Abstract
Rosa rugosa Thunb., is as a medicinal plant known for anti‐diabetic, and anti‐inflammatory activities. However, the specific active compounds responsible for the individual pharmacological effects of in R. rugosa extract (95% EtOH) remain unknown. Here, we hypothesized that terpenoid structure, the most abundant constituents in R. rugosa extract, are responsible for its anti‐inflammatory activity. We investigated the phytochemical substituents (compounds 1–13) and newly purified 11‐methoxy polisin A, and 13‐methoxy bisaborosaol F using NMR and ESI‐MS and to screened their effects on NO production in LPS‐induced macrophages. Rugosic acid A (RA) induced to ameliorate NO production, iNOS, and pro‐inflammatory cytokines associated with the NF‐κB. And, RA suppressed IL‐6 secretion and IL‐6‐mediated STAT3 activation in LPS‐mediated inflammation. In addition, RA was evaluated in LPS‐mediated acute lung injury (ALI) model similar to acute pneumonia. Our results suggested that RA was suppressed to translocate nuclear NF‐κB and IL‐6‐mediated STAT3 activation. Finally, RA led to amelioration of ALI by decreasing myeloperoxidase (MPO) and inhibiting phosphorylation of NF‐κB and STAT3. Our group originally found that R. rugosa extract had new methoxy compounds and RA may be alternative natural agent for acute pneumonia similar to severe acute respiratory syndrome by coronavirus.
Keywords: Acute pneumonia, IL‐6, NF‐κB, Rosa rugose Thunb, Rugosic acid A, STAT3
1. INTRODUCTION
Botanical character of Rosa rugosa Thunb., (R. rugosa) occurs naturally in Eastern Asia from ochotsk to Korea and the northern parts of Japan to China. Traditionally, R. rugosa extracts of the flowers or hips have been valued for Asian culinary, cosmetic, aromatherapy, and medicinal properties and used in herbal medicines for diabetes mellitus and osteoarthritis (Cho, Yokozawa, Kim, Shibahara, & Park, 2004; Yang et al., 2016). The several effects seemed to be involved in the presence of many phytochemicals in R. rugosa extracts, such as flavonoids, phenylpropanoid, bisabolanes, acoranes, tannins, fatty acid, and terpenoids (Hashidoko, 1996). Among these natural products, terpenoids were included in several medicinal plants such as Dysoxylum gotadhora, Cyperus rotaundus, Dodonaea viscosa, Onopordum Illyricum, and Gardenia sootepenis and reacted as anti‐inflammatory agent (Dang, Parekar, Kamat, Scindia, & Rege, 2011; Formisano et al., 2017; Jiang et al., 2015; Khalil, Sperotto, & Manfron, 2006; Youn et al., 2016). A concomitant feature of anti‐inflammatory activity among the exhibited medicinal plants was down‐regulation associated with nuclear factor kappa B (NF‐κB), or signal transducer and activator of transcription 3 (STAT3), as a transcription factor.
NF‐κB and STAT3 signaling mainly had known to regulate several biological interactions; development, proliferation, differentiation as well as cancer and inflammation (He & Karin, 2011). In perspectives of inflammatory action, NF‐κB acts on a family of inducible transcription factors that regulate a lot of genes associated with individual functional processes in inflammation in innate immune cells, including macrophages (Oeckinghaus & Ghosh, 2009). In addition, STAT3 is involved in a family of cytoplasmic protein with SH2 (Src Homology‐2) domains. They act as signal messengers and transcription factors and participate in normal cellular response to growth factors and cytokines (Guanizo, Fernando, Garama, & Gough, 2018). when activate both NF‐κB and STAT3 signaling, genes responsible for inflammation including IL‐6 (interleukin‐6), IL‐10 (interleukin‐10), IL‐11 (interleukin‐11), IL‐17 (interleukin‐17), IL‐23 (interleukin‐23), IL‐1b (interleukin‐1b), Tnf‐α (tumor necrosis factor‐α), and iNOS (inducible nitric oxide synthase) were induced and regulated in pathogenesis of inflammation diseases (Liu et al., 2017; He & Karin, 2011). Thus, the safe development of inhibitory agents of activation both NF‐κB, and STAT3 is crucial for clinical treatment of inflammatory diseases among natural constituents.
In present study, we newly investigated anti‐inflammatory effects of R. rugosa extracts (EtOH 95%) and developed effective compounds under inflammatory condition associated with lipopolysaccharides (LPS) and IL‐6‐mediated inflammation. Although R. rugosa flower extracts (EtOH 50%) and euscaphic acid, derived from R. rugosa root extracts performed anti‐inflammatory and effects in previous study (Um, Han, & Lee, 2018), there were no academic evaluation for information of ingredients and quantitative analysis among R. rugosa extracts. Here, we further studied that anti‐inflammatory agents of terpenoid structures (compound 1 to 13) and originally reported the identification of a new compound, 11‐methoxy polisin A and 13‐methoxy bisaborosaol F from the R. rugosa extracts (EtOH 95%) using spectroscopic data, including nuclear magnetic resonance (NMR) and electrospray ionization mass spectrometry (ESI‐MS). In several phytochemical constituents of R. rugosa extracts (EtOH 95%), our group originally reported that rugosic acid A, sesquiterpenoid structure, had anti‐inflammatory effects by amelioration of nitric oxide (NO) production and inhibition of nuclear‐translocation of NF‐κB under LPS‐mediated inflammatory conditions in RAW 264.7. In addition, rugosic acid A significantly disturbed IL‐6‐mediated STAT3 activation under IL‐6/STAT3 screening system. Finally, we tested RA‐mediated amelioration of inflammation in LPS‐mediated acute lung injury as an acute pneumonia model, which was similar to severe acute respiratory syndrome by coronavirus such as COVID‐19 pneumonia (Zhao et al., 2020; Bernheim et al., 2020). Through our results, RA expect to be alternative natural agent and treatment for acute pneumonia similar to COVID‐19 pneumonia.
2. MATERIALS AND METHODS
2.1. General experimental procedures
1H, 13C and 2D NMR spectra were acquired with instruments of a JEOL JNM‐ECA 400 and JEOL JNM‐ECA 600 instruments (JEOL, Tokyo, Japan) using TMS as an internal standard. Optical rotations were obtained on a Jasco P‐2000 polarimeter (Jasco Corp., Tokyo, Japan), and UV spectra were recorded using a SpectraMax M2 e spectrophotometer (Molecular Devices, Sunnyvale, CA). High resolution electrospray ionization mass spectrometer (HRESIMS) data were acquired using a Waters SYNAPT G2‐Si HDMS spectrometer (Waters, Milford, MA). Silica gel (Kieselgel 60, 230–400 mesh, Merck, Darmstadt, Germany) column chromatography (CC) and medium‐pressure liquid chromatography (MPLC, CombiFlash RF, Teledyne lsco, Lincoln, NE) were used to separate the extract fractions. Each fractions was monitored by TLC using silica gel 60 F254 and RP‐18 F254s (Merck, Burlington). Sephadex LH‐20 (GE Healthcare, Uppsala, Sweden) column chromatography (CC) and semipreparative high speed liquid chromatography (semipreparative HPLC) were used to acquire pure compounds. Semipreparative HPLC was performed on a Shimadzu LC‐6 AD instrument (Shimadzu Corp., Tokyo, Japan) equipped with an SPD‐20A detector and Phenomenex Luna C18 (21.2 mm × 150 mm, 5 μm), Phenomenex Luna C18 (21.2 mm × 250 mm, 5 μm) columns. All fractions and compounds were analyzed using an Agilent 1,260 series HPLC system (Agilent, Santa Clara).
