Abstract
Maternally transmitted Wolbachia bacteria infect about half of all insect species. They usually show imperfect maternal transmission and often produce cytoplasmic incompatibility (CI). Irrespective of CI, Wolbachia frequencies tend to increase when rare only if they benefit host fitness. Several Wolbachia, including wMel that infects Drosophila melanogaster, cause weak or no CI and persist at intermediate frequencies. On the island of São Tomé off West Africa, the frequencies of wMel-like Wolbachia infecting Drosophila yakuba (wYak) and Drosophila santomea (wSan) fluctuate, and the contributions of imperfect maternal transmission, fitness effects, and CI to these fluctuations are unknown. We demonstrate spatial variation in wYak frequency and transmission on São Tomé. Concurrent field estimates of imperfect maternal transmission do not predict spatial variation in wYak frequencies, which are highest at high altitudes where maternal transmission is the most imperfect. Genomic and genetic analyses provide little support for D. yakuba effects on wYak transmission. Instead, rearing at cool temperatures reduces wYak titer and increases imperfect transmission to levels observed on São Tomé. Using mathematical models of Wolbachia frequency dynamics and equilibria, we infer that temporally variable imperfect transmission or spatially variable effects on host fitness and reproduction are required to explain wYak frequencies. In contrast, spatially stable wSan frequencies are plausibly explained by imperfect transmission, modest fitness effects, and weak CI. Our results provide insight into causes of wMel-like frequency variation in divergent hosts. Understanding this variation is crucial to explain Wolbachia spread and to improve wMel biocontrol of human disease in transinfected mosquito systems.
Keywords: cytoplasmic incompatibility, Drosophila yakuba, Drosophila santomea, endosymbiosis, host–microbe interactions
MATERNALLY transmitted Wolbachia bacteria are the most widespread group of intracellular symbionts, infecting about half of all insect species, as well as other arthropods and nematodes (Werren et al. 2008; Zug and Hammerstein 2012; Weinert et al. 2015). Wolbachia often manipulate host reproduction to favor infected females (Rousset et al. 1992; Hoffmann and Turelli 1997; Hurst and Jiggins 2000). For example, Wolbachia like wRi, which infects Drosophila simulans, cause strong cytoplasmic incompatibility (CI) that reduces egg hatch when uninfected females mate with Wolbachia-infected males (Hoffmann and Turelli 1997)—infected females are protected from CI (Shropshire et al. 2018), providing them with a relative fitness advantage. Many other Wolbachia do not strongly manipulate host reproduction yet persist in nature. These include wMau in D. mauritiana (Meany et al. 2019), wAu in D. simulans (Hoffmann et al. 1996), wSuz in D. suzukii (Hamm et al. 2014; Cattel et al. 2016), wMel in D. melanogaster (Kriesner et al. 2016), and Wolbachia variants infecting D. yakuba-clade hosts (wYak in D. yakuba, wSan in D. santomea, and wTei in D. teissieri) that diverged from wMel in only the last 30,000 years (Charlat et al. 2004; Zabalou et al. 2004; Cooper et al. 2017, 2019).
Population frequency dynamics and equilibria of Wolbachia can be approximated with three parameters: (1) the proportion of uninfected ova produced by infected females (μ; i.e., imperfect maternal transmission), (2) the fitness of infected females relative to uninfected females (F; i.e., components of host fitness like fecundity), and (3) the relative egg hatch of uninfected eggs fertilized by infected males (H; i.e., the severity of CI) (Hoffmann et al. 1990). When initially rare, Wolbachia must generate F(1–μ) > 1 to spread deterministically from low frequencies, regardless of whether they cause CI. The specific fitness benefits underlying low-frequency spread are poorly understood, but potential candidates include fecundity effects (Weeks et al. 2007), viral protection (Hedges et al. 2008; Teixeira et al. 2008; Osborne et al. 2009; Martinez et al. 2014), and nutrient provisioning (Brownlie et al. 2009; Hosokawa et al. 2010; Nikoh et al. 2014; Moriyama et al. 2015). Once infections become sufficiently common, strong CI drives Wolbachia to high equilibrium frequencies balanced by imperfect maternal transmission (Hoffmann et al. 1990; Barton and Turelli 2011), as observed for rapid wRi spread through global D. simulans populations (Turelli and Hoffmann 1991, 1995; Carrington et al. 2011; Kriesner et al. 2013). Conversely, Wolbachia that do not cause strong CI tend to occur at intermediate frequencies that fluctuate through time and space (wSuz, Hamm et al. 2014; wMel, Kriesner et al. 2016; D. yakuba-clade Wolbachia, Cooper et al. 2017), suggesting the parameters influencing Wolbachia spread must vary.
Imperfect maternal transmission has been documented for wRi and wAu in D. simulans and wMel in D. melanogaster in the field (Hoffmann et al. 1990, 1998; Turelli and Hoffmann 1995; Carrington et al. 2011). In contrast, maternal transmission is nearly perfect under laboratory conditions (Hoffmann et al. 1990; Meany et al. 2019), indicating that environmental conditions influence the fidelity of Wolbachia transmission. Transmission is predicted to depend on Wolbachia titer and localization within developing female oocytes (Ferree et al. 2005; Serbus and Sullivan 2007; Casper-Lindley et al. 2011; Serbus et al. 2015). Host diet influences Wolbachia titer and localization during late oogenesis (Serbus et al. 2015; Camacho et al. 2017; Christensen et al. 2019), suggesting that seasonal or spatial availability of preferred fruits (e.g., marula fruit for D. melanogaster or figs for D. santomea) could potentially affect maternal transmission (Cariou et al. 2001; Mansourian et al. 2018; Sprengelmeyer et al. 2020). Thermal conditions may directly alter Wolbachia transmission. For instance, stressful temperatures disrupt wMel transmission in transfected Aedes aegypti mosquitoes (Ross et al. 2017, 2019a; Foo et al. 2019), potentially reducing the efficacy of wMel biocontrol of human disease transmission (Hoffmann et al. 2011; van den Hurk et al. 2012; Aliota et al. 2016; Caragata et al. 2016; O’Neill 2018). The frequency of wMel in its natural D. melanogaster host declines at temperate latitudes in Eastern Australia and Eastern North America, which may be due to a combination of Wolbachia fitness costs and reduced maternal transmission in cold environments (Kriesner et al. 2016). Together, these data suggest nutritional and thermal variation may perturb Wolbachia maternal transmission, particularly for wMel-like Wolbachia.
wMel-like Wolbachia infect the sister-species D. yakuba and D. santomea, which diverged from model wMel-infected D. melanogaster up to 13 million years ago (MYA; Tamura et al. 2004). D. yakuba is a human commensal distributed throughout sub-Saharan Africa that is generally found in open habitats, but absent in rainforests (Llopart et al. 2005a,b; Cooper et al. 2018). On Pico de São Tomé off West Africa, D. yakuba is found in open disturbed areas below 1450 m, whereas endemic D. santomea is found between 1153 and 1800 m in montane mist and rain forests. The distributions of D. yakuba and D. santomea overlap in the midlands of Pico de São Tomé where they hybridize (Lachaise et al. 2000; Comeault et al. 2016; Cooper et al. 2017; Turissini and Matute 2017). Phylogenomic analyses indicate that wMel-like Wolbachia spread among D. yakuba-clade host species via introgressive transfer in the last 2500 to 4500 years (Cooper et al. 2019). wYak and wSan share very high sequence similarity across their genomes (0.0017% third-position pairwise differences; 643 genes across 644,586 bp; Cooper et al. 2019), and, like wMel, they cause weak CI potentially modulated by host factors (Reynolds and Hoffmann 2002; Cooper et al. 2017). Relative to wMel (Beckmann et al. 2017; LePage et al. 2017), these Wolbachia have an additional set of loci implicated in CI that they acquired horizontally from divergent B-group Wolbachia (Cooper et al. 2019). Infection frequencies on São Tomé are temporally variable in idiosyncratic ways, such that wYak frequency increased from 2001 (0.40) to 2009 (0.76; P < 0.001) and wSan decreased from 2009 (0.77) to 2015 (0.37; P < 0.0001), during a period where wYak frequencies were stable. wYak frequencies in 2009 also varied spatially between São Tomé and the nearby island of Bioko (0.03; P < 0.001) (Cooper et al. 2017). This variation is similar to sporadic fluctuations of wMel observed in Africa (Kriesner et al. 2016), and suggests that wMel-like Wolbachia with high sequence similarity may behave differently across different abiotic conditions and host backgrounds.
The causes of wMel-like Wolbachia frequency fluctuations remain mostly unknown. Here, we use a combination of field, laboratory, and mathematical analyses to dissect the contributions of imperfect maternal transmission, Wolbachia effects on host fitness, and CI to spatiotemporal wYak and wSan frequency variation on São Tomé. In 2018, we generated concurrent estimates of imperfect maternal transmission (μ) and infection frequencies (p) of wYak and wSan along an altitudinal transect on Pico de São Tomé to test whether field estimates of μ predict spatial variation in p. In the laboratory, we then tested how environmental conditions and host genetic factors contribute to variation in imperfect maternal transmission. Finally, we used mathematical models of Wolbachia frequency dynamics and equilibria to better understand wMel-like frequency variation on São Tomé. Our results provide insight into the basis of wMel-like Wolbachia frequency fluctuations in two sister species that diverged from model D. melanogaster up to 13 MYA (Tamura et al. 2004).
Materials and Methods
Estimating Wolbachia frequencies and imperfect maternal transmission
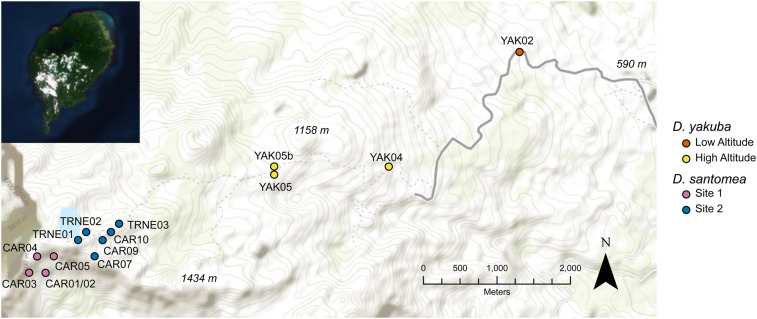
To determine spatial variation in wYak and wSan frequencies, we sampled female D. yakuba (N = 81) and D. santomea (N = 78) from 15 trapping sites on São Tomé in 2018 (Figure 1 and Supplemental Material, Table S1). Flies were sampled using fruit traps and by sweeping nets over piles of local jackfruit and bananas. Flies sampled from the four D. yakuba traps were assigned to a low altitude (N = 40) and high altitude (N = 41) group. The low altitude region (590 m) is characterized by relatively hot and dry conditions, whereas high altitude (900–1104 m) is generally cooler and more humid. The high altitude sites are contiguous with the known D. yakuba–D. santomea hybrid zone (Comeault et al. 2016; Turissini and Matute 2017). D. santomea individuals sampled from 11 traps were initially assigned into two regional groups based on geography such that site one (N = 24) and site two (N = 54) were collected on opposite sides of a small mountainous region (Figure 1). Subsequent analyses found no regional variation in p or μ for wSan (see below).
