Abstract
Organisms adapted to life in extreme habitats (extremophiles) can further our understanding of the mechanisms of genetic stability, particularly replication and repair. Despite the harsh environmental conditions they endure, these extremophiles represent a great deal of the Earth’s biodiversity. Here, for the first time in a member of the archaeal domain, we report a genome-wide assay of spontaneous mutations in the halophilic species Haloferax volcanii using a direct and unbiased method: mutation accumulation experiments combined with deep whole-genome sequencing. H. volcanii is a key model organism not only for the study of halophilicity, but also for archaeal biology in general. Our methods measure the genome-wide rate, spectrum, and spatial distribution of spontaneous mutations. The estimated base substitution rate of 3.15 × 10−10 per site per generation, or 0.0012 per genome per generation, is similar to the value found in mesophilic prokaryotes (optimal growth at ∼20–45°). This study contributes to a comprehensive phylogenetic view of how evolutionary forces and molecular mechanisms shape the rate and molecular spectrum of mutations across the tree of life.
Keywords: Haloferax volcanii, mutation accumulation, mutation rate, mutation spectrum, AT bias, genome copy number
SPONTANEOUS mutation is the ultimate source of novel genetic variation in nature and influences the genetic features of all populations. Thus, because mutation affects processes including evolution, inheritance, and genetic disorders, it is important to understand the intrinsic and extrinsic factors influencing the rate and molecular spectrum of mutations. Until recently, most strategies for determining the mutation rate and spectrum were primarily indirect, including interspecies comparison of putatively neutral sites in specific genes (Graur and Li 2000; Wielgoss et al. 2011) and analyses using reporter-construct genes (Drake 1991). However, because selection can affect synonymous sites (Lawrie et al. 2013), mutation rates can vary significantly across different genomic regions (Foster et al. 2013) and only a fraction of reporter-construct mutations are detectable (Drake 1991), both of these methods are likely to have significant biases.
In long-term mutation accumulation (MA) experiments, replicate lines are taken through regular population bottlenecks to minimize the efficiency of selection, allowing cell lines to accumulate mutations in an unbiased fashion (Kibota and Lynch 1996; Lynch et al. 2016). By applying high-throughput (WGS)], it has become possible to directly identify these mutations (Lynch et al. 2008; Keightley and Halligan 2009). As a result, unbiased estimates of the genome-wide rate and molecular spectrum of spontaneous mutations have been generated for a large number of eukaryotic and bacterial organisms (Lynch et al. 2016; Long et al. 2018). However, to understand how DNA replication and repair cooperate, and ultimately determine the genome-wide mutation rate across the entire tree of life, it is necessary to expand these experimental assays to the unexplored archaeal domain.
The moderately halophilic (optimal NaCl 1.7–2.5 M) and mesothermophilic (30–49°) archaeon Haloferax volcanii has a moderately GC-rich genome (65.6%), containing a primary 2.848-Mb chromosome, three smaller chromosomes (pHV1, 3, and 4), and the pHV2 plasmid (Hartman et al. 2010). H. volcanii can be cultured relatively easily, which is rare for an extremophile, and is a model organism not only for the study of halophilicity, but also for archaeal biology in general. In the last decade, H. volcanii has been widely studied in efforts to understand archaeal DNA replication/repair and transcription mechanisms. For example, it has been found that the cell machinery involved in replication and repair in eukaryotes is more closely related to that of archaea than bacteria (Barry and Bell 2006). Previous in vitro studies suggesting the possibility that the fidelity of archaeal DNA polymerases is higher than that of its bacterial counterparts (Kunkel 1992; Cline et al. 1996; Bloom et al. 1997; Grogan et al. 2001) provide further motivation for the current study.
As is characteristic in many clades of Euryarchaeota (e.g., Halobacteriales, Methanosarcinales, Thermococcales, and Methanococcales), H. volcanii—while genetically haploid—has been shown to carry a variable number of genome copies, with 2–45 genome copies per cell depending on the growth phase (Breuert et al. 2006; Zerulla et al. 2014). It has been proposed that high genome copy numbers may have selective advantages for prokaryotes, especially those living in extreme environments. The possible advantages of many genome copies in prokaryotes include efficient DNA repair by homologous recombination, higher rates of transcription/translation in nutrient-limiting conditions, restrained phenotypic expression of deleterious recessive mutations (Breuert et al. 2006; Zahradka et al. 2006), and the use of genomic DNA as a phosphate storage polymer (Zerulla et al. 2014). Some of these effects might either increase or decrease indirect mutation rate estimations.
Previous mutation rate estimates in a few Archaeal species have been derived using indirect methods, such as fluctuation tests, which rely on the production of a phenotype to indicate the random mutation of a known locus or loci, and are sometimes coupled with Sanger sequencing: H. volcanii (Mackwan et al. 2007), Halobacterium salinarum (Busch and DiRuggiero 2010), Sulfolobus acidocaldarius (Jacobs and Grogan 1997; Grogan et al. 2001), and Thermus thermophilus (Mackwan et al. 2008). In this study—for the first time to our knowledge—we directly determine the rate, spectrum, and distribution of genome-wide mutation in an archaeon, thereby enhancing understanding of how different phylogenetic backgrounds influence the mutation process.
Materials and Methods
Mutation accumulation
One hundred independent H. volcanii DS2 [American Type Culture Collection (ATCC) 29605] MA lines were initiated from a single colony. Halobacterium medium 974, as recommended by the ATCC, was used for the MA line transfers. Every week, a single colony from each MA line was transferred by streaking to a new plate, ensuring that each line regularly passed through a single-cell bottleneck. This bottlenecking procedure ensures that mutations accumulate in an effectively neutral fashion (Muller 1928; Bateman 1959; Mukai 1964; Kibota and Lynch 1996). MA lines were incubated at 30° under aerobic conditions and each line passed through ∼3000 cell divisions (Supplemental Material, Table S1). Frozen stocks of all lineages were prepared by growing a final colony per isolate in 1 ml Halobacterium broth incubated at 30°, and freezing in 20% glycerol at −80°.
