Abstract
Chromatin domain insulators are thought to help partition the genome into genetic units called topologically associating domains (TADs). In Drosophila, TADs are often separated by inter-TAD regions containing active housekeeping genes and associated insulator binding proteins. This raises the question of whether insulator binding proteins are involved primarily in chromosomal TAD architecture or gene activation, or if these two activities are linked. The Boundary Element-Associated Factor of 32 kDa (BEAF-32, or BEAF for short) is usually found in inter-TADs. BEAF was discovered based on binding to the scs’ insulator, and is important for the insulator activity of scs’ and other BEAF binding sites. There are divergent promoters in scs’ with a BEAF binding site by each. Here, we dissect the scs’ insulator to identify DNA sequences important for insulator and promoter activity, focusing on the half of scs’ with a high affinity BEAF binding site. We find that the BEAF binding site is important for both insulator and promoter activity, as is another sequence we refer to as LS4. Aside from that, different sequences play roles in insulator and promoter activity. So while there is overlap and BEAF is important for both, insulator and promoter activity can be separated.
Keywords: BEAF, insulators, chromatin domains, promoter function, Drosophila
CHROMATIN domain insulators have been defined based on their ability to block chromosomal position effects on transgene expression and to disrupt the communication of an enhancer with a promoter when inserted in between. This depends on insulator binding proteins, which are thought to somehow define the boundaries of genetic domains such that enhancer–promoter communication and the spreading of chromatin states can occur within domains, but not between adjacent domains. Evidence indicates these proteins can also play roles as positive or negative regulators of gene expression and in mediating intra- and interchromosomal interactions (Raab and Kamakaka 2010; Chetverina et al. 2014; Kyrchanova and Georgiev 2014; Cubeñas-Potts and Corces 2015, 2017; Ali et al. 2016). Because they are thought to influence the organization of chromosomes in nuclei, insulator proteins are also called architectural proteins (Gómez-Diaz and Corces 2014; Bouwman and de Laat 2015). The main architectural protein studied in vertebrates is CTCF, which has been shown to play a prominent role in maintaining chromatin loops (Rao et al. 2014). It is highly conserved, with 11 zinc finger domains that target CTCF to thousands of genomic sites (Kim et al. 2007; Xie et al. 2007). In Drosophila, several sequence-specific DNA-binding architectural proteins are known, in addition to a homolog of CTCF [dCTCF, Su(Hw), Pita, ZIPIC, Zw5, GAF, BEAF-32, Ibf1, Ibf2 and the Elba complex] (Pauli et al. 2016), doubtless with more yet to be described.
In Drosophila, most regions bound by insulator proteins correspond to enhancers and promoters, especially of housekeeping genes (Cubeñas-Potts et al. 2017). Interestingly, certain RNA polymerase III promoters in budding and fission yeast, as well as the budding yeast RNA polymerase II promoter of the CHA1 gene have been found to act as barriers to heterochromatin spreading (Donze and Kamakaka 2001; Simms et al. 2004; Scott et al. 2006). Topologically associated domains (TADs) in Drosophila are frequently separated by inter-TAD regions formed by active housekeeping genes together with associated insulator proteins (Ulianov et al. 2016; Cubeñas-Potts et al. 2017; Hug et al. 2017; Rowley et al. 2017), consistent with a role for insulator proteins in chromatin domain organization. This leads to the question of whether insulator proteins play a role primarily in gene activation or in chromosomal TAD architecture, or both. If both, are these two activities linked or separable?
We study the Drosophila Boundary Element-Associated Factor of 32 kDa, BEAF-32 (hereafter referred to as BEAF), as a model insulator binding protein (Zhao et al. 1995). There are two 32 kDa forms of BEAF made from one gene, BEAF-32A and BEAF-32B (Hart et al. 1997). They differ only in their ∼80 amino acid N-termini, which both encode a DNA-binding zinc finger, and interact via a C-terminal BESS domain (Avva and Hart 2016). Only BEAF-32B is essential (Roy et al. 2007a), and genome-wide mapping found that BEAF-32B has the dominant DNA binding activity (Jiang et al. 2009). It binds CGATA motifs and might bind as a trimer (Hart et al. 1997). BEAF-32A binds CGTGA motifs, and might play a role at a subset of possibly lower-affinity binding sites (Jiang et al. 2009). However, BEAF binding sites cannot be predicted easily from DNA sequences. CGATA motif clusters are not sufficient to guarantee binding by BEAF, motifs in bound clusters have highly variable spacing and relative orientations, and BEAF can bind sequences with a single CGATA (Jiang et al. 2009). No clear rules on CGATA clustering for BEAF binding, or ancillary motifs that might also play a role other than possibly CGTGA, have emerged.
BEAF was discovered based on its binding to one of the first described insulators, scs’ (Udvardy et al. 1985; Kellum and Schedl 1991). It has been shown to be important for the activity of several insulators in addition to scs’ (Cuvier et al. 1998; Cuvier et al. 2002; Sultana et al. 2011; Schwartz et al. 2012). Consistent with insulators playing a role in chromatin organization, interfering with BEAF function disrupts polytene chromosome structure and enhances position effect variegation (Gilbert et al. 2006; Roy et al. 2007a). Genome-wide mapping found that ∼85% of BEAF binding regions are centered within 300 bp of transcription start sites (Bushey et al. 2009; Jiang et al. 2009; Nègre et al. 2010), and ∼85% of these are housekeeping genes (Jiang et al. 2009; Shrestha et al. 2018). An example is scs’, which contains divergent promoters for housekeeping genes. One promoter (CG3281) has an adjacent low affinity BEAF binding site, and the other (aurA) has a high affinity site, both composed of three CGATA motifs (Hart et al. 1997). We recently reported that BEAF interacts with the transcription factor Serendipity-δ (Sry-δ), and interactions between promoter-proximal BEAF and Sry-δ bound either adjacent or 2.3 kb upstream can activate a reporter gene (Dong et al. 2020). There are differences between housekeeping and developmental promoters (Zabidi et al. 2015), and we also found that BEAF directly activated two housekeeping promoters but not a developmental promoter. One is the promoter from the scs’ insulator with the high affinity BEAF binding site. This raises the question of whether BEAF helps to establish or maintain active chromatin, and whether that is the reason it is the DNA-binding insulator protein most commonly found at inter-TAD regions (Ulianov et al. 2016). Indeed, BEAF colocalizes with active chromatin marks, including RNA polymerase II (Jiang et al. 2009). Another question is whether the role of BEAF at promoters is related to its role in insulator function. Here, we address these questions by dissecting scs’ to find sequences important for insulator and promoter function.
