The abundance of truncation mutations suggests that inactivation of the pol2-P301R allele allows cells to use WT POL2 as the sole source for Polε, thereby dramatically lowering their mutation rate. This mirrors how the combination of nonsense and antimutator mutations suppresses the mutator phenotype of some pol3-01/pol3-01 msh6Δ/msh6Δ cultures. In light of this, the observed missense mutations may either inactivate the mutator polymerase or function as antimutators.
The central role of the pol2 mutations in the evolution of these cultures helps to interpret the structure of the phylogenetic trees. Since the majority of pol2 mutations occurred after divergence of the subclones, most of the 300 ± 126 (SD) shared mutations in the trunks likely arose prior to zygote formation. The magnitude of the mutator phenotype in pol2-P301R msh2Δ haploid cells is likely similar to pol2-P301R/pol2-P301R msh2Δ/msh2Δ diploid cells, which formed small colonies (Figure 5B and Figure S8). In the mathematical model of diploid EEX, homozygous inactivation of essential genes limits growth (Herr et al. 2014). A terminal colony size of 1 × 105 cells corresponds to a mutation rate of ∼5 × 10−3 CanR mutants per division, which is 10 times greater than the observed pol2-P301R/POL2 msh2Δ/msh2Δ mutation rate (5.43 × 10−4 CanR mutants/division). We can deduce the genome-wide mutation rate of pol2-P301R/POL2 msh2Δ/msh2Δ cells from the minimal internodal distance between branch points in phylogenetic trees that rapidly diverged and acquired independent pol2 mutations (Dataset S2). The lowest number of shared SNVs between any two subclones was 24. Pairs of subclones from eight cultures (A5, A7, A8, C11, F10, G6, G8, and H6), with similar low increments of shared SNVs, averaged 30 ± 7 (SD) SNVs/diploid genome. This would place the mutation rate of pol2-P301R/pol2-P301R msh2Δ/msh2Δ diploids at ∼300 SNVs/division and the pol2-P301R msh2Δ haploids cells at 150 SNVs/division, easily accounting for the observed number of mutations in the trunks of the phylogenetic trees.
If a mutation rate of 30 SNVs/division continued unabated in pol2-P301R/POL2 msh2Δ/msh2Δ cells, the subclones would have accumulated ∼3600 mutations over 120 generations. However, the average branch length was 418 (± 188 SD). A truncating mutation in pol2-P301R that nullifies the mutator phenotype should lower the mutation rate to that observed in msh2Δ/msh2Δ cells (5 × 10−6 CanR mutants/division) (see H9-1, Figure 5C and Dataset S2), which we estimate would be ∼0.3 mutations/diploid genome/division. A 100 cellular generations at this suppressed mutation rate would produce 30 SNVs, equal to the number that occurs in just one division of the original mutator strain. Subtracting this value from the average branch length suggests that the average adaptative event occurred by ∼13 cellular generations, before the colony reached 104 cells (213). Thus, adaptive mutations in pol2-P301R occur early during the evolution of these strains, substantially moderating the mutation burden.
Since the bulk of mutations in branches with strong pol2 null alleles come from these early divisions, we pooled them to determine the mutation spectrum produced by the pol2-P301R/POL2 msh2Δ/msh2Δ mutator phenotype (Figure 5F). The spectrum from the branches matches the spectrum of the pooled mutations from the trunks (cosine similarity of 0.994) despite likely differences in the magnitudes of their mutator phenotypes (Figure 5F). We also compared the branch mutation spectrum to all mutational signatures from the Catalogue of Somatic Mutations in Cancer (Cosmic) database (Alexandrov et al. 2020) to test how well these yeast cells recapitulate the mutational processes occurring in human disease. We found the highest similarity with Cosmic mutational signature number 14 (cosine similarity of 0.793), which results from the synergy between POLE and MMR mutator alleles (Haradhvala et al. 2018; Alexandrov et al. 2020) (Table S1). Thus, our analysis of the phylogenic trees of evolved pol2-P301R/POL2 msh2Δ/msh2Δ strains reveal a severe mutator phenotype that resembles the mutagenesis in cancers with combined POLE and MMR deficiencies. In this model, we show that adaptation readily occurs through suppression of the pol2-P301R allele. In nearly every culture, the adaptation is polyclonal, resulting in a genetically diverse population of cells that survive extinction.
