Summary
The outbreak of coronavirus disease 2019 (COVID‐19) and pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has become a major concern globally. As of 14 April 2020, more than 1.9 million COVID‐19 cases have been reported in 185 countries. Some patients with COVID‐19 develop severe clinical manifestations, while others show mild symptoms, suggesting that dysregulation of the host immune response contributes to disease progression and severity. In this review, we have summarized and discussed recent immunological studies focusing on the response of the host immune system and the immunopathology of SARS‐CoV‐2 infection as well as immunotherapeutic strategies for COVID‐19. Immune evasion by SARS‐CoV‐2, functional exhaustion of lymphocytes, and cytokine storm have been discussed as part of immunopathology mechanisms in SARS‐CoV‐2 infection. Some potential immunotherapeutic strategies to control the progression of COVID‐19, such as passive antibody therapy and use of interferon αβ and IL‐6 receptor (IL‐6R) inhibitor, have also been discussed. This may help us to understand the immune status of patients with COVID‐19, particularly those with severe clinical presentation, and form a basis for further immunotherapeutic investigations.
Keywords: coronavirus, COVID‐19, immune response, immunotherapeutic, SARS‐CoV‐2
1. INTRODUCTION
The outbreak of coronavirus disease 2019 (COVID‐19) that started from the Hubei Province in China in December 2019 1 was declared as a Public Health Emergency of International Concern (PHEIC) by the World Health Organization (WHO) on 30 January 2020, 2 has now attained pandemic status affecting more than 200 countries. 3 , 4 , 5 The infection is caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) that shares 79.6% sequence identity to severe acute respiratory syndrome coronavirus (SARS‐CoV). 6 As of 14 April 2020, based on COVID‐19 Global Cases, 7 1 920 985 confirmed COVID‐19 cases and 119 686 deaths have been reported in 185 countries. Several studies have demonstrated that the virus is more transmissible than SARS‐CoV, with a mean reproductive number (R 0) ranging from 1.4 to 3.58, 8 , 9 , 10 which means that one infected person can transmit the virus to up to three susceptible people. This situation prompted the WHO to declare the current COVID‐19 outbreak as a global pandemic. 11
SARS‐CoV‐2 is a member of the coronavirus (CoV) family, a group of enveloped and positive‐sense single‐stranded RNA (ssRNA) viruses, with a genome ranging from 26 to 32 kb in size. 12 SARS‐CoV‐2 belongs to the family Coronaviridae and subfamily Coronavirinae; this subfamily has been subdivided into four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus, based on serology and phylogenetic analysis. 13 , 14 SARS‐CoV‐2 belongs to the genus Betacoronavirus together with SARS‐CoV and Middle East respiratory syndrome coronavirus (MERS‐CoV). The virus consists of a membrane and a nucleocapsid. The membrane comprises three proteins: (a) spike (S) type I glycoprotein, forming peplomers on the virion surface; (b) membrane (M) protein, spanning the membrane three times and containing a short N‐terminal ectodomain and cytoplasmic tail; and (c) small membrane protein (E), a highly hydrophobic protein. 15 S protein is trimeric and processed by host cellular proteases to form two functional subunits S1 and S2, which remain noncovalently bound in the prefusion conformation. 16 The S protein plays a major role in membrane fusion and viral entry into host cells through the cellular angiotensin‐converting enzyme 2 (ACE2) receptor. Moreover, the receptor‐binding domain (RBD) is located in the S1 subunit at the apex of the S protein of SARS‐CoV. 17 , 18
Although some risk factors, such as age and the presence of comorbidity, have been associated with disease severity and mortality of patients with COVID‐19, 19 there is still a lack of understanding of the role of the immune system on disease severity and mortality rate. An initial study suggested clear changes in immune responses during disease progression in a patient with COVID‐19. 20 Therefore, dysregulated host‐immune responses might be one of the important factors for the pathogenesis and disease severity of COVID‐19. This review aims to collate the recent immunological studies from patients with COVID‐19, which may give us an insight on (a) the interaction between the host immune system and the virus and (b) disease pathogenesis thereby, helping in developing effective immune‐based therapies for SARS‐CoV‐2 infection.