2.2. Extraction and isolation
The dried Rosa. rugosa Thunb. (2.5 kg) was purchased from the acote co., ltd in Gochang, Korea. R. rugosa Thunb. (2.5 kg) dry powder was extracted twice with 95% ethanol (3 × 25 L) at 70°C and the EtOH solution was concentrated under vacuum to yield a pinkish brown residue (375.9 g). The residue (375.9 g) was suspended in H2O (1 L), and the aqueous layer was partitioned with n‐hexane, chloroform, ethyl acetate, and n‐buthanol. The n‐hexane layer (30.8 g) was subjected to silica gel column eluted with n‐hexane: EtOAc (1:0 to 0:1, v/v) to obtain fifteen fractions (H1 ~ H15). Fraction H11 (2.78 g) was subjected to MPLC [column: C18 RediSepRf (130 g); mobile phase: H2O: MeOH (40 ~ 85%)] to yield 13 fractions (H11‐1 ~ 13). Fraction H11‐3 and H11‐5 were subjected to preparative HPLC (Phenomenex Luna C18 column (150 × 21.2 mm, 5 μm). Fraction H11‐3 (57.4 mg) was used isocratic elution 50% CH3CN in H2O to afford compound 3 (t R 25 min, 18.1 mg, 88.4% area). Fraction H11‐5 (68 mg) was used isocratic elution 40% CH3CN in H2O to afford compound 2 (t R 55 min, 15.6 mg, 85.7% area). Fraction H12 (3.65 g) was subjected to MPLC [column: C18 RediSepRf (130 g); mobile phase: H2O: MeOH (20 ~ 85%)] to yield 15 fractions (H12‐1 ~ 15). Fraction H12‐6 (13.4 mg) was used isocratic elution 55% CH3CN in H2O to afford compound 7 (t R 15 min, 2.7 mg, 92.2% area). Fraction H13 (1.21 g) was subjected to MPLC [column: C18 RediSepRf (130 g); mobile phase: H2O: MeOH (35 ~ 100%)] to yield eight fractions (H13‐1 ~ 8). The extracts contained appreciable levels of compound 7 content (0.8 mg/g extract). Fraction H13‐1 (53.6 mg) was used isocratic elution 48% CH3CN in H2O to afford compound 1 (t R 16 min, 2.0 mg, 85.1% area). Fraction H15 (520.4 mg) was subjected to chromatography a C18 MPLC [column: C18 RediSepRf (130 g)] with a gradient solvent composed of H2O and MeOH (1:0 to 0:1, v/v), (30–100%)] to generate 8 fractions (H15‐1 ~ 8). Compound 4 (3.4 mg, 86.8% area) and 6 (1.5 mg, 90.0% area) were purified from fraction H15‐2 (23 mg) by semi‐preparative HPLC (Phenomenex Luna 5 μm C18, 250 × 21.2 mm, 35% CH3CN) at 42 and 47 min.
The chloroform layer (18.6 g) was subjected to silica gel column eluted with CHCl3: MeOH to obtain 14 fractions (C1 ~ C14) and purify compound 9 (t R 13 min, 9.1 mg, 81.6% area). Fraction C6 (172.3 mg) was subjected to MPLC [column: C18 RediSepRf (26 g); mobile phase: H2O: MeOH (30 ~ 90%)] to yield 11 fractions (C6‐1 ~ 11). Fraction C6‐7 (14.9 mg) was subjected to preparative HPLC (Phenomenex Luna C18 column (150 × 21.2 mm, 5 μm), using isocratic elution 50% CH3CN in H2O to afford compound 5 (t R 43 min, 2.1 mg, 80.3% area). Compound 10 (3.0 mg, 79.2% area) was purified from fraction C6‐11 (30.1 mg) by semi‐preparative HPLC (Phenomenex Luna 5 μm C8, 150 × 21.2 mm, 80% CH3CN) at 27 min. Fraction C11 (1.8 g) was subjected to MPLC [column: Silica RediSepRf (120 g); mobile phase: n‐hexane: EtOAc (1:0 to 0:1, v/v)] to yield 13 fractions (C11‐1 ~ 13). Fraction C11‐12 (321.4 mg) was subjected to preparative HPLC (Phenomenex Luna C18 column [250 × 21.2 mm, 5 μm], using isocratic elution 70% CH3CN in H2O) to purify compound 8 (t R 14 min, 26.7 mg, 86.9% area), 11 (t R 48 min, 12.3 mg, 94.3% area). Fraction C11‐13 (190.5 mg) was subjected to preparative HPLC (Phenomenex Luna C18 column [250 × 21.2 mm, 5 μm], using isocratic elution 35, 80% CH3CN in H2O) to obtain two fractions (C11‐13‐1, C11‐13‐2). Fraction C11‐13‐2 (21.1 mg) was subjected Sephadex LH‐20 column (CHCl3: MeOH = 1:1) to afford compound 12 (5.4 mg, 87.3% area).
The ethyl acetate layer (53.2 g) was subjected to silica gel column eluted with n‐hexane: EtOAc (1:0 to 0:1, v/v) to obtain 10 fractions (E1 ~ E10). Compound 13 (1.9 mg, 63.6% area) was purified from fraction E1 (40.2 mg) by semi‐preparative HPLC (Phenomenex Luna 5 μm C18, 250 × 21.2 mm, 40% CH3CN) at 22 min.
11‐Methoxy polilsin A (1): White amorphous powder, + 4.8 (c 0.1 CH3OH), UV (CH3OH) λmax (log ε); 214 (3.01), HRESI‐MS ion peak at m/z 261.1103 [M + Na]+ (calcd for C13H18O4Na, 261.1097), 1H NMR data (Methanol‐d 4 , 600 MHz): δH 6.99 (1H, m, H‐2), 3.71 (3H, s, OCH3‐11), 2.71 (1H, m, H‐9a), 2.59 (1H, m, H‐9b), 2.52 (1H, m, H‐6a), 2.37 (1H, m, H‐3a), 2.23 (1H, m, H‐8a), 2.18 (1H, m, H‐6b), 2.10 (1H, m, H‐3b), 2.03 (1H, m, H‐8b), 1.88 (1H, m, H‐5a), 1.87 (1H, m, H‐4), 1.36 (3H, s, H‐12), 1.29 (1H, m, H‐5b), 13C NMR data (Methanol‐d 4 , 150 MHz): δC 179.5 (C‐10), 169.2 (C‐11), 140.1 (C‐2), 131.3 (C‐1), 90.5 (C‐7), 52.2 (OCH3‐11), 44.0 (C‐4), 32.0 (C‐8), 29.9 (C‐9), 28.1 (C‐3), 25.9 (C‐6), 24.3 (C‐5), 23.0 (C‐12).
13‐Methoxy Bisaborosaol F (5): Colorless syrup, + 0.1 (c 0.1 CH3OH), UV (CH3OH) λmax (log ε); 215 (3.47), HRESI‐MS ion peak at m/z 319.1885 [M + Na]+ (calcd for C17H28O4Na, 319.1880), 1H NMR data (DMSO‐d 6 , 600 MHz): δH 6.93 (1H, m, H‐2), 5.62 (1H, dt, J = 15.6, 7.2, H‐11), 5.39 (1H, d, J = 15.6, H‐12), 3.63 (3H, s, OCH3‐7), 3.01 (3H, s, OCH3‐13), 2.37 (1H, m, H‐6a), 2.26 (1H, m, H‐3a), 2.14 (2H, d, J = 7.2, H‐9), 2.01 (2H, m, H‐3b, 6b), 1.81 (1H, m, H‐5a), 1.44 (1H, m, H‐4), 1.16 (3H, s, H‐14, 15), 1.12 (1H, m, H‐5b), 1.00 (3H, s, H‐10) 13C NMR data (DMSO‐d 6 , 150 MHz): δC 166.7 (C‐7), 140.1 (C‐2), 137.4 (C‐12), 129.1 (C‐1), 126.0 (C‐11), 74.2 (C‐13), 72.0 (C‐8), 51.2 (OCH3‐7), 49.4 (OCH3‐13), 42.8 (C‐9), 41.5 (C‐4), 26.3 (C‐3), 25.8 (C‐14), 25.6 (C‐15), 24.8 (C‐6), 23.8 (C‐10), 22.6 (C‐5).