Materials and Methods
Plasmids and fly germline transformation
All plasmids used for testing insulator activity in flies were made from pC4scs (linker-scanning) or pC4-attB-scs (minimal scs’) (Maharjan et al. 2018). Test sequences were based on the 216 bp scs’ M fragment that encompasses the high affinity BEAF binding site, with M* having mutations in the binding site that eliminate BEAF binding (Cuvier et al. 1998). The S fragment has 102 bp of the 3′ end of the M fragment. The X fragment is a 43 bp sequence centered on the high affinity binding site that has been used to DNA-affinity purify BEAF (Zhao et al. 1995). The 6-linker-scanning mutations (LS1–LS6) change 22 bp of scs’ to a sequence from bacteriophage lambda with 3 bp mutated to create a SpeI site (5′ AGGTAATAacTAGtGCCTGCAT, mutations in lowercase). Adjacent LS mutations overlap by 3 bp. The 5′ deletion removes 52 bp from the 5′ end of the M fragment, while the 3′ deletion removes 51 bp from the 3′ end. The spacer mutation changes 42 bp of scs’ between the LS4 location and the BEAF binding site to lambda sequences with 3 bp mutated to create a SpeI site (5′ CAACAGGTAAGACAGTTCGCAGGTAATAacTAGtGCCTGCAT), while the spacer deletion deletes these 42 bp. Monomers were cloned as BglII–BamHI fragments that were dimerized and inserted into the BamHI site of pC4scs or pC4-attB-scs. For P-element-based integration, pC4scs-derived plasmids (0.4 mg/ml) were co-injected with the helper plasmid pπ25.7wc (0.1 mg/ml) into preblastoderm y1 w67c23 embryos to generate P[w+] transgenic flies. For integration into phiC31 attP sites, pC4-attB-scs-derived plasmids were injected by GenetiVision (Houston, TX) into three attP fly lines (attP-H-X; Int.4, attP-I-2; Int.X and attP-M-X; Int.4) that show strong chromosomal position effects in eyes (Maharjan et al. 2018). Insulator function was assessed by scoring the eye color of 2- to 3-day old heterozygous female flies; eye color was not observed to vary between staged individuals of a given transgenic line.
Luciferase plasmids were built in pBSKS- (Stratagene). A PCR-amplified 225 bp SV40 polyadenylation region from pEGFP-N3 (Clontech) was inserted into XbaI and SacI sites, followed by insertion of PCR-amplified firefly luciferase (pGEM-luc; Promega) or Renilla luciferase coding sequences (pGL4.70; Promega) into HindIII and BamHI sites. Using Gibson assembly (New England Biolabs) the scs’ M fragment (Cuvier et al. 1998) was inserted into the SalI site of the firefly plasmid as a promoter control, while test sequences were inserted into the SalI site of the Renilla plasmid.
Comparison of BEAF ChIP-seq to transcription start sites and AT content
ChIP-seq data were downloaded from the ChIP-Atlas database (Oki et al. 2018) as normalized bigwig files aligned to the dm3 genome (Kc167: SRX749021; S2: SRX386677; embryo: SRX119302). ChIP-seq signal was plotted in a 2-kb window centered on transcription start sites (TSSs) using the metaseq python package (Dale et al. 2014). Heatmaps were made using deepTools (Ramírez et al. 2016) after using bedtools (Quinlan 2014) to make bigwig files of the dm3 genome AT and GC contents using 10-bp sliding windows. Annotated TSSs were ordered from highest to lowest BEAF signal for Kc167 cells, and the top 3000 and bottom 3000 TSSs were plotted using a 2-kb window centered on TSSs. AT and GC content heatmaps have the same gene order as used for BEAF.
S2 cell culture
Drosophila S2 cells were grown in Shields and Sang M3 Insect Medium (Sigma S8398) supplemented with 10% FBS (Corning 35-010-CV) and antibiotic-antimycotic (Gibco 15240062) from 5 × 105 to 1 × 107 cells/ml. Cells were grown at 25° in 5 ml in 25 cm2 T flasks and were maintained with cell splitting every 4 days with 1:4 dilutions into new M3 medium and flasks.
Transfections and luciferase assay
Transfections were done in 24-well plates. Wells were seeded with 7.5 × 105 cells in a final volume of 1 ml M3 complete medium, and incubated for 24 hr at 25° to bring the cells to 70% confluency. DNA was prepared by mixing 300 ng control firefly luciferase plasmid (M fragment promoter) with 700 ng experimental Renilla luciferase plasmid, then adding 5 µl of Lipofectamine 2000 reagent (Invitrogen 11668-019) and 500 µl of serum-free M3 medium and incubating at room temperature for 15 min. Cells in the plate were carefully washed with serum free M3 medium, the DNA solution was added, and cells were incubated 4 hr. The transfection mix was then replaced by M3 complete medium and incubated another 48 hr.
Cells were lysed and assayed for luciferase activity following the dual-luciferase system instructions (Promega E1910). The experimental Renilla luciferase activity was divided by the control firefly luciferase activity to control for transfection efficiency, and values were then normalized to the Renilla M fragment promoter. Three biological replicates were done.
Data availability
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Fly lines and plasmids are available upon request. Insulator (dimers), promoter (monomers), and primer sequences used for cloning are in Supplemental Material, Table S1. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12473447.
