Abstract
Since its emergence in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has developed into a global pandemic within a matter of months. While subunit vaccines are one of the prominent options for combating coronavirus disease 2019 (COVID‐19), the immunogenicity of spike protein‐based antigens remains unknown. When immunized in mice, the S1 domain induced much higher IgG and IgA antibody levels than the receptor‐binding domain (RBD) and more efficiently neutralized SARS‐CoV‐2 when adjuvanted with alum. It is inferred that a large proportion of these neutralization epitopes are located in the S1 domain but outside the RBD and that some of these are spatial epitopes. This finding indicates that expression systems with posttranslational modification abilities are important to maintain the natural configurations of recombinant spike protein antigens and are critical for effective COVID‐19 vaccines. Further, adjuvants prone to a Th1 response should be considered for S1‐based subunit COVID‐19 vaccines to reduce the potential risk of antibody‐dependent enhancement of infection.
Keywords: antibody‐dependent enhancement, COVID‐19, receptor‐binding domain, S1, SARS‐CoV‐2 subunit vaccine, spike protein
Highlights
Antibodies induced by the S1 domain neutralized SARS‐Cov‐2 more efficiently than those induced by the receptor‐binding domain (RBD). Antibodies induced by the S1 domain produced from HEK293K cells neutralized SARS‐Cov‐2 more efficiently than those induced by the S1 domain produced from E. coli. Both the S1 domain and the RBD induced a highly Th2 response when adjuvanted with alum.
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has infected 10 million people and caused 0.5 million deaths around the globe as of 1 July 2020, 6 months after the first case was reported in December 2019. 1 , 2 , 3 Several medications, including hydroxychloroquine, remdesivir, and dexamethasone, have quickly been utilized as treatments for coronavirus disease 2019 (COVID‐19); however, none have showed significant benefits yet. 4 , 5 , 6 , 7 , 8 , 9 Considering its highly contagious character, vaccines may be the optimal choice to combat SARS‐CoV‐2.
Several platforms, including nucleic acids (DNA/RNA), viral vectors, live attenuated, inactivated and protein subunit vaccine candidates, are being evaluated as potential SARS‐CoV‐2 vaccines. 10 , 11 Subunit vaccines, which are one of the two most commonly adopted vaccine platforms in the market, have definitive advantages over whole‐virion vaccines (both attenuated and inactivated), particularly when considering the risk of exposure during vaccine production, which can only be avoided by high biosafety level factory buildings and strict training in operation processes.
Prior research focused on the development of vaccines against other coronaviruses (including SARS‐CoV‐1, which originated in 2002, and Middle East respiratory syndrome coronavirus (MERS‐CoV), which was identified in 2012) determined that spike (S) proteins are ideal targets for subunit vaccine antigens. S proteins are found on the surface of coronaviruses and are responsible for viral attachment to host cells (S1 domain) and virus‐cell membrane fusion (S2 domain). 12 , 13 , 14 , 15 Taking into account production difficulties for large recombinant proteins (the S protein extracellular domain is ~1300 amino acids) and the risk of antibody‐dependent enhancement (ADE) of infection, S1 (∼700 amino acids) and its receptor‐binding domain (RBD, ∼200 amino acids) are widely considered the most attractive potential coronavirus vaccine targets. 16 , 17 , 18 Unlike other S segments, the RBD of SARS‐CoV‐2 shows low similarity with those of other known coronaviruses. 3 Though neutralizing antibodies targeting the RBD have been reported in SARS‐CoV‐2 patients, similar to those found in SARS‐CoV‐1 and MERS patients, the immunogenicity of the SARS‐CoV‐2 RBD remains unknown. 19 , 20 , 21 , 22
2. MATERIALS AND METHODS
2.1. Antigen preparation
HEK293K cells expressed recombinant SARS‐CoV‐2 S1 (cat: 40591‐V08H, purity >90% as determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis [SDS‐PAGE] and more than 95% as determined by size exclusion chromatography high performance liquid chromatography) and RBD (Cat: 40592‐V08H, purity >95% as determined by SDS‐PAGE) proteins were purchased from Sino Biological Inc. (Beijing, China).