The number of generations undergone by each MA line per transfer was determined based on the average number of cells in a colony. Specifically, each month, one single colony randomly chosen from at least 10 MA lines was transferred to a sterile tube with basal salt solution (medium without carbon source), vortexed, serially diluted, and plated. After 6 days of incubation at 30°, colony-forming units (denoted as N here) of the diluted cultures were counted and averaged. The number of generations was calculated by
DNA extraction and sequencing
Lines surviving to the end of the MA experiment were prepared for WGS: DNA was extracted with the Wizard Genomic DNA Purification kit (Promega, Madison, WI) and DNA libraries for Illumina HiSeq 2500 sequencing (insert size 300 bp) were constructed using the Nextera DNA Sample Preparation kit (Illumina, San Diego, CA). Paired-end 150-nt read sequencing of MA lines was done by the Hubbard Center for Genome Studies, University of New Hampshire, with an average sequencing depth of 126× across all lines (Table S1).
Mutation identification and analyses
Adaptors of paired-end reads were removed with Trimmomatic 0.32 (Bolger et al. 2014); trimmed reads were mapped to the reference genome [National Center for Biotechnology Information (NCBI) accession numbers: NC_013964–NC_013968] using Burrows-Wheeler Aligner (BWA), version 0.7.12 (Li and Durbin 2009). The output was parsed with SAMTOOLS (Li et al. 2009); we also applied duplicate-read removal using picardtools-1.141, and read realignment around insertion/deletions (indels) using GATK 3.6, before performing SNP and indel discovery with standard hard-filtering parameters described in GATK Best Practices recommendations (Phred-scaled quality score QUAL > 100 and root mean square of mapping quality (MQ) > 59 for both variant and nonvariant sites; ploidy setting: diploid higher ploidy did not change mutation detection) (McKenna et al. 2010; DePristo et al. 2011). Base substitutions and small indels were called using the Haplotype Caller in GATK. In order to call a variant, a minimum of 10 reads was needed and 99% of reads in a line were required to call the line-specific consensus nucleotide at a site; a ∼1% cutoff was set to allow for aberrant reads originating from sequencing errors, contamination of pure indexes during library construction, or barcode degeneracy during sequence demultiplexing. All novel homozygous and heterozygous variants (mutations that are segregating within an individual) that were supported by at least three reads for both alleles were kept. Presences of structural variants (transpositions, deletions, inversions, and duplications) were called using GRASPER (Lee et al. 2016) and RetroSeq (Keane et al. 2013), while the relative ploidy level within an MA line was determined by creating sequencing coverage maps of 10,000-bp sliding windows with BEDTools (Quinlan and Hall 2010). All structural variants were confirmed by visual examination using Integrative Genomics Viewer (IGV) (Robinson et al. 2011).
Statistics and calculations
The per-site per-cell division mutation rate (μ) was calculated for each line as , where m is the number of observed mutations, n is the number of sites analyzed in the MA line, and T is the number of generations for the line. The SEM mutation rate was calculated with the equation , where SD is the SD of mutation rates of each line.
The expected GC content at mutation equilibrium was calculated as (Lynch 2007): , where μA/T→G/C is the rate of mutations at A:T sites resulting in an A/T → G/C change (including A/T → G/C transitions and A/T → C/G transversions), and μG/C→A/T is the rate of mutations at G:C sites resulting in a G/C → A/T change (including G/C → A/T transitions and G/C → T/A transversions) (Table S1). Mutation bias in the G/C direction was calculated by . We used R v3.1.0 (R Development Core Team 2014) for all statistical tests. The 95% Poisson C.I.s were calculated using the Poisson test in R. While testing if the observed conditional base substitution mutation spectra differed between chromosomes, the cutoff P-value was determined using Bonferroni correction, i.e., P = 0.05/n, where n equals 6 – the number of conditional base substitution types.
A consideration of genome copy number effect on MA experiment outcomes
Because H. volcanii is reported to maintain a large number of genome copies (2–45 genome copies/cell) (Zerulla et al. 2014), a newly arising neutral mutation needs time to reach fixation within a cell. In a single-cell lineage, the average time to fixation is around two times the ploidy for a newly arisen selectively neutral mutation (Kimura and Ohta 1969). Thus, if genomes are randomly transmitted to new cells and each generation starts from a single cell, conditional on drifting to fixation, a new mutation in H. volcanii should reach fixation in 4–90 generations. Considering that each H. volcanii MA line passed through an average of 3000 cell divisions in this study, heterozygous mutations should be a very small fraction of the total; even if genome copy number were as high as 45, heterozygous mutations should still be a small fraction [∼0.05 (45 genome copies × 3,866,751 nt sites per genome copy × 3.15 10−10 base pair substitutions per site)] and of negligible significance if they were undetected. Furthermore, even if the genome copy number increases at some stage of colony development, the two-genome stage acts as a bottleneck to accelerate the fixation of a mutation within a lineage. Therefore, the number of fixed mutations per generation is very close to the mutation rate per generation. However, as a conservative approach, we conducted a secondary estimation of mutation rate taking heterozygous variants into account. Here, heterozygous variants were scaled by their proportion of supporting reads where and mutation rate was then recalculated for each sample as .
Data availability
The workflow has been deposited at the GitHub repository (https://github.com/sibelkucukyildirim) and raw sequences are available at the Sequence Read Archive at the NCBI (Bioproject No.: PRJNA386190). Supplemental material available at figshare: https://doi.org/10.25386/genetics.12411035.
Results
To estimate the mutation rate in H. volcanii DS2, an MA experiment was carried out for 37 months (∼3000 generations) with 100 independent lineages, all derived from a common progenitor colony of H. volcanii. Every week, a single colony from each line was restreaked onto a fresh plate with transfers producing colonies from probably just a single cell. Such treatment minimizes the effective population size, such that all but lethal mutations should accumulate in an effectively neutral manner (Kibota and Lynch 1996). The number of generations undergone by each MA line per transfer was determined based on the number of cells in the colonies at the time of transfer (23.6 ± 6.4 generations per transfer). There was no significant change in the number of cells per colony throughout the experiment, suggesting that the number of generations per transfer stayed constant. The 57 lines that survived over 122 transfers—to the end of the MA experiment—were prepared for WGS. Of these lines, 54 had sufficient depth of coverage (> 156×) to calculate the mutation rate and spectrum. After sequencing and detecting mutations in those 54 MA lines, we noted that three mutations were shared by all the MA lines, indicating that those mutations were present in the ancestor line and were removed from further downstream analysis (Table S1).