2. INNATE IMMUNE RESPONSES AGAINST SARS‐COV‐2
As SARS‐CoV‐2 and SARS‐CoV are similar, 6 it is suggested that the biochemical interactions are likely to be similar. Like SARS‐CoV, SARS‐CoV‐2 enters the host cells through ACE‐2 receptors on type II pneumocytes in the lungs. 6 , 21 , 22 After breaching the first line of immune defence, such as mucus and ciliated cells, the pathogen‐associated molecular patterns (PAMPs) on the virus alert frontline innate immune cells such as monocytes, alveolar macrophages, natural killer (NK) cells, and neutrophils to the presence of the invading virus. 23 To mount the appropriate response, these innate immune cells express pattern recognition receptors (PRRs) such as toll‐like receptors (TLRs), retinoic acid‐inducible gene I (RIG‐I)‐like receptors (RLRs), and nucleotide‐binding and oligomerization domain (NOD)‐like receptors (NLRs) that recognize PAMPs. The interaction between PRR and PAMP induces phagocytosis and activates intracellular signaling pathways to stimulate the synthesis of proinflammatory cytokines, such as type I interferons (IFN‐I, ie, IFNα/β) and type II interferons (IFN‐II, ie, IFN‐γ), and chemokines, such as CXCL‐10 and CCL‐2. 12 , 23 , 24 IFN‐I inhibit viral replication via multiple mechanisms. 25 Infected cells with reduced major histocompatibility complex I (MHC‐I) expression also activate NK cells to produce significant amounts of IFN‐γ and induce apoptosis through cytotoxic granules and antibody‐dependent cellular cytotoxicity (ADCC). 26 , 27 In SARS‐CoV infection, dendritic cells (DCs) and macrophages are the key players in innate immune responses as they possess the capacity to kill through phagocytosis and IFN release (ie, plasmacytoid DCs utilize TLR‐7 to sense viral nucleic acid leading to IFN‐I secretion to lyse the viruses). 24 More importantly, they are capable of presenting viral peptides to bridge to adaptive immune responses. 28
IFN‐I, secreted by infected cells and innate immune cells, such as macrophages and DCs, is a potent cytokine in controlling viral infection and priming adaptive immune responses. 29 IFN‐I blocks viral replication by inducing hundreds of interferon stimulated genes (ISGs), including key components of the protein synthesis machinery, protein kinase R (PKR), and 2′‐5′‐oligoadenylate synthase (OAS)/RNAse L. 29 PKR halts general protein synthesis through phosphorylation of eukaryotic initiation factor 2 subunit‐α (eIF2α) and OAS/RNase L degrades viral ssRNA, thereby effectively impairing viral replication. 25 The key role played by PKR and OAS/RNAse L in the innate immune responses to CoV is emphasized by MERS‐CoV expressing protein NS4a to inhibit PKR activation 30 and NS4b to inhibit RNase L activation. 31 Moreover, IFN‐I stimulates CD8+ T proliferation, promotes B cell activation, antibody production, and class switching (ie, IgM to IgG), 32 and activates NK cells and macrophages to clear the viruses. 33 The role of IFN in SARS‐CoV‐2 infection, including other innate immune components and responses, is shown in Figure 1A. Pretreatment of nonlymphatic cells with IFN‐α before infection with SARS‐CoV induces substantial gene expression for IFN induction, IFN‐signaling pathways, and antiviral effector proteins. 34 An in vitro study shows that IFN‐α can inhibit SARS‐CoV as seen by the cytopathic effect, plaque assay, and immunoblot analysis. 35 SARS‐CoV‐2 is sensitive to IFN‐I pretreatment. 36 In this study, Vero cells pretreated with recombinant IFN‐I prior to infection with SARS‐CoV‐2 showed a significant reduction of SARS‐CoV‐2 replication—a 3 to 4 log drop in viral titer. 36 This suggests that IFN‐I has antiviral potential for SARS‐CoV‐2; therefore, further clinical trials are required to evaluate this potential.