Figure 1.
Trapping sites on the island of São Tomé (inset) off West Africa. Points denote individual trapping sites (Table S1), which are grouped into regions based on altitude and geography. Points are colored according to their regional grouping for each species. D. santomea sites were pooled for all analyses. Topographic contour lines delineate altitude along the Pico de São Tomé transect.
Wild-caught females were placed individually into vials and allowed to lay eggs in the field on instant media (Nutri-Fly Instant Formulation, Genesee Scientific) supplemented with active yeast. This enabled us to establish isofemale lines and to sample F1 offspring that we preserved in ethanol upon emergence in the field. We used PCR to assess the infection status of each line from each site. The proportion of infected isofemale lines served as our estimate of p for each site, region, and species. To estimate μ, we assayed the Wolbachia infection status of newly emerged F1 offspring, and then determined the proportion of uninfected adults produced by each wild-caught Wolbachia-infected female in the field. Importantly, because we sampled infected females from nature, the infection status of the male in these crosses is unknown. In crosses with infected males, uninfected ova produced by infected females could be lost if they are susceptible to CI [see Equation 2 from Turelli and Hoffmann (1995) for further dissection of this point]. All PCR assays used primers for the Wolbachia surface protein (wsp), and a second set of primers for the arthropod-specific 28S rDNA, which served as a positive control (Cooper et al. 2017; Meany et al. 2019). There is no evidence that D. yakuba-clade hosts are infected with other unaccounted for heritable symbionts, such as Spiroplasma (Mateos et al. 2006).
All statistical analyses were performed in R (R Core Team 2018). For each species, we first generated pooled estimates of p and μ that included all 2018 trapping sites. We then examined variation in p and μ by region and by trapping site. Assuming a binomial distribution, we estimated exact 95% binomial confidence intervals for p for each species/region/trapping site using the “binconf” function in the package Hmisc (Harrell and Dupont 2018). We used Fisher’s exact tests to assess pairwise differences in p among species and among regions. Estimates of p from 2018 were also compared to previously published data on São Tomé from Cooper et al. (2017); however, these prior estimates lacked adequate sampling to examine regional variation in wYak and wSan frequencies.
We estimated μ for each species/region/trapping site as the weighted mean of uninfected offspring produced by infected mothers, across all infected families, weighted by the number of F1s in each family. We then estimated 95% bias-corrected and accelerated bootstrap (BCa) confidence intervals using the “boot.ci” function and 5000 iterations in the boot package in R (Canty and Ripley 2017). BCa confidence intervals are calculated using the two-sample acceleration constant given by equation 15.36 of Efron and Tibshirani (1993). Prior work has demonstrated substantial heterogeneity in μ among wild-caught wRi-infected D. simulans (Turelli and Hoffmann 1995; Carrington et al. 2011) and wSuz-infected D. suzukii females (Hamm et al. 2014). Therefore, we visualized the full distribution of μ values for wYak and wSan, and used Kruskal–Wallis tests to assess heterogeneity in μ among wild-caught females for each species (Fox and Weisberg 2011). We used Kruskal–Wallis tests to assess differences in μ among trapping sites and Wilcoxon rank sum tests to assess pairwise differences in μ between species and between regions. Prior work also suggests maternal transmission of Wolbachia infecting D. pseudotakahashii is sex-biased, with perfect transmission to female progeny and reduced transmission to males (Richardson et al. 2019). Thus, we also evaluated whether imperfect transmission varied between female and male progeny for wYak or wSan.
Determining environmental effects on imperfect wYak maternal transmission
Our 2018 sampling revealed that transmission rates vary spatially on São Tomé for wYak but not wSan (see Results). Specifically, μ was significantly higher for wYak at high altitude, where conditions are relatively cool compared to low altitude. Wolbachia that are imperfectly transmitted in the field are perfectly, or near perfectly, transmitted under standard laboratory conditions (Hoffmann et al. 1990; Meany et al. 2019), suggesting that abiotic conditions may contribute to imperfect transmission in nature. Moreover, Turelli and Hoffmann (1995) found evidence that wRi maternal transmission becomes near perfect when isofemale lines of D. simulans are cultured in the laboratory for six months. Environmental conditions can also influence other traits, like cellular Wolbachia titer and localization in developing host oocytes, both of which are predicted to influence μ (Clancy and Hoffmann 1998; Serbus and Sullivan 2007; Ross et al. 2017; Christensen et al. 2019; Ross et al. 2019a). Thus, we manipulated environmental conditions in the laboratory in an attempt to determine the basis of variable imperfect wYak transmission on São Tomé.
We first tested whether changes to the standard fly food diet alter wYak transmission rates. Female D. melanogaster reared on a yeast-enriched diet for two days have reduced cellular wMel titer in developing stage 10a oocytes (Serbus et al. 2015; Christensen et al. 2019). Reductions in oocyte titer are predicted to generate imperfect transmission, although this hypothesis has not been tested. We reared an infected D. yakuba isofemale line (L5; Table S2) at 25° on a standard food diet and then placed newly emerged virgin females individually with two males of the same infected genotype in vials containing yeast-enriched food. Yeast-enriched food was prepared by mixing 1.5 ml of heat-killed yeast paste into 3.5 ml of standard food. Each subline was allowed to lay eggs for one week, and, in the following generation, newly emerged male and female F1 offspring were screened for wYak infection individually using PCR as described above.
Because imperfect maternal transmission was greatest at high altitude where conditions are generally cool (Table S1), we next tested whether rearing D. yakuba in cold conditions (20°) perturbs maternal transmission relative to standard laboratory conditions (25°). World Bioclim data (www.worldclim.org) indicate that 20° is well within the average temperature range of our trapping sites on São Tomé (Table S1), and previous work has demonstrated that D. yakuba experience reduced larval survival, egg hatch, and longevity at temperatures <20° (Matute et al. 2009; Cooper et al. 2018). Thus, we reared the L5 isofemale line in a 20° incubator under a 12L:12D light cycle (Pericival Model I-36LL). Virgin females were placed individually with two males into vials and allowed to lay eggs for one week. Newly emerged male and female F1s from each subline were screened for wYak infection using PCR as described above.
The 20° cold treatment generated a significant increase in imperfect maternal transmission, whereas yeast-enriched food did not (see Results). We therefore investigated whether rearing L5 females at 20° reduces wYak titer, a trait predicted to determine Wolbachia transmission. We used qPCR to compare titer in females reared at 20° to those reared under standard conditions at 25°. Females from each temperature treatment were aged to three days and then homogenized together in groups of 10. The final sample included 10 biological replicates of each temperature treatment. DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen). We used a Stratagene Mx3000P (Agilent Technologies) and qPCR primers designed for D. yakuba to amplify a Drosophila-specific locus (Rpl32; F: 5′-CCGCTTCAAGGGACAGTATC-3′, R: 5′-CGATCTCCTTGCGCTTCTTG-3′) and a Wolbachia-specific locus (ftsZ; F: 5′-ATCCTTAACTGCGGCTCTTG-3′, R: 5′-TTCATCACAGCAGGAATGGG-3′) with the following cycling conditions: 50° for two min, 95° for two min, and then 40 cycles of 95° for 15 sec, 58° for 15 sec, and 72° for one min. Efficiency curves were generated to confirm that primer efficiency was between 90 and 100% for Rpl32 (96.58%) and ftsZ (93.24%). We used the average cycle threshold (Ct) value of three technical replicates for each sample. We estimated relative Wolbachia density as where (Pfaffl 2001). We then used a Wilcoxon rank sum test to assess differences in titer between females reared at 20° and 25°.
Dissecting Wolbachia and host effects on wYak maternal transmission
We next investigated the contributions of wYak and D. yakuba genomes to imperfect wYak transmission. Prior work indicates very little differentiation among wYak genomes from West Africa (0.0007% third-position pairwise differences; Cooper et al. 2019), suggesting wYak genomic variation may have little influence on variation in imperfect wYak transmission. In contrast, host genomes have been shown to influence Wolbachia titer in developing host oocytes, which is predicted to determine transmission fidelity (Serbus and Sullivan 2007; Funkhouser-Jones et al. 2018). The genomes of D. yakuba sampled from low and high altitudes on São Tomé are weakly, but significantly, differentiated (FST = 0.0503, P < 0.001; Comeault et al. 2016; Turissini and Matute 2017). While we are ignorant of the type of host factors that modify Wolbachia titer and transmission in Drosophila, we characterized allele frequency and fixed differences between D. yakuba at low and high altitudes. In addition to this genomic characterization, we explicitly tested for host and Wolbachia contributions to variation in μ using genotypes with reciprocally introgressed D. yakuba and wYak genomes.
Genomic reads were obtained from the published data of Turissini and Matute (2017) for D. yakuba from low (N = 7) and high (N = 22) altitude on São Tomé. As described in Turissini and Matute (2017), reads were mapped to version 1.04 of the D. yakuba reference genome (Drosophila 12 Genomes Consortium et al. 2007) using bwa version 0.7.12 (Li and Durbin 2010). We merged BAM files with SAMtools version 0.1.19 (Li et al. 2009). Reads were then remapped locally in the merged BAM files using GATK version 3.2-2 using the RealignerTargetCreator and IndelRealigner functions (McKenna et al. 2010; DePristo et al. 2011). We used the GATK UnifiedGenotyper with the parameter het = 0.01, and applied QD = 2.0, FS_filter = 60.0, MQ_filter = 30.0, MQ_Rank_Sum_filter = −8.0 to the resulting vcf file. Sites were excluded if the coverage was <5 or the coverage was greater than the 99th quantile of the distribution of genomic coverage for each line, or if the SNP failed to pass any of the GATK filters. All polymorphic sites from the resulting vcf file were stored in a MySQL table for analyses.
For each SNP, we calculated the difference in allele frequencies between low vs. high altitude D. yakuba on São Tomé. We summarized mean allelic differences along each chromosome using a sliding widow with 5-kb increments. We calculated the mean and SD of the full distribution of allele frequency differences using the function “tapply” in R. We also identified fixed differences across the genome. Finally, we tested whether the mean allele frequency difference observed along the genome differed from zero using a parametric Welch test and a nonparametric Wilcoxon rank sum test, which both agreed. We generated a normal distribution with 6,528,464 observations (the number of polymorphic sites) using the R function “rnorm” matching the same SD observed in the empirical genome-wide data but centered on zero. We used the function “compare.2.vectors” in the afex package (Singmann et al. 2016) to compare the observed and simulated allele frequency distributions.