Mutation rate
Across 54 H. volcanii MA lines, we identified 167 single-base substitution changes (Table S2), yielding an overall base substitution mutation rate of 3.15 × 10−10 (SE = 0.27 × 10−10) per site per generation, or 0.0012 per genome per generation. In addition to single-base substitutions, we also identified 14 multinucleotide mutations (MNMs; defined as multiple mutations that occur within 50 nt in the same MA line) (Schrider et al. 2011) (Table S3). Including these 14 MNMs increases the overall base substitution mutation rate to 3.24 × 10−10 (SE = 0.28 × 10−10) per site per generation. As noted in the Material and Methods section, because the H. volcanii genome is known to be transiently maintained in a polyploid state of up to 45 genome copies, we repeated our analysis to include heterozygous mutations (only the heterozygous variants that were supported by at least three reads for both alleles). This analysis revealed 12 heterozygous mutations, all of which were MNMs (Table S3). These heterozygous mutations represent a very small fraction of our data (∼0.02) and account for a negligible difference in the estimated mutation rate. Thus, the mutation rate in H. volcanii (per haploid genome) is similar to mutation rates commonly observed in mesothermophilic bacteria (Long et al. 2018).
We also found 19 short indels 1–30-bp in length (2 insertions and 17 deletions), yielding an indel rate of 3.58 × 10−11 (SE = 0.82 × 10−11) per site per generation (Tables S2 and S4). These small indel events comprise 10.2% of all accumulated mutations in H. volcanii, consistent with results from many other microbial MA experiments (Sung et al. 2016). The deletion rate for H. volcanii is ∼8.5× greater than the insertion rate resulting in a small net rate of loss of genomic sites, consistent with a suggested universal prokaryotic deletion-bias hypothesis (Mira et al. 2001), and 21% of the small indels occur in simple sequence repeats, e.g., homopolymer runs (Table S4).
One of the goals of this study was to examine whether mutation rates and spectra differ between the main chromosome and the mini chromosomes (pHV1, pHV3, and pHV4), which differ in size and content. The heterogeneity of overall base substitution mutation rates of each chromosome was evaluated with a χ2 test. We were unable to reject the null expectation that the number of substitutions is proportional to the number of sites covered on each chromosome (χ2 = 7.34, P = 0.062) (Tables S5–S8 and Figure S1). This may not appear to be surprising, given that all chromosomes are expected to be replicated and repaired by the same mechanisms.
Using the annotated H. volcanii DS2 genome, we identified the functional context of each base substitution (Table S9). Across the 54 sequenced MA lines, 120 of the 167 (71.9%) substitutions occurred in coding regions (∼86% of the genome consists of coding sequence), while the remaining 47 were found at noncoding sites (Table S9). Given the codon usage and transition/transversion ratio in H. volcanii, the expected ratio of nonsynonymous to synonymous mutations is 2.77 in the absence of selection, which is not significantly different from the observed ratio of 2.24 (83/37) (χ2 = 1.06, P = 0.3). Thus, selection does not appear to have had a significant influence on the distribution of mutations in this experiment, as has been shown repeatedly for the MA-WGS method (Lee et al. 2012; Dillon et al. 2015; Long et al. 2015; Kucukyildirim et al. 2016; Senra et al. 2018).
Structural variants
We called medium/large deletions, transpositions, inversions, and tandem duplications using a combination of GRASPER (Lee et al. 2016), RetroSeq (Keane et al. 2013), and IGV (Robinson et al. 2011). We detected 24 total structural variants consisting of 3 large deletions on the main chromosome, and 3 inversions and 18 large deletions on the plasmid pHV4. Large deletions were the most common form of structural variation (21 events) accounting for a genome-wide large-scale deletion rate of 3.77 × 10−11 per site per generation (Table S10). Of these large-scale deletions, 19 were mediated by recombination between insertion sequence (IS) elements.
IS elements are short mobile elements in prokaryotic (bacterial and archaeal) genomes, initially identified by their capacity to generate mutations. Previous studies have identified IS element-related mutations as being mostly deleterious or selectively neutral (Kidwell and Lisch 2001; Chandler and Mahillon 2002). The H. volcanii genome has two major IS element types (ISH51 and the newly described IS4-type elements), both of which are members of the IS4 family (Hofman et al. 1986; Hartman et al. 2010). IS element-mediated genomic rearrangement has previously been reported in Ha. salinarum (Sapienza et al. 1982).
In H. volcanii, IS elements are unevenly distributed among the chromosomes with pHV4 harboring more than the other chromosomes (Hartman et al. 2010). Thus, we looked to see if a chromosomal bias existed for IS-mediated deletions. If IS-mediated deletions were distributed evenly between the main chromosome and pHV4 with respect to size, we would expect 85% of the deletions to occur on the main chromosome and 15% of the deletions on pHV4. However, we see the opposite relationship, i.e., a strong bias for IS-mediated deletion toward pHV4 (χ2 = 45.296, P = 1.23 × 10−11). Our results are consistent with the highly dynamic nature of archaeal genomes (Redder and Garrett 2006; Bridger et al. 2012), and indicate that a high deletion rate related to IS elements may be affected by genome composition and by the fact that various IS elements have different activities.