Rugosic acid A (7): Colorless syrup. + 4.7 (c 0.1 CH3OH), ESI‐MS ion peaks at m/z 265 [M – H] ‐, 1H NMR data (Chloroform‐d, 600 MHz): δH 9.52 (1H, s, H‐14), 6.83 (1H, dd, J = 6.0, 1.2, H‐3), 5.20 (1H, ddd, J = 5.4, 2.4, 1.2, H‐5), 4.51 (1H, dd, J = 11.4, 6.0, H‐2), 2.61 (1H, m, H‐11), 2.22 (1H, dd, J = 14.4, 5.4, H‐6a), 1.89 (1H, ddd, J = 10.8, 9.0, 2.4, H‐10), 1.82 (1H, m, H‐8a), 1.77 (1H, dd, J = 14.4, 2.4, H‐6b), 1.70 (1H, dd, J = 12.0, 6.0, H‐8b), 1.61 (1H, m, H‐9a), 1.43 (1H, m, H‐9b), 0.96 (3H, d, J = 6.6, H‐12), 0.92 (1H, d, J = 6.6, H‐13), 0.86 (1H, s, H‐15), 13C NMR data (Chloroform‐d, 150 MHz): δC 191.6 (C‐14), 150.0 (C‐3), 146.6 (C‐4), 95.1 (C‐1), 70.0 (C‐5), 69.0 (C‐2), 54.5 (C‐10), 42.1 (C‐8), 39.8 (C‐7), 38.6 (C‐6), 26.0 (C‐11), 25.0 (C‐12), 23.3 (C‐15), 20.2 (C‐9), 18.6 (C‐13).
2.3. Cell culture
RAW 264.7 (ATCC TIB‐71) cells and Hep3B (ATCC HB‐8064) cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin sulfate. Cells were maintained at 37°C in humidified air with 5% CO2.
2.4. Measurement of NO contents and cell cytotoxicity
RAW 264.7 (ATCC TIB‐71) cells were cultured in Dulbecco's modified Eagle medium (DMEM), and NO assay was carried out for measurements of NO release using previously reported method (Su et al., 2019). Briefly, RAW 264.7 cells were plated at 1 × 105 cell density in 96‐well microplate, and cultured for 24 hr. Compounds (1–13) were pretreated with increasing dose concentrations (‐10 and 30 μM), and then stimulated with LPS (1 μg/mL, Sigma‐Aldrich, St. Louis, MO) for 18hr. The mixture of cell supernatant (100 μL) and Griess reagent [1% sulfanilamide +0.1% N‐(1‐naphthyl)ethylenediamine (Sigma–Aldrich, St. Louis, MO)] in 5% phosphoric acid was recorded at 550 nm using a microplate reader (Varioskan LUX, Thermo Fisher Scientific Inc., Waltham, MA). RAW 264.7 cell cytotoxicity was evaluated using 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay (Lim et al., 2019).
2.5. Real‐time PCR using Taqman probe
Total RNA was extracted from RAW 264.7 cells using the TaKaRa MiniBEST Universal RNA extraction Kit following the manufacturer's instructions (TaKaRa Bio Inc. Japan). The complementary DNA (cDNA) was synthesized from 1 μg of the total RNA using a PrimeScript first strand cDNA synthesis kit (Takara Bio Inc. Japan). Quantitative real‐time PCR (qPCR) of Il1bβ (Mm00434228_m1), Tnf (Mm00443258_m1), and Il6 (Mm00446190_m1) was performed with a Taqman Gene Expression Assay Kit (Thermo Fisher Scientific, San Jose, CA). To normalize the gene expression, an 18S rRNA endogenous control (Applied Biosystems, Foster City, CA) was used. The qPCR was employed to verify the mRNA expression using a Step‐One Plus Real‐Time PCR system. To quantify mRNA expression, Taqman mRNA assay was performed according to the manufacturer's protocol (Applied Biosystems) (Bertucci et al., 2019). PCR amplification was analyzed using the comparative ΔΔCT method.
2.6. Immunoblots analysis
The whole cell lysate was extracted using Cell Lysis Buffer (Cell Signaling Technology, Beverly, MA). The cytosolic and nuclear extracts were prepared using an NE‐PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific) according to the manufacturer's protocol. Immunoblots analysis was performed as previously described method (Jang et al., 2017). After transfer to nitrocellulose (NC) membrane, the blocking membrane with 5% skimmed milk powder was incubated overnight at 4°C with primary antibody, including anti‐p65 NF‐κB (1:1000), anti‐IκBα (1:1000), anti‐iNOS (1:1000), anti‐lamin B (1:1000), and anti‐β‐actin antibodies (Cell Signaling Technology, Beverly, MA). The membranes were then incubated with a horseradish peroxide‐conjugated anti‐rabbit secondary antibody (1:5000) at room temperature. The band densities were calculated with Quantity One software (Bio‐Rad Laboratories, Hercules, CA).
2.7. Immunocytochemistry
RAW 264.7 (1 × 106 per well) cells were seeded on collagen‐coated coverslips in 6‐well plates and incubated overnight. Then, the cells were pretreated with compound 7 (30 μM) or dexamethasone (10 μM) for 2 hr. Then, the coverslips were washed with phosphate‐buffered saline (PBS) and fixed with ice‐cold methanol for 10 min at room temperature. After blocking with 1% bovine serum albumin (BSA) in PBS containing 0.1% Tween 20 (PBST) for 30 min, the coverslips were incubated overnight with an anti‐p65 NF‐κB antibody in a humidified chamber at 4°C, followed by coincubation with a FITC‐labeled anti‐rabbit IgG antibody in a 37°C humidified chamber for 60 min in the dark. The coverslips were sealed and stained with DAPI by using ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific, Waltham, MA). Immunofluorescence images were obtained using an Olympus IX73 microscope with cellSens software (Olympus, Center Valley, PA) (Lim et al., 2019).
2.8. pSTAT3 luciferase assay
Hep3B cells stably expressing pSTAT3‐Luc were established as described previously (Hwang et al., 2016; Oh et al., 2014). In brief, Hep3B cells stably expressing pSTAT3‐Luc were seeded in 96‐well culture plates at 2 × 104 cells/well, and cells were treated with samples for 1 hr before stimulation with IL‐6 (10 ng/mL) for 12 hr. Luciferase activity was measured according to the manufacturer's protocol (Promega Corp., Madison, WI).
2.9. Animals
Female BALB/c mice (6 weeks old) were purchased from Orient Bio (Gwangju, Korea). All animals had ad libitum access to a standard rodent chow and filtered water during the study. The animals were housed six per cage in a laminar air flow room maintained under a temperature of 24°C, relative humidity of 55%, and a 12 hr light/dark cycle throughout the study. The care and treatment of the animals were carried out in accordance with the guidelines established by the Public Health Service Policy on the Humane Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the Korea Research Institute of Bioscience and Biotechnology. Approved number of animal experiment was KRIBB‐AEC‐20033.
2.10. Isolation of bronchoalveolar lavage in LPS‐induced acute lung injury
BALB/C mice (female, 6 weeks) were randomly divided into 5 groups and each group contained six mice: control, LPS, LPS + RA (10 μg/kg and 1 μg/kg), and LPS + DX (5 mg/kg, positive control). RA and DX were orally administrated as a pretreatment for 24 hr. Then, acute lung injury (ALI) in all mice were induced by intratracheally injection of LPS (1 mg/mL, 25 μL) after anesthesia by intraperitoneal injection of tribromoethanol. After 5 hr of LPS treatment, the mice were euthanized by CO2 inhalation, and their bronchoalveolar lavage fluid (BALF) was collected.
2.11. Measurement of myeloperoxidase concentration
Myeloperoxidase (MPO) concentration in lung tissues represents pulmonary infiltration of inflammatory cells, such as neutrophils and macrophages. MPO concentration in BALF of lung tissues was measured using commercial kits (DY3667) purchased from R&D SYSTEMS INC (MN, USA).
3. RESULTS AND DISCUSSION
3.1. Identification of compounds
Fractionation and chromatographic separation of R. rugosa Thunb. led to the purification of 13 compounds, including eight sesquiterpenoids (1 ~ 8), five triterpenoids (9 ~ 13) from the n‐hexane, CHCl3 and EtOAc‐soluble fractions. By means of LC–MS analysis and 1D and 2D NMR, the structures of these compounds were identified to be 11‐methoxy polisin A (1) (Wang, Li, Dong, Liu, & Feng, 2013), bisaborosaol B1, 2 (2, 3) (Hashidoko, Tahara, & Mizutani, 1991), bisaborosaol F, E1/E2, 13‐methoxy bisaborosaol F (4, 5, 6) (Hashidoko, Tahara, & Mizutani, 1993), rugosic aicd A, D (7, 8) (Hashidoko et al., 1991), oleanolic acid acetate (9) (Hichri, Jannet, Cheriaa, Jegham, & Mighri, 2003), ursolic acid (10) (Seebacher, Simic, Weis, Saf, & Kunert, 2003), corosolic acid (11) (Hou et al., 2009), trans‐, sis‐jacumaric acid (12, 13) (Häberlein & Tschiersch, 1994).