FIGURE 1.
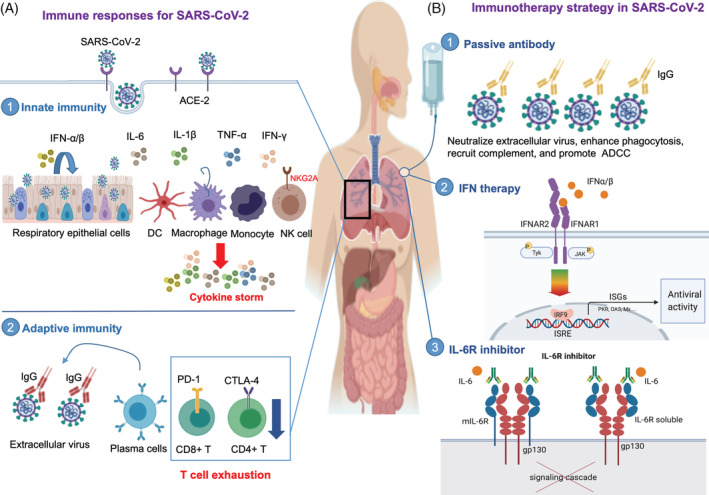
Immune responses and immunotherapy strategy in SARS‐CoV‐2 infection. Immune response to SARS‐CoV‐2 involving innate and adaptive immunity. A1, SARS‐CoV‐2 enters the host cells by binding the receptor for angiotensin converting enzyme 2 (ACE2). The infected cells then release interferon type I (IFN‐α/β); innate immune cells respond to the IFNs by establishing an antiviral state. In case of severe infection, the viruses are sensed by monocytes, tissue macrophages, and resident dendritic cells resulting in uncontrolled proinflammatory cytokine (IFN, TNF‐α, IL‐1β, and IL‐6) production, leading to a phenomenon called the cytokine storm, which damages the host’s respiratory epithelial cells. Exhausted natural killer cells with increased expression of the inhibitory receptor, NKG2A, are also seen in SARS‐CoV‐2. A2, In SARS‐CoV‐2 infection, increased antibody production and poor T cell responses are observed (A2). CD4+ T cell and CD8+ T cell numbers decrease, and the exhausted phenotype, which is characterized by a higher expression of inhibitory receptors, such programmed death receptor‐1 (PD‐1) and cytotoxic T lymphocyte associated antigen‐4 (CTLA‐4), is seen. Immunotherapy strategies for SARS‐CoV‐2 have been proposed. B1, Transferring convalescent sera with neutralizing antibodies from the recovered patients. The antibodies can directly bind to SARS‐CoV‐2 and prevent the virus from infecting new cells (neutralization), enhance phagocytosis (opsonization), recruit complement to lyse infected cells or neutralize the viruses, and promote NK cell mediated killing of infected cells through antibody dependent cellular cytotoxicity (ADCC). B2, IFN α/β bind to IFN receptors and induce an antiviral response by expressing several interferon stimulated genes (ISGs), such as PKR, OAS, and Mx. The protein product of ISGs controls viral infection. (B3) The IL‐6R inhibitor (such as tocilizumab) binds to the membrane bound IL‐6 receptor (mIL‐6R) and soluble IL‐6 receptor (soluble IL‐6R). Binding of tocilizumab to IL‐6R inhibits the IL‐6 signaling pathway
3. ANTIBODY RESPONSES AGAINST SARS‐COV‐2 INFECTION
The B cell response of humoral immunity is essential for controlling viral infection and replication of extracellular viruses through antibody production. B cell activation is achieved through the crosslinking of two or more membrane immunoglobulins (Ig) to multiple antigenic molecules (epitopes or antigenic determinants). Moreover, T‐cell‐mediated immune responses lead to the generation of antibody secreting plasma cells and memory B cells. 37 , 38 Antibodies control viral infection through several mechanisms, including neutralization, opsonization, phagocytosis, activation of the classical complement pathway, and ADCC (an NK cell mediated apoptosis), and therefore prevent extracellular viruses from replicating and further infecting healthy cells. 37 , 39 , 40
A study has found an increase in the number of antibody‐secreting cells (ASCs) during SARS‐CoV‐2 infection. 20 The median duration of detectable IgA and IgM is approximately 5 days (ranging from 2 to 6 days) and 14 days for IgG (ranging from 10 to 18 days) after the onset of symptoms. 41 Although no crossreactivity was observed between SARS‐CoV‐2 nucleocapsid protein and positive human plasma for coronaviruses NL63, 229E, OC43, and HKU1, there was a strong crossreactivity with SARS‐CoV positive plasma. 41 The increased production of IgM and IgG in SARS‐CoV‐2 infection was detected in blood before the recovery of symptoms and persisted for at least 7 days after the full resolution of symptoms. 