Mutation spectrum
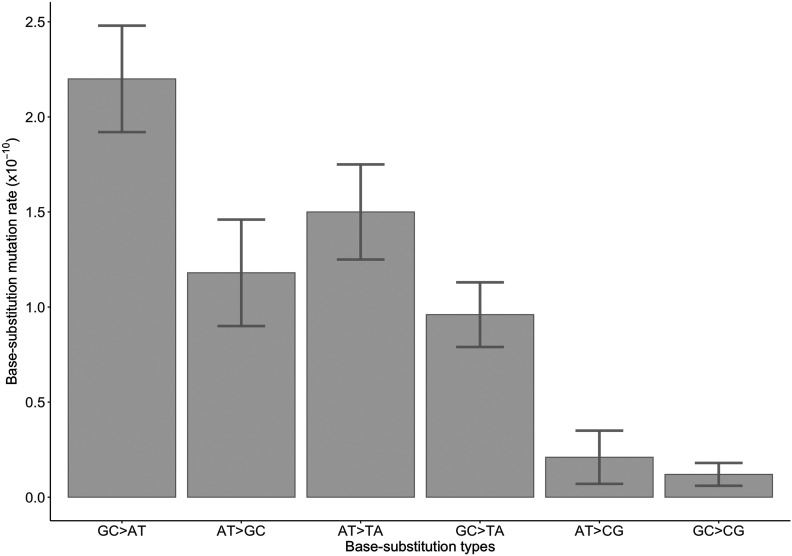
Across the 54 MA lines, there were 98 transitions and 69 transversions, resulting in a transition/transversion ratio of 1.42. There are 76 G:C→A:T transitions and 33 G:C→T:A transversions at GC sites, yielding a mutation rate in the AT direction of per G:C site per generation. In contrast, 22 A:T→G:C transitions and 4 A:T→C:G transversions yielded a mutation rate in the GC direction of per A:T site per generation (Table S2 and Figure 1), which is significantly lower than the rate (95% Poisson C.I.s for are 2.6−3.8 × 10−10 and, for , are 0.9−2.0 × 10−10). As such, the expected GC content under mutation–drift equilibrium is 0.31 ± 0.04 (SEM), lower than the genome-wide GC content (0.66).
Figure 1.
Conditional base substitution mutation rates for H. volcanii MA lines. Rates for each base substitution type normalized by genome base composition; error bars indicate the SE.
We performed a χ2 test to investigate whether the observed conditional base substitution mutation spectra differ between chromosomes (main chromosome/pHV1: χ2 = 4.73, d.f. = 5, P = 0.45; main chromosome/pHV3: χ2 = 5.72, d.f. = 5, P = 0.33; and main chromosome/pHV4: χ2 = 12.6, d.f. = 5, P = 0.03; after the Bonferroni correction, the cutoff P value = 0.05/6 = 0.0083). These comparisons show that there is no significant chromosomal variation in the overall base substitution spectra of H. volcanii (Figure S1 and Tables S5–S8).
Spontaneous DNA damage, such as deamination of cytosine to uracil and C5 methylcytosine to thymine, or oxidation of guanine to 7,8-dihidro-8-oxoguanine, can lead to mutational biases. Methylated bases are well-known mutational hotspots in bacteria (Coulondre et al. 1978; Duncan and Miller 1980; Lind and Andersson 2008; Lee et al. 2012). Ouellette et al. (2015) found that H. volcanii DS26 contains methyltransferases that modify two motifs in the genome: GCAm6BN6VTGC (B stands for C, G, or T, V stands for A, C, or G, and N stands for any base) and Cm4TAG. Although the percentage of methylation of these sites is high in the H. volcanii genome (77 and 28%, respectively) (Ouellette et al. 2015), we found no elevation of the mutation rate at the above-noted methylated sites. We also examined bacterial canonical and noncanonical sites (e.g., GATC, CTAG, GACC, and CACC), previously suggested as methylation sites (Clark et al. 2012; Long et al. 2015), but found no indication of mutation rate elevation.
Discussion
The biological processes related to mutation (prevention, production, and repair) vary among genetic contexts. This variation leads to biases in the types, locations, and numbers of genetic changes that occur. As a consequence, organisms with different genome compositions may experience different types of genetic variation. As such, determining variation in the mutation process is important to understand how genome composition affects neutral and adaptive sequence evolution.
Analyses of spontaneous mutation have shown that most mesothermophilic bacteria share two mutational characteristics: (1) total genome-wide mutation rates in the general range of 0.0025–0.0046 per genome per generation (Drake 1991; Drake et al. 1998; Lynch et al. 2016), with the variation potentially explained by the influence of the drift barrier (Kuo and Ochman 2009; Sung et al. 2012; Lynch et al. 2016), and (2) base substitutions comprising ∼83% of the total mutations (Sung et al. 2016). However, previous analyses of Archaea based on reporter constructs estimated a H. volcanii genomic mutation rate of 0.00045 per genome per generation, ∼10-fold lower than the consensus value of 0.004 observed in other mesothermophilic prokaryotes, as well as a predominance of small indel events (Grogan et al. 2001; Mackwan et al. 2007; Mackwan et al. 2008).
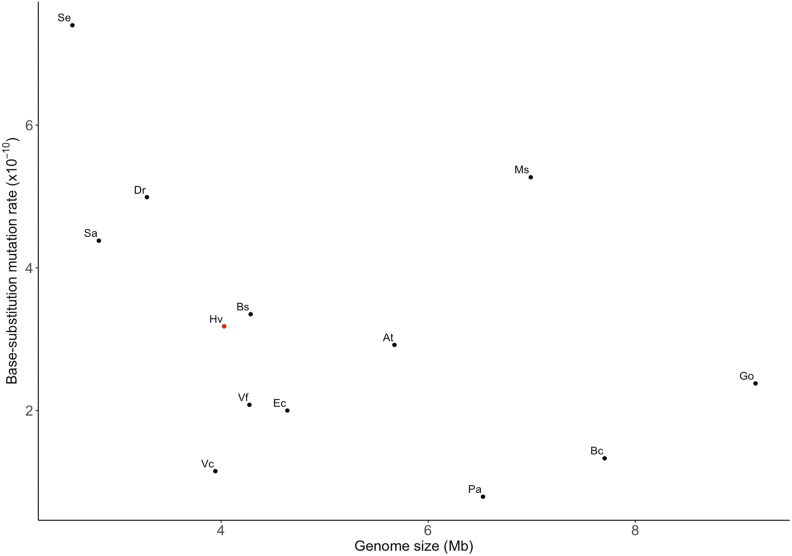
It has been observed that mutation rate estimates can differ between MA experiments and specific locus methods, even with carefully chosen reporter genes (Table 1), and these differences are often significant (paired Student’s t-test, two-tailed P = 0.029) (Drake 2012). These discrepancies likely arise in part because mutation rates vary among individual nucleotide types (Long et al. 2015, 2018; Kucukyildirim et al. 2016), depending on their neighboring content (Sung et al. 2012), and vary among chromosomal regions (Long et al. 2014; Dillon et al. 2015; Sung et al. 2015; Niccum et al. 2019). In addition, only a fraction of reporter-construct mutations are molecularly detectable (Drake 1991). Thus, for any organism, a single locus will hardly ever provide an unbiased estimate of the genomic mutation rate or molecular spectrum. As next-generation sequencing becomes increasingly less expensive, and now that higher-quality reference genomes are available for a large number of microbial species, MA combined with WGS provides a more comprehensive and nearly unbiased description of spontaneous mutations (for the given environmental setting) across the entire genome, as supported by our cross-checks on synonymous vs. nonsynonymous base substitutions. This reduces the risk of spurious conclusions based on a single gene and avoids other methodological issues associated with reporter gene approaches (e.g., mutation detectability) for mutation rate measurement. We report a base substitution mutation rate for H. volcanii of 3.15 × 10−10 per base pair per generation, nearly threefold higher than the previously reported rate of 1.08 × 10−10 based on the pyrE2 reporter locus (Mackwan et al. 2007), but still lower than the average mesophilic bacterial base substitution rate (Figure 2). Moreover, we do not observe an excess of indel mutations, contrary to a previous estimate for H. volcanii (Mackwan et al. 2007), which is the only study to date exhibiting this pattern in any organism. The reason for the observed difference between the two mutation rate estimates may be that not all mutations could be molecularly detected at the reporter locus (∼75%).