Compound 1 was obtained as a white amorphous powder, + 4.8 (c 0.1 CH3OH). The molecular formula of 1 is C13H18O4, as determined by HRESI‐MS analysis (m/z 261.1103 [M + Na]+, calcd for C13H18O4Na, 261.1097). The UV spectrum showed absorption peaks at 214 nm. 1H NMR data exhibited signals for a methyl group [δH 1.36 (s, H‐12)], and a methoxy group [δH 3.71 (s, 11‐OCH3)]. The 13C and DEPT 135 NMR data showed 13 signals corresponding to a methyl carbon [δC 22.9 (C‐12)], five methylene carbons [δC 24.2 (C‐5), 25.9 (C‐6), 28.0 (C‐3), 29.9 (C‐9), 32.0 (C‐8)], a methine carbon [δC 44.0 (C‐4)], a methoxy carbon [δC 52.2 (11‐OCH3)], a sp2 carbon [δC 140.1 (C‐2)], and two carbonyl carbons [δC 169.2 (C‐11), 179.5 (C‐10)]. The 1H‐1H COSY spectrum showed that the proton at H‐2 correlated with the methylene proton at δH 2.37, 2.10 (H‐3), H‐3 correlated with the methylene proton at δH 1.87 (H‐4), H‐6 correlated with the methylene proton at δH 1.88, 1.29, (H‐5) and H‐9 correlated with the methylene proton at δH 2.23, 2.03 (H‐8). Additionally, the HMBC spectrum showed correlations from δH 3.71 (11‐OCH3) to δC 169.2 (C‐11) indicating the position of methoxy group. The HMBC correlations from δH 3.71 (11‐OCH3) to δC 131.3 (C‐1) suggested that the 11‐OCH3 was connected to the C‐1. The connectivities of ring A were identified by HMBC correlations from δH 6.99 (H‐2) to δC 25.9 (C‐6), from δH 2.52, 2.18 (H‐6) to δC 131.3 (C‐1), from δH 1.88, 1.29 (H‐5) to δC 44.0 (C‐4) and 1H‐1H COSY correlations. In addition, the HMBC correlations from δH 2.23, 2.03 (H‐8) to δC 90.5 (C‐7), 29.9 (C‐9), 179.5 (C‐10) and correlations from δH 2.71, 2.59 (H‐9) to δC 90.5 (C‐7), 179.5 (C‐10) suggested the presence of a lactone ring B. The connection between ring A and ring B was constructed by the HMBC correlations from δH 1.36 (H‐12) to δC 44.0 (C‐4), 90.5 (C‐7), 32.0 (C‐8), from δH 1.87 (H‐4) to δC 90.5 (C‐7) and from δH 2.23, 2.03 (H‐8) to δC 44.0 (C‐4) as shown in Figure 1b. The compound 1 was finally identified as 11‐methoxy polisin A (Figure 1a).
FIGURE 1.
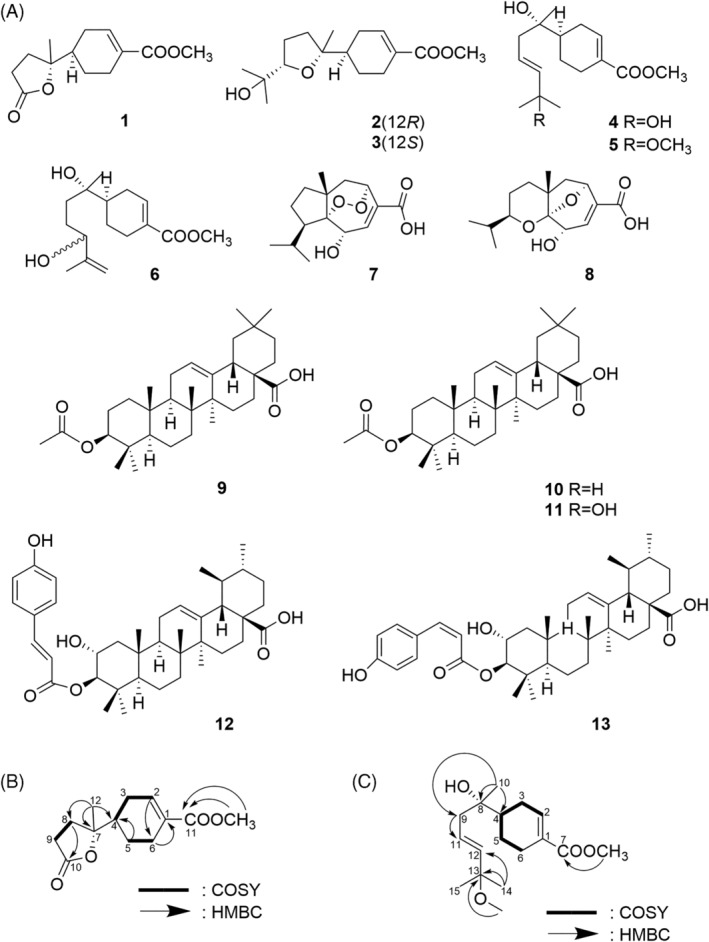
Terpenoid structures and new methoxy structures in R. rugosa extracts. (a) Structure of isolated compounds 1–13 of R. rugosa. (b) Key 1H‐1H COSY and HMBC correlations of 1 (c) Key 1H‐1H COSY and HMBC correlations of 5
Compound 5 was obtained as a Colorless syrup, + 1 (c 0.1 CH3OH). The molecular formula of 5 is C17H28O4, as determined by HRESI‐MS analysis (m/z 319.1885 [M + Na]+, calcd for C17H28O4Na, 319.1880). The UV spectrum showed absorption peaks at 215 nm. 1H NMR data exhibited signals for three methyl groups [δH 1.00 (s, H‐10), 1.16 (s, H‐14, H‐15)], two methoxy groups [δH 3.63 (s, 7‐OCH3), 3.01 (s, 13‐OCH3)], and two trans‐conjugated olefinic protons [δH 5.62 (dt, J = 15.6, 7.2 Hz, H‐11), 5.39 (d, J = 15.6 Hz, H‐12)]. Since the H‐11 were further coupled vicinally with H‐9(δH 2.14, d, J = 7.2 Hz), partial structure [—CH2—CH=CH—] became feasible. The 13C, DEPT 90 and DEPT 135 NMR data showed 17 signals corresponding to three methyl carbons [δC 23.8 (C‐10), 25.6 (C‐15), 25.8 (C‐14)], four methylene carbons [δC 22.6 (C‐5), 24.8 (C‐6), 26.3 (C‐3), 42.8 (C‐9)], a methine carbon [δC 41.5 (C‐4)], two methoxy carbons [δC 49.4 (13‐OCH3), 51.2 (11‐OCH3)], three sp2 carbons [δC 125.9 (C‐11), 137.4 (C‐12), 140.1 (C‐2)], and a carbonyl carbon [δC 166.7, (C‐7), 179.5, (C‐10)]. The 1H‐1H COSY spectrum showed that the proton at H‐2 correlated with the methylene proton at δH 2.26, 2.01 (H‐3), H‐3 correlated with the methine proton at δH 1.44 (H‐4), H‐4 correlated with the methylene proton at δH 1.81, 1.21 (H‐5) and H‐5 correlated with the methylene proton at δH 2.37, 2.01 (H‐6). The 1H‐1H COSY spectrum clearly revealed the presence of a cyclohexene. Additionally, the HMBC spectrum showed correlations from δH 3.63 (7‐OCH3) to δC 166.7 (C‐7) and from δH 3.01 (13‐OCH3) to δC 74.1 (C‐13) indicating the position of two methoxy groups. The HMBC correlations from δH 1.16 (H‐14, H‐15) to δC 137.4 (C‐12), 74.1 (C‐13), from δH 2.14 (H‐9) to δC 125.9 (C‐11) and correlations from δH 1.00 (H‐10) to δC 41.5 (C‐4), 71.9 (C‐8), 42.8 (C‐9) confirmed the side chain of the compound 5. (Figure 1c). The compound 5 was finally identified as 13‐methoxy bisaborosaol F. (Figure 1a).