Results
Linker-scanning analysis of scs’
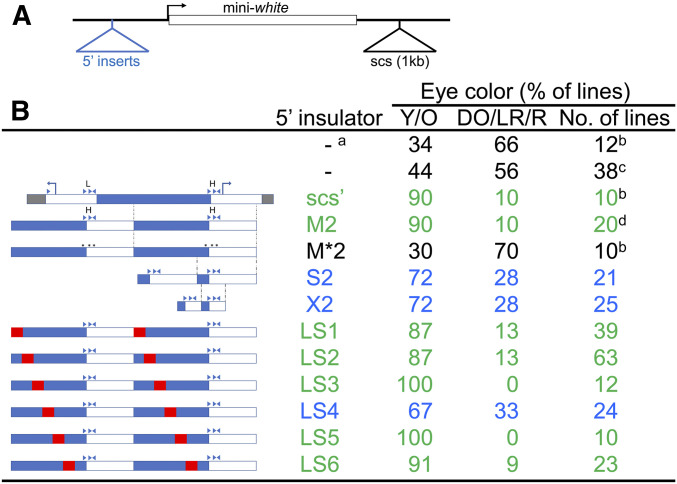
It was previously shown that a dimer of a 216 bp fragment from the 500 bp scs’ insulator is able to insulate a transgene from chromosomal position effects (CPE) as well as scs’ does (Cuvier et al. 1998), while a monomer is a weaker insulator (Maharjan et al. 2018). The assay was based on a P-element with an enhancerless mini-w gene with a 3′ scs insulator and variable sequences to test for insulator activity at its 5′ end (Figure 1A). Eye color is the readout, with light eye color (yellow to light orange) indicating either a lack of activating CPE at the site of transposon insertion or protection from CPE by the insulators. Thus a higher percentage of transgenic fly lines having lighter eye color indicates stronger insulation from activating CPE. Scs’ has two binding sites for the chromatin domain insulator protein BEAF, a low affinity site and a high affinity site (Hart et al. 1997). The 216 bp sequence, termed the M fragment, contains the high affinity BEAF binding site (hereafter referred to as the H site) such that the distance between the two H sites in the M dimer (M2) is the same as the distance between the H site and low affinity site in scs’. When the H site is mutated so that BEAF does not bind to the mutant M* dimer (M*2), insulator activity is lost (Cuvier et al. 1998).
Figure 1.
The LS4 sequence contributes to scs’ insulator activity. (A) Schematic of the mini-w reporter gene (white rectangle; bent arrow: TSS), showing the location of 5′ test sequences and the 3′ scs insulator. The upstream and downstream P-element sequences are not shown. (B) Assay results for each sequence tested in the 5′ insulator location, given as percent of fly lines with yellow/orange (Y/O) or dark-orange/light-red/red (DO/LR/R) eyes. Also indicated are the number of fly lines scored. Values in green indicate a functional insulator; in blue indicate impaired insulator activity; in black indicate a nonfunctional insulator. On the left are schematics of the scs’ sequences tested. Arrowheads represent CGATA motifs, stars represent mutated motifs, and H and L indicate the high and low affinity BEAF binding sites. Bent arrows represent TSSs. Open and filled rectangles roughly represent nuclease sensitive and resistant regions, respectively (Udvardy et al. 1985). Dotted lines indicate regions of scs’ present in M, S, and X monomers. Red boxes indicate the linker-scanning mutations present in LS1–LS6, which were tested as dimers. All constructs had a 3′ scs insulator except: a: no 3′ scs; b: data from Cuvier et al. (1998);. c: includes 18 fly lines from Cuvier et al. (1998); d: includes 10 fly lines from Cuvier et al. (1998).
Starting with the same P-element plasmid, we made dimers of shorter scs’ sequences to test for protection against CPE. The S fragment has 114 bp deleted from the 5′ end of the M fragment, while the X fragment is 43 bp encompassing the H site. The X fragment has been used to DNA-affinity purify BEAF (Zhao et al. 1995) and has 123 bp deleted from the 5′ end and 50 bp deleted from the 3′ end of the M fragment. Both the S2 and X2 dimers provide similar, reduced insulation from CPE relative to that obtained with M2 or scs’ (Figure 1B). Thus the BEAF binding site alone, as a dimer, has some insulator activity. The partial activity suggested that sequences in addition to the H site, spacing between H sites, or both contribute to insulator activity. Because deleting additional sequences to make the X2 dimer did not seem to reduce insulator activity more than the S2 dimer, we focused on the sequences deleted in the S fragment relative to the M fragment.
We performed a linker-scanning analysis on the 114 bp present in M but not in S, using six 22-bp steps that overlap by 3 bp (LS1–LS6, tested as dimers). From 10 to 63 fly lines were generated for each construct, including the M2 and no 5′ insulator controls. A dimer of the LS4 linker-scanning mutation impaired insulator activity similar to S2 and X2 (Figure 1B). This suggests that an accessory protein, or protein complex, binds at the wild-type LS4 sequence and cooperates with BEAF for stronger insulator activity. No clear protein binding motif is apparent in this AT-rich sequence (5′ GCACTATTCAATAATTCTCTTG). Sequence analysis using Jaspar (Khan et al. 2018) and the MEME suite (Bailey et al. 2009), detailed in the Discussion, did not identify promising protein candidates. Although sequence-specific binding cannot be excluded, it is possible that the DNA has some feature that is recognized, such as curvature or a narrow minor groove (Yoon et al. 1988; Carrera and Azorin 1994).
Further analysis of sequence requirements for scs’ insulator activity
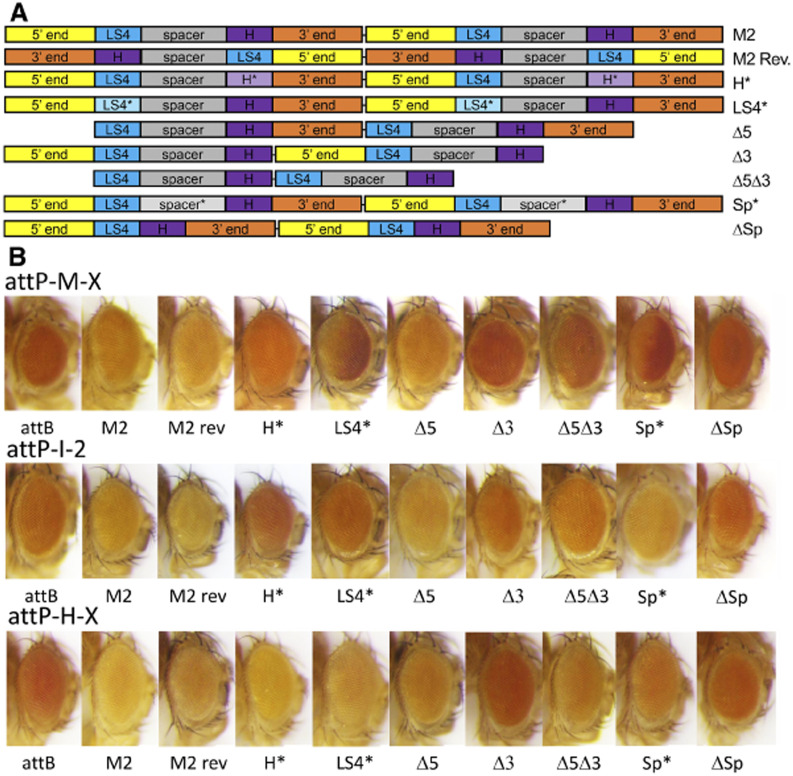
Based on the LS results, the M fragment was divided into five regions for further analysis: the 5′ end, LS4 region, spacer region, H site, and 3′ end. These regions were deleted or mutated, as indicated in Figure 2A. We found that a monomer of the M fragment does not insulate as well as a dimer (Maharjan et al. 2018), so all derivatives were tested as dimers. To simplify the analysis, we placed a phiC31 attB site just upstream of the 5′ insulator test site shown in Figure 1A, and used three fly lines we recently described that have attP sites at locations that show CPE (Maharjan et al. 2018). This allowed all constructs to be tested at the same genomic locations. Without an insulator, CPE activates mini-w to give a dark-orange eye color, while M2 provides insulation in both the forward or reverse orientation resulting in dark-yellow or light-orange eyes (Figure 2B).