Discussion
Cell lineages with very high mutation rates face a progressive loss of fitness, and even extinction, unless they adapt by becoming polyploid or acquiring antimutator mutations. The competition between polyploids and antimutators during the evolution of mutators is influenced by the rate at which they arise, and the extent to which they preserve fitness.
In our study of pol2-4 msh2Δ haploids, we found that two-thirds of cells sampled from the perimeter of the initial colonies (< 20 cellular generations) were inviable, illustrating the strong selection pressure for eex mutants. At this early stage, polyploids and antimutators were present at similar frequencies, which is in keeping with the comparable rates of spontaneous polyploidization of haploid yeast (7 × 10−5 per division) (Harari et al. 2018) and gene inactivation of pol2-4 msh2Δ cells (1 × 10−4 CanR mutants/division). Diploids frequently emerge in WT haploid evolution studies, with a fitness advantage of ∼3.5% per generation (Gerstein et al. 2006; Venkataram et al. 2016; Fisher et al. 2018). The fitness advantage of polyploids during the evolution of haploid mutators is likely much larger and constantly increasing as mutations accumulate, as suggested by studies with chemically mutagenized haploid yeast (Mable and Otto 2001). As the mutator parent strain loses fitness, and polyploids and antimutators compete for supremacy, continued mutation accumulation in antimutators may place them at a competitive disadvantage. Even more consequential, the antimutator mutations may inherently compromise fitness by diminishing polymerase processivity. In some cases, antimutators may even cause DNA replication stress and a need for homologous recombination, which would favor the emergence of polyploid variants within the antimutator population.
While spontaneous polyploids and antimutators also arise during the evolution of strong diploid mutators, antimutators dominate nearly every culture. This may reflect stark differences in how frequently the two types of mutants arise. The rate of tetraploidization of diploid yeast, to our knowledge, has not been reported, but our data provide an estimate based on the following logic. Sequencing shows that a single adapted clone rose to prominence in each pol3-01/pol3-01 msh6Δ/msh6Δ culture. We observed that 1% of these cultures became tetraploid during their evolution. Mutator suppression of pol3-01/pol3-01 msh6Δ/msh6Δ cells requires two independent pol3 mutations, whereas polyploidization is a singular event. Biallelic attenuation of the pol3-01 mutator phenotype will occur at a rate no greater than the square of the per gene mutation rate, which for pol3-01/pol3-01 msh6Δ/msh6Δ cells is ∼(1 × 10−3)2 or 10−6. If we assume that tetraploids and antimutators have a similar stabilizing effect on the loss of fitness, the rate of tetraploid formation may be no greater than 10−8. Of course, the rate of tetraploid formation could potentially be higher if tetraploids were inherently less fit than diploid antimutator strains. Growth competition studies between WT tetraploids and diploids suggest that this may be true (Gerstein et al. 2006). Tetraploids suffer higher rates of genomic instability and depend more heavily on factors involved in homologous recombination (Storchová et al. 2006). In the context of a mutator phenotype, a higher rate of homologous recombination in tetraploids may limit buffering of deleterious mutations by promoting loss of heterozygosity. Diploid antimutators are already buffered from mutations and may retain that protection better than tetraploids if they indeed have lower recombination rates.