3.2. Inhibitory effect of nitric oxide production in R. rugosa extract
NO is a major pro‐inflammatory mediator, involved in the pathogenesis of inflammation (Sharma, Al‐Omran, & Parvathy, 2007). During an inflammatory response, the large amount of NO formed by the action of iNOS surpasses the standard physiological amount of NO (Xie, Kashiwabara, & Nathan, 1994). A previous study reported that R. rugosa extracts of 50% EtOH had main active substituents; gallic acid and ellagic acid, and anti‐inflammatory effects by inhibiting NF‐κB and MAPK activation (Tursun et al., 2016). However, the efficacy of R. rugosa extracts of 95% EtOH and the specific active compound responsible for the inflammatory effects became unclear. Then, we evaluated NO production in LPS‐induced RAW 264.7, a mouse macrophage cell line, upon treatment with R. rugosa of 95% EtOH extract and its terpenoid compounds (Figure 2). R. rugosa of 95% EtOH extract dose‐dependently decreased NO production under LPS stimulation (Figure 2a). In addition, to find active compound of inhibitory effect of NO production, we tested 13 compounds of terpenoid structure derived from R. rugosa of 95% EtOH extract (Figure 2b). Through screening terpenoid compounds in R. rugosa of 95% EtOH extract, we observed highly inhibitory effect of NO production and selected rugosic acid A, oleanolic acid acetate, and ursolic acid. These results suggested that R. rugosa of 95% EtOH extract had inhibitory effect of NO production similar to dexamethasone, and its major active compounds were rugosic acid A, oleanolic acid acetate, and ursolic acid.
FIGURE 2.
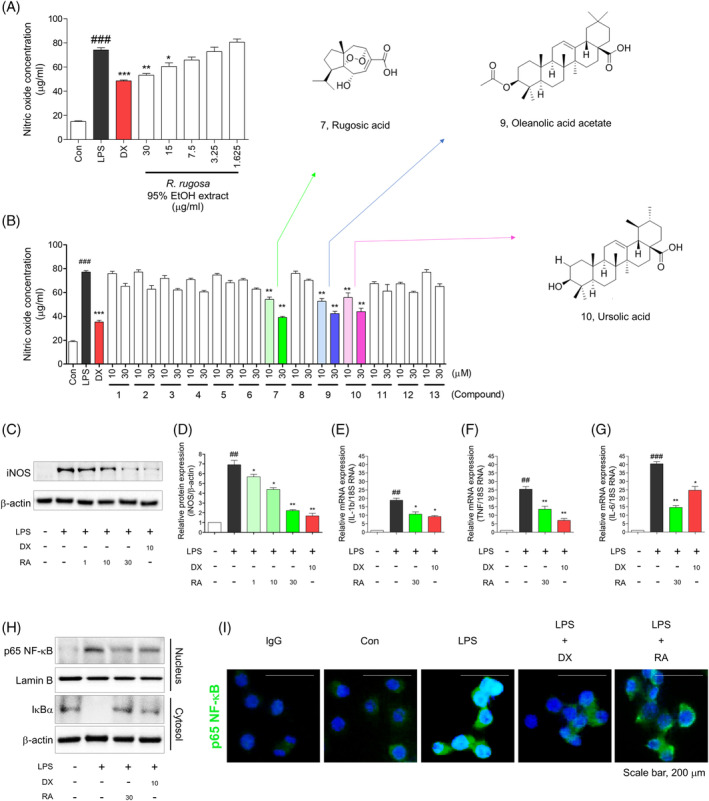
Anti‐inflammatory effect of R. rugosa extract and Rugosic acid A in RAW 264.7 cell line. (a) NO concentration was evaluated in R. rugosa extract. (b) The inhibition screening of NO production was evaluated among 13 constituents in R. rugosa extract. NO evaluation performed triplicate test, and results described as Mean ± SE. An unpaired Student's t test was used for statistical analysis. ###p < .001 versus Con. **p < .01, ***p < .001, versus LPS. Con; control, LPS; lipopolysaccharide, Dx; dexamethasone. (c) Rugosic acid Adecreased iNOS expression levels in LPS‐induced RAW 264.7 cells. (d) Relative ratio of iNOS versus β‐actin were measured using densitometry, and dexamethasone was used as positive control. These graphs represented that rugosic acid A dose‐dependently inhibited iNOS at concentration of 1, 10, and 30 μM using Immunoblots analysis. (e‐g) The mRNA expression levels of IL‐1β, TNF‐α, and IL‐6 were measured using quantitative real‐time PCR experiment, and these pro‐inflammatory cytokines were significantly diminished by rugosic acid A. Cells were pre‐incubated for 2 hr with rugosic acid A at concentration of 30 μM, and activated by LPS (1 μg/mL) for 2 hr. Results represent as mean ± SE, and dexamethasone was used as a positive control. ##p < .01, ###p < .001 versus Con, *p < .05, **p < .01, and ***p < .001 versus LPS. Con; control, LPS; lipopolysaccharide, Dx; dexamethasone, RA; rugosic acid. (h) Immunoblots analysis displayed that translocation of NF‐κB into nucleus and degradation of cytosolic IκBα were suppressed by rugosic acid (30 μM) in RAW 264.7 cells. (i) Rugosic acid (30 μM) suppressed nuclear translocation of NF‐κB. After 2 hr of compound 20 treatment, the cells were fixed and permeabilized. NF‐κB (green) was immunostained with rabbit‐anti‐NF‐κB followed by FITC‐conjugated secondary antibodies and the nuclei (blue) were stained with DAPI. The results shown were representative of two independent experiments [Colour figure can be viewed at wileyonlinelibrary.com]
3.3. Inhibitory activities of iNOS, pro‐inflammatory cytokines, and NF‐κB by rugosic acid A
iNOS, which is involved in the production of NO, is a novel signaling molecule associated with the MAPK pathway and NF‐κB activity in both microglia and macrophage cells (Tripathi, Tripathi, Kashyap, & Singh, 2007). Subsequently, pro‐inflammatory cytokines such as IL‐1β, TNF‐α, and IL‐6 are induced by activating nuclear translocation of NF‐κB (Liu et al., 2017).
To investigate intracellular biological evidence of reduced NO level by rugosic acid A, we evaluated the effect of rugosic acid A on the pro‐inflammatory mediators such as iNOS, IL‐1β, TNF‐α, IL‐6, and in LPS‐induced macrophages (Figure 2c‐g). In our results, rugosic acid A was newly developed inhibition of iNOS under LPS‐induced macrophages based on western blotting (Figure 2c). The graph described significantly inhibition of iNOS (Figure 2d). The inhibitory effect of rugosic acid A (30 μM) was similar to that of dexamethasone (positive control, 10 μM). Moreover, we tested pro‐inflammatory cytokines after treatment with rugosic acid A. Rugosic acid A (30 μM) significantly inhibited the pro‐inflammatory cytokine mRNA of IL‐1β, and TNF‐α, and IL‐6. Specially, rugosic acid A had the more effective prevention of mRNA of IL‐6 than dexamethasone (Figure 2e‐g).
Subsequently, we evaluated preventive effect of nuclear translocation of NF‐κB by pretreatment with rugosic acid A quantitatively and qualitatively (Figure 2h,i). To determine whether rugosic acid A suppressed the nuclear translocation of NF‐κB, we evaluated nuclear NF‐κB and cytosolic IκBα using western blotting (Figure 2h). Immunocytochemistry clearly demonstrated that rugosic acid A reduced the translocation of NF‐κB into the nucleus in RAW 264.7 cells (Figure 2i). Rugosic acid A remarkably ameliorated to translocate nuclear NF‐κB by suppressing the degradation of IκBα, similar to dexamethasone. These results suggested that, rugosic acid A was significant anti‐inflammatory compound by inhibition of nuclear translocation of NF‐κB among terpenoid structures in R. rugosa extracts (95% EtOH).