To explicitly dissect wYak and D. yakuba contributions to variation in μ, we reciprocally introgressed wYak and D. yakuba genomes using two infected D. yakuba isofemale lines as starting material (L42 and L48; Table S2). We first crossed L42 females with L48 males, and then backcrossed F1 females to L48 males. We repeated this cross for five generations to generate the L48L42 genotype (wYak variant denoted by superscript) composed of ∼97% of the L48 nuclear background and the introgressed L42 cytoplasm. We then generated the reciprocal genotype (L42L48) using the same approach. We measured μ for the two naturally sampled genotypes (L42 and L48) and for the two reciprocally introgressed genotypes (L42L48 and L48L42) reared under standard laboratory conditions (25°, 12L:12D). Virgin females were placed individually with two males of the same genotype into vials and allowed to lay eggs for one week. Newly emerged male and female F1 offspring were screened for wYak infection individually using PCR (as described above). We first tested whether μ varied among the four genotypes (L42, L48, L42L48, L48L42) using a Kruskal-Wallis test. Because μ takes the form of a frequency, we also used a generalized linear model (GLM) to assess the contributions of the host and Wolbachia genomes to imperfect transmission. We used the “glm” function in R to fit a GLM with a Poisson error structure and used the raw count data of uninfected F1s from each family as the dependent variable. We then included the host nuclear genome, the Wolbachia genome, and their interaction as independent variables. Both the Kruskal–Wallis test and the GLM revealed no evidence for host or Wolbachia effects on μ, so only the Kruskal–Wallis results are presented herein.
Modeling infection equilibria with field estimates of p and μ
To explore the relationship between our field estimates of p and μ on São Tomé, we considered an idealized discrete-generation model for Wolbachia frequency dynamics proposed by Hoffmann et al. (1990). This model incorporates μ, F (Wolbachia effects on host fitness), and H (the severity of CI) (Hoffmann and Turelli 1997)—we previously estimated the latter two parameters in the laboratory (Cooper et al. 2017). In cases of imperfect transmission, the model assumes that uninfected ova are equally susceptible to CI regardless of the infection status of their mother, which is supported by results from wRi-infected D. simulans (Turelli and Hoffmann 1995; Carrington et al. 2011). Embryos produced by uninfected mothers mated with infected fathers hatch with frequency , relative to the other three possible fertilizations, which are all considered equally compatible (Cooper et al. 2017). Thus, represents the severity of CI, or the frequency of unhatched eggs in pairings between uninfected females and infected males.
Prior estimates of CI indicate that wYak and wSan reduce egg-to-adult viability of uninfected females mated to infected males by about 10–15%, relative to compatible crosses (Cooper et al. 2017). We first ignored this weak CI and considered a stable equilibrium balanced by imperfect transmission (μ > 0) and positive fitness effects (F > 1). Benefits to host fitness have yet to be directly connected to low frequency Wolbachia spread in these systems or any others, but, like Hoffmann and Turelli (1997) conjectured for non-CI-causing Wolbachia, we assume F(1 – μ) must be >1 given the spread and persistence of wYak and wSan in nature (Cooper et al. 2017). When F(1 – μ) > 1, the stable equilibrium frequency is
| (1) |
In this case, increases from 0 toward 1 – μ as F increases from 1/(1 – μ). We incorporated our field estimates of imperfect maternal transmission to explore how μ and a range of positive Wolbachia fitness effects might explain observed estimates of p for wYak and wSan.
Next, we considered equilibria that incorporate CI (i.e., ) (Turelli and Hoffmann 1995; Kriesner et al. 2016). CI does not contribute to the initial spread of Wolbachia, yet strong-CI-causing strains like wRi spread rapidly to high equilibrium frequencies (Turelli and Hoffmann 1995; Kriesner et al. 2013). Thus, we assume CI-causing strains must also increase host fitness to initially spread from low frequency such that F(1 – μ) > 1. CI-causing infections that generate F(1 – μ) > 1 and Fμ < 1 produce a single stable equilibrium between 0 and 1 given by
| (2) |
Laboratory estimates of CI in the D. yakuba clade support for both wYak and wSan (Cooper et al. 2017). In contrast, the relatively high infection frequencies of wYak and wSan in 2018 (Figure 2 and Table 1) are a hallmark of strong CI (e.g., Turelli and Hoffmann 1995), implying these Wolbachia may cause stronger CI in the field. Thus, we considered stronger CI ( and ), in addition to our laboratory estimates of for each Wolbachia.
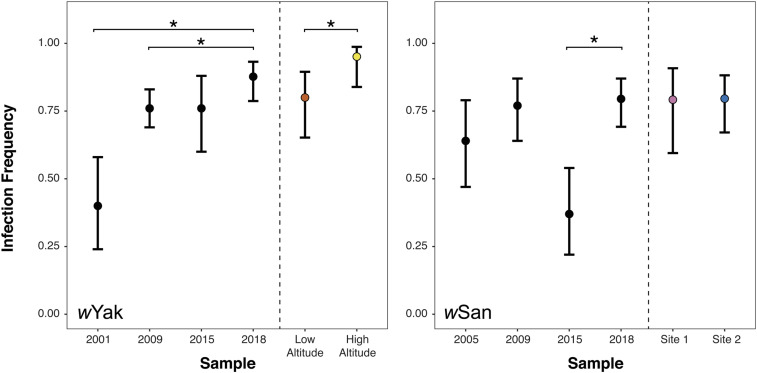
Figure 2.
Wolbachia infection frequencies on São Tomé. Data from the present study (2018) are compared to previous estimates of infection frequencies from 2001 to 2015 (Cooper et al. 2017). Error bars represent 95% binomial confidence intervals. To the right of vertical dashed lines, 2018 infection frequencies are separated by region for each species. Points are color-coded according to Figure 1. Asterisks indicate statistically significant differences between 2018 and prior years, or between regions in the 2018 sample at P < 0.05.
Table 1. Wolbachia infection frequencies in D. yakuba and D. santomea on São Tomé.
| Species | Year | N | Infected | p (confidence interval) |
|---|---|---|---|---|
| D. yakuba | 2001 | 35 | 14 | 0.40 (0.24, 0.58) |
| 2009 | 155 | 118 | 0.76 (0.69, 0.83) | |
| 2015 | 41 | 31 | 0.76 (0.60, 0.88) | |
| 2018 | 81 | 71 | 0.88 (0.79, 0.93) | |
| Low altitude | 40 | 32 | 0.80 (0.65, 0.90) | |
| High altitude | 41 | 39 | 0.95 (0.84, 0.99) | |
| D. santomea | 2005 | 39 | 25 | 0.64 (0.47, 0.79) |
| 2009 | 57 | 44 | 0.77 (0.64, 0.87) | |
| 2015 | 38 | 14 | 0.37 (0.22, 0.54) | |
| 2018 | 78 | 62 | 0.80 (0.69, 0.87) | |
| Site 1 | 24 | 19 | 0.79 (0.60, 0.91) | |
| Site 2 | 54 | 43 | 0.80 (0.67, 0.88) |
Sample sizes (N), infection frequencies (p), and exact 95% binomial confidence intervals are shown for each year. Data from 2001 to 2015 are from Cooper et al. (2017). For each species, infection frequencies are calculated for the pooled 2018 dataset and for each region.
Our analysis of wYak infection equilibria at high altitude revealed that biologically unrealistic parameter values (e.g., F > 4, ) are required to explain the observed combination of high wYak frequency [p = 0.95 (95% confidence interval: 0.84, 0.99)] and very imperfect wYak transmission [μ = 0.20 (95% BCa interval: 0.087, 0.364)]. We explored two additional processes the might contribute to this counterintuitive pattern at high altitude: (1) temporal variation in imperfect maternal transmission and (2) stochastic infection frequency fluctuations.
First, we consider temporal variation in μ. Discrete generation models use estimates of μ, F, and H to infer in the next host generation. We estimated μ and p concurrently in the field, assuming that our estimates of μ in the current generation reflect μ in the prior host generation. Turelli and Hoffmann (1995) found that imperfect wRi transmission decreased significantly from 0.044 (0.029, 0.090) in April to 0 (0.000, 0.007) in November of 1993 (P < 0.001) in an Ivanhoe, California population of D. simulans (Carrington et al. 2011), but it remains unknown whether Wolbachia transmission rates vary over shorter timescales within the same host population (e.g., across a single host generation). We repeated our mathematical analyses to consider the full range of μ point estimates across all trapping sites on São Tomé (Table S1). This range exceeds the seasonal variation in μ estimated for wRi in Ivanhoe, CA, and we reasoned that any generation-to-generation variation in imperfect transmission within a single location is likely less than the range of point estimates across all trapping sites. Thus, this analysis conservatively considers how different values of μ in the prior host generation would alter .
Next, we consider the potential for stochastic wYak frequency fluctuations to influence the patterns we observed. We explored the effect of host population size on wYak frequency fluctuations by modifying the model of Hoffmann and Turelli (1997) as described in Kriesner and Hoffmann (2018) to incorporate randomly variable outcomes for μ, F, and H, as well as for male mating success, crossing types arising from matings, female reproductive success, and the chance of viable embryos surviving to adulthood. We selected plausible combinations of μ, F, and H for wYak at low and high altitude (based on the abovementioned mathematical analysis; Table S5) to illustrate how stochasticity contributes to variation in wYak frequencies at . Monte Carlo simulations with 10,000 replicates of population events were enacted using functions in the PopTools package (Hood 2011) and the parameters described above (see Supplemental Methods for a detailed description). The initial generation for each replicate trial comprised a set number of adults with two infection status types: uninfected and wYak-infected. Each infection type consisted of an equal number of females and males, and we assumed females had premated with a male of the same infection type. Trials were initiated with the host population at and were run for 100 host generations. To evaluate how host population size affects stochasticity, we tested three different host census population sizes (N = 5,000, 50,000, and 500,000). Every 20 host generations, we calculated the mean infection frequency of the Monte Carlo simulations and associated 95% confidence intervals.
Data accessibility
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All supplemental materials, data, and scripts are available on figshare: https://doi.org/10.25386/genetics.12275513. Genomic data from Turissini and Matute (2017) are available at NCBI (Bioproject: PRJNA395473).