The ease with which pol3-01/pol3-01 msh6Δ/msh6Δ cells adapted to EEX despite being homozygous for the mutator alleles suggests that similar events may be common in cancers with heterozygous POLE mutations. We modeled this scenario using pol2-P301R/POL2 msh2Δ/msh2Δ cells, which incur 2.7 SNVs/Mb/division (30 SNVs/11 Mb). This level of mutagenesis imposes a strong selection for suppressor mutations, and we found a wide variety of suppressor mutations that reduced the mutation rate as much as a 100-fold. The average mutation burden of these adapted subclones (60 SNVs/Mb) is less than the clonal mutation burden of many ultramutated tumors (> 100 SNVs/Mb). One-half of POLE-P286R tumors curated on cBioPortal (Cerami et al. 2012) carry clonal, secondary mutations in POLE (Figure 5G and Dataset S1). Intriguingly, four of these carry truncating POLE mutations including TCGA-IB-7651 (X1335_splice), whose mutation spectrum reflects synergy between POLE and MMR defects (Haradhvala et al. 2018). The short-read sequencing used for these samples does not reveal whether the mutations are colinear with P286R, but there are only two possibilities: the mutations either suppress the mutator or the WT allele. Yeast cells that only express pol2-P301R have a 27-fold-higher mutation rate (1.4 × 10−4 CanR mutants/division) than cells that are heterozygous for the allele (Kane and Shcherbakova 2014). Thus, truncating POLE mutations in the WT allele may enhance the POLE-P286R mutator phenotype. The TCGA-IB-7651 tumor carries four other mutations in POLE besides the truncating mutation, making it a reasonable candidate for mutator suppression regardless of which POLE allele carries the truncating mutation. The mutational signatures of the other three tumors with truncating POLE mutations (TCGA-AX-A05Z, P0010967, and P0005824) have high mutation loads but lack a mutational signature of combined POLE and MMR defects (Haradhvala et al. 2018). Thus, these truncating mutations may enhance the P286R mutator phenotype. Given their high mutation loads, we predict that these tumors may also experience selection pressure for P286R suppressor mutations. However, since POLE is essential, adaptations in these types of tumors may be limited to missense mutations that restore Polε fidelity.
The adaptive missense mutations in our study encode amino acid changes that may inactivate the polymerase or attenuate the mutator phenotype. They group together in different locations within the Pol2p structure (Swan et al. 2009; Parkash et al. 2019), suggesting discrete mechanisms of suppression (see Figure S9 for details). The most striking antimutator candidate is D306Y, which is adjacent to P301R in the exonuclease structure (Figure S9B) and may even be allele-specific. Together, the truncating and missense mutations argue that POLE cancer cells have numerous ways to escape EEX. The frequent emergence of independent eex mutants in the same evolved culture indicates that polyclonal adaptation may be common in POLE tumors, potentially obscuring the critical role of suppressor mutations in preventing EEX of the cancer and preserving genetic diversity for tumor evolution.
Our studies also indicate that there may be other changes that attenuate the mutator phenotype in diploid cells. In yeast, deletion of DUN1 suppresses Polδ (Datta et al. 2000; Mertz et al. 2015) and Polε mutator phenotypes (Williams et al. 2015) in haploid MMR-proficient cells. The dun1Δ antimutator effect involves a reduction in dNTP pools, since restoration of dNTPs by deletion of SML1, a Dun1 target, restores the mutator phenotype (Williams et al. 2015). Like intragenic mutations in polymerase genes predicted to compromise DNA binding, mutations that lower dNTP pools may enhance polymerase dissociation from the primer•template, permitting extrinsic proofreading (Williams et al. 2015). Although heterozygous variants in DUN1 are observed in some pol3-01/pol3-01 msh6Δ/msh6Δ lineages, they do not explain the observed variation in mutation rates between subclones. While MMR-proficient pol3-01 cells appear to activate the checkpoint (Datta et al. 2000), this activation may depend, in part, on processing of mismatches by MMR (Reha-Krantz et al. 2011). Therefore, in the absence of MMR, the contribution of Dun1p to the pol3-01 mutator phenotype may be substantially reduced. Thus, we may need to look elsewhere for antimutators. Recent evolution studies in diploid yeast have pointed to unconventional mutator phenotypes caused by heterozygous mutations in housekeeping genes (Coelho et al. 2019). The vast phenotypic space created by heterozygous mutations and their epistatic interactions remains largely unexplored. If mutation rate can be enhanced by such mutations, perhaps it may be attenuated as well.
Acknowledgments
We thank Maitreya Dunham for use of her flow cytometer and horn sonicator; Iñigo Martincorena for help implementing dNdscv; and Maitreya Dunham and Scott Kennedy for critically reading the manuscript. We acknowledge funding from the National Institute of General Medical Sciences (NIGMS) via grant R01GM118854, a Cellular and Molecular Biology training grant to M.B.L. (T32GM7270-39), and supported for the University of Washington Molecular Medicine Training grant for I.T.D. (T32GM095421). M.B.L. was also supported by the Howard Hughes Medical Institute (HHMI) Gilliam Fellowship for Advanced Study and the University of Washington (UW) Graduate Opportunities and Minority Achievement Program (UW GO-MAP Bank of America Fellowship). I.D.T was also supported by a National Institute on Aging (NIA) Genetic Approaches to Aging Training grant (T32AG000057). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the NIA, the NIGMS, or the HHMI. The authors declare that there are no competing financial or nonfinancial interests.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12276461.