E. coli expressed SARS‐CoV‐2 S1 and RBD proteins were prepared as follows. DNA sequences encoding either the S1 subunit (YP_009724390.1, Met1‐Tyr695) or the RBD domain (YP_009724390.1, Arg328‐Pro521) were synthesized by Genscript Inc. (Nanjing, China) downstream of a DNA sequence encoding the norovirus shell domain (to enhance immunity). These sequences were then inserted between BamHI and NotI restriction sites in a pET‐28b plasmid (Novagen), transformed into E. coli (BL21, DE3) and induced overnight at room temperature (~25°C) using 0.4 mM isopropyl‐β‐d‐thiogalactopyranoside. 23 , 24 Cells were collected by centrifugation, resuspended in PBS, sonicated and centrifuged again. Precipitates were resuspended in PBS and sonicated again. Inclusion bodies were dissolved in PBS containing 8M urea and dialyzed against 20 mM Tris‐HCl (pH 7.4) containing sequential dilutions of urea (6, 8, 2, 1, and 0M). Recombinant proteins were analyzed by 10% SDS‐PAGE followed by Western blot analysis (primary antibody: rabbit anti‐SARS‐CoV‐1 S1 subunit polyclonal antibody from Sino Biological Inc.; secondary antibody: horse radish peroxidase (HRP)‐conjugated goat anti‐rabbit polyclonal antibody from ThermoFisher Scientific). Proteins were quantified by both SDS‐PAGE in 10% gels loaded with known concentrations of serially diluted bovine serum albumin (Bio‐Rad) and by bicinchoninic acid protein assay kit (Beyotime Inc., Beijing China). Finally, proteins were observed by transmission electron microscopy (Hitachi, Japan). 25 , 26 , 27
2.2. Immunization of mice
Six‐week‐old female specific pathogen‐free BALB/c mice (14‐17 g) were supplied and maintained by the Central Services of the Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College (IMB, CAMS). Animals were randomly divided into five groups with six mice in each group (N = 6). Antigens were diluted to 10 μg/mouse/dose in 25 μL of phosphate buffered saline (PBS) and mixed with the same volume of alum adjuvant (Thermo Fisher Scientific) before immunization. Thus, 50 μL of immunogens were administered intramuscularly into the thigh muscle three times at 2 week intervals. Two weeks after the final immunization, mice were anesthetized with ketamine, and blood was collected via cardiac puncture. After clotting at 4°C overnight, serum was collected by centrifugation at 3 000 rpm for 10 minutes and pooled by group.
All experiments were performed in compliance with the Guiding Principles for the Care and Use of Laboratory Animals of the Animal Ethics Committee of the IMB, CAMS (permit number: SCXK (dian) K2017‐0002).
2.3. Enzyme‐linked immunosorbent assay of antibody titers
Ninety‐six‐well plates were coated with 2 μg/mL HEK293K cell‐expressed recombinant SARS‐CoV‐2 S1 or RBD proteins overnight at 4°C. Plates were washed one time with wash buffer (PBS containing 0.05% (v/v) polysorbate 20) and then blocked with 5% (w/v) skim milk dissolved in wash buffer for 1 hour at 37°C. Plates were then washed four times and incubated with serially diluted mouse sera for 1 hour at 37°C. Next, plates were washed five times and incubated with goat anti‐mouse IgG/IgA/IgG1/IgG2a HRP‐conjugated secondary antibodies (ThermoFisher Scientific) for 1 hour at 37°C. Following five additional washes, 3,3′,5,5′‐tetramethylbenzidine (TMB, BD Bioscience) substrate was added. The plate was incubated at room temperature in the dark for 10 minutes, and reactions were stopped by the addition of 2M sulfuric acid. Absorbance (450 nm) was detected using a microplate reader (Bio‐Tek Instruments, Inc). Antibody titers were defined by end‐point dilution with a cut‐off signal of OD450 = 0.1. Sera samples that did not produce an optical density of more than 0.1 at 1:500 were determined as 0.