Results
wYak and wSan infection frequencies vary on São Tomé
wYak and wSan frequencies vary through time on São Tomé, and wYak frequencies vary spatially within São Tomé and between São Tomé and Bioko (Figure 2 and Table 1; Cooper et al. 2017). Across all D. yakuba trapping sites, our pooled estimate of wYak frequency in 2018 (p = 0.88) is significantly higher than our estimates from 2001 (p = 0.40; Fisher’s exact P < 0.0001) and 2009 (p = 0.76; Fisher’s exact P = 0.040), although 2018 did not differ from our 2015 estimate (p = 0.76; Fisher’s exact P = 0.120) (Cooper et al. 2017). In 2018, wYak frequencies varied regionally, such that wYak frequency was significantly higher at high altitude (p = 0.95; yellow sites in Figure 1) than at low altitude (p = 0.80; orange sites in Figure 1) along the Pico de São Tomé transect (Fisher’s exact P = 0.048). While this is the first demonstration of within-island spatial (altitudinal) variation in p, it is consistent with previous observations of geographic variation in wYak frequencies in 2009 between São Tomé (p = 0.76) and the neighboring island of Bioko (p = 0.03; P < 0.001) (Cooper et al. 2017).
Across all D. santomea trapping sites, the pooled frequency of wSan in 2018 was relatively high (p = 0.80), and did not differ statistically from past estimates on the island in 2005 (p = 0.64; Fisher’s exact P = 0.115) and 2009 (p = 0.77; Fisher’s exact P = 0.833). The 2018 estimate was significantly greater than the most recent 2015 estimate (p = 0.37; Fisher’s exact P < 0.0001), providing further support for temporal variation in wSan frequencies on São Tomé (Cooper et al. 2017). wSan frequencies varied between years when wYak frequencies were relatively stable, and vice versa, suggesting these nearly identical Wolbachia behave differently across host species backgrounds and/or abiotic environments. Unlike wYak, we did not find evidence for regional variation in wSan frequencies, such that site one (p = 0.79; purple sites in Figure 1) and site two (p = 0.80; blue sites in Figure 1) were statistically indistinguishable (Fisher’s exact P = 1).
Imperfect maternal transmission varies spatially for wYak, but not wSan
Imperfect maternal transmission was heterogeneous among wild-caught D. yakuba and D. santomea females. Wolbachia were usually transmitted with high fidelity, but some females exhibited substantial imperfect transmission (Figure 3). Accordingly, we found significant heterogeneity in μ among wild-caught females for D. yakuba (, P < 0.001) and D. santomea (, P < 0.001). Pooled μ values across all trapping sites (Table 2) did not differ between wYak (μ = 0.126) and wSan (μ = 0.068; W = 2355.5, P = 0.283). Contrary to our predictions, regional estimates of μ for wYak and wSan were not negatively correlated with p (F1,2 = 5.333, P = 0.147). This remained true when families were grouped by individual trapping sites (e.g., YAK02, CAR01, etc.) rather than region (F1,15 = 0.078, P = 0.784).
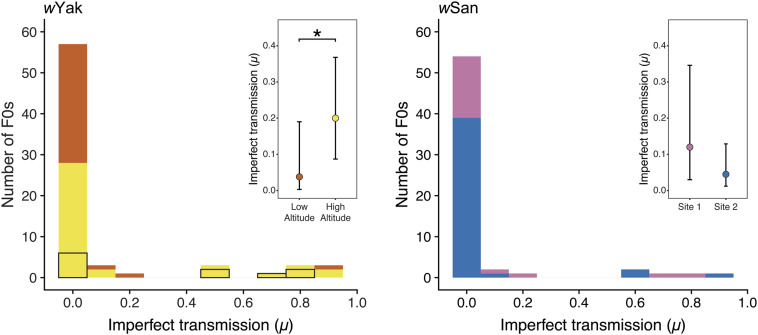
Figure 3.
Histogram of μ for wYak-infected D. yakuba (left) and wSan-infected D. santomea (right) females. The bars are color-coded by region as in Figure 1. For wYak, the contributions of F0 females from the anomalous site YAK05b are outlined in black (see main text). Insets show mean estimates of μ and associated 95% BCa confidence intervals for each region. Asterisk indicates a statistically significant difference in wYak μ between low and high altitude at P < 0.05.
Table 2. Imperfect maternal Wolbachia transmission by D. yakuba and D. santomea on São Tomé.
| Species | Year | Mean N F1s | μ (confidence interval) |
|---|---|---|---|
| D. yakuba | 2018 | 10.05 | 0.126 (0.062, 0.238) |
| Low altitude | 9.85 | 0.038 (0.003, 0.184) | |
| High altitude | 10.24 | 0.20 (0.087, 0.364) | |
| D. santomea | 2018 | 9.80 | 0.068 (0.027, 0.154) |
| Site 1 | 9.96 | 0.120 (0.032, 0.346) | |
| Site 2 | 8.64 | 0.045 (0.011, 0.130) |
Mean number of F1 offspring per family (Mean N F1s), weighted mean imperfect transmission (μ), and 95% BCa confidence intervals are shown for 2018. For each species, rates of maternal transmission are calculated for the pooled 2018 dataset and for each region.
Differences in wYak μ among individual trapping sites were not statistically significant ( P = 0.055; Table S1); however, when traps where grouped into altitudinal regions, we detected regional variation in wYak μ, such that maternal transmission was more imperfect at high altitude (μ = 0.20) relative to low altitude (μ = 0.038; W = 503, P = 0.045; Figure 3 and Table 2). This is surprising given that wYak frequencies were significantly higher at high altitude compared to low altitude. To explain this counterintuitive relationship between wYak μ and p, either μ must vary on short timescales within regions or F and/or H must vary between regions (see below). Finally, we found no evidence for differences in wYak transmission to male [μ = 0.192 (0.087, 0.346)] and female [μ = 0.204 (0.084, 0.386)] offspring at high altitude (W = 722.5, P = 0.994), or to males [μ = 0.033 (0, 0.172)] and females [μ = 0.043 (0, 0.216)] at low altitude (W = 512.5, P = 0.551).
Imperfect wSan transmission did not vary among trapping sites ( P = 0.098; Table S1) or between regional groupings, such that site one (μ = 0.120) and site two (μ = 0.045) were statistically indistinguishable (W = 456.5, P = 0.213; Figure 3 and Table 2). This is not surprising given only a mountainous ridge separates these sites (Figure 1). As with wYak, we observed no difference between rates of wSan transmission to male [μ = 0.129 (0.027, 0.341)] and female [μ = 0.114 (0.03, 0.378)] offspring at site one (W = 190, P = 0.698) or to male [μ = 0.054 (0.013, 0.144)] and female [μ = 0.038 (0.004, 0.127)] offspring at site two (W = 866, P = 0.779). Because we found no evidence for variation in μ or p for wSan, we disregarded regional groupings and used the pooled 2018 parameter estimates (μ = 0.068, p = 0.795) when modeling infection equilibria (see below). The pooled estimate of μ = 0.068 for wSan was statistically indistinguishable from wYak at low (W = 1028.5, P = 0.607) and at high altitude (W = 1018, P = 0.0513).
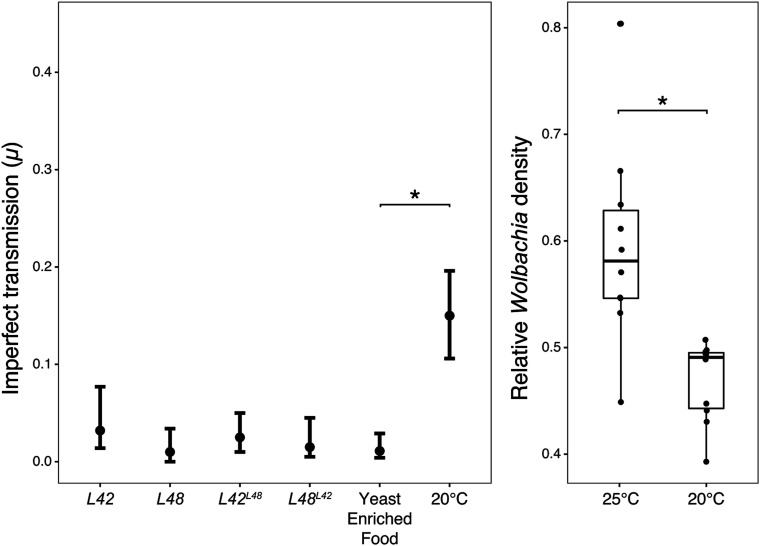
Cool rearing temperature increases imperfect maternal transmission
In the laboratory, maternal transmission was near perfect when infected females were placed on yeast-enriched food (μ = 0.011; Figure 4 and Table S3). μ values from the yeast treatment were homogenous among individual mothers ( P = 0.523; Figure S1). Compared to our field data, μ on yeast-enriched food was not significantly different from μ at low altitude (W = 440, P = 0.557), but was significantly lower than μ at high altitude (W = 440, P = 0.030) (Figures 3 and 4). Imperfect transmission on yeast-enriched food also did not vary between males (μ = 0) and females [μ = 0.021 (0.007, 0.058); W = 434, P = 0.081].
Figure 4.
Laboratory estimates of imperfect wYak transmission and titer. Left: Mean μ and 95% confidence intervals for wYak from laboratory experiments dissecting genetic and environmental contributions to imperfect maternal transmission. μ values are shown for naturally sampled (L42, L48) and for reciprocally introgressed genotypes (L42L48, L48L42) reared under standard conditions (25°), and for L5 reared on yeast-enriched food at 25° and on standard food at 20°. Right: Estimates of wYak titer (relative Wolbachia density) for L5 females reared at 25° and 20°. Asterisks indicate statistically significant differences at P < 0.05.
In contrast, we detected substantial imperfect wYak transmission when infected D. yakuba females were reared at relatively cool 20° (μ = 0.15; Figure 4 and Table S3). μ values at 20° were homogenous among individual mothers ( P = 0.627; Figure S1). μ from the cold treatment was significantly greater than our field estimate at low altitude on São Tomé (W = 548, P < 0.001), but it was significantly lower than μ at high altitude (W = 535, P = 0.011) (Figures 3 and 4). wYak transmission under cold exposure was also more imperfect than in the yeast-enriched food treatment (W = 74, P < 0.001; Figure 4). When reared at 20°, D. yakuba females also had a lower relative wYak density [ (0.441, 0.496)] than females developed at 25° [ (0.527, 0.663); W = 6, P < 0.001; Figure 4]. We predict that reductions in wYak titer at cool temperatures contribute to imperfect maternal transmission. Interestingly, maternal transmission to female progeny in our 20° treatment was an order of magnitude more imperfect [μ = 0.277 (0.118, 0.376)] than to males [μ = 0.02 (0, 0.065); W = 342, P < 0.001], a pattern opposite that observed for the Wolbachia infecting D. pseudotakahashii (Richardson et al. 2019). This pattern is also opposite to our theoretical expectation that selection should favor faithful transmission to female offspring since their uninfected ova are susceptible to CI (Prout 1994; Turelli 1994). While transmission to females and males did not differ in the field on São Tomé, or in our yeast-enriched food treatment, in all cases our point estimates for wYak μ were higher for females than males.
