Abstract
Turmeric, or Curcuma longa, is commonly consumed in the South East Asian countries as a medical product and as food due to its therapeutic properties. However, with increasing demand for turmeric powder, adulterated turmeric powders mixed with other cheap starch powders, such as from corn or cassava, are being distributed by food suppliers for economic benefit. Here, we developed molecular markers using quantitative real-time PCR to identify adulteration in commercial turmeric powder products. Chloroplast genes, such as matK, atpF, and ycf2, were used to design species-specific primers for C. longa and Zea mays. Of the six primer pairs designed and tested, the correlation coefficients (R2) were higher than 0.99 and slopes were −3.136 to −3.498. The efficiency of the primers was between 93.14 and 108.4%. The specificity of the primers was confirmed with ten other species, which could be intentionally added to C. longa powders or used as ingredients in complex turmeric foods. In total, 20 blind samples and 10 commercial C. longa food products were tested with the designed primer sets to demonstrate the effectiveness of this approach to detect the addition of Z. mays products in turmeric powders. Taken together, the real-time PCR assay developed here has the potential to contribute to food safety and the protection of consumer’s rights.
Keywords: anti food fraud, Curcuma longa, DNA markers, species identification, SYBR-GREEN real-time PCR, Zea mays
1. Introduction
Turmeric (Curcuma longa) belongs to the ginger family, Zingiberaceae, and is native to Southern Asia and India. Turmeric rhizomes, which have brown skin and a unique flavor, are commonly used as a coloring and flavoring agent in Asian cuisines. Due to its fragrant aroma and slightly bitter taste, turmeric is a common culinary spice in Indian cuisines, especially curry. Additionally, beyond food products, turmeric is commonly consumed as a medical product in South East Asian countries due to its therapeutic properties [1]. The market size of curcumin was valued at USD 58.4 million in 2019 and is expected to experience a CAGR (compound annual growth rate) of 12.7% from 2020 to 2027 [2]. Globally, the demand for turmeric has grown due to its therapeutic functions and low toxicity. Curcumin, (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione), also known as diferuloylmethane, is the main natural polyphenol found in rhizomes of C. longa (turmeric) and in other Curcuma spp. [3]. It has been shown to target multiple signaling molecules while also demonstrating activity at the cellular level, which has helped support its multiple health benefits [4]. It has beneficial effects in inflammatory conditions [3], metabolic syndrome [5], and pain [6], as well as helps in the management of inflammatory and degenerative eye conditions [7]. While there appear to be countless therapeutic benefits of curcumin supplementation, most of them may be due to its antioxidant and anti-inflammatory effects [3].
Reports on the medicinal value of turmeric in treating a variety of ailments have further increased the global demand for turmeric [8]. In the United States, the largest market for turmeric supplements, turmeric was the top-selling herbal supplement, with sales exceeding US $47.6 million in 2016 [8,9]. In addition, turmeric-based dietary supplements, which also include standardized extracts with high concentrations of curcumin, have seen a steady increase in popularity in the United States and elsewhere [10,11]. However, with the increasing demand for turmeric powder, adulterated turmeric powders mixed with other cheap starch powders, such as from corn or cassava, have been distributed by food supplies for economic benefit [12]. According to the United States Grocery Manufacturers Association, food fraud costs $10–15 billion annually in the global food industry and affects approximately 10% of all commercial foods sold [13].
To detect fraudulent ingredients in complicated mixed foods, various technologies, such as sensory-, physicochemical-, chromatographic-, spectroscopic-, and DNA-based assays have been developed. DNA is generally believed to be stable enough to withstand various chemical treatments and high temperatures, and small quantities of DNA can be detected with specific primers using PCR-based methods [14]. DNA-based methods, such as quantitative real-time PCR (real-time PCR), multiplex PCR, and PCR-RFLP have been successfully applied to detect food fraud and adulteration due to their economical and time-saving advantages over other approaches [15,16].
Specifically, real-time PCR (real-time PCR) assay presents with high specificity and sensitivity, capable of detecting very small amounts of target DNA in complex foods. General types of real-time PCR approaches, probe-based real-time PCR (TaqMan assay), and DNA intercalating dye-based real-time PCR (SYBR Green I assay) have been employed for the detection and identification of DNA [17]. Probe-based real-time PCR detects the target sequence with specificity using probes designed to be complementary to a target sequence [18]; however, this approach requires many SNPs or indels to differentiate species, and it is difficult to design probes and optimize real-time PCR conditions [19,20]. Alternatively, SYBR Green I, an intercalating dye that binds to double-stranded DNA in a sequence independent manner, can provide a more flexible, convenient, and inexpensive method over probe-based methods [19].
It is generally believed that the nuclear genome of a cell has a single copy of a particular gene along with a few sequences in low copy numbers; hence, it is difficult to obtain high uniformity in PCR amplification. Especially, DNA extracted from processed commercial foods is of low quality, possibly because of degradation caused by the processes of drying, heating, fermentation, and addition of ingredients. Therefore, markers designed on the extracted nuclear DNA from processed foods exhibit a low ability to discriminate between species because of the low quality of nuclear DNA that has either a single gene or low-copies of genes [21,22]. The chloroplast genome size varies among species, ranging from 107 to 208 kb and consisting of a single circular molecule of DNA that is generally present in hundreds of copies per cell [23]. Chloroplasts are composed of two layers of membranes that enable chloroplasts to persist through decomposition during food processing [24]. The chloroplast genome is generally believed to contain 120–130 genes [22]. Some genes, such as matK, ndhF, ycf2, and ccsA, exhibit higher frequencies of single-nucleotide polymorphisms (SNPs) and insertion/deletions (indels) than other chloroplast genes [23]. A variety of chloroplast markers, including atpF-atpH spacer, matK gene, rbcL gene, rpoB gene, rpoC1 gene, psbK-psbl spacer, and trnH-psbA spacer, have been employed for species identification [25,26].
As described above, cheap corn powder with a similar color to turmeric has been wildly used in adulterated turmeric powders by food suppliers for illegal economic benefit. In this study, we developed SYBR Green-based quantitative real-time PCR assay to identify adulteration in commercial turmeric powder products using turmeric and corn species-specific primer sets. The real-time PCR methodology was optimized for both species-specific primers to correctly identify target species in complex powder products. Subsequently, the designed primers were applied to commercial turmeric products.