The IgG1‐to‐IgG2a titer ratio was calculated to evaluate Th1‐Th2 balance. 28 , 29
2.4. SARS‐CoV‐2 neutralization
First, 100 μL of Vero cells cultured in Dulbecco's modified Eagle's medium (DMEM, Corning, NY) supplemented with 5% fetal bovine serum (FBS, HyClone, Logan) were seeded into 96‐well plates (Corning) at 2.5 × 105 cells per well and cultured at 37°C in 5% CO2 overnight. The KMS‐1 strain of SARS‐CoV‐2 (GenBank No: MT226610.1) was diluted 10‐fold in DMEM and added at 100 μL/well. After 6 to 7 days of incubation at 37°C in 5% CO2, cytopathic effect (CPE) was assessed with an inverted microscope. Viral load was calculated based on the dilutions at which more than 50% of the cells exhibited lesions using the cell culture infective dose (CCID50) formula. 30
For neutralization, mouse sera were diluted with DMEM in a two‐fold series. Then, 50 μL of SARS‐CoV‐2 diluted with DMEM to 3.3 lg CCID50 was added to 50 μL of diluted serum, incubated at 37°C for 1 hour, and then added to 100 μL of Vero cells suspended in DMEM and 10% FBS. After culturing cells at 37°C in 5% CO2 for 4 days, the neutralization titer was reported as the serum dilution at which SARS‐CoV‐2 infection was inhibited by 50%.
All SARS‐CoV‐2 manipulations were carried out in a biosafety level 3 (BSL‐3) laboratory at IMB, CAMS.
2.5. Statistical analysis
Data are shown as the mean and standard deviation. GraphPad Prism 7.0 (San Diego, CA) was used for statistical analyses.
3. RESULTS
3.1. SARS‐CoV‐2 S1 and RBD recombinant proteins are expressed as inclusion bodies in E. coli
Here, we fused S1 and RBD to the carboxyl terminus of the norovirus shell domain, which has been reported to present recombinant expressed proteins on the surface of virus‐like particles to enhance the immunity of recombinant proteins. 23 , 24 While both RBD and S1 were expressed well with the norovirus shell domain (S‐RBD and S‐S1, respectively), as certified by the corresponding band (~52 kDa for S‐RBD and ~108 kDa for S‐S1) by SDS‐PAGE (Figure 1A) and Western blot analysis (Figure 1B), both were expressed as inclusion bodies. Following sonication, washing and dialysis, while S‐RBD showed quite high purity (lane S‐RBD in Figure 1A), S‐S1 showed only ∼60% purity by SDS‐PAGE (lane S‐S1 in Figure 1A). Transmission electron microscopy showed that after dialysis, only a small portion of the S‐RBD and S‐S1 fusion proteins formed similar but not identical virus‐like particles with diameters of ∼30 to 60 nm (showed by arrows in Figure 1C,D), while the majority of these recombinant proteins formed irregular aggregates (Figure 1C,D).
Figure 1.
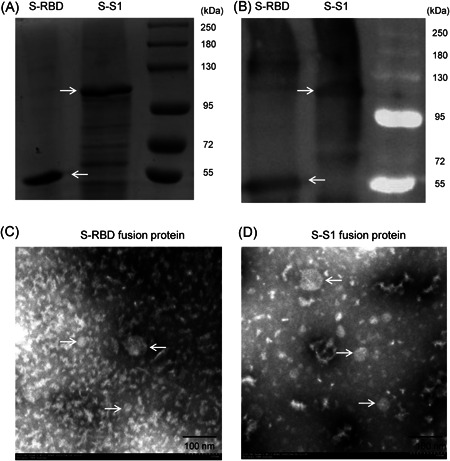
Production and characterization of E. coli‐expressed SARS‐CoV‐2 S1 and RBD proteins. (A) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) of norovirus shell domain‐RBD (S‐RBD, lane 1, indicated by arrows pointing to the left) and norovirus shell domain‐S1 (S‐S1, lane 2, indicated by arrows pointing to the right). (B) Western blot characterization of S‐RBD (lane 1, indicated by arrows pointing to the left) and S‐S1 (lane 2, indicated by arrows pointing to the right). (C, D) Transmission electron microscopy of the (C) S‐RBD and (D) S‐S1 fusion proteins. Particles with diameters ∼30 to 60 nm are indicated by arrows. RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
3.2. SARS‐CoV‐2 S1 induces higher IgG and IgA titers than RBD
2 weeks after the third intramuscular immunization (Figure 2A), both S1‐specific (S1‐coated plate in Figure 2) and RBD‐specific (RBD‐coated plate in Figure 2) antibodies were analyzed.
Figure 2.