Figure 2.
CPE analysis of M fragment derivatives. (A) Schematics of the dimeric insulator constructs tested. The M fragment was divided into the 5′ end, 3′ end, the LS4 sequence, the H (high affinity BEAF binding site) sequence, and spacer sequence between LS4 and H. Sequences were deleted or mutated as indicated. Mutant sequences are denoted by a * and lighter rectangle shading. Dimers were placed between an attB site and the 5′ end of mini-w for integration into chromosomal attP sites. As in Figure 1A, there is an scs insulator downstream of mini-w. (B) Eye color of 2–3 day old heterozygous transgenic female flies with the indicated 5′ insulator sequences. Release 6 locations of the attP sites are: attP-M-X: chrX:10,366,253; attP-I-2: chr2:7,040,089; attP-H-X: chrX:13,128,844. attB: no 5′ insulator. Results show that 5′ end and spacer sequences are dispensable, while the 3′ end, LS4, H and spacing between LS4 and H play a role in insulator function.
Consistent with earlier results, mutating the LS4 sequence or H site reduced insulator activity in 2 attP lines. It is not clear why the mutated insulators functioned well in the attP-H-X line. According to high-throughput expression data on FlyBase the divergent genes bracketing this site (CG32638 and CG1517) are expressed at low-to-moderate levels in heads (Gramates et al. 2017), so maybe local enhancers that drive the CPE are more sensitive to blocking. In contrast, the insertion sites in the other two lines are just upstream of genes with high levels of expression in heads (attP-M-X: l(1)G0289; attP-I-2: Tsp42Ej), so maybe local enhancers that drive the CPE are stronger and more resistant to blocking.
Also consistent with earlier results, deleting the 5′ end or mutating the spacer sequence between LS4 and the H site does not affect insulator function. This agrees with the linker-scanning analysis, since the 5′ end corresponds to LS1 to LS3, and the spacer corresponds to LS5 and LS6. The 5′ deletion changes the spacing between H sites by 52 bp, from 226 to 174 bp, showing that spacing between the two H sites is not critical for insulator function.
Deleting the spacer sequence or the 3′ end impaired insulator function. Consistent with the latter result, deleting the 5′ and 3′ regions together also impaired insulator activity. The effect of the spacer deletion, in contrast to the spacer mutation, most likely indicates that spacing between LS4 and H site sequences is important. Spacing between H sites in the dimer is also affected, but this spacing is also affected by the 5′ end deletion. The spacer deletion is 42 bp while the 5′ end deletion is 52 bp, so DNA helical phasing is unlikely to be a factor. On the other hand, the 3′ end deletion likely deletes important sequences. This deletion is 55 bp, or 3 bp more than the 5′ end deletion, so helical phasing between BEAF binding sites cannot be ruled out.
With the attP-M-X line, flies with insulators with LS4 or spacer mutations exhibited an unexpected phenotype. Eyes had an anterior-dark to posterior-light pigment gradient. For the LS4 insulator, two separate injections were done with different plasmid preparations and the same result was obtained. For both insulators, correct insertion into the attP site was confirmed by PCR for two independent transgenic lines (data not shown). Differential CPE on different transgenes inserted into the same attP site has been previously reported, presumably due to differential susceptibility of different promoters to local enhancers (Mikhaylichenko et al. 2018). However, in our case, the difference between transgene constructs is confined to LS4 or spacer mutations present in the insulator dimer. The cause of the effect we see is not clear.
Analysis of sequence requirements for scs’ aurA promoter activity
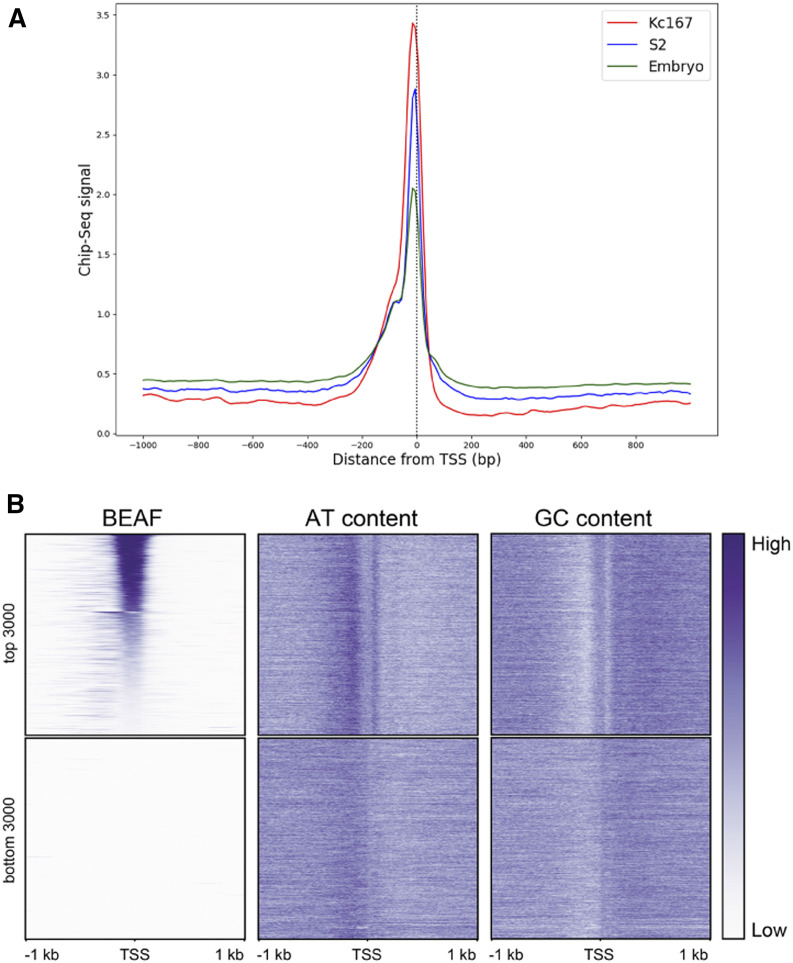
We previously mapped genome-wide BEAF binding in embryos and found that over 85% of BEAF peaks are centered within 300 bp of a TSS (Jiang et al. 2009). This is true of other genome-wide datasets as well (Figure 3A). The sequences around BEAF peaks are AT-rich (Figure 3B). Many BEAF peaks are between head-to-head divergent gene pairs, representing at least one-third of the genes organized in this fashion (Jiang et al. 2009). Some of these could fit the dual-core model where BEAF binds near both TSSs of divergent gene pairs (Emberly et al. 2008). An example of this is scs’, which has two promoters with BEAF binding sites next to both TSSs. The M fragment half of scs’ contains the TSS for aurA, a gene encoding a kinase needed during the cell cycle (Glover et al. 1995). As a model of a BEAF-associated promoter, we were curious to compare the sequence requirements for M fragment insulator and promoter activity, particularly with respect to BEAF binding. To this end we used luciferase assays to test the M fragment and its derivatives for promoter activity after transient transfection of S2 cells (Figure 4).