20 Analysis of serum samples collected 14 days or longer after the onset of COVID‐19 symptoms revealed 94% and 88% seropositivity rate for IgG and IgM, respectively, against the SARS‐CoV‐2 internal nucleoprotein (NP) antigen. 42 For IgG and IgM against surface spike protein receptor binding domain (RBD), the seropositivity rates were higher, 100% and 94%, respectively. 42 The level of antibodies against NP and RBD correlated with their neutralizing activities. 42 Taken together, humoral responses were clearly activated with diverse isotypes of antibodies during SARS‐CoV‐2 infection (Figure 1A). An increased IgG response with a higher titer of total immunoglobulins in the patients with severe COVID‐19 resulted in worse outcome, which suggests the possible role of antibody‐dependent enhancement (ADE) during SARS‐CoV‐2 infection. 43 , 44 , 45 However, how effective these antibodies are to neutralize extracellular viruses and to activate phagocytosis, classical complement pathway, and ADCC needs to be evaluated and further studies are warranted. In addition, impaired adaptive immune responses along with uncontrolled inflammatory innate responses were reported to be harmful, leading to local and systemic tissue damage. 45 However, one study found that the level of antibodies detected in patients with COVID‐19 is associated with virus neutralization titer, 42 so highlighting the potential use of convalescent plasma of COVID‐19 survivors or therapeutic monoclonal antibodies for patients with COVID‐19.
4. IMMUNOPATHOLOGY IN SARS‐COV‐2 INFECTION
4.1. Immune evasion of SARS‐CoV‐2: Lessons from SARS‐CoV
Groups of patients with COVID‐19, mostly elderly and those having comorbidities, develop severe clinical manifestations and succumb to death 46 , 47 , 48 ; however, some young healthy adults, 20‐40 years of age, have also developed severe clinical manifestations that require their admittance to the intensive care unit (ICU) and a small proportion (0.1% to 0.2%) has been reported to have died. 49 This might suggest that the host immune responses in those groups were unable to control viral replication, leading to severe disease progression. The exact immunological reasons why some patients cannot deal with SARS‐CoV‐2 remain largely unknown.
During SARS‐CoV infection, the virus employs strategies to block IFN production. 50 , 51 In this context, the production of IFN‐I induced by polyinosinic:polycytidylic acid (poly I:C) or Sendai virus was suppressed by the nucleocapsid (N) protein of the SARS‐CoV. 50 The SARS‐CoV membrane protein also inhibits the formation of the TRAF3·TANK·TBK1/IKK‐ɛ complex, leading to decreased interferon regulatory factor (IRF)3/IRF7 transcription factor and hence reduces IFN production. 51 The TRAF3·TANK·TBK1/IKK‐ɛ complex is required for the activation of IFN‐gene promoters. SARS‐CoV infection also downregulates gene expression of cathepsins and proteasomes, proteins required for antigen processing for MHC‐I and II pathways, and reduces MHC‐I expression in monocytes. 52 SARS‐CoV can infect and replicate in DCs, thereby inhibiting the maturation and antigen presentation capacity of DC, leading to impaired T cell activation for effective anti‐viral cell‐mediated immunity. 53 SARS‐CoV infection can induce production of cytokines, such as IL‐6 and IL‐8, from macrophages and DCs and upregulate CD40 and CD86 expression on DCs; IL‐6 and IL‐8 induction impaired the ability of DCs and macrophages to activate naïve T cells. 54 Taken together, SARs‐CoV blocks critical antiviral pathways and these evading mechanisms may also be utilized by SARS‐CoV‐2. However, extensive studies are required to elucidate the immune evasion of SARS‐CoV‐2.