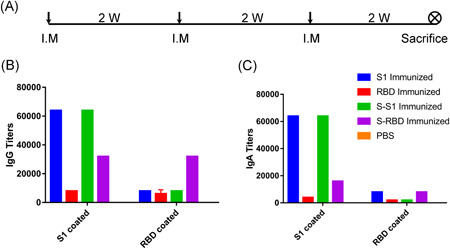
Immunization schedule and humoral responses of various immunogens in mice. A, Immunization schedule. I.M.: intramuscular. W: week. B, S1‐ and RBD‐specific IgG titers. C, S1‐ and RBD‐specific IgA titers. RBD, receptor‐binding domain
HEK293K cell‐expressed recombinant S1 (S1 immunized) and E.coli‐expressed norovirus shell domain‐S1 fusion protein (S‐S1 immunized) induced similar S1‐specific IgG titers (64 000) and similar RBD‐specific titers (8 000) (Figure 2B). HEK293K cell‐expressed recombinant RBD (RBD immunized) induced low S1‐specific IgG titers (8000) and RBD‐specific IgG titers (6 000), implying low immunogenicity of the RBD alone. Unlike S1 (similar IgG titers between S1 and S‐S1 may be attributed to low purity and thus low S1 content in S‐S1), fusion of the RBD with the norovirus shell domain (S‐RBD immunized) elevated both RBD‐specific IgG titers (from 6 000 to 32 000) and S1‐specific IgG titers (from 8 000 to 32 000).
As SARS‐CoV‐2 is a respiratory virus, mucosal immunity is important to fight infection and we therefore detected IgA titers (Figure 2C). While both HEK293K cell‐expressed recombinant S1 (S1 immunized) and E.coli‐expressed norovirus shell domain‐S1 fusion protein (S‐S1 immunized) induced equivalent levels of S1‐specific IgA titers and IgG titers (64 000), both HEK293K cell‐expressed recombinant RBD (RBD immunized) and E.coli‐expressed norovirus shell domain‐RBD fusion proteins (S‐RBD immunized) induced half the level of S1‐specific IgA titers as IgG titers (4000 vs 8000 for RBD and 16 000 vs 32 000 for S‐RBD, respectively). While RBD‐specific IgA titers were the lowest of all the IgG and IgA titers tested, there was a tendency for norovirus shell domain to elevate RBD immunogenicity on RBD‐specific IgA titers (RBD coated, S‐RBD immunized vs RBD immunized in Figure 2C), as was observed for the S1‐specific IgA titers (S1‐coated, S‐RBD immunized vs RBD immunized in Figure 2C).
3.3. SARS‐CoV‐2 S1 induced more balanced Th1‐Th2 responses than the RBD
Similar to S1‐specific total IgG titers (Figure 2B, S1‐coated), both HEK293K cell‐expressed recombinant S1 (S1 immunized) and E.coli‐expressed norovirus shell domain‐S1 fusion protein (S‐S1 immunized) induced the highest S1‐specific IgG1 (Figure 3A, S1 coated) and IgG2a (Figure 3B, S1 coated) titers and comparably low RBD‐specific IgG1 (Figure 3A, RBD coated) and IgG2a (Figure 3B, RBD coated) titers. HEK293K cell‐expressed recombinant RBD (RBD immunized) and E.coli‐expressed norovirus shell domain‐RBD fusion proteins (S‐RBD immunized) induced low levels of IgG1 and IgG2a titers specific to both S1 (S1 coated in Figure 3A,B) and the RBD (RBD coated in Figure 3A,B).
Figure 3.
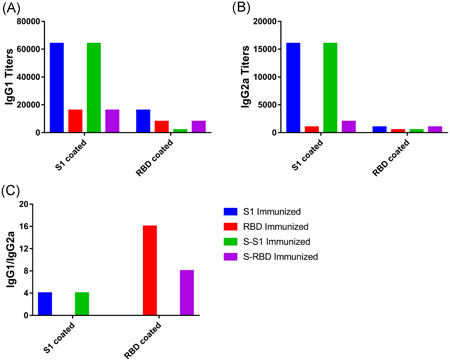
Th1‐Th2 balance analysis. A, S1‐ and RBD‐specific IgG1 titers. B, S1‐ and RBD‐specific IgG2a titers. C, IgG1/IgG2a ratios. RBD, receptor‐binding domain
Notably, the IgG1 titers in each group in Figure 3A are considerably higher than the IgG2a titers in Figure 3B. To enable direct comparisons between groups, we compared the IgG1/IgG2a ratios in each group induced by their own antigens (Figure 3C). While both HEK293K cell‐expressed recombinant S1 (S1 immunized) and E.coli‐expressed norovirus shell domain‐S1 fusion proteins (S‐S1 immunized) induced an IgG1/IgG2a ratio of 4, HEK293K‐expressed recombinant RBD (RBD immunized) induced an IgG1/IgG2a ratio as high as 16. This ratio could be lowered by ligation to the E.coli‐expressed norovirus shell domain (the S‐RBD immunized IgG1/IgG2a ratio was 8) but was still higher than in that in the S1 and S‐S1 immunized groups. Higher IgG1‐to‐IgG2a ratios, including the those for the S1 and S1‐RBD groups, imply a Th2‐biased immune response for these antigens.