Table 1. Examples for base substitution mutation rate (μ) estimates by using WGS of MA experiments and reporter-construct estimates.
Figure 2.
The relationship of the base substitution mutation rate per site per generation with total haploid genome size. Data points correspond to the following species: At: Agrobacterium tumefaciens (Sung et al. 2016), Bs: Bacillus subtilis (Sung et al. 2015), Bc: Bulkholderia cenocepacia (Dillon et al. 2015), Dr: Deinococcus radiodurans (Long et al. 2015a), Ec: E. coli (Lee et al. 2012), Go: Gemmata obscuriglobus (Long et al. 2018), Hv: H. volcanii (this study), Ms: Mycobacterium smegmatis (Kucukyildirim et al. 2016), Pa: Pseudomonas aeruginosa (Dettman et al. 2016), Sa: Staphylococcus aureus (Long et al. 2018), Se: S. epidermidis (Sung et al. 2016), Vf: Vibrio fischeri, and Vc: Vibrio cholera (Dillon et al. 2017). All data are derived from mutation accumulation-whole-genome sequencing projects.
Possible effects of temperature
In principle, mutation rates can be influenced by a number of factors including growth conditions, growth medium, number of generations, temperature, and genome copy number (ploidy level). Accordingly, growth temperature may contribute to the differences observed in the results for our study, conducted at 30°, and the previous study of the H. volcanii mutation rate, conducted at 37° and 41° (Mackwan et al. 2007). Extensive literature, with a wide range of organisms including viruses, bacteria, microbial eukaryotes, insects, and plants, has shown that extremely high temperatures are stressful and may result in very low growth/survival rates and have mutagenic effects (Zamenhof and Greer 1958; Drake 1966; Lindgren 1972). Recent work by Chu et al. (2018) has shown that for Escherichia coli, within a range of unstressful temperatures, the genome-wide spontaneous base pair substitution rate is 1.5-fold higher at 37° than at the cooler, suboptimal growth temperatures of 25° and 28°. If this relationship between temperature and base pair substitution rate extends to H. volcanii then we would expect to observe a lower base pair substitution rate at 30° than at 37°/41°, not higher. Therefore, it seems unlikely that temperature is the main factor contributing to the differences in mutation rate estimates in H. volcanii reported here and in the previous study (Mackwan et al. 2007).
Possible effects of genome copy number
Another feature with the potential to affect the mutation process is genome copy number, which can vary among prokaryotes as a function of phyla and growth rate. As in other haloarchaeal species (such as Ha. salinarum and H. mediterranei), H. volcanii can maintain multiple copies (2–45) of its genome, sometimes at high copy numbers (Zerulla et al. 2014), which may result in detection of fewer mutations in short-term surveys that depend on phenotype and Sanger sequencing to estimate base pair substitution rates (Mackwan et al. 2007). All else being equal, the number of mutations accumulated in diploid cells is expected to be twofold higher than in haploid cells, as they have two sets of homologous chromosomes and more targets for mutation (Crow and Kimura 1965). However, unlike haploid cells, diploids have continual access to homologous DNA. This access may reduce the rate of DNA damage, but the presence of homologous DNA may also increase the probability of spontaneous structural variants and rearrangements. As recently shown in yeast, diploid cells have greater replication fidelity (for single-nucleotide changes) but are more prone to large-scale mutations with deleterious fitness effects (Sharp et al. 2018). Thus, ploidy level may affect the mutational profile.
In an MA experiment, newly arising mutations in organisms with many genome copies will be present at low frequency and may be incorrectly filtered by the hard-mapping criteria. However, we found 12 heterozygous mutations (Table S10) that represent a very small fraction of our data (∼0.02 even if we consider them independent events) and account for a negligible difference in the estimated mutation rate. This negligible contribution of heterozygotes is expected based on population-genetic theory: if equals the copy number of a chromosome, then is both the frequency of a mutation on the chromosome when it initially occurs as well as the probability of the random fixation of that mutation within the genome. When these values are combined with the per-site mutation rate, μ, then the product is the total fixation probability of a mutation or the mutation rate. Even if the genome copy number increase is as high as 45 during colony growth, the single-cell bottleneck that occurs during streaking will effectively fix mutations on average within 45 generations, assuming random segregation of chromosomes to progeny, so the number of heterozygous base pair substitutions per MA line at any time is ∼0.05.
Taken together, the time need for intracellular fixation of any novel base pair mutation is short relative to the total length of the MA experiment.
Mutation spectrum
Our study demonstrates for the first time the spectrum of spontaneous mutations in an archaeon. A fundamental question that arises is how do intrinsic forces normally shape the GC content of genomes? Here, in the moderately GC-rich H. volcanii, we observed that mutations are biased in the AT direction (vs. ) (Table S2), consistent with near-universal AT mutation bias (Hershberg and Petrov 2010). As recently discussed in Long et al. (2018), genome-wide nucleotide composition variation among species is a consequence of both interspecific differences in mutation bias and the efficiency of selection or biased gene conversion for different nucleotides, and a GC-rich genome in species with AT mutational bias is not particularly surprising.