4.2. Functional exhaustion in SARS‐CoV‐2 infection
Lymphocytes such as T cells (CD4+ and CD8+ T cells) and NK cells are important for viral clearance. 55 T cells, particularly cytotoxic T cells (CD8+ T cells) target infected cells through the interaction of the T cell receptor (TCR) with MHC‐I bound viral peptides. MHC‐I and TCR ligation enables CD8+ T cells to degranulate cytotoxic factors, such as perforins and granzymes, to induce apoptosis of infected cells 37 , 56 , 57 ; thereby eliminating viral reservoirs. NK cells recognize distressed cells or cells with reduced surface MHC‐I expression 58 and control viral replication through the release of IFN‐γ and activation of ADCC. 59 Impaired responses of functional CD8+ T cells and NK cells are associated with disease progression. 60 , 61 , 62
The COVID‐19 patients revealed leucopenia, lymphopenia, and thrombocytopenia on blood examination. 47 Both cellular and humoral immune responses are believed to be responsible for COVID‐19 pathogenesis. 63 , 64 In addition, T‐cell‐ and B‐cell‐mediated immunological mechanisms such as B cell activation along with detection and presentation of the antigen are initiated against SARS‐CoV‐2 to direct lymphocyte and macrophage stimulated responses. 63 , 65 , 66 , 67 The probable involvement of helper T cells and cytotoxic T cells needs thorough evaluation. In this context, helper T cells may present virus antigens to B cells for production of antibodies and subsequent neutralization, whereas cytotoxic T cells may directly kill the virus. 63 , 68 The SARS‐CoV‐2 is reported to attack respiratory epithelial cells followed by spread to other cells. Moreover, the virus infects peripheral leucocytes and lymphocytes particularly T cells. 69 The damage caused by SARS‐CoV‐2 to lymphocytes including T cells leads to lymphopenia and predisposes the individuals to secondary bacterial infections and increased severity of the disease. 69 The increase in proinflammatory and decrease in anti‐inflammatory cytokines may indicate the T‐cell‐mediated response against SARS‐CoV‐2, resulting in cytokine storm associated hyperinflammation and subsequent severe pneumonia in COVID‐19. 67 , 68 , 69 The increased levels of proinflammatory cytokines may result in shock and severe tissue damage in heart, kidney, and liver along with respiratory and multiple organ failure. 45 The severe and fatal cases of COVID‐19 are characterized by lymphopenia and sustained inflammation. 66
A recent study has found a significant decrease of total T lymphocytes, CD8+ T cells, and NK cells in patients with COVID‐19 and this reduction was more prominent among those with severe disease compared with those with a mild one. 70 Moreover, NK cells become exhausted during SARS‐CoV‐2 infection, characterized by increased expression of inhibitory receptor, NKG2A (NK group 2 member A), 70 which induces NK cell exhaustion in chronic viral infection (Figure 1A). In patients with COVID‐19, significant decrease of CD107a+NK, IFN‐γ+ NK, IL‐2+ NK, TNF‐α+NK, and granzyme B+ NK cells has also been observed, most likely due to the overexpression of NKG2A. 70 A study confirmed that the number of NK and CD8+ T cells was restored when the expression of NKG2A was reduced among COVID‐19 survivors after treatment. 70
IFN‐γ from CD4+ T cells is required to activate macrophages as potent killers of intracellular pathogens through the Th1 pathway; IFN‐γ is also required for robust CD8+ T cell responses. 71 TNF‐α, secreted by CD4+ T cells also has direct antiviral effects, resulting in reduced viral replication. 72 In patients with SARS‐CoV‐2, there was a significant reduction of IFN‐γ+CD4+ T cells and TNF‐α+CD4+ T cells in those with severe disease compared with those with a mild clinical manifestation. 