Figure 3.
BEAF binds near TSSs and in AT-rich sequences. (A) Distribution of BEAF binding around TSSs in Kc167 cells (red), S2 cells (blue), and embryos (green). The data are from ChIP-seq experiments (Kc167: SRX749021; S2: SRX386677; embryo: SRX119302). (B) Heatmaps showing, from left to right: BEAF binding signal; AT content; and GC content around TSSs. Top panels: 3000 TSSs arranged from strongest to weakest BEAF signal. Bottom panels: 3000 TSSs lacking significant BEAF signal. Data are for Kc167 cells (SRX749021).
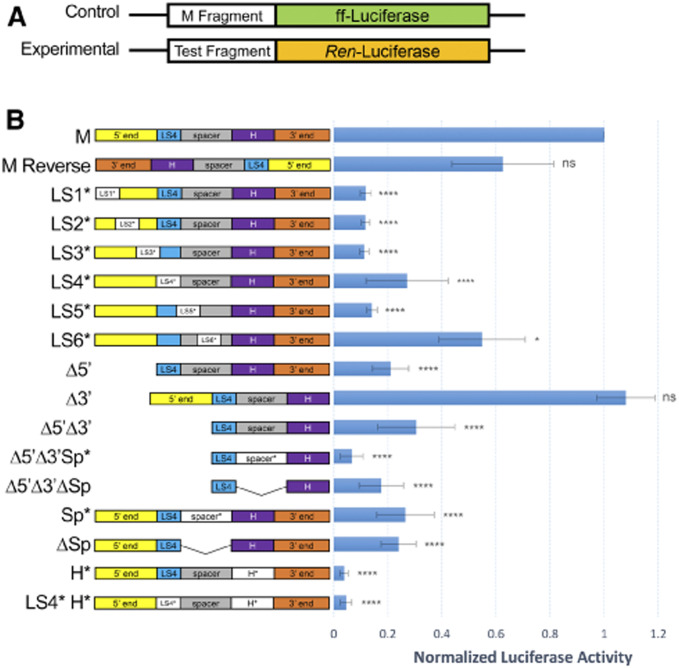
Figure 4.
Promoter analysis of the M fragment in transfected S2 cells. (A) Schematic of the control (M-firefly) and experimental (Test-Renilla) luciferase constructs. (B) Left: Schematics of the M fragment and derivatives used as test sequences in the promoter assay. See Figure 2A for details. Right: Luciferase activities of the indicated test sequences, normalized to the M-firefly luciferase activity. The M fragment has promoter activity in both orientations. Deletion of the 3′ end does not affect promoter activity. All other deletions or mutations affect promoter activity, with mutating the BEAF binding site having the strongest effect (H*: ∼50-fold) and LS6* having the weakest effect (∼2-fold). Three biological replicates were done, with SD indicated by the whiskers. One-way ANOVA P-values are indicated by asterisks (*P < 0.05; ****P < 0.0001; ns: not significant).
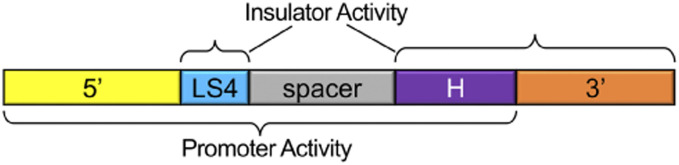
The M fragment has promoter activity, and every deletion or mutation that we tested resulted in decreased promoter activity except for deletion of the 3′ end (Figure 4B). The 3′ deletion starts 2 bp downstream of the annotated TSS, indicating that no promoter elements are located downstream of the TSS. Consistent with other studies showing bidirectional transcription in Drosophila, we found that the M fragment directs bidirectional transcription (Henriques et al. 2018; Meers et al. 2018). The LS6 mutation caused only around a twofold decrease in promoter activity, while most other deletions or mutations led to 3- to 10-fold reductions. The exception was mutation of the H site, which decreased promoter activity ∼50-fold. This strongly implicates BEAF in the function of this promoter. Thus, the sequence requirements for insulator and promoter activity overlap but are distinct (Figure 5). BEAF binding and the LS4 sequence play a role in both. However, the 5′ end plays a role in promoter function, while the 3′ end plays a role in insulator function. In addition, the sequence between LS4 and the H site, particularly the LS5 sequence, plays a role in promoter function. However, the spacer sequence is not important for insulator function, while spacing between LS4 and the H site is.
Figure 5.
Insulator and promoter activities of the scs’ M fragment can be separated despite overlapping. LS4 and the BEAF binding site (H) are important for both insulator and promoter function of the scs’ M fragment. The 3′ end plays a role in insulator function, but not promoter function. The 5′ end plays a role in promoter function, but not insulator function. Spacing between LS4 and H is important for insulator function while the spacer sequence is not, whereas the spacer sequence does play a role in promoter function.