Communicating editor: J. Nickoloff
Literature Cited
- Adams W. T., and Skopek T. R., 1987. Statistical test for the comparison of samples from mutational spectra. J. Mol. Biol. 194: 391–396. 10.1016/0022-2836(87)90669-3 [DOI] [PubMed] [Google Scholar]
- Agresti A., Wackerly D., and Boyett J. M., 1979. Exact conditional tests for cross-classifications: approximation of attained significance levels. Psychometrika 44: 75–83. 10.1007/BF02293786 [DOI] [Google Scholar]
- Albertson T. M., Ogawa M., Bugni J. M., Hays L. E., Chen Y. et al. , 2009. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. USA 106: 17101–17104. 10.1073/pnas.0907147106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L. B., Kim J., Haradhvala N. J., Huang M. N., Tian Ng A. W. et al. , 2020. The repertoire of mutational signatures in human cancer. Nature 578: 94–101. 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbari S. R., Kane D. P., Moore E. A., and Shcherbakova P. V., 2018. Functional analysis of cancer-associated DNA polymerase epsilon variants in Saccharomyces cerevisiae. G3 (Bethesda) 8: 1019–1029. 10.1534/g3.118.200042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulock C. R., Xing X., and Shcherbakova P. V., 2020. DNA polymerase δ proofreads errors made by DNA polymerase ε. Proc. Natl. Acad. Sci. USA 117: 6035–6041. 10.1073/pnas.1917624117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B. B., Light N., Fabrizio D., Zatzman M., Fuligni F. et al. , 2017. Comprehensive analysis of hypermutation in human cancer. Cell 171: 1042–1056.e10. 10.1016/j.cell.2017.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network , 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network; Kandoth C., Schultz N., Cherniack A. D., Akbani R. et al. , 2013. Integrated genomic characterization of endometrial carcinoma. Nature 497: 67–73 (erratum: Nature 500: 242). 10.1038/nature12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O. et al. , 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church D. N., Briggs S. E. W., Palles C., Domingo E., Kearsey S. J. et al. , 2013. DNA polymerase ɛ and δ exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 22: 2820–2828. 10.1093/hmg/ddt131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M. C., Pinto R. M., and Murray A. W., 2019. Heterozygous mutations cause genetic instability in a yeast model of cancer evolution. Nature 566: 275–278. 10.1038/s41586-019-0887-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Schmeits J. L., Amin N. S., Lau P. J., Myung K. et al. , 2000. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3–01 mutants. Mol. Cell 6: 593–603. 10.1016/S1097-2765(00)00058-7 [DOI] [PubMed] [Google Scholar]
- Dennis D. G., McKay-Fleisch J., Eitzen K., Dowsett I., Kennedy S. R. et al. , 2017. Normally lethal amino acid substitutions suppress an ultramutator DNA Polymerase δ variant. Sci. Rep. 7: 46535 10.1038/srep46535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska I. J., and Schaaper R. M., 1995. Effects of Escherichia coli dnaE antimutator alleles in a proofreading-deficient mutD5 strain. J. Bacteriol. 177: 5979–5986. 10.1128/JB.177.20.5979-5986.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska I. J., and Schaaper R. M., 1996. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc. Natl. Acad. Sci. USA 93: 2856–2861. 10.1073/pnas.93.7.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K. J., Buskirk S. W., Vignogna R. C., Marad D. A., and Lang G. I., 2018. Adaptive genome duplication affects patterns of molecular evolution in Saccharomyces cerevisiae. PLoS Genet. 14: e1007396 10.1371/journal.pgen.1007396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood C. L., Rodriguez G. P., Bao G., Shockley A. H., Kow Y. W. et al. , 2015. Replicative DNA polymerase δ but not ε proofreads errors in cis and in trans. PLoS Genet. 11: e1005049 10.1371/journal.pgen.1005049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Storchova Z., and Pellman D., 2007. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17: 157–162. 10.1016/j.gde.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Gerstein A. C., Chun H. J., Grant A., and Otto S. P., 2006. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2: e145 10.1371/journal.pgen.0020145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsby R. E., Lawrence N. A., Hays L. E., Olmsted E. A., Chen X. et al. , 2001. Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat. Med. 7: 638–639. 10.1038/88963 [DOI] [PubMed] [Google Scholar]