73 Moreover, the level of coinhibitory receptors, such as programmed death receptor‐1 (PD‐1), cytotoxic T lymphocyte associated antigen‐4 (CTLA‐4), and T‐cell immunoglobulin and ITIM domain (TIGIT) increased in patients with COVID‐19 compared with healthy controls. 73 Increased expression of PD‐1 and CTLA‐4 reduces the function of CD4+ T cells and CD8+ T cells through the ligation of these molecules to inhibitory ligands, such as PD‐L1 and CD80, on DC cells. 74 This mechanism might be important to reduce the activation and proliferation of T cells against SARS‐CoV‐2. T‐cell subset analysis from 44 patients with COVID‐19 revealed that the levels of helper T cells (CD3+CD4+ T cells) and cytotoxic T cells (CD3+CD8+ T cells) were below the normal range. 75 In addition, an analysis of the blood sample from a patient with COVID‐19 found increased follicular helper T cells (TFH cells) and activated CD4+ T cells and CD8+ T cells. 20 Taken together, the evidence suggests that the number of lymphocytes, such as NK cells and T cells, in patients with COVID‐19 decreased and were exhausted with increased expression of inhibitory receptors such as NKG2A, PD‐1, and CTLA‐4 (Figure 1A). Apart from the increase in the expression of inhibitory receptors, reduction of activation and proliferation capacities of lymphocytes may also be associated with low costimulatory receptor expression (CD28) and reduced growth cytokines, such as IL‐2. 75 The characteristics of impaired lymphocyte responses in SARS‐CoV‐2 infection, which may contribute to disease progression, are presented in Table 1.
TABLE 1.
Summary of lymphocyte response in patients with COVID‐19
| Immune component | COVID‐19 compared with normal | Severe compared with mild | Ref. |
|---|---|---|---|
| NKG2A expression on NK cells | Increase | [70] | |
| NKG2A+ NK cells | Increase | [70] | |
| NKG2A+ CD8+ T cells | Increase | [70] | |
| PD‐1 | Increase | [73] | |
| CTLA‐4 | Increase | [73] | |
| TIGIT | Increase | [73] | |
| Total T lymphocytes | Decrease | Decrease | [70] |
| CD8+ T cells | Decrease | Decrease | [70] |
| Total NK cells | Decrease | Decrease | [70] |
| CD107a+ NK cells | Decrease | [70] | |
| IFN‐γ+ NK cells | Decrease | [70] | |
| IL‐2+ NK cells | Decrease | [70] | |
| TNF‐α+NK cells | Decrease | [70] | |
| Granzyme B+ NK cells | Decrease | [70] | |
| IFN‐γ+CD4+ T cells | Decrease | [73] | |
| TNF‐α+CD4+ T cells | Decrease | [73] | |
| Helper T cells (CD3+CD4+ T cells) | Decrease | [75] | |
| Cytotoxic T cells (CD28+CD8+ T cells) | Decrease | [75] |
4.3. Cytokine storm
The COVID‐19 associated deaths in severe forms were mostly attributed to respiratory failure 68 , 69 probably caused by lethal pneumonia following hyperinflammation. 64 , 68 , 69 The cytokine storm, dysregulated and uncontrolled release of proinflammatory cytokines, inducing systemic immune responses and inflammation, can lead to severe clinical manifestations, such as sepsis and multiple organ failure. 76 A cytokine storm has been documented in several viral infections including SARS‐CoV. 77 Increased neutrophilic and monocytic inflammatory responses and cytokines, such as IL‐1β, are the main characteristics of the cytokine storm. 78 , 79 Acute lung injury is a common repercussion of the cytokine storm and is mostly associated with infection in the lungs and other organs. 77
The clinical signs associated with SARS‐CoV‐2 infection are fever, dry cough, sneezing, chest pain, and pneumonia along with ground glass opacities and bilaterally consolidated lungs on computed tomography (CT) scan. 68 , 80 , 81 , 82 Moreover, clinical manifestations of COVID‐19 include mild‐flu‐like symptoms in the beginning; a proportion of patients develop dyspnea, acute respiratory distress syndrome (ARDS), and death. 68 These clinical characteristics bear some resemblance to those for SARS‐CoV and MERS‐CoV. 83 , 84 A growing body of evidence suggests that a proportion of patients with severe COVID‐19 may have the cytokine storm. 