2. Materials and Methods
2.1. Plant and Food Sample Preparation
Turmeric (Curcuma longa) rhizomes and corn (Zea mays) seeds were kindly provided by Gangwondo Agriculture Research and Extension Services (Chuncheon, Korea). Both plants were grown in a stable temperature greenhouse for four weeks with horticulture soil. Samples for DNA isolation were extracted from the leaves of each plant. All C. longa commercial products used for the analysis of food complexes were purchased from local markets and stored at room temperature.
2.1.1. Reference Binary Mixtures
To generate a quantitative reference binary mixture model, binary mixtures containing different amounts (2 mg, 0.1%; 20 mg, 1%; 200 mg, 10%; and 2 g, 100%) of turmeric rhizome powders were mixed to prepare a final mixture of 2 g with corn powder, wheat flour, or rice flour purchased from a local market. Additionally, different amounts of corn powder were mixed (2 mg, 0.1%; 20 mg, 1%; 200 mg, 10%; and 2 g, 100%) to prepare final mixtures of 2 g with turmeric rhizome powders. Turmeric rhizomes and corn seeds were dried in a 55 °C dry oven for 48 hours and then ground with a mixing machine.
2.1.2. Blind Samples
Blind powder samples (n = 20) were provided by the National Institute of Food and Drug Safety Evaluation of the Ministry of Food and Drug Safety (Cheongju, Korea). The blind samples consisted of different percentages of corn and turmeric rhizome powders. The corn powders were added to turmeric rhizome powders at concentrations of 0–10% w/w, to prepare final mixtures of 150 mg.
2.2. DNA Extraction
For the efficiency of the designed primer sets, genomic DNA used for standard curves was extracted from C. longa and Z. mays leaves using the Dneasy Plant Pro Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. Genomic DNA used to plot standard curves of reference binary mixtures was isolated from the binary mixture samples (2 g each) using a large scale CTAB-based genomic DNA isolation method [27]. Genomic DNA from the commercial turmeric products was extracted using the Dneasy Plant Pro Kit according to the manufacturer’s protocol. To obtain high quality genomic DNA, DNA extracted with the large scale CTAB method was purified using the Wizard DNA Clean-Up system (Promega, Madison, USA). DNA quantity and purity were measured using a SPECTROstar Nano reader (BMG Labtech, Ortenberg, Germany). Purity of the DNA extracts was in the range of 1.7–2.
2.3. Sequence Analysis and Primer Design
Sequences of target chloroplast genes such as matK, atpF, and ycf2 of two species (C. longa for NC_042886.1 and Z. mays for NC_001666.2) were downloaded from the National Center for Biotechnology Information (NCBI) and used to design target-specific primers. The nucleotide sequences of the both species were aligned using ClustalW2 (EMBL-EBI, Hinxton, Cambridgeshire, UK) and BioEdit 7.2 (Ibis Biosciences, Carlsbad, CA, USA). Species-specific primer sets were designed based on the variable region between C. longa and Z. mays using Beacon DesignerTM (PRIMER Biosoft, Palo Alto, CA, USA). Species-specific primers were commercially synthesized (Macrogen, Seoul, Korea).
2.4. Quantitative Real-Time PCR
Real-time PCR was performed in a final volume of 20 µL using AccuPower® 2× GreenStar™ real-time PCR Master Mix with SYBR Green (Bioneer, Daejeon, Korea). The real-time PCR reaction mixture consisted of 10 µL 2× GreenStar Master Mix, 0.5 µL 10 pmol each primer, 1 µL of 10 ng/µL genomic DNA, and 0.25 µL ROX Dye. A QuantStudio 3 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) was used for real-time PCR amplification. The real-time PCR conditions were as follows: pre-denaturation (10 min at 95 °C), followed by 40 cycles of denaturation for 30 s at 95 °C, annealing for 20 s at 55–60 °C (depending on each targeting primer sequence), and extension for 30 s at 72 °C. All real-time PCRs were performed in technical triplicates for three biological replicates.
2.5. Cloning of PCR Amplicons and Sequencing
Conventional PCR was carried out using TaKaRa Ex TaqTM DNA polymerase (TaKaRa Bio Company, Kusatsu, Shiga, Japan) mixture with 10 ng DNA and 10 pmol each primer using a C1000 Thermal Cycler (BIO-RAD, California, USA). PCR conditions were as follows: pre-denaturation for 5 min at 95 °C, followed by 35 cycles of annealing and denaturation for 30 s at 95 °C, annealing for 20 s at 55–60 °C (depending on primer sequences), and extension 30 s at 72 °C, and final extension for 5 min at 72 °C. PCR products were amplified using target specific primers (CL_matK, CL_atpF, CL_ycf2, ZM_matK, ZM_atpF, and ZM_ycf2) and cloned using the RBC T&A Cloning Vector (Real Biotech Corporation, Taipei, Taiwan). Plasmid DNA was extracted from recombinant plasmids using the DokDo-Prep Plasmid Mini-Kit (ELPISBIOTECH, DaeJeon, South Korea) and sequenced by a commercial service (Macrogen, Seoul, Korea).
2.6. Standard Curve Construction and Data Analysis
The efficiency of the designed primer sets was evaluated using two approaches. First, species-specific PCR products were cloned into the RBC T&A Cloning Vector (Real Biotech Corporation, Taipei, Taiwan), and recombinant clones were then diluted serially (107, 106, 105, 104, and 103 copies) and used to quantify and confirm the efficiency of equivalent amplification [28,29]. Second, real-time PCR assays were applied to genomic DNA using target and non-target gDNA diluted ten-fold into five series (10 ng to 1 pg).
Each binary mixture with genomic DNAs extracted from the leaves or powder products of each species was diluted to a final concentration of 10 ng/μL. A baseline and a threshold were set for further analysis. The cycle number at the threshold level of log-based fluorescence was defined as the Ct (cycle threshold) number, which was the observed value in the conventional real-time PCR experiments [30]. Correlations between diluted DNAs and cycle threshold (Ct) standard curves were evaluated using a default parameter. The standard curve was calculated as y = −ax + b (a refers to the standard curve slope and b refers to the y-intercept). The efficiency of the reaction (E) was calculated as E = (10−1/a), and the percent efficiency was evaluated as (E − 1) × 100% [29,30]. For all analyses, three technical replicates of each biological replicate were performed.