3.4. Inhibition of IL‐6/STAT3 activation by rugosic acid A
To further conduct strong inhibition of IL‐6 after rugosic acid treatment in RAW 264.7, we tested to discover potent STAT3 inhibitor among major substituents; rugosic acid (compound 7), oleanolic acid acetate (compound 9), and ursolic acid (compound 10) in R. rugosa extracts under IL‐6 mediated STAT3 activation in Hep3B cell line, which was co‐transfected with pSTAT3‐Luc and pcDNA3.1/Hygro vector (Oh et al., 2014). Using IL‐6/STAT3 screening system of the transfected Hep3B cell line, we already reported that oleanolic acid acetate was major inhibitor of IL‐6/STAT3 activation (Hwang et al., 2016). In previous study, ursolic acid had prevention of hepatic carcinoma by inhibition of STAT3 signaling pathway (Liu, Li, Li, & Lin, 2010). In this study, we originally observed that rugosic acid significantly inhibited IL‐6‐mediated STAT3 activation (Figure 3a). The IC50 value of IL‐6/STAT3 activation was represented on Figure 3b‐d. These results suggested that rugosic acid similar to oleanolic acid acetate and ursolic acid had significant effect of IL‐6‐mediated STAT3 activation.
FIGURE 3.
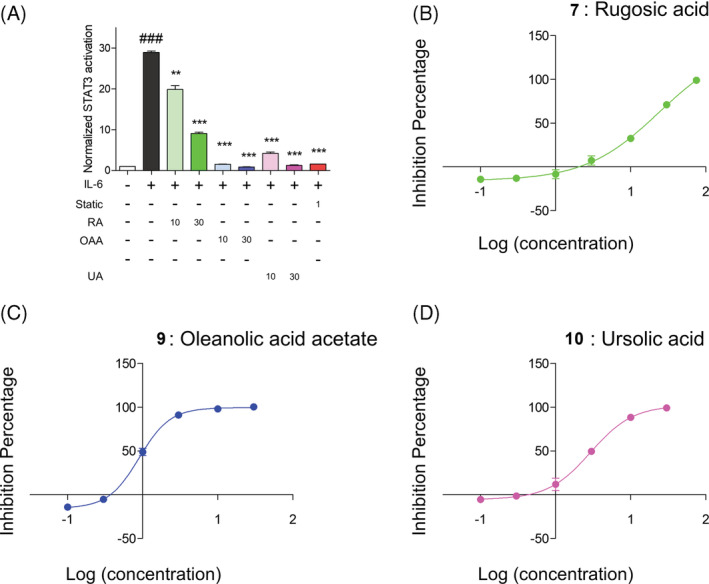
Rugosic acid ameliorated IL‐6‐mediated STAT3 activation under transfected Hep3B cell line. (a) Inhibitory evaluation of IL6‐mediated STAT3 activation was evaluated by treatment with rugosic acid A, oleanolic acid acetate, and ursolic acid the transfected Hep3B cell line. (b‐d) Dose dependent curves about rugosic acid A (b), oleanolic acid acetate (c), and ursolic acid (d) were exhibited [Colour figure can be viewed at wileyonlinelibrary.com]
3.5. Inhibition of phosphorylation of NF‐κB and STAT3 by rugosic acid A under acute lung injury model
To further evaluate inhibitory effect of NF‐κB and STAT3 by rugosic acid A, we established LPS‐mediated acute lung injury (ALI) model and tested oral treatment with rugosic acid A (Figure 4a). Because recent severe acute respiratory syndrome of coronavirus disease 2019 (COVID‐19) has rapidly spread in worldwide and has similar symptom with acute pneumonia accompanied by cytokine storm (Mehta et al., 2020). If rugosic acid A may ameliorate symptom of acute pneumonia by inhibition of NF‐κB and STAT3, it will be alternative treatment with COVID‐19 similar to tocilizumab, which is an agent of recombinant humanized anti‐human IL‐6 receptor monoclonal antibody, and used to cure severe COVID‐19 patients by inhibiting IL‐6 signaling transduction, before development of vaccine of COVID‐19 (Xu et al., 2020). Firstly, we performed evaluation of myeloperoxidase (MPO) concentration in BALF on ALI model, because MPO was used as biomarker of activation and accumulation of neutrophils in lung tissues and clinical signature of pneumonia patients (Abul et al., 2001). We observed that pretreatment of rugosic acid A (10 μg/mL) significantly ameliorated LPS‐mediated MPO concentration (Figure 4b). Subsequently, we evaluated preventive effect of phosphorylation of STAT3 and p65 NF‐κB by pretreatment with rugosic acid A using western blotting in lung tissue (Figure 4c). In our results, the administration of rugosic acid A (10 μg/mL) had significantly inhibition of phosphorylation of STAT3 (Figure 4d) and p65 NF‐κB (Figure 4e) in lung tissues. These results suggested that rugosic acid A ameliorated LPS‐mediated ALI by decreasing MPO concentration and inhibiting phosphorylation of STAT3 and p65 NF‐κB.
FIGURE 4.
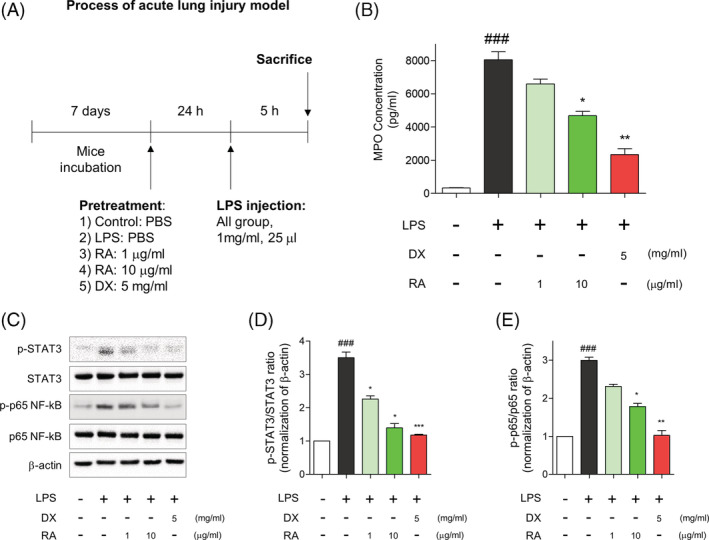
Effect of rugosic acid A on MPO activity in lung tissues of LPS‐stimulated ALI mice. (a) Schematic figure of induction of acute lung injury (ALI) mice. (b) After orally pretreatment with agents: RA 10 μg/kg, RA 1 μg/kg, and DX 5 mg/kg for 24 hr, mice intratracheally were injected with LPS (1 mg/mL, 25 μL). Vehicle group were orally administrated with PBS. The lung myeloperoxidase (MPO) concentration was determined on bronchoalveolar lavage fluid (BALF) after LPS injection. The values are presented as mean ± SE (n = 5 in each group). Multifactor ANOVA with Tukey's multiple comparison was used for statistical analyses; ###p < .001, versus control group. *p < .05, and **p < .01, versus LPS group. (c) Expression of phosphorylated STAT3, p65 NF‐κB, total STAT3, total p65 NF‐κB and β‐actin in lung tissue. (d) Quantitative densitometry analysis of the ratio of phos‐STAT3 to total STAT3. (e) Quantitative densitometry analysis of the ratio of phos‐p65 NF‐κB to total p65 NF‐κB. ###p < .001, versus control group. *p < .05, **p < .01 and ***p < .001, versus LPS group [Colour figure can be viewed at wileyonlinelibrary.com]
3.6. Quantitative analysis of rugosic acid A in R. rugosa extracts
To investigate yield value of rugosic acid A as an anti‐inflammatory agent, we performed quantitative analysis (Figure 5). The sesquiterpenoid structure rugosic acid A was exhibited (Figure 5a) and evaluated using calibration curve as a standard (Figure 5b). In quantitative study, R. rugosa extracts (375.9 g) contained 0.8 mg/g of rugosic acid A (Figure 5c and 2.3 Extraction and isolation).
FIGURE 5.