3.4. S1 but not RBD induces effective SARS‐CoV‐2‐neutralizing antibodies
Among all the immunized groups, only serum from the HEK293K cell‐expressed recombinant S1‐immunized group showed obvious neutralization of the KMS‐1 strain of SARS‐CoV‐2 (Figure 4, S1‐immunized mean titer: 48). Unsurprisingly, serum from the HEK293K cell‐expressed recombinant RBD‐immunized group (Figure 4, RBD‐immunized mean titer: 6) showed a low neutralization effect. This effect was 1/8 of that in the S1 immunized group, comparable with their S1‐specific IgG (8 000:64 000 in Figure 2B) and IgA (4 000:64 000 in Figure 2C) titers. Moreover, RBD‐specific IgG and IgA titers are not indicative of neutralization effects (6 000:8 000 and 2 000:8 000, respectively, for RBD‐immunized and S1‐immunized serum, as shown in Figure 2). These results imply that antibodies induced by domains within S1 but outside the RBD can also play decisive roles in neutralization. It is the immunogenicity rather than the functional role of the S1 segment that matters in the induction of effective SARS‐CoV‐2‐neutralizing antibodies. As both S1‐specific IgG and IgA titers are very high for S1‐immunized serum, it is unclear which plays a more important role in virus neutralization.
Figure 4.
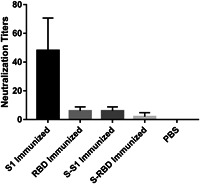
SARS‐CoV‐2 neutralization titers of serum from mice vaccinated with various immunogens. PBS: serum from mice immunized with PBS. PBS, phosphate buffered saline; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Notably, while E.coli‐expressed norovirus shell domain‐S1 fusion protein (S‐S1 immunized) induced both S1‐specific IgG and IgA titers comparable to those induced in the HEK293K cell‐expressed recombinant S1 immunized groups (Figure 2), only 1/8 neutralization titers were detected (6:48 in Figure 4). Similar phenomena were observed between the RBD‐ and S‐RBD‐immunized groups: while ligation of norovirus shell domain elevated the S1‐specific IgG (from 8 000 to 32 000) and IgA (from 4 000 to 16 000) titers, neutralization declined from six to two (Figure 4). These results showed that mammalian systems are more suitable than prokaryotic systems for the production of SARS‐CoV‐2 spike protein antigens that can induce effective neutralization antibodies.
4. DISCUSSION
Since its emergence in December 2019, SARS‐CoV‐2 has evolved into a global pandemic. Though public health measures have been taken, no effective medications or vaccines have been developed yet. Subunit vaccines, which are one of the two most commonly adopted vaccine forms in the market, are safer than whole‐virion vaccines due to the decreased risk of exposure during manufacturing.