No evidence for Wolbachia or host effects on maternal transmission
We found very little differentiation in allele frequencies along the genomes of D. yakuba from low and high altitudes (Figure S2). The mean difference in allele frequencies between low and high altitude D. yakuba was 0.0027 (lower 25% quartile = 0, upper quartile = 0.0455), which is significantly higher than zero (Welch two sample t-test: t = 51.42, df = 13,056,636, P < 0.0001); however, the simulated distribution centered on 0 and our observed distribution are almost completely overlapping (Figure S2). This is consistent with two prior studies showing very low differentiation between these two regions (Comeault et al. 2016; Turissini and Matute 2017). Only 8 of the 6,528,464 SNPs across the genome were fixed for different nucleotides between the low and high D. yakuba populations. None of these fixed differences were located in protein-coding sequence, but all eight SNPs occurred within 10 kb of genes (Table S4). All nearby genes were either functionally undescribed or had no obvious connection to interactions with Wolbachia. We are ignorant of the types of genes that might modify wYak transmission, and have no reason to think these fixed differences between low and high altitude D. yakuba influence the variation in transmission we observe.
To explicitly test for host modulation of wYak transmission, we reciprocally introgressed wYak and D. yakuba genomes and quantified μ for the starting (L42 and L48) and reciprocally introgressed (L42L48 and L48L42) genotypes. Maternal transmission was near perfect for all four genotypes when reared under standard 25° conditions (Table S3). We found no evidence for D. yakuba effects on wYak transmission, such that μ did not differ among the four genotypes ( P = 0.478). For the L42 genotype (μ = 0.032), μ was homogeneous among mothers ( P = 0.171; Figure S1), and wYak transmission to female progeny was significantly more imperfect [μ = 0.042 (0.016, 0.105)] than to males (μ = 0; W = 286, P = 0.041). For the L48 genotype (μ = 0.01), μ was also homogeneous among mothers ( P = 0.535), but transmission to female [μ = 0.009 (0, 0.049)] and male progeny [μ = 0.01 (0, 0.061)] did not differ (W = 220, P = 1). For the reciprocally introgressed genotype L42L48 (μ = 0.024), μ was homogeneous among mothers ( P = 0.709), and transmission to female progeny [μ = 0.05 (0.02, 0.103)] was again significantly more imperfect than to males (μ = 0; W = 250, P = 0.020). Finally, for the reciprocally introgressed genotype L48L42 (μ = 0.015), μ values were homogeneous among mothers ( P = 0.523), but transmission to female [μ = 0.01 (0, 0.059)] and male progeny [μ = 0.01 (0, 0.061)] did not differ (W = 200, P = 1). Interestingly, we detected female-biased imperfect transmission only for genotypes with the L42 host nuclear genome (L42 and L42L48), motivating future analysis of D. yakuba factors with sex-specific effects on wYak transmission.
Temporally variable μ or spatially variable F and/or H is required to explain wYak infection equilibria
We modeled , incorporating our field estimates of μ on São Tomé and laboratory estimates of F and H (Cooper et al. 2017). Beneficial Wolbachia effects of F > ∼1.2 have not been documented in any system (Weeks et al. 2007; Meany et al. 2019), and prior efforts found no evidence for positive Wolbachia effects on D. yakuba-clade host fecundity (Cooper et al. 2017). Wolbachia effects on other components of host fitness have not been examined, but we assume F(1 – μ) must be >1 given the spread and persistence of wYak and wSan in nature (Cooper et al. 2017). We generally consider values of F > 1.5 as biologically unrealistic (Meany et al. 2019). Finally, while wYak and wSan cause weak CI on average in the laboratory we considered equilibria ranging from no CI up to very strong CI . Laboratory crosses and field infection frequencies suggest CI strength may vary in this clade (Cooper et al. 2017).
The results of our mathematical analyses are summarized in Figure 5. We conservatively evaluated infection equilibria across the full credible intervals for estimates of μ for each region/species (Figure S3 and Table S5). Because μ could vary within sites across host generations, we also evaluated equilibria across the full range of μ point estimates for all trapping sites on São Tomé (Table S6). We reasoned that temporal variation in μ within any one site is likely lower than the full range of μ point estimates for all trapping sites on São Tomé, but future analysis of short-term variation in μ within field sites is required to test this assumption. Finally, we evaluated how host population size affects stochastic fluctuations in Wolbachia frequencies in host populations at infection equilibrium (Figure S4 and Table S7).
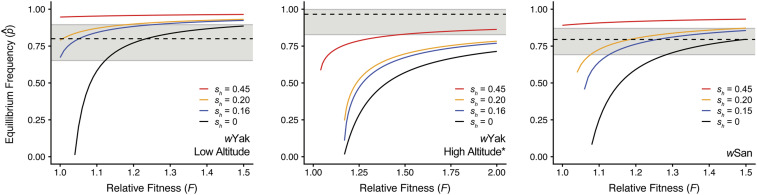
Figure 5.
Equilibrium wYak and wSan infection frequencies plotted against a range of F values, assuming field estimates of μ. Dotted lines indicate observed infection frequencies for each region, and the gray area denotes 95% binomial confidence intervals for observed frequencies. Black lines denote no CI , followed by laboratory estimates of weak CI in blue for wYak and wSan (Cooper et al. 2017), moderate CI in orange ( = 0.20), and strong CI in red ( = 0.45). The asterisk signifies that the YAK05b trapping site was removed from the high altitude wYak dataset due to an anomalously high value of μ (see Table S1, Figure 3). Similar plots showing parameter estimates across the full credible intervals of μ are shown in Figure S3.
We first modeled infection equilibria for wYak at low altitude. Given our estimate of μ = 0.038 (0.003, 0.184), no-to-moderate CI (sh = 0–0.20), and modest host fitness effects (F = 1.01–1.23), we can plausibly explain our wYak frequency estimate of p = 0.80 (0.65, 0.90) (Figure 5 and Table S5). Assuming our laboratory estimate of weak CI and our point estimate of μ = 0.038, F = 1.25 is required to explain the upper credible interval for p (0.90) (Figure S3 and Table S5). This suggests that our field estimate of imperfect transmission, combined with laboratory estimates of CI and modest-to-strong (positive) Wolbachia effects on host fitness, can reasonably explain the credible interval for p at low altitude. However, very unrealistic values of F combined with strong CI are almost always required to explain p if we assume the upper credible interval for μ at low altitude (0.184).
Next, we modeled infection equilibria for wYak at high altitude, where we observed the counterintuitive pattern of a high infection frequency [p = 0.95 (0.84, 0.99)] despite very imperfect maternal transmission [μ = 0.20 (0.087, 0.364)]. Using our point estimate for μ, stronger CI than previously estimated and biologically unrealistic positive fitness effects (F > 4) are required to explain p (Figure S3 and Table S5; Cooper et al. 2017). Even considering the full credible interval for μ, unrealistic values of F > 1.5 are required to approach the credible interval of p, regardless of CI strength. If we assume wYak transmission in the prior D. yakuba generation was substantially lower, such that μ = 0.038 (i.e., the lowest wYak μ point estimate for all trapping sites on São Tomé), we can plausibly explain the lower credible limit of p (0.84) using our laboratory estimate of CI and F = 1.1 (Table S6).
Further exploration of our data indicates that imperfect transmission at one particular trapping site at high altitude (YAK05b, altitude = 1104 m) was more than three times higher than prior field estimates in any system (μ = 0.354; see Figure 3 and Tables S1 and S8). This is surprising given that the YAK05b site does not differ noticeably from the nearby trap YAK05, which is within eyesight of YAK05b (Figure 1). Excluding the anomalously high YAK05b site from the high altitude region, p = 0.97 (0.83, 0.998) (N = 29) and μ = 0.143 (0.036, 0.356). Here, the lower credible interval of p can plausibly be explained by strong CI and F = 1.48 if we assume our point estimate of μ = 0.143, or by weak CI and F = 1.07 if we assume the lower credible interval of μ = 0.036 (Figure 5 and Table S5). Similar values of and F are required to explain p if we assume differences in imperfect maternal transmission in the prior host generation (Table S6).
We found no evidence that stochastic fluctuations in wYak frequencies could influence the patterns we observe. We chose three different combinations of parameter estimates from Table S5 and evaluated how host population size affects stochasticity (Supplemental Methods, Figure S4 and Table S7). In all cases, even small host populations are expected to yield deterministic infection dynamics. As an example, at high altitude our lower credible limit of p = 0.84 can be explained by assuming the lower credible limit of μ = 0.087, moderate CI of and very strong fitness benefits of F = 1.55 (Table S5). Assuming an unrealistically small host population of 5000 individuals and wYak we inferred mean p = 0.84 (0.82, 0.86) after 100 simulated host generations (Figure S4 and Table S7). When we removed the anomalous YAK05b site and assumed an infection equilibrium of with μ = 0.036, sh = 0.45, and F = 1.47 (Table S5), we found even less variance in infection frequencies with p = 0.966 (0.960, 0.972) after 100 generations. This is consistent with simulations and cage experiments using wRi- and wAu-infected D. simulans showing that small experimental host populations tend to yield deterministic infection dynamics (Kriesner and Hoffmann 2018). These results imply that host population size fluctuations are unlikely to explain why concurrent estimates of μ do not predict wYak infection frequencies at high altitude. Plausible explanations for wYak at high altitude generally require us to infer (1) a higher fidelity of transmission in the D. yakuba generation prior to our concurrent estimation of μ and p and/or (2) more positive wYak effects on D. yakuba fitness and/or stronger CI at high relative to low altitude. We predict that some combination of these two possibilities determines wYak frequency variation on São Tomé.
We found no evidence for regional variation in wSan p or μ on São Tomé, so we modeled infection equilibria for our pooled sample of D. santomea. Generally, we can explain p = 0.80 (0.69, 0.87) assuming our point estimate of μ = 0.068, weak-to-moderate CI and F < 1.5 (Figure 5 and Table S5). For example, assuming μ = 0.068 and our laboratory estimate of CI strength F must equal 1.26 to explain p = 0.80. Assuming more imperfect wSan transmission (e.g., μ = 0.154, the upper credible interval) we can still explain p = 0.80 with F < 1.5, but we must invoke strong CI Thus, unlike wYak frequencies on São Tomé, we can plausibly explain much of our credible interval for wSan p with our point estimate for μ, weak-to-moderate CI, and biologically reasonable values of F.