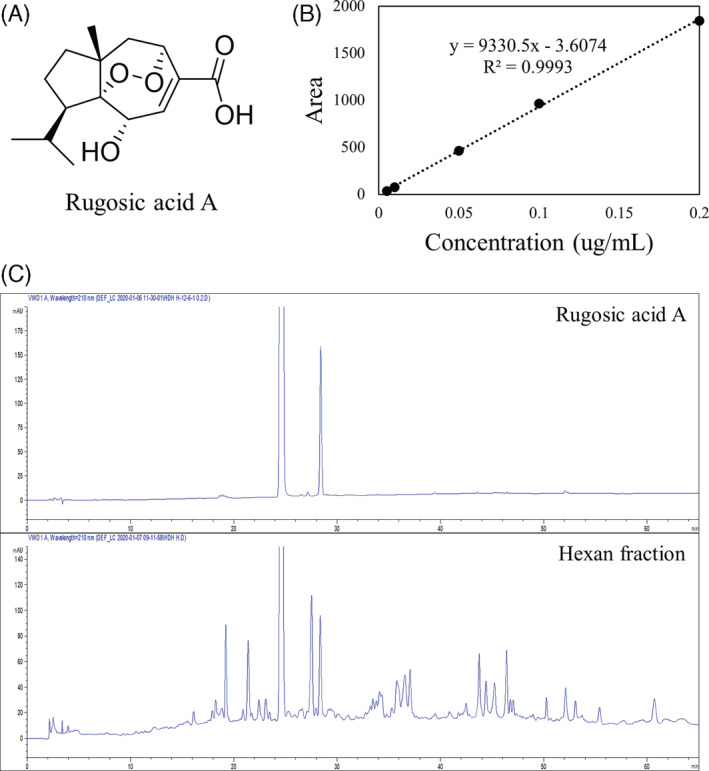
Quantitative analysis of rugosic acid A in R. rugosa extract. (a) Structure of rugosic acid A. (b) Calibration curve of rugosic aicd A standard. (c) HPLC chromatogram pattern of rugosic acid A in extract from hexan fraction of R.rugosa.; HPLC model: Agilent 1100series (Agilent), column: Phenomenex Luna C18 (5 μm, 4.6 × 250 mm), column temperature: 25°C, mobile phase: (a) H2O, (b) ACN, gradient: 0–0, 5–0, 45–100, 65(min)‐100(B%) [Colour figure can be viewed at wileyonlinelibrary.com]
4. CONCLUSIONS
Our group originally found that rugosic acid A, which has sesquiterpenoid structure has bioactivity responsible for the anti‐inflammatory activity of R. rugosa extract (EtOH 95%) by inhibition of the nuclear translocation of NF‐κB, by reduction of pro‐inflammatory mediators such as iNOS, TNF‐α, and IL‐6. Rugosic acid A significantly showed similar to major STAT3 inhibitory phytochemicals; oleanolic acid acetate and ursolic acid. In addition, our group measured quantitative analysis of rugosic acid A in R. rugosa extract (EtOH 95%). These results indicate that R. rugosa extract and its sesquiterpenoid compound, rugosic acid, may be safe and potential therapeutic agent for inflammatory diseases associated with NF‐κB and IL‐6/STAT3 signaling.
CONFLICTS OF INTEREST
The authors declare no potential conflict of interest.
Supporting information
Figure S1 HRESIMS spectrum of 1, Figure S2. 1H NMR (600 MHz, methanol‐d 4) spectrum of 1, Figure S3. 13C NMR (150 MHz, methanol‐d 4) and dept 135 spectrum of 1Figure S4. COSY (600 MHz, methanol‐d 4) spectrum of 4, Figure S5. HMQC (600 MHz, methanol‐d 4) spectrum of 4, Figure S6. HMBC (600 MHz, methanol‐d 4) spectrum of 4, Figure S7. HRESIMS spectrum of 5 Figure S8. 1H NMR (600 MHz, methanol‐d 4) spectrum of 5, Figure S9. 13C NMR (150 MHz, methanol‐d 4) and dept 135 spectrum of 5, Figure S10. COSY (600 MHz, methanol‐d 4) spectrum of 5, Figure S11. HMQC (600 MHz, methanol‐d 4) spectrum of 5, Figure S12. HMBC (600 MHz, methanol‐d 4) spectrum of 5.
ACKNOWLEDGMENTS
This research was supported by a grant from the KRIBB Research Initiative Program (KGS1052012 and KGS1002012).
Kim K‐H, Park Y‐J, Jang H‐J, et al. Rugosic acid A, derived from Rosa rugosa Thunb., is novel inhibitory agent for NF‐κB and IL‐6/STAT3 axis in acute lung injury model. Phytotherapy Research. 2020;34:3200–3210. 10.1002/ptr.6767
Kang‐Hoon Kim and Ye‐Ji Park contributed equally to this study.
Contributor Information
Seung Woong Lee, Email: lswdoc@kribb.re.kr.
Mun‐Chual Rho, Email: rho-m@kribb.re.kr.
REFERENCES
- Abul, H. , Abul, A. , Khan, I. , Mathew, T. C. , Ayed, A. , & Al‐Athary, E. (2001). Levels of IL‐8 and myeloperoxidase in the lungs of pneumonia patients. Molecular and Cellular Biochemistry, 217(1–2), 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim, A. , Mei, X. , Huang, M. , Yang, Y. , Fayad, Z. A. , Zhang, N. , … Chung, M. (2020). Chest CT Findings in Coronavirus Disease‐19 (COVID‐19): Relationship to Duration of Infection. Radiology, 295(3), 200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci, A. , Kim, K. H. , Kang, J. , Zuidema, J. M. , Lee, S. H. , Kwon, E. J. , … Sailor, M. J. (2019). Tumor‐targeting, MicroRNA‐silencing porous silicon nanoparticles for ovarian cancer therapy. ACS Applied Materials & Interfaces, 11(27), 23926–23937. [DOI] [PubMed] [Google Scholar]
- Cho, E. J. , Yokozawa, T. , Kim, H. Y. , Shibahara, N. , & Park, J. C. (2004). Rosa rugosa attenuates diabetic oxidative stress in rats with streptozotocin‐induced diabetes. The American Journal of Chinese Medicine, 32(4), 487–496. [DOI] [PubMed] [Google Scholar]
- Dang, G. K. , Parekar, R. R. , Kamat, S. K. , Scindia, A. M. , & Rege, N. N. (2011). Antiinflammatory activity of Phyllanthus emblica, Plumbago zeylanica and Cyperus rotundus in acute models of inflammation. Phytotherapy Research: PTR, 25(6), 904–908. [DOI] [PubMed] [Google Scholar]
- Formisano, C. , Sanna, C. , Ballero, M. , Chianese, G. , Sirignano, C. , Rigano, D. , … Taglialatela‐Scafati, O. (2017). Anti‐inflammatory sesquiterpene lactones from Onopordum illyricum L. (Asteraceae), an Italian medicinal plant. Fitoterapia, 116, 61–65. [DOI] [PubMed] [Google Scholar]
- Guanizo, A. C. , Fernando, C. D. , Garama, D. J. , & Gough, D. J. (2018). STAT3: a multifaceted oncoprotein. Growth Factors, 36(1–2), 1–14. [DOI] [PubMed] [Google Scholar]
- Häberlein, H. , & Tschiersch, K.‐P. (1994). Triterpenoids and flavonoids from Leptospermum scoparium . Phytochemistry, 35(3), 765–768. [Google Scholar]
- Hashidoko, Y. (1996). The phytochemistry of Rosa rugosa . Phytochemistry, 43(3), 535–549. [Google Scholar]
- Hashidoko, Y. , Tahara, S. , & Mizutani, J. (1991). Isolation of four novel carotanoids as possible metabolites of Rugosic acid A in Rosa rugosa leaves. Agricultural and Biological Chemistry, 55(4), 1049–1053. [Google Scholar]
- Hashidoko, Y. , Tahara, S. , & Mizutani, J. (1993). Sesquiterpenoids from Rosa rugosa leaves. Phytochemistry, 32(2), 387–390. [Google Scholar]
- He, G. , & Karin, M. (2011). NF‐κB and STAT3—Key players in liver inflammation and cancer. Cell Research, 21(1), 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri, F. , Jannet, H. B. , Cheriaa, J. , Jegham, S. , & Mighri, Z. (2003). Antibacterial activities of a few prepared derivatives of oleanolic acid and of other natural triterpenic compounds. Comptes Rendus Chimie, 6(4), 473–483. [Google Scholar]
- Hou, W. , Li, Y. , Zhang, Q. , Wei, X. , Peng, A. , Chen, L. , & Wei, Y. (2009). Triterpene acids isolated from Lagerstroemia speciosa leaves as alpha‐glucosidase inhibitors. Phytotherapy Research: PTR, 23(5), 614–618. [DOI] [PubMed] [Google Scholar]