Spike (S) proteins, which are found on the surface of coronavirus particles, are responsible for entrance into host cells and are widely considered prophylactic and treatment targets. 12 , 14 Similar to results using inactivated whole‐virus vaccines, immunopathology reports show eosinophilic infiltration and Th2‐mediated inflammatory alveolar damage when mice are challenged with infectious virus after immunization with full‐length S proteins. 31 , 32 , 33 , 34 , 35 To reduce this risk, S protein segments were evaluated as subunit vaccine antigens. While the S2 domain, which contains two heptad repeat segments, participates in virus‐cell membrane fusion and is a potential target for the development of antiviral inhibitors, the S1 domain, which contains the RBD, is responsible for attachment to host receptors (SARS‐CoV‐1&‐2 with human angiotensin‐converting enzyme 2 and MERS‐CoV with human dipeptidyl peptidase‐4) and has been widely accepted as a target for SARS‐CoV‐1 and MERS‐CoV vaccine development. 13 , 36 , 37 , 38 , 39 , 40 , 41
The SARS‐CoV‐2 RBD alone is not as immunogenic as S1, as evidenced by S1‐specific and RBD‐specific IgG (Figure 2B) and IgA (Figure 2C) titers induced in immunized mouse serum. This phenomenon may be attributed partly to the smaller size of the RBD (223 aa) than of S1 (670 aa). In fact, when the RBD was ligated to the norovirus shell domain (for a total molecular weight of ∼52 kDa), both S1‐specific and RBD‐specific IgG and IgA titers were elevated prominently (Figure 2B,C). Previous reports have demonstrated that RBD immunogenicity can be enhanced via presentation on virus‐like particles (VLPs) and multimerization of RBD monomers. 42 , 43 We originally intended to enhance RBD immunogenicity via presentation on the norovirus shell domain, but unfortunately, neither the S‐RBD nor S‐S1 recombinant proteins formed typical VLPs (Figure 1) as expected. 23 , 24 We are not sure whether these results should be attributed to the characteristics of the RBD and S1 or to difficulties of reconfiguration from inclusion bodies.
To our surprise, although both S1 and S‐S1 induced comparably high S1‐specific IgG and IgA titers (Figure 2), only S1‐immunized mouse serum showed prominent SARS‐CoV‐2 neutralization (Figure 4). This pattern was also observed with RBD‐ and S‐RBD‐immunized mouse serum. These results implied that antibody spectra induced by RBD and S1 from different expression systems are quite different. Compared with prokaryotic cells, mammalian expression systems efficiently incorporate RBD and S‐RBD posttranslational modifications, including glycosylation, which might be critical for the natural configuration of these proteins. 44 Conversely, inclusion bodies produced by E. coli cells may increase reconfiguration difficulties. Whatever the truth is, these results demonstrate that conformation‐dependent neutralizing antibodies induced by immunized S1 proteins play key roles in virus neutralization and subsequent effective protection. 20 , 45 As S1‐induced RBD‐specific IgG and IgA titers are very low and similar titers induced by RBD showed no prominent neutralization, we deduced that other S1 domains besides the RBD are also very important antigens for the induction of neutralizing antibodies.
Notably, those non‐neutralizing antibodies may inhibit neutralization. As has been observed in Dengue, respiratory syncytial virus, etc., ADE of infection phenomena were also reported with antibodies against SARS‐CoV‐1 and MERS‐CoV spike proteins (including antibodies against the RBD). 46 , 47 , 48 Interestingly, ADE‐inducing sera did not contain SARS‐CoV‐1 spike‐specific IgG2a subtype antibodies, which are present in neutralizing sera. 49 This result is consistent with the observed eosinophilic infiltration following vaccination and virus exposure, a typical characteristic of Th2 immune responses with elevated IgG1/IgG2a proportions. 28 , 29 Unfortunately, both SARS‐CoV‐2 S1 and RBD showed a Th2‐like immune response with high proportions of IgG1 when immunized with alum as adjuvant (Figure 3), implying a similar immunopathological risk to those reported for other coronaviruses. Though no ADE has been reported in animal models re‐exposed to SARS‐CoV‐2 or exposed following vaccine immunization, special attention should be paid to Th1‐biased adjuvants, as reported in the development of SARS‐CoV‐1 and MERS‐CoV vaccines. 31 , 32
In conclusion, the SARS‐CoV‐2 S1 domain is more immunogenic than the RBD domain, inducing higher IgG and IgA antibodies and also efficient virus neutralization antibodies. We infer that a large proportion of these neutralization epitopes exist within the S1 domain but outside of the RBD and that some of these are spatial epitopes. While S1 induced a more balanced Th1/Th2 response than the RBD when adjuvanted with alum, increased levels of IgG1 antibodies still indicate a potential risk of ADE, and adjuvants prone to a Th1 response should be considered for S1 subunit‐based COVID‐19 vaccines.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
YW and LW performed the experiments and analyzed the data. HC helped to perform part of the experiments. CL designed the study, drafted and finalized the manuscript.
ACKNOWLEDGMENTS
This work was financially supported by the CAMS Initiative for Innovative Medicine (grant number 2017‐I2M‐3‐022) and the National Natural Science Foundation of China (grant number 81503117).