Interestingly, we did not observe increased mutation rates for nucleotide sequences known to be methylation targets in H. volcanii. Mutation rates at these sites are expected to be higher, especially if there is mutational bias toward AT, as G/C nucleotides are more prone to DNA damage due to oxidative stress and spontaneous deamination. However, uracil DNA glycosylases have been noted to protect the genome against deamination in GC-rich bacteria (Venkatesh et al. 2003) and may explain why we found no elevation of the mutation rate at methylation sites in this organism.
In conclusion, by using the MA approach combined with WGS, we have shown that H. volcanii has a mutation rate similar to the consensus value of other prokaryotic organisms, as well as a commonly observed A/T-biased mutational spectrum. More precise genetic and biochemical assays are necessary to clarify the mechanisms maintaining genomic stability in Archaea.
Acknowledgments
We thank Samuel F. Miller for technical support and Hongan Long for helpful discussions. This study is funded by a Multidisciplinary University Research Initiative award from the US Army Research Office (W911NF-09-1-0444) and National Institutes of Health awards (R01-GM-036827, R35-GM-122566, and F32-GM-123703).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12411035.
Communicating editor: M. Nachman
Literature Cited
- Barry E. R., and Bell S. D., 2006. DNA replication in the Archaea. Microbiol. Mol. Biol. Rev. 70: 876–887. 10.1128/MMBR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. J., 1959. The viability of near-normal irradiated chromosomes. Int. J. Radiat. Biol. 1: 170–180. [Google Scholar]
- Behringer M. G., and Hall D. W., 2016. Genome-wide estimates of mutation rates and spectrum in Schizosaccharomyces pombe indicate CpG sites are highly mutagenic despite the absence of DNA methylation. G3 (Bethesda) 6: 149–160. 10.1534/g3.115.022129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom L. B., Chen X., Fygenson D. K., Turner J., O’Donnell M. et al. , 1997. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J. Biol. Chem. 272: 27919–27930. 10.1074/jbc.272.44.27919 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinfomatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuert S., Allers T., Spohn G., and Soppa J., 2006. Regulated polyploidy in halophilic archaea. PLoS One 1: e92 10.1371/journal.pone.0000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger S. L., Lancaster W. A., Poole F. L., Schut G. J., and Adams M. W., 2012. Genome sequencing of genetically tractable Pyrococcus furiosus strain reveals a highly dynamic genome. J. Bacteriol. 194: 4097–4106. 10.1128/JB.00439-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch C. R., and DiRuggiero J., 2010. MutS and MutL are dispensable for maintenance of the genomic mutation rate in the halophilic archaeon Halobacterium salinarum NRC-1. PLoS One 5: e9045 10.1371/journal.pone.0009045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., and Mahillon J., 2002. Insertion sequences revisited, pp. 305–366 in Mobile DNA II, edited by Craig N. L., Craigie R., Gellert M., and Lambowitz A.. ASM Press, Washington, D.C. [Google Scholar]
- Chang D.-Y., Gu Y., and Lu A.-L., 2001. Fission yeast (Schizosaccharomyces pombe) cells defective in the MutY-homologous glycosylase activity have a mutator phenotype and are sensitive to hydrogen peroxide. Mol. Genet. Genomics 266: 336–342. 10.1007/s004380100567 [DOI] [PubMed] [Google Scholar]
- Chu X.-L., Zhang B.-W., Zhang Q.-G., Zhu B.-R., Lin K. et al. , 2018. Temperature responses of mutation rate and mutational spectrum in an Escherichia coli strain and the correlation with metabolic rate. BMC Evol. Biol. 18: 126 10.1186/s12862-018-1252-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T. A., Murray I. A., Morgan R. D., Kislyuk A. O., and Spittle K. E., 2012. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 40: e29 10.1093/nar/gkr1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline J., Braman J. C., and Hogrefe H. H., 1996. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24: 3546–3551. 10.1093/nar/24.18.3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., and Gilbert W., 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775–780. 10.1038/274775a0 [DOI] [PubMed] [Google Scholar]
- Crow J. F., and Kimura M., 1965. Evolution in sexual and asexual populations. Am. Nat. 99: 439–450. 10.1086/282389 [DOI] [Google Scholar]
- DePristo M. B. E., Poplin R., Garimella K., Maguire J., Hartl C. et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman J. R., Sztepanacz J. L., and Kassen R., 2016. The properties of spontaneous mutations in the opportunistic pathogen Pseudomonas aeruginosa. BMC Genomics 17: 27 10.1186/s12864-015-2244-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon M. M., Sung W., Lynch M., and Cooper V. S., 2015. The rate and molecular spectrum of spontaneous mutations in the GC-rich multichromosome genome of Burkholderia cenocepacia. Genetics 200: 935–946. 10.1534/genetics.115.176834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon M. M., Sung W., Sebra R., Lynch M., and Cooper V. S., 2017. Genome-wide biases in the rate and molecular spectrum of spontaneous mutations in Vibrio cholerae and Vibrio fischeri. Mol. Biol. Evol. 34: 93–109. 10.1093/molbev/msw224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88: 7160–7164. 10.1073/pnas.88.16.7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., 1966. Spontaneous mutations accumulating in bacteriophage T4 in the complete absence of DNA replication. Proc. Natl. Acad. Sci. USA 55: 738–743. 10.1073/pnas.55.4.738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., 2012. Contrasting mutation rates from specific-locus and long-term mutation-accumulation procedures. G3 (Bethesda) 2: 483–485. 10.1534/g3.111.001842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Charlesworth B., Charlesworth D., and Crow J. F., 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., and Miller J. H., 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287: 560–561. 10.1038/287560a0 [DOI] [PubMed] [Google Scholar]
- Farlow A., Long H., Arnoux S., Sung W., Doak T. G. et al. , 2015. The spontaneous mutation rate in the fission yeast Schizosaccharomyces pombe. Genetics 201: 737–744. 10.1534/genetics.115.177329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Hanson A. J., Lee H., Popodi E. M., and Tang H., 2013. On the mutational topology of the bacterial genome. G3 (Bethesda) 3: 399–407. 10.1534/g3.112.005355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. L., Neill E., and Davey S., 2003. Fission yeast Uve1 and Apn2 function in distinct oxidative damage repair pathways in vivo. DNA Repair (Amst.) 2: 1253–1267. 10.1016/j.dnarep.2003.08.005 [DOI] [PubMed] [Google Scholar]