To evaluate amplification efficiency and sensitivity, two criteria were used to define an acceptable real-time PCR assay based on previous reports [28,29]: linear dynamic range and amplification efficiency. The linear dynamic range should ideally extend over four log10 concentrations, with the coefficient of determination (R2) being greater than 0.98, and the amplification efficiency should be in the range of 110-90%, corresponding to a slope between −3.1 and −3.6 [29].
To validate the specificity and sensitivity of the designed target-specific primers, interlaboratory validation was performed in two independent laboratories. Validation was performed in two laboratories using the same PCR conditions and with either an Applied Biosystems 7500 Fast Real-Time PCR Instrument System (Applied Biosystems, Foster City, CA, USA) or a CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA).
3. Results and Discussion
3.1. Design of Species-Specific Primers
To verify authenticity of C. longa commercial food products, we designed species-specific primer pairs for C. longa and Z. mays. Chloroplast genes, such as matK, atpF, and ycf2, with high frequencies of SNPs and indels between the two species [25] were targeted to design the species-specific primer sets. For designing species-specific primers, chloroplast genes of both species, as well as those of other starch crops (Oryza sativa and Triticum aestivum), were aligned using a software program with ClustalW2 (EMBL-EBI, Hinxton, Cambridgeshire, UK) and BioEdit 7.2 (Ibis Biosciences, Carlsbad, CA, USA; Supplementary Figures S1 and S2). We identified a variety of SNPs within three chloroplast genes among four species (Supplementary Figure S1). Food processing, such as heating, drying, and mixing, is known to damage and degrade DNA [31]. If the length of PCR amplicons is long, real-time PCR would be decreased in various food products. Therefore, based on species-specific SNPs, target-specific primers were designed to amplify short products ranging from 80 to 194 bp (Table 1).
Table 1.
Primer sets designed for species-specific targeting.
| Target Species | Target Gene | Primer | Length (bp) | Sequence (5′→3′) | Size (bp) | Tm (°C) |
|---|---|---|---|---|---|---|
| All plants | 18s rRNA region |
18s rRNA_F | 25 | TCTGCCCTATCAACTTTCGATGGTA | 137 | 58 |
| 18s rRNA_R | 25 | AATTTGCGCGCCTGCTGCCTTCCTT | ||||
| Curcuma longa | matK | CL_matK_F | 19 | CAATCCTATATGGTTGAGA | 171 | 55 |
| CL_matK_R | 18 | GTCAGAAGACTCTATGGA | ||||
| atpF | CL_atpF_F | 20 | GCATTATTGGTTGATAGAGA | 194 | 58 | |
| CL_atpF_R | 22 | GTTTATTTCAAGAATAGGATGG | ||||
| ycf2 | CL_ycf2_F | 20 | GAAGAAGAGGAAGAGGACAT | 80 | 60 | |
| CL_ycf2_R | 20 | CATATTCTAGGAGCCCAAAC | ||||
| Zea mays | matK | ZM_matK_F | 19 | TTGATATCGAACATAATGC | 135 | 55 |
| ZM_matK_R | 16 | ACATCTTCTGGAACCT | ||||
| atpF | ZM_atpF_F | 19 | TGGAAGCAGATGAGTATCG | 160 | 60 | |
| ZM_atpF_R | 18 | TGTTGTCGGACCTGATTC | ||||
| ycf2 | ZM_ycf2_F | 20 | AAGAGGATGAGTTGTCAGAG | 99 | 59 | |
| ZM_ycf2_R | 18 | GCAAGAAGTCCGAATCAG |
3.2. Amplification Efficiency of the Designed Primer Sets
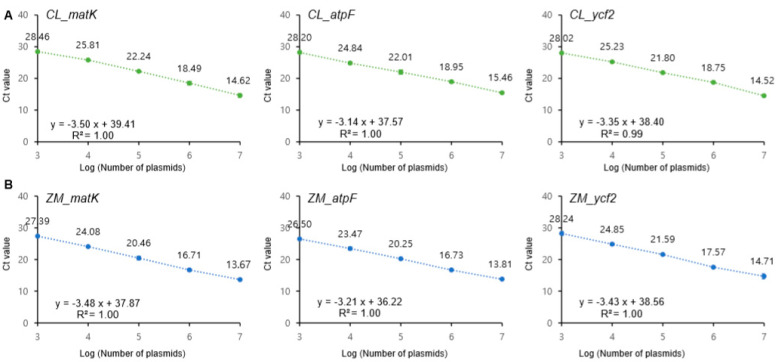
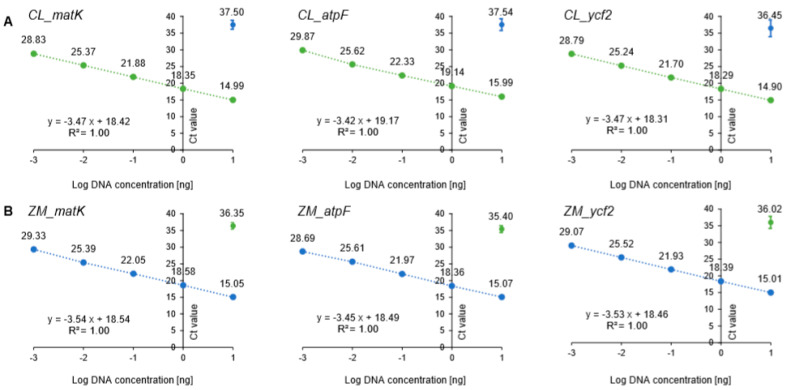
Amplification efficiency of the six primer sets (CL_matK, CL_atpF, CL_ycf2, ZM_matK, ZM_atpF, and ZM_ycf2) was evaluated by constructing standard curves using 10-fold serial dilutions (107 to 103) of each recombinant plasmid DNA, and regression analyses were performed (Figure 1, Supplementary Figure S3). The correlation coefficients (R2) of the six primer pairs were higher than 0.99, and slopes ranged from −3.14 to −3.50. The efficiency of the primers was between 93.14 and 108.40% (Supplementary Table S1). All values fit the ENGL (European Network of GMO Laboratories) guidelines, with the coefficient of determination (R2) being greater 0.98 and the amplification efficiency ranging from 110 to 90%, which corresponds to a slope between −3.1 and −3.6 [29]. Subsequently, we evaluated the efficiency of the primers using the 10-fold serially diluted genomic DNAs (from 10 ng to 1 pg) extracted from plant samples (Figure 2, Supplementary Figure S4). Similarly to the results of recombinant plasmid DNAs, standard curves in the gDNA samples also ranged from −3.42 to −3.54, exhibited R2> 0.99, and efficiency values of 91.78–95.92%, which also conformed to the ENGL guidelines (Supplementary Table S1) [29].