- Haber J. E., 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. 10.1534/genetics.111.134577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haradhvala N. J., Kim J., Maruvka Y. E., Polak P., Rosebrock D. et al. , 2018. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat. Commun. 9: 1746 10.1038/s41467-018-04002-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari Y., Ram Y., Rappoport N., Hadany L., and Kupiec M., 2018. Spontaneous changes in ploidy are common in yeast. Curr. Biol. 28: 825–835.e4. 10.1016/j.cub.2018.01.062 [DOI] [PubMed] [Google Scholar]
- Herr A. J., Ogawa M., Lawrence N. A., Williams L. N., Eggington J. M. et al. , 2011a Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 7: e1002282 10.1371/journal.pgen.1002282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A. J., Williams L. N., and Preston B. D., 2011b Antimutator variants of DNA polymerases. Crit. Rev. Biochem. Mol. Biol. 46: 548–570. 10.3109/10409238.2011.620941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A. J., Kennedy S. R., Knowels G. M., Schultz E. M., and Preston B. D., 2014. DNA replication error-induced extinction of diploid yeast. Genetics 196: 677–691. 10.1534/genetics.113.160960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel K. P., de Borja R., Henninger E. E., Campbell B. B., Ungerleider N. et al. , 2018. Explosive mutation accumulation triggered by heterozygous human Pol ε proofreading-deficiency is driven by suppression of mismatch repair. Elife 7: e32692 10.7554/eLife.32692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., and Mitchell A., 1994. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Kane D. P., and Shcherbakova P. V., 2014. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 74: 1895–1901. 10.1158/0008-5472.CAN-13-2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S. R., Schultz E. M., Chappell T. M., Kohrn B., Knowels G. M. et al. , 2015. Volatility of mutator phenotypes at single cell resolution. PLoS Genet. 11: e1005151 10.1371/journal.pgen.1005151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. B., Dowsett I. T., Carr D. T., Wasko B. M., Stanton S. G. et al. , 2019. Defining the impact of mutation accumulation on replicative lifespan in yeast using cancer-associated mutator phenotypes. Proc. Natl. Acad. Sci. USA 116: 3062–3071. 10.1073/pnas.1815966116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.-D., Cuevas I., Zhang M., Lu C., Alam M. M. et al. , 2018. Polymerase-mediated ultramutagenesis in mice produces diverse cancers with high mutational load. J. Clin. Invest. 128: 4179–4191. 10.1172/JCI122095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A., 2016. Human cancers express a mutator phenotype: hypothesis, origin, and consequences. Cancer Res. 76: 2057–2059. 10.1158/0008-5472.CAN-16-0794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A., Springgate C. F., and Battula N., 1974. Errors in DNA replication as a basis of malignant changes. Cancer Res. 34: 2311–2321. [PubMed] [Google Scholar]
- Lynch H. T., Lynch P. M., Lanspa S. J., Snyder C. L., Lynch J. F. et al. , 2009. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 76: 1–18. 10.1111/j.1399-0004.2009.01230.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable B. K., and Otto S. P., 2001. Masking and purging mutations following EMS treatment in haploid, diploid and tetraploid yeast (Saccharomyces cerevisiae). Genet. Res. 77: 9–26. 10.1017/S0016672300004821 [DOI] [PubMed] [Google Scholar]
- Martincorena I., Raine K. M., Gerstung M., Dawson K. J., Haase K. et al. , 2017. Universal patterns of selection in cancer and somatic tissues. Cell 171: 1029–1041.e21 [corrigenda: Cell 173: 1823 (2018)]. 10.1016/j.cell.2017.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz T. M., Sharma S., Chabes A., and Shcherbakova P. V., 2015. Colon cancer-associated mutator DNA polymerase δ variant causes expansion of dNTP pools increasing its own infidelity. Proc. Natl. Acad. Sci. USA 112: E2467–E2476. 10.1073/pnas.1422934112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., and Sugino A., 1994. The 3′→5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 242: 289–296. 10.1007/BF00280418 [DOI] [PubMed] [Google Scholar]