Discussion
wMel-like Wolbachia in Drosophila tend to persist at intermediate frequencies that vary (Kriesner et al. 2016; Cooper et al. 2017), but the contributions to frequency variation of imperfect maternal transmission, Wolbachia fitness effects, and CI remain unknown. Our field analysis found that both wYak and wSan are imperfectly transmitted on São Tomé, and wYak transmission varies spatially. Contrary to our prediction, concurrent field estimates of imperfect maternal transmission do not predict spatial variation in wYak frequencies, which are highest at high altitudes where maternal transmission is the most imperfect. Genomic and genetic analyses suggest little contribution of host genomes to variation in wYak maternal transmission. Instead, rearing D. yakuba females at a cool temperature significantly decreases wYak titer and increases imperfect transmission to levels observed in nature. Using mathematical models, we infer that temporal variation in wYak transmission within sites, and/or spatial variation in wYak effects on host fitness and CI among sites, is required to explain wYak frequencies. In contrast, while wSan frequencies continue to vary across years (Cooper et al. 2017), they were spatially stable within São Tomé in 2018, and plausibly explained by imperfect transmission, modest fitness effects, and weak CI. We discuss the implications of our findings below.
Environmental effects on maternal Wolbachia transmission
Altitudinal differences in wYak frequency (p = 0.80–0.95) and imperfect maternal transmission (μ = 0.038–0.20) occur over short geographic distances, with low and high altitude D. yakuba sites separated by only 2.4 km and an elevation gain of 310 m (Table S1). Conditions are relatively cool at high altitude on Pico de São Tomé (Table S1). While maternal transmission is near perfect under standard laboratory conditions (25°), wYak transmission at relatively cool 20° (μ = 0.15) is imperfect and intermediate to our low (μ = 0.038) and high (μ = 0.20) altitude field estimates. The pooled wYak frequency on São Tomé in 2018 was relatively high (p = 0.88), suggesting most matings are between infected females and infected males. Because uninfected ova produced by infected females can be eliminated by CI in this cross (Turelli and Hoffmann 1995), we chose to pair infected females with infected males for all laboratory transmission experiments to mimic the most likely cross in nature. Therefore, it is unlikely that differences in the susceptibility of uninfected ova to CI can account for differences between our estimates of μ in the field and the laboratory. Instead, we predict that unknown factors in addition to temperature contribute to the relatively imperfect wYak transmission we observe at high altitude.
Relative wYak titer is reduced at 20° compared to 25°, suggesting temperature effects on titer contribute to variation in imperfect maternal transmission. wMel titer and maternal transmission are also reduced in transinfected Ae. aegypti when mosquitoes are exposed to heat stress in the lab (26–40°) and hot temperatures in the field (>39°) (Ulrich et al. 2016; Ross et al. 2017, 2019a; Foo et al. 2019). Interestingly, Foo et al. (2019) found a sex-specific effect where Ae. aegypti exposed to heat stress produce female progeny with relatively low wMel titer. This is similar to our laboratory results at 20° and for wYak variants paired with the L42 D. yakuba genome where transmission to female progeny was more imperfect than to males. Selection for faithful transmission to females should be relatively intense, given that the uninfected offspring produced by infected mothers may be susceptible to CI. The fact that we detected sex-biased transmission at 20° and for the L42 nuclear genome, but not for a yeast-enriched diet or the L48 nuclear genome suggests that biases in transmission may depend on abiotic and genetic environments.
Host diet can perturb cellular traits predicted to influence maternal transmission (Serbus et al. 2015; Christensen et al. 2019). Adult female D. melanogaster reared on a yeast-enriched diet for 2 days exhibited a 72% decrease in cellular wMel titer in stage 10a oocytes (Serbus et al. 2015). Titer at this stage of oogenesis is predicted to determine Wolbachia transmission to the developing offspring (Hadfield and Axton 1999; Serbus and Sullivan 2007; Serbus et al. 2015; Camacho et al. 2017; Russell et al. 2018), which led us to predict that yeast-enriched food could generate imperfect transmission. Instead, maternal wYak transmission is near perfect on a yeast-enriched diet (μ = 0.011). Despite much analysis of Wolbachia titer and localization in developing host oocytes (Hadfield and Axton 1999; Serbus and Sullivan 2007; Serbus et al. 2015; Camacho et al. 2017; Christensen et al. 2019), there have been no direct tests for covariance between these cellular traits and imperfect transmission. Our results and others (Kriesner et al. 2016; Ulrich et al. 2016; Ross et al. 2017, 2019a; Foo et al. 2019) motivate such analyses, particularly for wMel-like Wolbachia reared at different temperatures.
Little evidence for host or Wolbachia effects on imperfect transmission
wYak variants in West Africa are extremely similar (0.0007% third-position pairwise differences; Cooper et al. 2019), having differentiated only a few thousand years ago. Indeed, wYak, wSan, and wTei possess a more recent common ancestor (2500–4500 years) than do wMel variants within D. melanogaster (4900–7200 years; Cooper et al. 2019). Our whole-genome analysis also suggests little differentiation between D. yakuba from low and high altitudes on São Tomé (mean difference = 0.0027; Figure S2). This is consistent with past genomic and phenotypic analyses of these regions by Comeault et al. (2016) and Turissini and Matute (2017). We found no evidence of Wolbachia or host effects on wYak maternal transmission in our genetic analysis of reciprocally introgressed genotypes (L42, L48, L48L42, L42L48). Interspecific crosses in Drosophila (Serbus and Sullivan 2007) and Nasonia (Funkhouser-Jones et al. 2018) revealed host factors that modify Wolbachia titer in host embryos, which could ultimately influence maternal transmission. We observed substantial heterogeneity in imperfect transmission among individual wild-caught females of D. yakuba and D. santomea ranging from μ = 0 to 0.929 (Figure 3), which resembles patterns observed for wRi (Turelli and Hoffmann 1995; Carrington et al. 2011) and wSuz (Hamm et al. 2014). This could indicate that host genotypes vary in their ability to transmit Wolbachia, but it seems more likely that variation in host development or adult environments (e.g., temperature) underlies heterogeneity in Wolbachia transmission.
The contributions of imperfect transmission, fitness effects, and CI to Wolbachia frequency dynamics
Our field estimates of imperfect wYak and wSan transmission do not predict Wolbachia frequencies on São Tomé. In particular, imperfect wYak transmission was highest at high altitude where wYak frequency was also the highest. We estimated μ and p concurrently, assuming that μ does not vary significantly between host generations. If we assume that imperfect wYak transmission in the D. yakuba generation prior to our sampling at high altitude was equal to our lowest μ point estimate on São Tomé (μ = 0.038; Table S1), we can plausibly explain the credible interval of wYak frequency at high altitude with our laboratory estimate of weak CI (sh = 0.16) and modest positive fitness effects (F = 1.1) (Cooper et al. 2017; Table S6). While it remains unknown whether wYak transmission varies temporally within sites on São Tomé, three observations suggest it could. First, wYak titer and μ are altered by temperature in the laboratory (Figure 4). If thermal environments experienced by D. yakuba females in the field vary between generations, estimating μ in the current D. yakuba generation may not reflect μ in the prior generation. Second, we observed significant heterogeneity in wYak transmission rates among D. yakuba females (Figure 3), suggesting μ may be particularly labile and depend on local conditions. Third, wRi transmission varied seasonally within a single population of D. simulans at Ivanhoe, CA between April and November 1993 (Carrington et al. 2011), indicating temporal variation in μ within sites is possible. Future analysis of fine-scale variation in host developmental environments in the field, in concert with field and laboratory estimation of μ, will help elucidate the basis of Wolbachia frequency fluctuations.
Stronger CI and/or more positive wYak effects on D. yakuba fitness at high relative to low altitude could potentially explain high wYak frequency despite very imperfect transmission. However, even if we consider the full credible interval for μ at high altitude (0.087, 0.364), F must be >1.5 to approach the credible interval of p, regardless of CI strength (Figure S3 and Table S5). Excluding the putatively anomalous YAK05b site, we still must invoke very strong CI and F approaching 1.5 to plausibly explain p using our point estimate of μ = 0.143. Assuming μ = 0.036 (the lower credible interval), our laboratory estimate of weak CI and F = 1.07 plausibly explain the lower credible interval of p (Figure 5 and Table S5). While wYak causes weak CI on average in the laboratory (Cooper et al. 2017), CI strength of some Wolbachia can vary across environmental conditions (Clancy and Hoffmann 1998; Ross et al. 2017, 2019a), host backgrounds (Reynolds and Hoffmann 2002; Cooper et al. 2017), and male ages (Reynolds and Hoffmann 2002). Strong positive fitness effects have not been estimated directly in this system or most others (Cooper et al. 2017; Shi et al. 2018; Meany et al. 2019), although wRi evolved from causing a fecundity cost of F = 0.8–0.9 to generating a benefit of F = 1.1 over the course of 20 years (Weeks et al. 2007). Few data exist for other components of fitness, but protection from viruses and nutrient provisioning remain candidates for wMel-like and other Wolbachia (Hedges et al. 2008; Teixeira et al. 2008; Brownlie et al. 2009; Osborne et al. 2009; Martinez et al. 2014; Nikoh et al. 2014; Newton and Rice 2020). For example, Wolbachia are known to provide protection from a limited number of RNA viruses under experimental conditions in the laboratory (Hedges et al. 2008; Teixeira et al. 2008; Osborne et al. 2009; Martinez et al. 2014), but there is currently no evidence that Wolbachia provide viral protection in natural Drosophila populations (Webster et al. 2015; Shi et al. 2018). Future work must focus on how wYak and other Wolbachia strains benefit components of host fitness.
Given that we found no evidence that stochastic fluctuations in wYak frequencies could influence the patterns we observe (Figure S4 and Table S7), we predict that some combination of variable imperfect maternal transmission, wYak effects on D. yakuba fitness, and CI strength underlie wYak frequency variation on São Tomé. In contrast, spatially stable wSan frequencies can plausibly be explained by our field estimate of imperfect transmission (μ = 0.068), our laboratory estimate of weak CI and modest positive fitness effects on D. santomea. The differences in the dynamics and equilibria of these wMel-like Wolbachia in different host species is particularly interesting given their extreme sequence similarity across the genome.
Conclusion
Our results add to the growing number of examples of Wolbachia frequency fluctuations in nature (Shoemaker et al. 2003; Ahrens and Shoemaker 2005; Toju and Fukatsu 2011; Hamm et al. 2014). Similar fluctuations have also been observed in another facultative symbiont; Rickettsia bellii spread to near fixation in an Arizona population of the sweet potato whitefly (Bemisia tabaci) in 2011, but then declined in frequency (p = 0.36) in 2017 (Bockoven et al. 2020). Our 2018 sampling on São Tomé revealed that wYak frequencies vary between low and high altitudes, and wSan varied temporally from 2015 to 2018. Wolbachia fluctuations are common in the D. yakuba clade over the last two decades (Figure 2; Cooper et al. 2017), and, more broadly, these fluctuations seem to be a general property of wMel-like Wolbachia. wMel frequencies vary greatly among D. melanogaster populations across the globe (Hoffmann et al. 1994, 1998; Ilinsky and Zakharov 2007; Richardson et al. 2012; Early and Clark 2013; Webster et al. 2015; Kriesner et al. 2016). In Eastern Australia, wMel frequencies decline clinally with latitude due in part to wMel fitness costs in cold environments (Kriesner et al. 2016). Our results suggest relatively more imperfect wMel transmission in cold environments could contribute to the wMel frequency cline in D. melanogaster. Additional work evaluating the basis of spatially varying Wolbachia frequencies, in addition to clinal host variation (Adrion et al. 2015), is needed.