Figure 1.
Standard curve of cycle threshold (Ct) values were obtained on the basis of efficiency and correlation of coefficient (R2) in serial dilution series recombinant plasmids (C. longa and Z. mays) using species-specific primer sets. The x-axis represents log number of plasmids and the y-axis represents means of Ct value ± SD. (A) C. longa targeting primer sets (CL_matK, CL_atpF, and CL_ycf2). Green dots represent serial dilution series of recombinant plasmids (107–103) containing C. longa specific target gene (matK, atpF and ycf2) sequences; (B) Z. mays targeting primer sets (ZM_matK, ZM_atpF, and ZM_ycf2). Blue dots represent serial dilution series of recombinant plasmids (107–103) containing Z. mays specific target gene (matK, atpF and ycf2) sequences. The real-time PCRs were carried out in triplicate (n = 3).
Figure 2.
Standard curve of cycle threshold (Ct) values were obtained on the basis of efficiency and correlation of coefficient (R2) in serial dilution series genomic DNA (C. longa and Z. mays) using species-specific primer sets. The x-axis represents log DNA concentration (ng) and the y-axis represents means of Ct value ± SD. (A) C. longa targeting primer sets (CL_matK, CL_atpF, and CL_ycf2). Green dots represent serial dilution series of genomic DNA in C. longa leaves (10ng–1pg) and blue dots represent genomic DNA of Z. mays (10ng); (B) Z. mays targeting primer sets (ZM_matK, ZM_atpF, and ZM_ycf2). Blue dots represent serial dilution series of genomic DNA in Z. mays leaves (10ng–1pg) and green dot represents genomic DNA of C. longa (10ng). The real-time PCRs were carried out in triplicate (n = 3).
In addition, to evaluate the adaptability of the primes across machines, amplification efficiency was evaluated by two independent laboratories. As a result, the primer sets were found to meet the ENGL criteria (R2 > 0.98 and efficiency ranges of 91.78–108.40; Supplementary Table S2). Based on the evaluation of amplification efficiency of the designed primer sets through three approaches, with recombinant plasmids, genomic DNA, and interlaboratory evaluation, the designed primer sets could be suitable to detect the target species.
3.3. Sensitivity and Specificity of the Assay
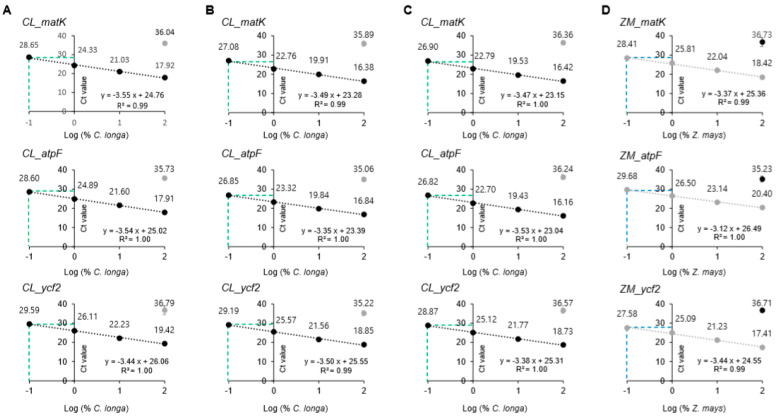
Globally, most C. longa-containing foods are prepared with rhizomes and dry powders. Therefore, we tested the sensitivity and specificity of the designed C. longa primer sets with binary mixtures (0.1–100% (w/w)) of C. longa dry rhizome powders containing each of three starch crops, including corn, rice, and wheat (Figure 3A–C). All three C. longa primer sets with slopes ranging from −3.35 to −3.550 exhibited R2 > 0.99 and efficiency values of 91.29–98.84% when used on mixed powders of C. longa and each starch crop, supporting the high sensitivity of the primer sets for verifying the presence of C. longa in mixtures sets (Supplementary Table S3). Subsequently, sensitivity of the three Z. mays primer sets was tested with binary mixtures of Z. mays and C. longa (0.1–100% (w/w)). Similarly, the three Z. mays primer sets with slope ranging from −3.12 to −3.44 exhibited R2 > 0.99 and efficiency values of 95.30–109.18% when used on mixed powders of C. longa and Z. mays, supporting the high sensitivity of the primer sets for verifying the presence of Z. mays in mixtures. Next, we determined the cut-off of Ct values based on the binary mixture standard collinearity equation of each primer set (Supplementary Table S3) to identify intended additions of cheap starch ingredients, such as Z. mays, in the C. longa powders. Ct values of 0.1% target species were determined as cut-off values for each primer set because additions of less than 0.1% of non-target species were not considered to be intended for illegal economic profit. The cut-off Ct values (0.1% target species in binary mixtures) were established to verify the presence of the target species from the calibration curves (Figure 3). The cut-off Ct values ranged from 26.82 to 29.59 cycles for each primer set targeting C. longa and 27.58 to 29.68 cycles for those targeting Z. mays (Supplementary Table S3).
Figure 3.