Wang Y, Wang L, Cao H, Liu C. SARS‐CoV‐2 S1 is superior to the RBD as a COVID‐19 subunit vaccine antigen. J Med Virol. 2021;93:892–898. 10.1002/jmv.26320
Yunfei Wang and Lichun Wang contributed equally to this study.
DATA AVAILABILITY STATEMENT
All data used during the study are available from the corresponding author by request.
REFERENCES
- 1. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of covid‐19 ‐ preliminary report. N Engl J Med. 2020;NEJMoa2007764. 10.1056/NEJMoa2007764 [DOI] [PubMed] [Google Scholar]
- 5. Geleris J, Sun Y, Platt J, et al. Observational study of hydroxychloroquine in hospitalized patients with Covid‐19. N Engl J Med. 2020;382(25):2411‐2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582:469. [DOI] [PubMed] [Google Scholar]
- 7. Rosenberg ES, Dufort EM, Udo T, et al. Association of treatment with hydroxychloroquine or azithromycin with in‐hospital mortality in patients with COVID‐19 in New York State. JAMA. 2020.323(24):2493–2502. 10.1001/jama.2020.8630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID‐19: a randomised, double‐blind, placebo‐controlled, multicentre trial. Lancet. 2020;395(10236):1569‐1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Williamson BN, Feldmann F, Schwarz B, et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS‐CoV‐2. Nature. 2020.585(7824):273–276. 10.1038/s41586-020-2423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghaebi M, Osali A, Valizadeh H, Roshangar L, Ahmadi M. Vaccine development and therapeutic design for 2019‐nCoV/SARS‐CoV‐2: challenges and chances. J Cell Physiol. 2020. 10.1002/jcp.29771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharpe HR, Gilbride C, Allen E, et al. The early landscape of COVID‐19 vaccine development in the UK and rest of the world. Immunology. 2020;160:223‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Enjuanes L, Zuniga S, Castano‐Rodriguez C, Gutierrez‐Alvarez J, Canton J, Sola I. Molecular basis of coronavirus virulence and vaccine development. Adv Virus Res. 2016;96:245‐286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang S, He Y, Liu S. SARS vaccine development. Emerg Infect Dis. 2005;11(7):1016‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou Y, Jiang S, Du L. Prospects for a MERS‐CoV spike vaccine. Expert Rev Vaccines. 2018;17(8):677‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu M. SARS immunity and vaccination. Cell Mol Immunol. 2004;1(3):193‐198. [PubMed] [Google Scholar]
- 16. He Y, Li J, Heck S, Lustigman S, Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus‐expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J Virol. 2006;80(12):5757‐5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Q, Zhang L, Kuwahara K, et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non‐human primates. ACS Infect Dis. 2016;2(5):361‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang N, Shang J, Jiang S, Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brouwer PJM, Caniels TG, van der Straten K, et al. Potent neutralizing antibodies from COVID‐19 patients define multiple targets of vulnerability. Science. 2020.369(6504):643–650. 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature. 2020.584(7819):115–119. 10.1038/s41586-020-2380-z [DOI] [PubMed] [Google Scholar]
- 21. Premkumar L, Segovia‐Chumbez B, Jadi R, et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS‐CoV‐2 patients. Sci Immunol. 2020;5(48):eabc8413. 10.1126/sciimmunol.abc8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tai W, He L, Zhang X, et al. Characterization of the receptor‐binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia M, Huang P, Jiang X, Tan M. Immune response and protective efficacy of the S particle presented rotavirus VP8* vaccine in mice. Vaccine. 2019;37(30):4103‐4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xia M, Huang P, Sun C, et al. Bioengineered norovirus S60 nanoparticles as a multifunctional vaccine platform. ACS Nano. 2018;12(11):10665‐10682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu C, Yang X, Yao Y, Huang W, Sun W, Ma Y. Diverse expression levels of two codon‐optimized genes that encode human papilloma virus type 16 major protein L1 in Hansenula polymorpha. Biotechnol Lett. 2014;36(5):937‐945. [DOI] [PubMed] [Google Scholar]
- 26. Liu C, Yao Y, Yang X, et al. Production of recombinant human papillomavirus type 52 L1 protein in Hansenula polymorpha formed virus‐like particles. J Microbiol Biotechnol. 2015;25(6):936‐940. [DOI] [PubMed] [Google Scholar]