Understanding factors that govern Wolbachia spread is crucial to explain the global Wolbachia pandemic and to improve the efficacy of wMel biocontrol (Ross et al. 2019b). The latter requires establishing and maintaining virus-blocking wMel in mosquito-vector populations to reduce human disease transmission (e.g., dengue and Zika) (McMeniman et al. 2009; Hoffmann et al. 2011; Ross et al. 2019b). Our discovery that cool rearing temperature generates imperfect wYak transmission opens the door to future analysis of Wolbachia transmission in the laboratory. Particularly promising avenues of research include the timing of infection loss during host development and the specific changes to Wolbachia titer and localization in host oocytes that result in imperfect maternal transmission. Ultimately, understanding how abiotic conditions and host genetic backgrounds influence all three determinants of Wolbachia spread—imperfect maternal transmission, fitness effects, and CI—is crucial to explain the spread and maintenance of Wolbachia in nature.
Acknowledgments
We thank all members of the 2018 São Tomé field crew that assisted with sampling D. yakuba and D. santomea. Tim Wheeler and Paighton Noel assisted in the laboratory. Dave Begun, Leonie Moyle, and two anonymous reviewers provided comments that improved our manuscript. The Cooper laboratory group and Aaron Comeault also provided valuable feedback. We especially thank Michael Turelli for very critical comments on an earlier draft that greatly improved our manuscript. We thank the Genomics Core and the Environmental Control for Organismal Research Laboratories at the University of Montana for their support. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award numbers R01GM121750 to D.R.M. and R35GM124701 to B.S.C.
Footnotes
Communicating editor: L. Moyle
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12275513.
Literature Cited
- Adrion J. R., Hahn M. W., and Cooper B. S., 2015. Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. 31: 434–444. 10.1016/j.tig.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens M. E., and Shoemaker D., 2005. Evolutionary history of Wolbachia infections in the fire ant Solenopsis invicta. BMC Evol. Biol. 5: 35 10.1186/1471-2148-5-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliota M. T., Peinado S. A., Velez I. D., and Osorio J. E., 2016. The wMel strain of Wolbachia reduces transmission of Zika virus by Aedes aegypti. Sci. Rep. 6: 28792 10.1038/srep28792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N., and Turelli M., 2011. Spatial waves of advance with bistable dynamics: cytoplasmic and genetic analogues of Allee effects. Am. Nat. 178: E48–E75. 10.1086/661246 [DOI] [PubMed] [Google Scholar]
- Beckmann J. F., Ronau J. A., and Hochstrasser M., 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2: 17007 10.1038/nmicrobiol.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockoven A. A., Bondy E. C., Flores M. J., Kelly S. E., Ravenscraft A. M. et al. , 2020. What goes up might come down: the spectacular spread of an endosymbiont is followed by its decline a decade later. Microb. Ecol. 79: 482–494. 10.1007/s00248-019-01417-4 [DOI] [PubMed] [Google Scholar]
- Brownlie J. C., Cass B. N., Riegler M., Witsenburg J. J., Iturbe-Ormaetxe I. et al. , 2009. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 5: e1000368 10.1371/journal.ppat.1000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho M., Oliva M., and Serbus L. R., 2017. Dietary saccharides and sweet tastants have differential effects on colonization of Drosophila oocytes by Wolbachia endosymbionts. Biol. Open 6: 1074–1083. 10.1242/bio.023895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty C., and B. Ripley, 2017 boot: Bootstrap R (S-Plus) functions. R Package version 1.3–19.
- Caragata E. P., Dutra H. L. C., and Moreira L. A., 2016. Inhibition of Zika virus by Wolbachia in Aedes aegypti. Microb. Cell 3: 293–295. 10.15698/mic2016.07.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou M.-L., Silvain J.-F., Daubin V., Da Lage J.-L., and Lachaise D., 2001. Divergence between Drosophila santomea and allopatric or sympatric populations of D. yakuba using paralogous amylase genes and migration scenarios along the Cameroon volcanic line. Mol. Ecol. 10: 649–660. 10.1046/j.1365-294x.2001.01225.x [DOI] [PubMed] [Google Scholar]
- Carrington L. B., Lipkowitz J. R., Hoffmann A. A., and Turelli M., 2011. A re-examination of Wolbachia-induced cytoplasmic incompatibility in California Drosophila simulans. PLoS One 6: e22565 [corrigenda: PLoS One 10: e0138050 (2015)]. 10.1371/journal.pone.0022565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper-Lindley C., Kimura S., Saxton D. S., Essaw Y., Simpson I. et al. , 2011. Rapid fluorescence-based screening for Wolbachia endosymbionts in Drosophila germ line and somatic tissues. Appl. Environ. Microbiol. 77: 4788–4794. 10.1128/AEM.00215-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattel J., Kaur R., Gibert P., Martinez J., Fraimout A. et al. , 2016. Wolbachia in European populations of the invasive pest Drosophila suzukii: regional variation in infection frequencies. PLoS One 11: e0147766 (erratum: PLoS One 11: e0150050). 10.1371/journal.pone.0147766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlat S., Ballard J., and Mercot H., 2004. What maintains noncytoplasmic incompatibility inducing Wolbachia in their hosts: a case study from a natural Drosophila yakuba population. J. Evol. Biol. 17: 322–330. 10.1046/j.1420-9101.2003.00676.x [DOI] [PubMed] [Google Scholar]
- Christensen S., Camacho M., Sharmin Z., Momtaz A. Z., Perez L. et al. , 2019. Quantitative methods for assessing local and bodywide contributions to Wolbachia titer in maternal germline cells of Drosophila. BMC Microbiol. 19: 206 10.1186/s12866-019-1579-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy D. J., and Hoffmann A. A., 1998. Environmental effects on cytoplasmic incompatibility and bacterial load in Wolbachia-infected Drosophila simulans. Entomol. Exp. Appl. 86: 13–24. 10.1046/j.1570-7458.1998.00261.x [DOI] [Google Scholar]
- Comeault A. A., Venkat A., and Matute D. R., 2016. Correlated evolution of male and female reproductive traits drive a cascading effect of reinforcement in Drosophila yakuba. Proc. Biol. Sci. 283: 20160730 10.1098/rspb.2016.0730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B. S., Ginsberg P. S., Turelli M., and Matute D. R., 2017. Wolbachia in the Drosophila yakuba complex: pervasive frequency variation and weak cytoplasmic incompatibility, but no apparent effect on reproductive isolation. Genetics 205: 333–351. 10.1534/genetics.116.196238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B. S., Sedghifar A., Nash W. T., Comeault A. A., and Matute D. R., 2018. A maladaptive combination of traits contributes to the maintenance of a Drosophila hybrid zone. Curr. Biol. 28: 2940–2947.e6. 10.1016/j.cub.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B. S., Vanderpool D., Conner W. R., Matute D. R., and Turelli M., 2019. Wolbachia acquisition by Drosophila yakuba-clade hosts and transfer of incompatibility loci between distantly related Wolbachia. Genetics 212: 1399–1419. 10.1534/genetics.119.302349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R. et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., Oliver B. et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- Early A. M., and Clark A. G., 2013. Monophyly of Wolbachia pipientis genomes within Drosophila melanogaster: geographic structuring, titre variation and host effects across five populations. Mol. Ecol. 22: 5765–5778. 10.1111/mec.12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B., and R. J. Tibshirani, 1993 An introduction to the bootstrap. Monographs on Statistics and Applied Probability, vol 57. Chapman and Hall/CRC, London.
- Ferree P. M., Frydman H. M., Li J. M., Cao J., Wieschaus E. et al. , 2005. Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 1: e14 10.1371/journal.ppat.0010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo I. J.-H., Hoffmann A. A., and Ross P. A., 2019. Cross-generational effects of heat stress on fitness and Wolbachia density in Aedes aegypti mosquitoes. Trop. Med. Infect. Dis. 4: 13 10.3390/tropicalmed4010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J., and Weisberg S., 2011. An R companion to applied regression, Sage, Thousand Oaks, CA. [Google Scholar]
- Funkhouser-Jones L. J., van Opstal E. J., Sharma A., and Bordenstein S. R., 2018. The maternal effect gene Wds controls Wolbachia titer in Nasonia. Curr. Biol. 28: 1692–1702.e6. 10.1016/j.cub.2018.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield S. J., and Axton J. M., 1999. Germ cells colonized by endosymbiotic bacteria. Nature 402: 482 10.1038/45002 [DOI] [PubMed] [Google Scholar]
- Hamm C. A., Begun D. J., Vo A., Smith C. C., Saelao P. et al. , 2014. Wolbachia do not live by reproductive manipulation alone: infection polymorphism in Drosophila suzukii and D. subpulchrella. Mol. Ecol. 23: 4871–4885. 10.1111/mec.12901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell F. E. Jr, and Dupont C., 2018. Hmisc: Harrell Miscellaneous https://CRAN.R-project.org/package=Hmisc.
- Hedges L. M., Brownlie J. C., O’Neill S. L., and Johnson K. N., 2008. Wolbachia and virus protection in insects. Science 322: 702 10.1126/science.1162418 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., and M. Turelli, 1997 Cytoplasmic incompatibility in insects. In: O’Neill SL, Hoffman AA, Werren JH (eds) Influential passengers. Oxford University Press: New York, pp. 42–80.