Standard curve of cycle threshold (Ct) values obtained on the basis of efficiency and correlation of coefficient (R2) by reference binary mixtures. The x-axis represents log percentage of the target species (%) and the y-axis represents means of Ct value ± SD. plotted against the logarithm of the target species concentration (100, 10, 1, and 0.1%). (A–C); each C. longa rhizome powders were mixed with three different plant powders (Z. mays, O. sativa, and T. aestivum) by ten-fold dilutions (0.1, 1, 10 and 100%, final mass of 2g) and the each mixture gDNA(10 ng/uL) was amplified using the C. longa targeting primer sets (CL_matK, CL_atpF, and CL_ycf2). The green dotted line means the 0.1% binary mixture Cts amplified using the C. longa targeting primer sets, CL_matK, CL_atpF and CL_ycf2) (A) binary mixture of C. longa and Z. mays; (B) binary mixture of C. longa and O. sativa; (C) binary mixture of C. longa and T. aestivum. (D) Z. mays powders were mixed with C. longa rhizome powders by ten-fold dilutions (0.1, 1, 10 and 100%, final mass of 2g) and each mixture gDNA(10 ng/uL) was amplified using the Z. mays targeting primer sets (ZM_matK, ZM_atpF, and ZM_ycf2). The blue dotted line means the 0.1% binary mixture Cts amplified using the Z. mays targeting primer sets, ZM_matK, ZM_atpF and ZM_ycf2). The real-time PCRs were carried out in triplicate (n = 3).
Subsequently, we conducted a specificity test using the species-specific primer sets. A total of 10 species of cereals and vegetables were examined to assess cross-reactivity (Table 2). The cheap starch crops such as barley, wheat, oats, rice, sweat potato, and cassava, which are likely to be intentionally mixed as ingredients in complex turmeric foods for illegal economic profits, were included for the specificity test. In addition, one vegetable crop such as cabbage and one oilseed crop such as peanuts were used for the specificity test as out groups. 18S plant rRNA primer sets were used as a positive control [32], which exhibited lower Ct values than the cut-off. As shown in Table 2, Cl_matK, CL_atpF, and CL_ycf2 exhibited C. longa-specific amplification but did not amplify the DNA of other species. Similarly, ZM_matK, ZM_atpF, and ZM_ycf2 exhibited Z. mays specific amplification did not amplify the DNA of other species. The specificity test demonstrates that the primer sets could be useful for detecting the target species in unknown-ingredient powders and in complex food products.
Table 2.
Results of the specificity test with other plants.
| NO | Species | Plant Systems | Curcuma longa | Zea mays | |||||
|---|---|---|---|---|---|---|---|---|---|
| 18s rRNA | CL_matK | Cl_atpF | CL_ycf2 | ZM_matK | ZM_atpF | ZM_ycf2 | |||
| 28 a (Cycles) | 28 (Cycles) | 29 (Cycles) | 28 (Cycles) | 29 (Cycles) | 28 (Cycles) | ||||
| 1 |
Curcuma longa (Turmeric) |
+ | + b | + | + | - | - | - | |
| 2 |
Hodeum vulgare (Barley) |
+ | - c | - | - | - | - | - | |
| 3 |
Avena sativa (Oats) |
+ | - | - | - | - | - | - | |
| 4 |
Triticum aestivum (Wheat) |
+ | - | - | - | - | - | - | |
| 5 |
Zea mays (Corn) |
+ | - | - | - | + | + | + | |
| 6 |
Oryza sativa (Rice) |
+ | - | - | - | - | - | - | |
| 7 | Brassica oleracea var. capitate (Cabbage) | + | - | - | - | - | - | - | |
| 8 |
Ipomoea batatas (Sweat potato) |
+ | - | - | - | - | - | - | |
| 9 |
Arachis hypogaea (Peanuts) |
+ | - | - | - | - | - | - | |
| 10 | Manihot esculenta (Cassava) | + | - | - | - | - | - | - | |
a Cycles, conventional PCR cycles based on cut-off (Ct) values of each specific primer sets (Ct values of 0.1% binary mixture ); b +, detected at less than Ct values of primers; c -, not detected before the primers′ Ct values.
3.4. Application of the Developed Real-Time PCR Assay to Blind Samples
A blind test was conducted to estimate the reliability of the developed real-time PCR assays. Twenty unknown powder samples of C. longa and Z. mays were mixed randomly by an independent research group. The 18S rRNA plant primer sets were used as positive amplification controls [32], which exhibited low Cts (13.27–16.21; Table 3).
Table 3.
Results of the blind mixture test for evaluating the reliability of the developed primer sets.
| No. | Ingredient | PACa | Eb (%) | Z. mays Specific Primer Ct ± SD | A/D | |||
|---|---|---|---|---|---|---|---|---|
| C. longa (%) | Z. mays (%) | ZM_matK | ZM_atpF | ZM_ycf2 | ||||
| 1 | 99 | 1 | 13.57 ± 0.04 | 0.5–1.5 | 25.02 ± 0.06 | 25.61 ± 0.20 | 26.44 ± 0.21 | Ad |
| 2 | 98 | 2 | 14.73 ± 0.09 | 1–5 | 25.09 ± 0.02 | 24.78 ± 0.12 | 22.8 ± 0.18 | A |
| 3 | 100 | 0 | 14.03 ± 0.01 | NDc | 31.51 ± 0.15 | 31.32 ± 0.26 | 35.15 ± 0.05 | A |
| 4 | 98 | 2 | 14.31 ± 0.01 | 1–5 | 24.40 ± 0.01 | 24.75 ± 0.15 | 23.92 ± 0.09 | A |
| 5 | 95 | 5 | 14.05 ± 0.01 | 1–5 | 23.72 ± 0.09 | 24.34 ± 0.09 | 21.94 ± 0.22 | A |
| 6 | 97 | 3 | 14.11 ± 0.06 | 1–5 | 23.45 ± 0.05 | 24.90 ± 0.07 | 22.97 ± 0.25 | A |
| 7 | 99.5 | 0.5 | 14.21 ± 0.02 | 0.5–1.5 | 25.09 ± 0.05 | 26.95 ± 0.07 | 26.34 ± 0.15 | A |
| 8 | 98.5 | 1.5 | 14.12 ± 0.05 | 0.5–1.5 | 25.59 ± 0.03 | 26.32 ± 0.10 | 24.69 ± 0.19 | A |
| 9 | 100 | 0 | 14.33 ± 0.08 | ND | 31.67 ± 0.20 | 31.01 ± 0.80 | 34.89 ± 0.10 | A |
| 10 | 98.5 | 1.5 | 14.23 ± 0.03 | 0.3–2 | 25.13 ± 0.09 | 25.69 ± 0.10 | 24.21 ± 0.02 | A |