- Morrison A., Johnson A. L., Johnston L. H., and Sugino A., 1993. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 12: 1467–1473. 10.1002/j.1460-2075.1993.tb05790.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palles C., Cazier J.-B., Howarth K. M., Domingo E., Jones A. M. et al. , 2012. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 45: 136–144 (erratum: Nat. Genet. 45: 713). 10.1038/ng.2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkash V., Kulkarni Y., ter Beek J., Shcherbakova P. V., Kamerlin S. C. L. et al. , 2019. Structural consequence of the most frequently recurring cancer-associated substitution in DNA polymerase ε. Nat. Commun. 10: 373 10.1038/s41467-018-08114-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N., Gakhar L., Winistorfer S. C., Sunshine A. B., Rich M. et al. , 2013. The yeast Alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev. Cell 25: 520–533. 10.1016/j.devcel.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz L. J., 1988. Amino acid changes coded by bacteriophage T4 DNA polymerase mutator mutants. Relating structure to function. J. Mol. Biol. 202: 711–724. 10.1016/0022-2836(88)90552-9 [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Siddique M. S. P., Murphy K., Tam A., O’Carroll M. et al. , 2011. Drug sensitive DNA polymerase δ reveals a role for mismatch repair in checkpoint activation in yeast. Genetics 189: 1211–1224. 10.1534/genetics.111.131938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., 1993. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 268: 23762–23765. [PubMed] [Google Scholar]
- Sherman F., 2002. Getting started with yeast, pp. 3–41 in Part B: Guide to Yeast Genetics and Molecular and Cell Biology, edited by Guthrie C., and Fink G. R.. Academic Press, San Diego: 10.1016/S0076-6879(02)50954-X [DOI] [Google Scholar]
- Shlien A., Campbell B. B., de Borja R., Alexandrov L. B., Merico D. et al. , 2015. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet. 47: 257–262. 10.1038/ng.3202 [DOI] [PubMed] [Google Scholar]
- Storchová Z., Breneman A., Cande J., Dunn J., Burbank K. et al. , 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443: 541–547. 10.1038/nature05178 [DOI] [PubMed] [Google Scholar]
- Strathern J., Hicks J., and Herskowitz I., 1981. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J. Mol. Biol. 147: 357–372. 10.1016/0022-2836(81)90488-5 [DOI] [PubMed] [Google Scholar]
- Swan M. K., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K., 2009. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 16: 979–986. 10.1038/nsmb.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataram S., Dunn B., Li Y., Agarwala A., Chang J. et al. , 2016. Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell 166: 1585–1596.e22. 10.1016/j.cell.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L. N., Herr A. J., and Preston B. D., 2013. Emergence of DNA polymerase ε antimutators that escape error-Induced extinction in yeast. Genetics 193: 751–770. 10.1534/genetics.112.146910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L. N., Marjavaara L., Knowels G. M., Schultz E. M., Fox E. J. et al. , 2015. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants. Proc. Natl. Acad. Sci. USA 112: E2457–E2466. 10.1073/pnas.1422948112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing X., Kane D. P., Bulock C. R., Moore E. A., Sharma S. et al. , 2019. A recurrent cancer-associated substitution in DNA polymerase ε produces a hyperactive enzyme. Nat. Commun. 10: 374 10.1038/s41467-018-08145-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R., Miyashita K., Inoue M., Shimamoto A., Yan Z. et al. , 2011. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur. J. Hum. Genet. 19: 320–325. 10.1038/ejhg.2010.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., 2015. A new practical guide to the Luria–Delbrück protocol. Mutat. Res. Fundam. Mol. Mech. Mutagen. 781: 7–13. 10.1016/j.mrfmmm.2015.08.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. All whole-genome sequencing data are available from the Sequence Read Archive (BioProject identifier: PRJNA629499). Supplemental Dataset S1 contains flow cytometry results, mutation rates, and modeling of diploid mutator evolution. Supplemental Dataset S2 describes whole-genome sequencing results of diploid mutator strains. Supplemental Dataset S1 and S2, along with all computer scripts used in this work, can also be found at GitHub (https://github.com/mutatorUW/MutatorEvolution). Supplemental material available at figshare: https://doi.org/10.25386/genetics.12276461.