- 27. Tan M, Huang P, Xia M, et al. Norovirus P particle, a novel platform for vaccine development and antibody production. J Virol. 2011;85(2):753‐764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lefeber DJ, Benaissa‐Trouw B, Vliegenthart JF, et al. Th1‐directing adjuvants increase the immunogenicity of oligosaccharide‐protein conjugate vaccines related to Streptococcus pneumoniae type 3. Infect Immun. 2003;71(12):6915‐6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mountford AP, Fisher A, Wilson RA. The profile of IgG1 and IgG2a antibody responses in mice exposed to Schistosoma mansoni . Parasite Immunol. 1994;16(10):521‐527. [DOI] [PubMed] [Google Scholar]
- 30. Liao Y, Li X, Mou T, et al. Distinct infection process of SARS‐CoV‐2 in human bronchial epithelial cells line. J Med Virol. 2020. 10.1002/jmv.26200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Honda‐Okubo Y, Barnard D, Ong CH, Peng BH, Tseng CT, Petrovsky N. Severe acute respiratory syndrome‐associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology. J Virol. 2015;89(6):2995‐3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwata‐Yoshikawa N, Uda A, Suzuki T, et al. Effects of Toll‐like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV‐inactivated severe acute respiratory syndrome‐related coronavirus vaccine. J Virol. 2014;88(15):8597‐8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tseng CT, Sbrana E, Iwata‐Yoshikawa N, et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7(4):e35421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agrawal AS, Tao X, Algaissi A, et al. Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum Vaccin Immunother. 2016;12(9):2351‐2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sekimukai H, Iwata‐Yoshikawa N, Fukushi S, et al. Gold nanoparticle‐adjuvanted S protein induces a strong antigen‐specific IgG response against severe acute respiratory syndrome‐related coronavirus infection, but fails to induce protective antibodies and limit eosinophilic infiltration in lungs. Microbiol Immunol. 2020;64(1):33‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS‐CoV‐‐a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS‐CoV spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21(2):131‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsunetsugu‐Yokota Y, Ohnishi K, Takemori T. Severe acute respiratory syndrome (SARS) coronavirus: application of monoclonal antibodies and development of an effective vaccine. Rev Med Virol. 2006;16(2):117‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Du L, Zhao G, He Y, et al. Receptor‐binding domain of SARS‐CoV spike protein induces long‐term protective immunity in an animal model. Vaccine. 2007;25(15):2832‐2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lan J, Yao Y, Deng Y, et al. Recombinant receptor binding domain protein induces partial protective immunity in rhesus macaques against Middle East Respiratory syndrome coronavirus challenge. EBioMedicine. 2015;2(10):1438‐1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y, Tai W, Yang J, et al. Receptor‐binding domain of MERS‐CoV with optimal immunogen dosage and immunization interval protects human transgenic mice from MERS‐CoV infection. Hum Vaccin Immunother. 2017;13(7):1615‐1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tai W, Zhao G, Sun S, et al. A recombinant receptor‐binding domain of MERS‐CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS‐CoV infection. Virology. 2016;499:375‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang C, Zheng X, Gai W, et al. Novel chimeric virus‐like particles vaccine displaying MERS‐CoV receptor‐binding domain induce specific humoral and cellular immune response in mice. Antiviral Res. 2017;140:55‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shajahan A, Supekar NT, Gleinich AS, Azadi P. Deducing the N‐ and O‐ glycosylation profile of the spike protein of novel coronavirus SARS‐CoV‐2. Glycobiology. 2020:cwaa042. 10.1093/glycob/cwaa042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Du L, Zhao G, Yang Y, et al. A conformation‐dependent neutralizing monoclonal antibody specifically targeting receptor‐binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol. 2014;88(12):7045‐7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smatti MK, Al Thani AA, Yassine HM. Viral‐induced enhanced disease illness. Front Microbiol. 2018;9:2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wan Y, Shang J, Sun S, et al. Molecular mechanism for antibody‐dependent enhancement of coronavirus entry. J Virol. 2020;94(5):e02015–e02019. 10.1001/jama.2020.8630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang SF, Tseng SP, Yen CH, et al. Antibody‐dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun. 2014;451(2):208‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jaume M, Yip MS, Cheung CY, et al. Anti‐severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH‐ and cysteine protease‐independent FcgammaR pathway. J Virol. 2011;85(20):10582‐10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used during the study are available from the corresponding author by request.