| 11 | 99.5 | 0.5 | 14.27 ± 0.01 | 0.1–1 | 26.06 ± 0.09 | 27.08 ± 0.10 | 26.19 ± 0.09 | A |
| 12 | 100 | 0 | 13.61 ± 0.12 | ND | 31.33 ± 0.28 | 30.98 ± 0.11 | 34.46 ± 0.26 | A |
| 13 | 97 | 3 | 14.31 ± 0.10 | 1–5 | 24.97 ± 0.09 | 24.37 ± 0.07 | 23.86 ± 0.13 | A |
| 14 | 98 | 2 | 13.22 ± 0.07 | 1–5 | 24.19 ± 0.09 | 26.14 ± 0.12 | 23.22 ± 0.09 | A |
| 15 | 96 | 4 | 13.33 ± 0.06 | 1–5 | 23.92 ± 0.15 | 24.26 ± 0.03 | 22.72 ± 0.27 | A |
| 16 | 99 | 1 | 14.26 ± 0.05 | 0.1–1 | 25.75 ± 0.10 | 26.75 ± 0.14 | 25.11 ± 0.10 | A |
| 17 | 93 | 7 | 13.95 ± 0.07 | 5–10 | 22.11 ± 0.11 | 23.99 ± 0.13 | 21.72 ± 0.11 | A |
| 18 | 90 | 10 | 13.74 ± 0.02 | 10–15 | 22.11 ± 0.10 | 21.99 ± 0.18 | 21.23 ± 0.13 | A |
| 19 | 100 | 0 | 16.21 ± 0.07 | ND | 30.82 ± 0.20 | 31.09 ± 0.01 | 34.06 ± 0.56 | A |
| 20 | 97 | 3 | 13.27 ± 0.1 | 1–5 | 23.96 ± 0.05 | 24.28 ± 0.10 | 23.22 ± 0.12 | A |
a Positive amplification control (18s rRNA); b, expected ratio of the Z. mays; C, not detected; d accordance.
Next, we determined whether Z. mays powder was present in the samples based on the cut-off Ct values of the designed primer sets (0.1% Z. mays in binary mixtures). As a result, we identified four samples (sample 3, 9, 12, and 19) with Ct values exceeding the cut-off Ct values, indicating that the samples did not contain Z. mays powder mixed in C. longa powder. The other 16 samples were exhibited lower Cts than the cut-off Ct values, indicating that those samples contained Z. mays powder. In addition, the ratio of Z. mays powder mixed into the 16 samples was predicted using the developed binary mixture assay of the three primer sets (ZM_matK, ZM_atpF, and ZM_ycf2). The predicted percentage of Z. mays in each blind sample was extrapolated by inserting the Cts into the standard collinearity equation of each primer set (ZM_matK, ZM_atpF, and ZM_ycf2). As a result, the predicted percentages of Z. mays present in each sample were consistent with those of the mixed samples (Table 3). Therefore, the real-time PCR methodologies developed in this study demonstrated high accuracy for detecting the addition of Z. mays in C. longa rhizome powders.
3.5. Application of the Developed Assay in Commercial Products
To verify adulteration with corn powder of C. longa food products, we performed the developed real-time PCR assays on 10 C. longa commercial food products (Supplementary Table S4, Table 4). First, the quality of genomic DNA isolated from the food products was evaluated using a spectrometer. As depicted in Table 4, the 18S rRNA primer sets exhibited low Cts (14.01–19.82), indicating that the gDNA from all the commercial products was sufficient to provide amplifiable gDNA. We found that all C. longa commercial food products (samples 1–10) were amplified with lower Ct values (from 14.1 to 21.971 cycles) using the C. longa species-specific primers (CL_matK, CL_atpF, and CL_ycf2) than the cut-off Ct values (Ct values of 0.1% C. longa-specific primer set in binary mixtures) for each primer set (CT values of CL_matK, CL_atpF, and CL_ycf2 were 28.65, 28.60, and 29.59 cycles, respectively; Figure 3, Supplementary Table S3). Additionally, all samples were amplified with higher Ct values (from 30.23 cycles to not detected before 40 cycles) with Z. mays targeting primers (ZM_matK, ZM_atpF, and ZM_ycf2) than the cut-off Ct values (Ct values of 0.1% Z. mays-specific primer sets in binary mixtures) for each primer set (28.41, 29.68, and 27.58 cycles, respectively; Supplementary Table S2). As a result, the commercial products purchased from local markets did not contain Z. mays, suggesting that the developed real-time PCR assays could be successfully applied to detect the presence of Z. mays in commercial complex C. longa products.
Table 4.
Result of the real-time PCR assay using 10 commercial products.
| Real Commercial Products Tested that Labeled as 100% Curcuma longa | |||||||
|---|---|---|---|---|---|---|---|
| Sample Number | Plant System (18s rRNA) | CL_matK | CL_atpF | CL_ycf2 | ZM_matK | ZM_atpF | ZM_ycf2 |
| 1 | 14.49 | 14.83 | 14.10 | 14.24 | NDa | 34.00 | ND |
| ±0.15 | ±0.13 | ±0.17 | ±0.14 | ±0.07 | |||
| 2 | 15.23 | 15.27 | 15.70 | 14.50 | 38.26 | 33.59 | ND |
| ±0.06 | ±0.18 | ±0.09 | ±0.08 | ±1.1 | ±2.01 | ||
| 3 | 19.82 | 22.62 | 22.65 | 20.16 | 36.28 | 32.32 | 34.79 |
| ±0.08 | ±0.27 | ±0.04 | ±0.06 | ±0.61 | ±0.54 | ±0.47 | |
| 4 | 15.25 | 15.95 | 16.33 | 15.25 | 33.60 | 30.05 | 34.35 |
| ±0.06 | ±0.07 | ±0.03 | ±0.14 | ±0.20 | ±0.07 | ±0.21 | |
| 5 | 14.01 | 15.29 | 15.46 | 15.29 | 31.53 | 31.41 | ND |
| ±0.05 | ±0.07 | ±0.01 | ±0.07 | ±0.54 | ±0.56 | ||
| 6 | 19.19 | 17.68 | 21.97 | 15.28 | 32.34 | 31.14 | ND |
| ±0.03 | ±0.12 | ±0.04 | ±0.04 | ±0.76 | ±0.14 | ||
| 7 | 16.44 | 16.10 | 16.39 | 14.90 | 34.89 | 34.57 | ND |
| ±0.18 | ±0.03 | ±0.03 | ±0.09 | ±0.35 | ±0.65 | ||
| 8 | 18.87 | 15.35 | 20.24 | 14.10 | 32.82 | 34.55 | ND |
| ±0.08 | ±0.02 | ±0.12 | ±0.02 | ±0.33 | ±0.95 | ||
| 9 | 15.31 | 14.63 | 15.15 | 13.26 | 31.05 | 34.00 | ND |
| ±0.09 | ±0.08 | ±0.06 | ±0.02 | ±4.55 | ±0.08 | ||
| 10 | 16.10 | 15.84 | 16.57 | 14.95 | 32.02 | 30.23 | 33.77 |
| ±0.01 | ±0.03 | ±0.02 | ±0.06 | ±0.24 | ±0.06 | ±0.92 | |
a ND indicates not detected at less than 40 cycles.