- Graur D., and Li W. H., 2000. Fundamentals of molecular evolution, Sinauer Associates, Massachusetts. [Google Scholar]
- Grogan D. W., Carver G. T., and Drake J. W., 2001. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 98: 7928–7933. 10.1073/pnas.141113098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W. L., Claessens A., Otto T. D., Kekre M., Fairhurst R. M. et al. , 2017. Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic Acids Res. 45: 1889–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A. L., and Norais C., Badger J. H., Delmas S., Haldenby S., et al. , 2010. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5: e9605 10.1371/journal.pone.0009605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., and Petrov D. A., 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6: e1001115 10.1371/journal.pgen.1001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman J. D., Schalkwyk L. C., and Doolittle W. F., 1986. ISH51: a large, degenerate family of insertion sequence-like elements in the genome of the archaebacterium, Halobacterium volcanii. Nucleic Acids Res. 14: 6983–7000. 10.1093/nar/14.17.6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C., Fleck O., Hansen H. A., Liu C., Slaaby R. et al. , 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 19: 853–862. 10.1101/gad.329905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs K. L., and Grogan D. W., 1997. Rates of spontaneous mutation in an archaeon from geothermal environments. J. Bacteriol. 179: 3298–3303. 10.1128/JB.179.10.3298-3303.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Wong K., and Adams D. J., 2013. RetroSeq: transposable element discovery from next-generation sequencing data. Bioinformatics 29: 389–390. 10.1093/bioinformatics/bts697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., and Halligan D. L., 2009. Analysis and implications of mutational variation. Genetica 136: 359–369. 10.1007/s10709-008-9304-4 [DOI] [PubMed] [Google Scholar]
- Kibota T. T., and Lynch M., 1996. Estimate of the genomic mutation rate deleterious to overall fitness in E. coli. Nature 381: 694–696. 10.1038/381694a0 [DOI] [PubMed] [Google Scholar]
- Kidwell M. G., and Lisch D. R., 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1–24. 10.1111/j.0014-3820.2001.tb01268.x [DOI] [PubMed] [Google Scholar]
- Kim M., Wolff E., Huang T., Garibyan L., Earl A. M. et al. , 2004. Developing a genetic system in Deinococcus radiodurans for analyzing mutations. Genetics 166: 661–668. 10.1534/genetics.166.2.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., and Ohta T., 1969. The average number of generations until fixation of a mutant gene in a finite population. Genetics 61: 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukyildirim S., Long H., Sung W., Miller S. F., Doak T. G. et al. , 2016. The rate and spectrum of spontaneous mutations in Mycobacterium smegmatis, a bacterium naturally devoid of the postreplicative mismatch repair pathway. G3 (Bethesda) 6: 2157–2163. 10.1534/g3.116.030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., 1992. DNA replication fidelity. J. Biol. Chem. 267: 18251–18254. [PubMed] [Google Scholar]
- Kunz C., and Fleck O., 2001. Role of the DNA repair nucleases Rad13, Rad2 and Uve1 of Schizosaccharomyces pombe in mismatch correction. J. Mol. Biol. 313: 241–253. 10.1006/jmbi.2001.5054 [DOI] [PubMed] [Google Scholar]
- Kuo C.-H., and Ochman H., 2009. Deletional bias across the three domains of life. Genome Biol. Evol. 1: 145–152. 10.1093/gbe/evp016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., and Murray A. W., 2008. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178: 67–82. 10.1534/genetics.107.071506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie D. S., Messer P. W., Hershberg R., and Petrov D. A., 2013. Strong purifying selection at synonymous sites in D. melanogaster. PLoS Genet. 9: e1003527 10.1371/journal.pgen.1003527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Popodi E., Tang H., and Foster P. L., 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 109: E2774–E2783. 10.1073/pnas.1210309109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Doak T. G., Popodi E., Foster P. L., and Tang H., 2016. Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli. Nucleic Acids Res. 44: 7109–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind P. A., and Andersson D. I., 2008. Whole-genome mutational biases in bacteria. Proc. Natl. Acad. Sci. USA 105: 17878–17883. 10.1073/pnas.0804445105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren D., 1972. The temperature influence on the spontaneous mutation rate. I. Literature review. Hereditas 70: 165–170. 10.1111/j.1601-5223.1972.tb01377.x [DOI] [PubMed] [Google Scholar]
- Long H., Sung W., Miller S. F., Ackerman M. S., Doak T. G. et al. , 2014. Mutation rate, spectrum, topology, and context-dependency in the DNA mismatch repair-deficient Pseudomonas fluorescens ATCC948. Genome Biol. Evol. 7: 262–271. 10.1093/gbe/evu284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Kucukyildirim S., Sung W., Williams E., Lee H. et al. , 2015. Background mutational features of the radiation-resistant bacterium Deinococcus radiodurans. Mol. Biol. Evol. 32: 2383–2392. 10.1093/molbev/msv119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Sung W., Kucukyildirim S., Williams E., Miller S. F. et al. , 2018. Evolutionary determinants of genome-wide nucleotide composition. Nat. Ecol. Evol. 2: 237–240. 10.1038/s41559-017-0425-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2006. The origins of eukaryotic gene structure. Mol. Biol. Evol. 23: 450–468. 10.1093/molbev/msj050 [DOI] [PubMed] [Google Scholar]
- Lynch M., 2007. The Origins of Genome Architecture, Sinauer Associates, Inc., Sunderland, MA. [Google Scholar]
- Lynch M., 2010. Evolution of the mutation rate. Trends Genet. 26: 345–352. 10.1016/j.tig.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R. et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. 10.1073/pnas.0803466105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Ackerman M. S., Gout J. F., Long H., Sung W. et al. , 2016. Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet. 17: 704–714. 10.1038/nrg.2016.104 [DOI] [PubMed] [Google Scholar]
- Mackwan R. R., Carver G. T., Drake J. W., and Grogan D. W., 2007. An unusual pattern of spontaneous mutations recovered in the halophilic archaeon Haloferax volcanii. Genetics 176: 697–702. 10.1534/genetics.106.069666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackwan R. R., Carver G. T., Kissling G. E., Drake J. W., and Grogan D. W., 2008. The rate and character of spontaneous mutation in Thermus thermophilus. Genetics 180: 17–25. 10.1534/genetics.108.089086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. H. M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennecier S., Coste G., Servant P., Bailone A., and Sommer S., 2004. Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol. Genet. Genomics 272: 460–469. 10.1007/s00438-004-1077-6 [DOI] [PubMed] [Google Scholar]