Discussion
Previous studies have shown that BEAF binding sites can contribute to insulator function (Cuvier et al. 1998, 2002; Sultana et al. 2011; Schwartz et al. 2012), and this depends on a functional BEAF gene (Roy et al. 2007a). One of the first characterized insulators, scs’ (Kellum and Schedl 1991), has a low affinity and a high affinity BEAF binding site (Zhao et al. 1995). Also, most BEAF sites are found near TSSs (Bushey et al. 2009; Jiang et al. 2009; Nègre et al. 2010). For example, both ends of scs’ have a TSS with a BEAF binding site by each. The M fragment is half of scs’ containing the high affinity BEAF binding site and the aurA TSS. A dimer of M insulates against CPE as well as scs’ does (Cuvier et al. 1998), although a single copy is not as effective (Maharjan et al. 2018), indicating that two copies of at least some binding proteins enhances insulator function. We dissected the M fragment for insulator and promoter activity. We confirmed the importance of BEAF binding for insulator function, and found that additional sequences contribute. We also found that mutating the BEAF binding site reduced the activity of the aurA promoter ∼50-fold. We recently reported that BEAF also activates a minimal housekeeping promoter from the RpS12 ribosomal protein gene, but not a minimal developmental promoter from the y gene (Dong et al. 2020). Other sequences contribute to promoter activity, but to a lesser degree. Of interest is that while sequences involved in insulator and promoter function overlap and include the BEAF binding site, insulator and promoter function can be separated, as illustrated in Figure 5.
While the X2 dimer of the BEAF binding site alone has insulator activity, sequences important for stronger insulator activity could only be pared down to 164 bp from the original 216 bp M fragment. This includes the spacer sequence between LS4 and the H site, where spacing is important but not the sequence. Perhaps the sequences could be reduced by further analysis of the 50 bp 3′ end sequences and the spacing constraints between LS4 and the H site. Similarly, sequences important for promoter activity could only be reduced to 165 bp. This includes the 5′ end and spacer sequences, and linker-scanning analysis indicates that all are important with the possible exception of part of LS6 in the spacer region. This indicates that multiple proteins in addition to BEAF contribute to both insulator and promoter activity of M, and the only overlap in these additional proteins could be at LS4.
A goal of defining minimal sequences was to use them to identify proteins that work with BEAF, with our initial interest being insulator activity. However, the only binding to the M fragment other than BEAF that we detected did not seem sequence-specific (Zhao et al. 1995). Also, the binding activity was present using Kc proteins extracted from nuclei with 350 mM KCl, but was much weaker in nuclear extracts prepared from S2 cells and absent from embryonic nuclear extracts. These properties cast doubt on the relevance of the binding activity to insulator or promoter function. Finally, the activity was not recovered when we tried to purify it. Maybe different results would be obtained using an alternative nuclear extraction method (Parker and Topol 1984; Kamakaka and Kadonaga 1994), or, since insulator activity was detected by w expression in eyes, using nuclei isolated from adult heads or an eye disc-derived cell line.
In an alternative approach, we used programs such as JASPAR (Khan et al. 2018) to search for protein binding motifs in LS4 and MEME, DREME, and FIMO (Bailey et al. 2009) to search for sequences that are conserved in other BEAF binding regions. BEAF peak regions are AT-rich (Figure 3B), so potentially have homeodomain binding sites. We previously found genetic interactions between BEAF and several homeodomain transcription factors (Roy et al. 2007b), although no convincing physical interactions were observed using yeast two-hybrid assays (Dong et al. 2020). The best binding site motif matches we found in LS4 are for vvl, cad (CG4328 binds the same motif), and HHEX, all of which have homeodomains. These transcription factors are not expressed, or are expressed at very low levels, in Kc cells, so are unlikely to be responsible for the low mobility binding complex we detected. Also, they are developmental transcription factors, while BEAF binds predominantly near housekeeping promoters. Still, homeodomain proteins cannot be ruled out and could be pursued in the future. As previously reported (Jiang et al. 2009), we did not find any promising binding motifs common to a large number of BEAF peak regions using MEME or DREME.
Proteins that work with BEAF on the M fragment could be somewhat similar to the MSL (male specific lethal) dosage compensation complex and the LBC (late binding complex), two protein complexes that bind extended DNA sequences with poor sequence conservation between binding sites. The MSL complex is initially targeted to ∼150 chromosome entry sites (CESs), or high affinity sites, that contain an MSL recognition sequence (MRE) or a related pioneer X chromosome (PionX) site (Gelbart and Kuroda 2009; Villa et al. 2016). This targeting can be done by the protein CLAMP (Soruco et al. 2013) or the MSL2 subunit of MSL (Villa et al. 2016). DNA shape as well as sequence is important for binding to at least PionX sites. After initial binding, MSL spreads along active chromatin of the X chromosome. Since these sequences are diverse, MSL likely recognizes some feature of the active chromatin. Perhaps proteins that bind to the M fragment to work with BEAF recognize some shape feature of the AT-rich DNA, or even a nucleosome with specific modifications.
The LBC might be more relevant to our results. It is important for the insulator and insulator-bypass activities of Fab-7 and Fab-8 in the Bithorax Complex (Kyrchanova et al. 2019a,b), and binds various sequences that lack similarities other than usually having a GAGA motif (Wolle et al. 2015; Cleard et al. 2017; Kyrchanova et al. 2018, 2019b). This includes X chromosome CESs with multiple MREs where it helps recruit MSL for dosage compensation, although MSL is not recruited to LBC binding sites not on the X chromosome (Kaye et al. 2017). Binding sites are a minimum of 65 bp, although the LBC binds better to even longer DNA fragments (Kyrchanova et al. 2018). Evidence suggests the LBC is a preassembled complex of over 1000 kDa that has variable protein composition (Wolle et al. 2015; Kaye et al. 2017; Kyrchanova et al. 2018). LBC subunits include GAGA factor, Mod(mdg4), E(y)2 and CLAMP, although it has not been fully characterized. Mod(mdg4) has 31 predicted isoforms (Dorn and Krauss 2003) and forms octamers (Bonchuk et al. 2011), so probably accounts for some LBC variability. The flexible sequence recognition properties of the LBC combined with the extended length of DNA required for binding and the apparent variability in subunit composition indicate that the LBC is complex. The lack of essential binding motifs suggests some feature of the DNA other than (or in addition to) sequence could play a role in recognition. This could be similar to the situation with the M fragment, where the only obvious motif is for BEAF binding. However, unlike the LBC, maybe the proteins that bind with BEAF to the M fragment are not preassembled. Maybe they bind sequentially, and cannot assemble in the nuclear extracts that we used.