4. Conclusions
A real-time PCR assay is a highly sensitive, rapid, and specific method to detect target-species in processed food complexes. We designed three chloroplast gene targeted primer sets for both C. longa and Z. mays. To assess the quantities of the target-species present, standard curves were constructed using recombinant plasmid DNA and binary DNA mixtures. The specificities of the designed primers were confirmed with ten other species. Blind sample analysis and the application to commercial C. longa food products supported the effectiveness of the real-time PCR assays to detect Z. mays products added for illegal economic profits. Therefore, the developed real-time PCR assay could contribute to food safety and the protection of consumer’s rights.
Acknowledgments
This research was supported by a grant (17162MFDS065) from the Ministry of Food and Drug Safety.
Supplementary Materials
The following are available online at https://www.mdpi.com/2304-8158/9/7/882/s1, Figure S1: Alignment of the target chloroplast gene (matK, atpF and ycf2) nucleotide sequences of C. longa, Z. mays and starch crops (O. sativa and T. aestivum) mainly eating as powders amplified by C. longa specific primer sets (CL_matK CL_atpF and CL_ycf 2), Figure S2: Alignment of the target chloroplast gene(matK, atpF and ycf2)nucleotide sequences of Z. mays, C. longa and starch crops(O. sativa, and T. aestivum) mainly eating as powders, amplified by Z. mays s pecific primer sets (ZM_matK, ZM_atpF and ZM_ycf2), Figure S3: Real time PCR with SYBR Green and DNA melting curve analyses.(A) Serial dilution series recombinant plasmids (107–103) containing C. longa specific gene (matK, atpF and ycf 2) sequence were amplified using C. longa specific primer sets. (B) Serial dilution series recombinant plasmids (107–103) containing Z. mays specific gene (matK, atpF and ycf 2) sequence were amplified using Z. mays specific primer sets. The real time PCRs were performed on a QuantStudio 3 Real Time PCR System (Applied Biosystems, Foster City, CA, USA) and carried out in triplicate (n = 3), Figure S4: Real time PCR with SYBR Green and DNA melting curve analyses green lanes mean the C. longa blue lanes mean Z. mays and pink lanes mean NTC. (A) Serial dilution series of C. longa genomic DNA (10 ng–1 pg) was amplified using C. longa specific primer sets. (B) Serial dilution series of Z. mays (10 ng–1 pg) was amplified using Z. mays specific primer sets. The real time PCRs were performed on a QuantStudio 3 Real Time PCR System (Applied Biosystems, Foster City, CA, USA) and carried out in triplicate (n = 3), Table S1: Evaluation of slope, R2, and efficiency using the developed primer sets, Table S2: Result of the real-time PCR assay in an interlaboratory experiment, Table S3: Evaluation of the slope, R2, and efficiency using binary mixtures containing three different intentionally added powders, Table S4: Information on the commercial food products.
Author Contributions
C.S.J. conceived of the overall study; S.H.O. carried out the experiment; S.H.O and C.S.J. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Priyadarsini K.I. The chemistry of curcumin: From extraction to therapeutic agent. Molecules. 2014;19:20091–20112. doi: 10.3390/molecules191220091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curcumin Market Size, Share & Trends Analysis Report by Application (Pharmaceutical, Food, Cosmetics), by Region (North America, Europe, Asia Pacific, Central & South America, Middle East & Africa), and Segment Forecasts, 2020–2027. [(accessed on 26 April 2020)]; Available online: https://www.grandviewresearch.com/industry-analysis/turmeric-extract-curcumin-market.
- 3.Aggarwal B.B., Kumar A., Bharti A.C. Anticancer potential of curcumin: Preclinical and clinical studies. Anticancer Res. 2003;23:363–398. [PubMed] [Google Scholar]
- 4.Gupta S.C., Patchva S., Aggarwal B.B. Therapeutic Roles of Curcumin: Lessons Learned from Clinical Trials. AAPS J. 2013;15:195–218. doi: 10.1208/s12248-012-9432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panahi Y., Hosseini M.S., Khalili N., Naimi E., Simental-Mendia L.E., Majeed M., Sahebkar A. Effects of curcumin on serum cytokine concentrations in subjects with metabolic syndrome: A post-hoc analysis of a randomized controlled trial. Biomed Pharmacother. 2016;82:578–582. doi: 10.1016/j.biopha.2016.05.037. [DOI] [PubMed] [Google Scholar]
- 6.Kuptniratsaikul V., Dajpratham P., Taechaarpornkul W., Buntragulpoontawee M., Lukkanapichonchut P., Chootip C., Saengsuwan J., Tantayakom K., Laongpech S. Efficacy and safety of Curcuma domestica extracts compared with ibuprofen in patients with knee osteoarthritis: A multicenter study. Clin. Interv. Aging. 2014;9:451–458. doi: 10.2147/CIA.S58535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazzolani F., Togni S. Oral administration of a curcumin-phospholipid delivery system for the treatment of central serous chorioretinopathy: A 12-month follow-up study. Clin. Ophthalmol. 2013;7:939–945. doi: 10.2147/OPTH.S45820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith T., Kawa K., Eckl V., Morton C., Stredney R. Herbal supplement sales in US increase 7.7% in 2016. HerbalGram. 2017;115:56–65. [Google Scholar]
- 9.Pina P. The Rise of Functional Foods. Think with Google Website. [(accessed on 19 July 2017)];2016 Apr; Available online: www.thinkwithgoogle.com/consumer-insights/2016-food-trends-google/
- 10.Food Trends 2016. [(accessed on 19 July 2017)]; Available online: https://think.storage.googleapis.com/docs/FoodTrends-2016.pdf.