- Mira A., Ochman H., and Moran N. A., 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17: 589–596. 10.1016/S0168-9525(01)02447-7 [DOI] [PubMed] [Google Scholar]
- Mukai T., 1964. The genetic structure of natural populations of Drosophila melanogaster. I. Spontaneous mutation rate of polygenes controlling viability. Genetics 50: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1928. The measurement of gene mutation rate in Drosophila, its high variability, and its dependence upon temperature. Genetics 13: 279–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niccum B. A., Lee H., MohammedIsmail W., Tang H., and Foster P. L., 2019. The symmetrical wave pattern of base-pair substitution rates across the Escherichia coli chromosome has multiple causes. mBio 10: e01226–e01219. 10.1128/mBio.01226-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W. L., and Maughan H., 2002. The spectrum of spontaneous rifampin resistance mutations in the rpoB gene of Bacillus subtilis 168 spores differs from that of vegetative cells and resembles that of Mycobacterium tuberculosis. J. Bacteriol. 184: 4936–4940. 10.1128/JB.184.17.4936-4940.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M., Jackson L., Chimileski S., and Papke T. R., 2015. Genome-wide DNA methylation analysis of Haloferax volcanii H26 and identification of DNA methyltransferase related PD-(D/E)XK nuclease family proteinHVO_A0006. Front. Microbiol. 6: 1–11. 10.3389/fmicb.2015.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget-McNicol S., and Saul A., 2001. Mutation rates in the dihydrofolate reductase gene of Plasmodium falciparum. Parasitology 122: 497–505. 10.1017/S0031182001007739 [DOI] [PubMed] [Google Scholar]
- Perkins A. E., Schuerger A. C., and Nicholson W. L., 2008. Isolation of rpoB mutations causing rifampicin resistance in Bacillus subtilis spores exposed to simulated Martian surface conditions. Astrobiology 8: 1159–1167. 10.1089/ast.2007.0224 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., and Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2014. R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Redder P., and Garrett R. A., 2006. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 188: 4198–4206. 10.1128/JB.00061-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldstottir H., Winckler W., Guttman M., Lander E. S. et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rojas A., and Blázquez J., 2009. The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. J. Bacteriol. 191: 844–850. 10.1128/JB.01081-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph C., Kunz C., Parisi S., Lehmann E., Hartsuiker E. et al. , 1999. The msh2 gene of Schizosaccharomyces pombe is involved in mismatch repair, mating-type switching, and meiotic chromosome organization. Mol. Cell. Biol. 19: 241–250. 10.1128/MCB.19.1.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., Rose M. R., and Doolittle W. F., 1982. High-frequency genomic rearrangements involving archaebacterial repeat sequence elements. Nature 299: 182–185. 10.1038/299182a0 [DOI] [PubMed] [Google Scholar]
- Schrider D. R., Hourmozdi J. N., and Hahn M. W., 2011. Pervasive multinucleotide mutational events in eukaryotes. Curr. Biol. 21: 1051–1054. 10.1016/j.cub.2011.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senra M. V. X., Sung W., Ackerman M. S., Miller S. F., Lynch M. et al. , 2018. An unbiased genome-wide view of the mutation-accumulation rate and spectrum of the endosymbiotic bacterium Teredinibacter turnerae. Genome Biol. Evol. 10: 723–730. 10.1093/gbe/evy027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp N. P., Sandell L., James C. G., and Otto S. P., 2018. The genome-wide rate and spectrum of spontaneous mutations differs between haploid and diploid yeast. Proc. Natl. Acad. Sci. USA 115: E5046–E5055. 10.1073/pnas.1801040115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doak T. G., and Lynch M., 2012. Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. USA 109: 18488–18492. 10.1073/pnas.1216223109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Gout J.-F., Miller S. F., Williams E. et al. , 2015. Asymmetric context-dependent mutation patterns revealed through mutation-accumulation experiments. Mol. Biol. Evol. 32: 1672–1683. 10.1093/molbev/msv055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Dillon M. M., Platt T. G., Fuqua C. et al. , 2016. Evolution of the insertion-deletion mutation rate across the tree of life. G3 (Bethesda) 6: 2583–2591. 10.1534/g3.116.030890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh J., Kumar P., Krishna P. S. M., Manjunath R., and Varshney U., 2003. Importance of uracil DNA glycosylase in Pseudomonas aeruginosa and Mycobacterium smegmatis, G+C-rich bacteria, in mutation prevention, tolerance to acidified nitrite, and endurance in mouse macrophages. J. Biol. Chem. 278: 24350–24358. 10.1074/jbc.M302121200 [DOI] [PubMed] [Google Scholar]
- Wielgoss S., Barrick J. E., Tenaillon O., Cruveiller S., Chane-Woon-Ming B. et al. , 2011. Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3 (Bethesda) 1: 183–186. 10.1534/g3.111.000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradka K., Slade D., Bailone A., Sommer S., Averbeck D. et al. , 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443: 569–573. 10.1038/nature05160 [DOI] [PubMed] [Google Scholar]
- Zamenhof G., and Greer S., 1958. Heat as an agent producing high frequency of mutations and unstable genes in Escherichia coli. Nature 182: 611–613. 10.1038/182611a0 [DOI] [PubMed] [Google Scholar]
- Zerulla K., Chimileski S., Nather D., Gophna U., Papke R. T. et al. , 2014. DNA as a phosphate storage polymer and the alternative advantages of polyploidy for growth or survival. PLoS One 9: e94819 10.1371/journal.pone.0094819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. O., Siegal M. L., Hall D. W., and Petrov D. A., 2014. Precise estimates of mutation rate and spectrum in yeast. Proc. Natl. Acad. Sci. USA 111: E2310–E2318. 10.1073/pnas.1323011111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The workflow has been deposited at the GitHub repository (https://github.com/sibelkucukyildirim) and raw sequences are available at the Sequence Read Archive at the NCBI (Bioproject No.: PRJNA386190). Supplemental material available at figshare: https://doi.org/10.25386/genetics.12411035.