The results presented here indicate that BEAF has both insulator and promoter functions, and these can be separated. A common set of proteins might be involved at LS4, but other than that it is likely that different proteins are involved in the two activities. It is possible that DNA shape or a nucleosome with a specific modification, rather than, or in addition to, DNA sequence, plays a role in both functions. It is also possible that variable protein complexes, as seems to be the case for the LBC, could be involved. These two possibilities could account for the lack of obvious binding motifs other than for BEAF in the M fragment and other BEAF peak regions. Our results set the stage for the identification of proteins involved in these activities, which will provide insight into the role of BEAF and perhaps also other promoter-associated insulator proteins in genome architecture and gene regulation.
Acknowledgments
The authors would like to thank past and present members of the Hart laboratory for helpful discussions, Jamie Wood for advice on luciferase assays, and FlyBase as an essential Drosophila resource. This work was supported by National Science Foundation (NSF) grant 1244100 from the Division of Molecular and Cellular Biosciences (www.nsf.gov) to CMH.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12473447.
Communicating editor: M. Kuroda
Literature Cited
- Ali T., Renkawitz R., and Bartkuhn M., 2016. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 37: 17–26. 10.1016/j.gde.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Avva S. V., and Hart C. M., 2016. Characterization of the Drosophila BEAF-32A and BEAF-32B insulator proteins. PLoS One 11: e0162906 10.1371/journal.pone.0162906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E. et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonchuk A., Denisov S., Georgiev P., and Maksimenko O., 2011. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J. Mol. Biol. 412: 423–436. 10.1016/j.jmb.2011.07.052 [DOI] [PubMed] [Google Scholar]
- Bouwman B. A., and de Laat W., 2015. Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol. 16: 154 10.1186/s13059-015-0730-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey A. M., Ramos E., and Corces V. G., 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23: 1338–1350. 10.1101/gad.1798209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera P., and Azorin F., 1994. Structural characterization of intrinsically curved AT-rich DNA sequences. Nucleic Acids Res. 22: 3671–3680. 10.1093/nar/22.18.3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D., Aoki T., Erokhin M., Georgiev P., and Schedl P., 2014. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays 36: 163–172. 10.1002/bies.201300125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F., Wolle D., Taverner A. M., Aoki T., Deshpande G. et al. , 2017. Different evolutionary strategies to conserve chromatin boundary function in the bithorax complex. Genetics 205: 589–603. 10.1534/genetics.116.195586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C., and Corces V. G., 2015. Architectural proteins, transcription, and the three-dimensional organization of the genome. FEBS Lett. 589: 2923–2930. 10.1016/j.febslet.2015.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C., Rowley M. J., Lyu X., Li G., Lei E. P. et al. , 2017. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 45: 1714–1730. 10.1093/nar/gkw1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O., Hart C. M., and Laemmli U. K., 1998. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 18: 7478–7486. 10.1128/MCB.18.12.7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O., Hart C. M., Kas E., and Laemmli U. K., 2002. Identification of a multicopy chromatin boundary element at the borders of silenced chromosomal domains. Chromosoma 110: 519–531. 10.1007/s00412-001-0181-1 [DOI] [PubMed] [Google Scholar]
- Dale R. K., Matzat L. H., and Lei E. P., 2014. metaseq: a Python package for integrative genome-wide analysis reveals relationships between chromatin insulators and associated nuclear mRNA. Nucleic Acids Res. 42: 9158–9170. 10.1093/nar/gku644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Avva S., Maharjan M., Jacobi J., and Hart C. M., 2020. Promoter-proximal chromatin domain insulator protein BEAF mediates local and long-range communication with a transcription factor and directly activates a housekeeping promoter in Drosophila. Genetics 215: 89–101. 10.1534/genetics.120.303144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., and Kamakaka R. T., 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20: 520–531. 10.1093/emboj/20.3.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R., and Krauss V., 2003. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica 117: 165–177. 10.1023/A:1022983810016 [DOI] [PubMed] [Google Scholar]
- Emberly E., Blattes R., Schuettengruber B., Hennion M., Jiang N. et al. , 2008. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 6: 2896–2910 [corrigenda: PLoS Biol. 7 (2009)] 10.1371/journal.pbio.0060327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M. E., and Kuroda M. I., 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136: 1399–1410. 10.1242/dev.029645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. K., Tan Y. Y., and Hart C. M., 2006. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173: 1365–1375. 10.1534/genetics.106.056002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M., Leibowitz M. H., McLean D. A., and Parry H., 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81: 95–105. 10.1016/0092-8674(95)90374-7 [DOI] [PubMed] [Google Scholar]
- Gómez-Diaz E., and Corces V. G., 2014. Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol. 24: 703–711. 10.1016/j.tcb.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Santos G. D., Urbano J. M., Antonazzo G. et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. 10.1093/nar/gkw1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart C. M., Zhao K., and Laemmli U. K., 1997. The scs’ boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 17: 999–1009. 10.1128/MCB.17.2.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T., Scruggs B. S., Inouye M. O., Muse G. W., Williams L. H. et al. , 2018. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32: 26–41. 10.1101/gad.309351.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C. B., Grimaldi A. G., Kruse K. and Vaquerizas J. M., 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Jiang N., Emberly E., Cuvier O., and Hart C. M., 2009. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 29: 3556–3568. 10.1128/MCB.01748-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., and Kadonaga J. T., 1994. The soluble nuclear fraction, a highly efficient transcription extract from Drosophila embryos. Methods Cell Biol. 44: 225–235. 10.1016/S0091-679X(08)60916-4 [DOI] [PubMed] [Google Scholar]
- Kaye E. G., Kurbidaeva A., Wolle D., Aoki T., Schedl P. et al. , 2017. Drosophila dosage compensation loci associate with a boundary-forming insulator complex. Mol. Cell. Biol. 37: e00253-17 10.1128/MCB.00253-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., and Schedl P., 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950. 10.1016/0092-8674(91)90318-S [DOI] [PubMed] [Google Scholar]
- Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J. A. et al. , 2018. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46: D260–D266. 10.1093/nar/gkx1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I. et al. , 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245. 10.1016/j.cell.2006.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., and Georgiev P., 2014. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 588: 8–14. 10.1016/j.febslet.2013.10.039 [DOI] [PubMed] [Google Scholar]
- Kyrchanova O., Kurbidaeva A., Sabirov M., Postika N., Wolle D. et al. , 2018. The bithorax complex iab-7 Polycomb response element has a novel role in the functioning of the Fab-7 chromatin boundary. PLoS Genet. 14: e1007442 10.1371/journal.pgen.1007442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Sabirov M., Mogila V., Kurbidaeva A., Postika N. et al. , 2019a Complete reconstitution of bypass and blocking functions in a minimal artificial Fab-7 insulator from Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA 116: 13462–13467. 10.1073/pnas.1907190116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Wolle D., Sabirov M., Kurbidaeva A., Aoki T. et al. , 2019b Distinct elements confer the blocking and bypass functions of the bithorax fab-8 boundary. Genetics 213: 865–876. 10.1534/genetics.119.302694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan M., Maeda R. K., Karch F., and Hart C. M., 2018. Using a phiC31 “Disintegrase” to make new attP sites in the Drosophila genome at locations showing chromosomal position effects. PLoS One 13: e0205538 10.1371/journal.pone.0205538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers M. P., Adelman K., Duronio R. J., Strahl B. D., McKay D. J. et al. , 2018. Transcription start site profiling uncovers divergent transcription and enhancer-associated RNAs in Drosophila melanogaster. BMC Genomics 19: 157 10.1186/s12864-018-4510-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylichenko O., Bondarenko V., Harnett D., Schor I. E., Males M. et al. , 2018. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32: 42–57. 10.1101/gad.308619.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A. et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O. et al. , 2018. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19: e46255 10.15252/embr.201846255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. S., and Topol J., 1984. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell 36: 357–369. 10.1016/0092-8674(84)90229-0 [DOI] [PubMed] [Google Scholar]
- Pauli T., Vedder L., Dowling D., Petersen M., Meusemann K. et al. , 2016. Transcriptomic data from panarthropods shed new light on the evolution of insulator binding proteins in insects: Insect insulator proteins. BMC Genomics 17: 861 10.1186/s12864-016-3205-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., 2014. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 47: 11.12.1-34. 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab J. R., and Kamakaka R. T., 2010. Insulators and promoters: closer than we think. Nat. Rev. Genet. 11: 439–446. 10.1038/nrg2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D. P., Gruning B., Bhardwaj V., Kilpert F. et al. , 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D. et al. , 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680 [corrigenda: Cell 162: 687–688 (2015)]. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M. J., Nichols M. H., Lyu X., Ando-Kuri M., Rivera I. S. M. et al. , 2017. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol Cell 67: 837–852.e7. 10.1016/j.molcel.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Gilbert M. K., and Hart C. M., 2007a Characterization of BEAF mutations isolated by homologous recombination in Drosophila. Genetics 176: 801–813. 10.1534/genetics.106.068056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Tan Y. Y., and Hart C. M., 2007b A genetic screen supports a broad role for the Drosophila insulator proteins BEAF-32A and BEAF-32B in maintaining patterns of gene expression. Mol. Genet. Genomics 277: 273–286. 10.1007/s00438-006-0187-8 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Linder-Basso D., Kharchenko P. V., Tolstorukov M. Y., Kim M. et al. , 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22: 2188–2198. 10.1101/gr.138156.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. C., Merrett S. L., and Willard H. F., 2006. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 16: 119–129. 10.1016/j.cub.2005.11.065 [DOI] [PubMed] [Google Scholar]
- Shrestha S., Oh D. H., McKowen J. K., Dassanayake M., and Hart C. M., 2018. 4C-seq characterization of Drosophila BEAF binding regions provides evidence for highly variable long-distance interactions between active chromatin. PLoS One 13: e0203843 10.1371/journal.pone.0203843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms T. A., Miller E. C., Buisson N. P., Jambunathan N., and Donze D., 2004. The Saccharomyces cerevisiae TRT2 tRNAThr gene upstream of STE6 is a barrier to repression in MATalpha cells and exerts a potential tRNA position effect in MATa cells. Nucleic Acids Res. 32: 5206–5213. 10.1093/nar/gkh858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soruco M. M., Chery J., Bishop E. P., Siggers T., Tolstorukov M. Y. et al. , 2013. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 27: 1551–1556. 10.1101/gad.214585.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana H., Verma S., and Mishra R. K., 2011. A BEAF dependent chromatin domain boundary separates myoglianin and eyeless genes of Drosophila melanogaster. Nucleic Acids Res. 39: 3543–3557. 10.1093/nar/gkq1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A., Maine E., and Schedl P., 1985. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 185: 341–358. 10.1016/0022-2836(85)90408-5 [DOI] [PubMed] [Google Scholar]
- Ulianov S. V., Khrameeva E. E., Gavrilov A. A., Flyamer I. M., Kos P. et al. , 2016. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 26: 70–84. 10.1101/gr.196006.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa R., Schauer T., Smialowski P., Straub T., and Becker P. B., 2016. PionX sites mark the X chromosome for dosage compensation. Nature 537: 244–248. 10.1038/nature19338 [DOI] [PubMed] [Google Scholar]
- Wolle D., Cleard F., Aoki T., Deshpande G., Schedl P. et al. , 2015. Functional requirements for fab-7 boundary activity in the bithorax complex. Mol. Cell. Biol. 35: 3739–3752. 10.1128/MCB.00456-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Mikkelsen T. S., Gnirke A., Lindblad-Toh K., Kellis M. et al. , 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. USA 104: 7145–7150. 10.1073/pnas.0701811104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C., Prive G. G., Goodsell D. S., and Dickerson R. E., 1988. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proc. Natl. Acad. Sci. USA 85: 6332–6336. 10.1073/pnas.85.17.6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi M. A., Arnold C. D., Schernhuber K., Pagani M., Rath M. et al. , 2015. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559. 10.1038/nature13994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Hart C. M., and Laemmli U. K., 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81: 879–889. 10.1016/0092-8674(95)90008-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Fly lines and plasmids are available upon request. Insulator (dimers), promoter (monomers), and primer sequences used for cloning are in Supplemental Material, Table S1. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12473447.