- Hoffmann A. A., Turelli M., and Harshman L. G., 1990. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics 126: 933–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Clancy D. J., and Merton E., 1994. Cytoplasmic incompatibility in Australian populations of Drosophila melanogaster. Genetics 136: 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Clancy D., and Duncan J., 1996. Naturally-occurring Wolbachia infection in Drosophila simulans that does not cause cytoplasmic incompatibility. Heredity 76: 1–8. 10.1038/hdy.1996.1 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Hercus M., and Dagher H., 1998. Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics 148: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Montgomery B. L., Popovici J., Iturbe-Ormaetxe I., Johnson P. H. et al. , 2011. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476: 454–457. 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- Hood G. M., 2011 PopTools. http://www.poptools.org
- Hosokawa T., Koga R., Kikuchi Y., Meng X.-Y., and Fukatsu T., 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 107: 769–774. 10.1073/pnas.0911476107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst G. D., and Jiggins F. M., 2000. Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg. Infect. Dis. 6: 329–336. 10.3201/eid0604.000402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilinsky Y. Y., and Zakharov I., 2007. The endosymbiont Wolbachia in Eurasian populations of Drosophila melanogaster. Russ. J. Genet. 43: 748–756. 10.1134/S102279540707006X [DOI] [PubMed] [Google Scholar]
- Kriesner P., and Hoffmann A. A., 2018. Rapid spread of a Wolbachia infection that does not affect host reproduction in Drosophila simulans cage populations. Evolution 72: 1475–1487. 10.1111/evo.13506 [DOI] [PubMed] [Google Scholar]
- Kriesner P., Hoffmann A. A., Lee S. F., Turelli M., and Weeks A. R., 2013. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 9: e1003607 10.1371/journal.ppat.1003607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriesner P., Conner W. R., Weeks A. R., Turelli M., and Hoffmann A. A., 2016. Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy. Evolution 70: 979–997. 10.1111/evo.12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D., Harry M., Solignac M., Lemeunier F., Benassi V. et al. , 2000. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from Sao Tome. Proc. R. Soc. Lond. B Biol. Sci. 267: 1487–1495. 10.1098/rspb.2000.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePage D. P., Metcalf J. A., Bordenstein S. R., On J., Perlmutter J. I. et al. , 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543: 243–247. 10.1038/nature21391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A., Lachaise D., and Coyne J. A., 2005a Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea. Genetics 171: 197–210. 10.1534/genetics.104.033597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A., Lachaise D., and Coyne J. A., 2005b An anomalous hybrid zone in Drosophila. Evolution 59: 2602–2607. 10.1111/j.0014-3820.2005.tb00972.x [DOI] [PubMed] [Google Scholar]
- Mansourian S., Enjin A., Jirle E. V., Ramesh V., Rehermann G. et al. , 2018. Wild African Drosophila melanogaster are seasonal specialists on marula fruit. Curr. Biol. 28: 3960–3968.e3. 10.1016/j.cub.2018.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Longdon B., Bauer S., Chan Y.-S., Miller W. J. et al. , 2014. Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog. 10: e1004369 10.1371/journal.ppat.1004369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos M., Castrezana S. J., Nankivell B. J., Estes A. M., Markow T. A. et al. , 2006. Heritable endosymbionts of Drosophila. Genetics 174: 363–376. 10.1534/genetics.106.058818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D. R., Novak C. J., and Coyne J. A., 2009. Temperature-based extrinsic reproductive isolation in two species of Drosophila. Evolution 63: 595–612. 10.1111/j.1558-5646.2008.00588.x [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K. et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman C. J., Lane R. V., Cass B. N., Fong A. W. C., Sidhu M. et al. , 2009. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323: 141–144. 10.1126/science.1165326 [DOI] [PubMed] [Google Scholar]
- Meany M. K., Conner W. R., Richter S. V., Bailey J. A., Turelli M. et al. , 2019. Loss of cytoplasmic incompatibility and minimal fecundity effects explain relatively low Wolbachia frequencies in Drosophila mauritiana. Evolution 73: 1278–1295. 10.1111/evo.13745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama M., Nikoh N., Hosokawa T., and Fukatsu T., 2015. Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. MBio 6: e01732-15 10.1128/mBio.01732-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton I. L., and Rice D. W., 2020. The Jekyll and Hyde symbiont: could Wolbachia be a nutritional mutualist? J. Bacteriol. 202: e00589-19 10.1128/JB.00589-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N., Hosokawa T., Moriyama M., Oshima K., Hattori M. et al. , 2014. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA 111: 10257–10262. 10.1073/pnas.1409284111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S. L., 2018. The Use of Wolbachia by the World Mosquito Program to interrupt transmission of Aedes aegypti transmitted viruses, pp. 355–360 in Dengue and Zika: Control and Antiviral Treatment Strategies, Springer, New York. [DOI] [PubMed] [Google Scholar]
- Osborne S. E., Leong Y. S., O’Neill S. L., and Johnson K. N., 2009. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 5: e1000656 10.1371/journal.ppat.1000656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29: e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout T., 1994. Some evolutionary possibilities for a microbe that causes incompatibility in its host. Evolution 48: 909–911. 10.1111/j.1558-5646.1994.tb01371.x [DOI] [PubMed] [Google Scholar]
- R Core Team , 2018. R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Reynolds K. T., and Hoffmann A. A., 2002. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet. Res. 80: 79–87. 10.1017/S0016672302005827 [DOI] [PubMed] [Google Scholar]
- Richardson M. F., Weinert L. A., Welch J. J., Linheiro R. S., Magwire M. M. et al. , 2012. Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet. 8: e1003129 10.1371/journal.pgen.1003129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson K. M., Griffin P. C., Lee S. F., Ross P. A., Endersby-Harshman N. M. et al. , 2019. A Wolbachia infection from Drosophila that causes cytoplasmic incompatibility despite low prevalence and densities in males. Heredity 122: 428–440. 10.1038/s41437-018-0133-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. A., Wiwatanaratanabutr I., Axford J. K., White V. L., Endersby-Harshman N. M. et al. , 2017. Wolbachia infections in Aedes aegypti differ markedly in their response to cyclical heat stress. PLoS Pathog. 13: e1006006 10.1371/journal.ppat.1006006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. A., Ritchie S. A., Axford J. K., and Hoffmann A. A., 2019a Loss of cytoplasmic incompatibility in Wolbachia-infected Aedes aegypti under field conditions. PLoS Negl. Trop. Dis. 13: e0007357 10.1371/journal.pntd.0007357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. A., Turelli M., and Hoffmann A. A., 2019b Evolutionary ecology of Wolbachia releases for disease control. Annu. Rev. Genet. 53: 93–116. 10.1146/annurev-genet-112618-043609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F., Bouchon D., Pintureau B., Juchault P., and Solignac M., 1992. Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc. Biol. Sci. 250: 91–98. 10.1098/rspb.1992.0135 [DOI] [PubMed] [Google Scholar]
- Russell S. L., Lemseffer N., and Sullivan W. T., 2018. Wolbachia and host germline components compete for kinesin-mediated transport to the posterior pole of the Drosophila oocyte. PLoS Pathog. 14: e1007216 [corrigenda: PLoS Pathog. 15: e1007557 (2019)]. 10.1371/journal.ppat.1007216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R., and Sullivan W., 2007. A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog. 3: e190 10.1371/journal.ppat.0030190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R., White P. M., Silva J. P., Rabe A., Teixeira L. et al. , 2015. The impact of host diet on Wolbachia titer in Drosophila. PLoS Pathog. 11: e1004777 (erratum: PLoS Pathog. 11: e1004777). 10.1371/journal.ppat.1004777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., White V. L., Schlub T., Eden J.-S., Hoffmann A. A. et al. , 2018. No detectable effect of Wolbachia wMel on the prevalence and abundance of the RNA virome of Drosophila melanogaster. Proc. Biol. Sci. 285: 20181165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker D. D., Ahrens M., Sheill L., Mescher M., Keller L. et al. , 2003. Distribution and prevalence of Wolbachia infections in native populations of the fire ant Solenopsis invicta (Hymenoptera: Formicidae). Environ. Entomol. 32: 1329–1336. 10.1603/0046-225X-32.6.1329 [DOI] [Google Scholar]
- Shropshire J. D., On J., Layton E. M., Zhou H., and Bordenstein S. R., 2018. One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 115: 4987–4991. 10.1073/pnas.1800650115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singmann H., Bolker B., Westfall J., and Aust F., 2016. afex: Analysis of Factorial Experiments. R package version 0.16–1. https://cran.r-project.org/web/packages/afex/index.html
- Sprengelmeyer Q. D., Mansourian S., Lange J. D., Matute D. R., Cooper B. S. et al. , 2020. Recurrent collection of Drosophila melanogaster from wild African environments and genomic insights into species history. Mol. Biol. Evol. 37: 627–638. 10.1093/molbev/msz271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., and Kumar S., 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. 10.1093/molbev/msg236 [DOI] [PubMed] [Google Scholar]
- Teixeira L., Ferreira Á., and Ashburner M., 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6: e1000002 10.1371/journal.pbio.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju H., and Fukatsu T., 2011. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: relevance of local climate and host plants. Mol. Ecol. 20: 853–868. 10.1111/j.1365-294X.2010.04980.x [DOI] [PubMed] [Google Scholar]
- Turelli M., 1994. Evolution of incompatibility-inducing microbes and their hosts. Evolution 48: 1500–1513. 10.1111/j.1558-5646.1994.tb02192.x [DOI] [PubMed] [Google Scholar]
- Turelli M., and Hoffmann A. A., 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353: 440–442. 10.1038/353440a0 [DOI] [PubMed] [Google Scholar]
- Turelli M., and Hoffmann A. A., 1995. Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics 140: 1319–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turissini D. A., and Matute D. R., 2017. Fine scale mapping of genomic introgressions within the Drosophila yakuba clade. PLoS Genet. 13: e1006971 10.1371/journal.pgen.1006971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich J. N., Beier J. C., Devine G. J., and Hugo L. E., 2016. Heat sensitivity of wMel Wolbachia during Aedes aegypti development. PLoS Negl. Trop. Dis. 10: e0004873 10.1371/journal.pntd.0004873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk A. F., Hall-Mendelin S., Pyke A. T., Frentiu F. D., McElroy K. et al. , 2012. Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl. Trop. Dis. 6: e1892 10.1371/journal.pntd.0001892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C. L., Waldron F. M., Robertson S., Crowson D., Ferrari G. et al. , 2015. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 13: e1002210 10.1371/journal.pbio.1002210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks A. R., Turelli M., Harcombe W. R., Reynolds K. T., and Hoffmann A. A., 2007. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 5: e114 10.1371/journal.pbio.0050114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert L. A., Araujo-Jnr E. V., Ahmed M. Z., and Welch J. J., 2015. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. Biol. Sci. 282: 20150249 10.1098/rspb.2015.0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Baldo L., and Clark M. E., 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6: 741–751. 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- Zabalou S., Charlat S., Nirgianaki A., Lachaise D., Merçot H. et al. , 2004. Natural Wolbachia infections in the Drosophila yakuba species complex do not induce cytoplasmic incompatibility but fully rescue the wRi modification. Genetics 167: 827–834. 10.1534/genetics.103.015990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug R., and Hammerstein P., 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7: e38544 10.1371/journal.pone.0038544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All supplemental materials, data, and scripts are available on figshare: https://doi.org/10.25386/genetics.12275513. Genomic data from Turissini and Matute (2017) are available at NCBI (Bioproject: PRJNA395473).