- Hwang, J. T. , Kim, Y. , Jang, H.‐J. , Oh, H.‐M. , Lim, C.‐H. , Lee, S. W. , & Rho, M. C. (2016). Study of the UV light conversion of Feruloyl amides from Portulaca oleracea and their inhibitory effect on IL‐6‐induced STAT3 activation. Molecules, 21(7), 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, H. J. , Lee, S. , Lee, S. J. , Lim, H. J. , Jung, K. , Kim, Y. H. , … Rho, M. C. (2017). Anti‐inflammatory activity of Eudesmane‐type Sesquiterpenoids from Salvia plebeia. Journal of Natural Products, 80(10), 2666–2676. [DOI] [PubMed] [Google Scholar]
- Jiang, K. , Chen, L.‐L. , Wang, S.‐F. , Wang, Y. , Li, Y. , & Gao, K. (2015). Anti‐inflammatory Terpenoids from the leaves and twigs of Dysoxylum gotadhora. Journal of Natural Products, 78(5), 1037–1044. [DOI] [PubMed] [Google Scholar]
- Khalil, N. M. , Sperotto, J. S. , & Manfron, M. P. (2006). Antiinflammatory activity and acute toxicity of Dodonaea viscosa . Fitoterapia, 77(6), 478–480. [DOI] [PubMed] [Google Scholar]
- Lim, H. J. , Jang, H. J. , Bak, S. G. , Lee, S. , Lee, S. W. , Lee, K. M. , … Rho, M. C. (2019). In vitro inhibitory effects of cirsiliol on IL‐6‐induced STAT3 activation through anti‐inflammatory activity. Bioorganic & Medicinal Chemistry Letters, 29(13), 1586–1592. [DOI] [PubMed] [Google Scholar]
- Liu, T. , Zhang, L. , Joo, D. , & Sun, S.‐C. (2017). NF‐κB signaling in inflammation. Signal Transduction and Targeted Therapy, 2(1), 17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Li, P.‐K. , Li, C. , & Lin, J. (2010). Inhibition of STAT3 signaling blocks the anti‐apoptotic activity of IL‐6 in human liver cancer cells. The Journal of Biological Chemistry, 285(35), 27429–27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, P. , McAuley, D. F. , Brown, M. , Sanchez, E. , Tattersall, R. S. , & Manson, J. J. (2020). COVID‐19: Consider cytokine storm syndromes and immunosuppression. The Lancet, 395(10229), 1033–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus, A. , & Ghosh, S. (2009). The NF‐kappaB family of transcription factors and its regulation. Cold Spring Harbor Perspectives in Biology, 1(4), a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, H. M. , Lee, S. W. , Yun, B. R. , Hwang, B. S. , Kim, S. N. , Park, C. S. , … Rho, M. C. (2014). Vigna angularis inhibits IL‐6‐induced cellular signalling and ameliorates collagen‐induced arthritis. Rheumatology (Oxford, England), 53(1), 56–64. [DOI] [PubMed] [Google Scholar]
- Seebacher, W. , Simic, N. , Weis, R. , Saf, R. , & Kunert, O. (2003). Complete assignments of 1H and 13C NMR resonances of oleanolic acid, 18α‐oleanolic acid, ursolic acid and their 11‐oxo derivatives. Magnetic Resonance in Chemistry, 41(8), 636–638. [Google Scholar]
- Sharma, J. N. , Al‐Omran, A. , & Parvathy, S. S. (2007). Role of nitric oxide in inflammatory diseases. Inflammopharmacology, 15(6), 252–259. [DOI] [PubMed] [Google Scholar]
- Su, X. D. , Jang, H. J. , Wang, C. Y. , Lee, S. W. , Rho, M. C. , Kim, Y. H. , & Yang, S. Y. (2019). Anti‐inflammatory potential of Saponins from Aster tataricus via NF‐kappaB/MAPK activation. Journal of Natural Products, 82(5), 1139–1148. [DOI] [PubMed] [Google Scholar]
- Tripathi, P. , Tripathi, P. , Kashyap, L. , & Singh, V. (2007). The role of nitric oxide in inflammatory reactions. FEMS Immunology & Medical Microbiology, 51(3), 443–452. [DOI] [PubMed] [Google Scholar]
- Tursun, X. , Zhao, Y. , Talat, Z. , Xin, X., Tursun , A., Abdulla, R. , & AkberAisa, H. (2016). Anti‐Inflammatory Effect of Rosa rugosa Flower Extract in Lipopolysaccharide‐Stimulated RAW264.7 Macrophages. Biomolecules & Therapeutics, 24(2), 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um, M. , Han, T. H. , & Lee, J. W. (2018). Ultrasound‐assisted extraction and antioxidant activity of phenolic and flavonoid compounds and ascorbic acid from rugosa rose (Rosa rugosa Thunb.) fruit. Food Science and Biotechnology, 27(2), 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Li, Z. H. , Dong, Z. J. , Liu, J. K. , & Feng, T. (2013). Norbisabolane and eremophilane sesquiterpenoids from cultures of the Basidiomycete Polyporus ellisii. Fitoterapia, 91, 194–198. [DOI] [PubMed] [Google Scholar]
- Xie, Q. W. , Kashiwabara, Y. , & Nathan, C. (1994). Role of transcription factor NF‐kappa B/Rel in induction of nitric oxide synthase. The Journal of Biological Chemistry, 269(7), 4705–4708. [PubMed] [Google Scholar]
- Xu, X. , Han, M. , Li, T. , Sun, W. , Wang, D. , Fu, B. , … Wei, H. (2020). Effective treatment of severe COVID‐19 patients with tocilizumab. Proceedings of the National Academy of Sciences, 117(20), 10970–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Wang, Y. , Wang, Y. , Zhao, M. , Jia, H. , Li, B. , & Xing, D. (2016). Tormentic Acid Inhibits IL‐1β‐Induced Inflammatory Response in Human Osteoarthritic Chondrocytes. Inflammation, 39(3), 1151–1159. [DOI] [PubMed] [Google Scholar]
- Youn, U. J. , Park, E.‐J. , Kondratyuk, T. P. , Sripisut, T. , Laphookhieo, S. , Pezzuto, J. M. , & Chang, L. C. (2016). Anti‐inflammatory triterpenes from the apical bud of Gardenia sootepensis. Fitoterapia, 114, 92–97. [DOI] [PubMed] [Google Scholar]
- Zhao, D. , Yao, F. , Wang, L. , Zheng, L. , Gao, Y. , Ye, J. , … Gao, R. (2020). A comparative study on the clinical features of COVID‐19 pneumonia to other pneumonias. Clinical Infectious Diseases. ciaa247, 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 HRESIMS spectrum of 1, Figure S2. 1H NMR (600 MHz, methanol‐d 4) spectrum of 1, Figure S3. 13C NMR (150 MHz, methanol‐d 4) and dept 135 spectrum of 1Figure S4. COSY (600 MHz, methanol‐d 4) spectrum of 4, Figure S5. HMQC (600 MHz, methanol‐d 4) spectrum of 4, Figure S6. HMBC (600 MHz, methanol‐d 4) spectrum of 4, Figure S7. HRESIMS spectrum of 5 Figure S8. 1H NMR (600 MHz, methanol‐d 4) spectrum of 5, Figure S9. 13C NMR (150 MHz, methanol‐d 4) and dept 135 spectrum of 5, Figure S10. COSY (600 MHz, methanol‐d 4) spectrum of 5, Figure S11. HMQC (600 MHz, methanol‐d 4) spectrum of 5, Figure S12. HMBC (600 MHz, methanol‐d 4) spectrum of 5.