- 11.Turmeric Jumps from the Spice Rack to the Coffee Cup at Peet’s. Forbes. [(accessed on 3 July 2018)]; Available online: www.forbes.com/sites/michelinemaynard/
- 12.Balakrishnan K.V. Postharvest technology and processing of turmeric. In: Ravindran P.V., Nirmal Babu K., Sivaraman K., editors. Turmeric: The Genus Curcuma. Volume 45. CRC Press; Boca Raton, FL, USA: 2007. pp. 193–256. [Google Scholar]
- 13.Johnson R. Food Fraud and “Economically Motivated Adulteration” of Food and Food Ingredients, Report, January 10, 2014; Washington DC. [(accessed on 27 May 2020)]; Available online: https://digital.library.unt.edu/ark:/67531/metadc276904/
- 14.Hong E.Y., Lee S.Y., Jeong J.Y., Park J.M., Kim B.H., Kwon K.S., Chun H.S. Modern analytical methods for the detection of food fraud and adulteration by food category. J. Sci. Food Agric. 2017;97:3877–3896. doi: 10.1002/jsfa.8364. [DOI] [PubMed] [Google Scholar]
- 15.Kane D.E., Hellberg R.S. Identification of species in ground meat products sold on the US commercial market using DNA-based methods. Food Control. 2016;59:158–163. doi: 10.1016/j.foodcont.2015.05.020. [DOI] [Google Scholar]
- 16.An J., Moon J.C., Jang C.S. Markers for distinguishing Orostachys species by SYBR Green-based real-time PCR and verification of their application in commercial O. japonica food products. Appl. Biol. Chem. 2018;61:499–508. doi: 10.1007/s13765-018-0383-3. [DOI] [Google Scholar]
- 17.Arya M., Shergill I.S., Williamson M., Gommersall L., Arya N., Patel H.R. Basic principles of real-time quantitative PCR. Expert Rev. Mol. Diagn. 2005;5:209–219. doi: 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- 18.López-Calleja I., González I., Fajardo V., Martín I., Hernández P., García T., Martin R. Real-time TaqMan PCR for quantitative detection of cows’ milk in ewes’ milk mixtures. Int. Dairy J. 2007;17:729–736. doi: 10.1016/j.idairyj.2006.09.005. [DOI] [Google Scholar]
- 19.Ergün Ş., Ahmet K. Practical Molecular Detection Method of Beef and Pork in Meat and Meat Products by Intercalating Dye Based Duplex Real-Time Polimerase Chain Reaction. Int. J. Food Prop. 2016;19:31–40. doi: 10.1080/10942912.2015.1017049. [DOI] [Google Scholar]
- 20.Muhammad S., Yasmeen J. Development and validation of fast duplex real-time PCR assays based on SYBER Green florescence for detection of bovine and poultry origins in feedstuffs. Food Chem. 2015;173:660–664. doi: 10.1016/j.foodchem.2014.10.088. [DOI] [PubMed] [Google Scholar]
- 21.Chase M.W., Cowan R.S., Hollingsworth P.M., van den Berg C., Madriñán S., Petersen G., Seberg O., Jorgsensen T., Cameron K.M., Carine M.A., et al. A proposal for a standardized protocol to barcode all land plants. Taxon. 2007;56:295–299. doi: 10.1002/tax.562004. [DOI] [Google Scholar]
- 22.Vijayan K., Tsou C.H. DNA Barcoding in Plants: Taxonomy in a New Perspective. Curr. Sci. 2010;99:1530–1541. [Google Scholar]
- 23.Daniell H., Lin C.S., Yu M., Chang M.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garino C., De Paolis A., Coisson J.D., Bianchi D.M., Decastelli L., Arlorio M. Sensitive and specific detection of pine nut (Pinus spp.) by real-time PCR in complex food products. Food Chem. 2016;194:980–985. doi: 10.1016/j.foodchem.2015.08.114. [DOI] [PubMed] [Google Scholar]
- 25.Hollingsworth P.M., Forrest L.L., Spouge J.L., Hajibabaei M., Ratnasingham S., van der Bank M., Chase M.W., Cowan R.S., Erickson D.L., CBOL Plant Working Group et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caroline P.L., Anne C.E. Development and Evaluation of a Real-Time PCR Multiplex Assay for the Detection of Allergenic Peanut Using Chloroplast DNA Markers. J. Agric. Food Chem. 2018;66:8623–8629. doi: 10.1021/acs.jafc.8b02053. [DOI] [PubMed] [Google Scholar]
- 27.Sasikumar B., Remya R., John Zachariah T. PCR Based Detection of Adulteration in the Market Samples of Turmeric Powder. Food Biotechnol. 2004;18:299–306. doi: 10.1081/FBT-200035022. [DOI] [Google Scholar]
- 28.Bustin S.A., Benes V., Garson J.A., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 29.ENGL (European Network of GMO Laboratories) Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing. [(accessed on 20 October 2015)]; Available online: http://gmo-crl.irc.ec.ecrl.jrc.ec.europa.eu/doc/MPR%20Report%20Application%2020_10_2015.pdf.
- 30.Yuan J.S., Reed A., Chen F., Stewart C.N. Statistical analysis of real-time PCR data. BMC Bioinform. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo Y., Shaw P. DNA-based techniques for authentication of processed food and food supplements. Food Chem. 2018;240:767–774. doi: 10.1016/j.foodchem.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 32.Allmann M., Candrian U., Höfelein C., Lüthy J. Polymerase chain reaction (PCR): A possible alternative to immunochemical methods assuring safety and quality of food Detection of wheat contamination in non-wheat food products. Z. Lebensm. Unters. Forch. 1993;196:248–251. doi: 10.1007/BF01202741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.