77 , 85 Data from a retrospective multicenter study of 68 deaths and 82 discharged cases in Wuhan revealed that the level of IL‐6 was significantly increased among patients in the death group compared with the survivor group. 86 Compared with healthy controls, the level of plasma cytokines, such as IL‐1B, IL‐1RA, IL‐7, IL‐8, IL‐9, IL‐10, IFN‐γ, and TNFα; chemokines, such as macrophage inflammatory protein 1‐alpha (MIP1A), MIP1B, and interferon gamma‐induced protein 10 (IP10); chemokine ligands, such as monocyte chemoattractant protein 1 (MCP1), growth factors and other proteins, such as granulocyte colony stimulating factor (GCSF) were higher in patients with both severe and nonsevere disease than healthy controls. 68 The levels of cytokines, such as TNF‐α, IL‐2, and IL‐7 and chemokines, such as GCSF, IP10, MCP1, and MIP1A were also higher in severe compared with nonsevere COVID‐19. 68 Release of higher amounts of proinflammatory cytokines, such as IL‐1β and TNF‐α, have a detrimental effect on the host cells as these molecules can induce a high influx of new immune cells to the site of infection and mount an excessive response, leading to severe tissue damage through the release of toxic granules. The advancement in lung damage due to hyperinflammation results into hypoxemia and, to compensate, shortness of breath along with cardiac arrest may take place leading to death. Therefore, immediate clinical care after the onset of clinical signs is crucial to prevent further COVID‐19 associated pathology. 87 In addition, histological examination performed on biopsy tissues from lung revealed desquamation of pneumocytes, diffuse alveolar damage, and presence of cellular fibromyxoid exudate along with formation of the hyaline membrane. 68 Altogether, evidence indicates that the intensity of the cytokine storm in patients with COVID‐19 increases with disease severity. Therefore, a strategy to inhibit the cytokine storm (ie, to control the immune response in patients with COVID‐19) is a promising immunotherapeutic approach for SARS‐CoV‐2 infection.
5. IMMUNOTHERAPEUTIC STRATEGIES FOR SARS‐COV‐2 INFECTION
Disease outcome of viral infection is determined by the balance between the virulence of the virus and the host immune response against the infection. 88 Although the host proinflammatory response is required for viral clearance in the early stages of infection, an excessive proinflammatory response at late stages of infection can exacerbate clinical manifestations. 89 Immunotherapy strategies that involve promoting viral clearance or reducing hyperinflammatory responses could potentially be used to treat patients with COVID‐19. Most of the efforts for developing therapeutics against coronavirus were targeted against the S protein which is the major inducer of neutralizing antibodies. 90 , 91 Use of immunotherapies, such as passive antibody, IFN‐I, and IL‐6 receptor (IL‐6R) inhibitor, that have been applied to treat SARS‐CoV and MERS‐CoV, either alone or in combination with other drugs are potentially being considered to treat patients with COVID‐19 (Figure 1B). The immunotherapies attempted against SARS‐CoV‐2 include plasma therapy (polyclonal antibody), hormones for T cells maturation, neutralizing antibodies, ACE2 immunoadhesins, and monoclonal antibodies against IL‐6. Moreover, other interventions like viral‐vectors, inactivated whole virus, nanoparticles and DNA as vaccines along with monoclonal antibody were reported to be used for SARS‐CoV and may prove promising against the SARS‐CoV‐2. 90 , 92
5.1. Passive antibody therapy
Passive antibody therapy involves the administration of antibodies from recovered patients to new patients who suffer from the same infection. 93 Historically, the use of passive antibody therapy to treat patients during the 1918 flu pandemic, Ebola hemorrhagic fever, influenza A (H5N1) infection, and SARS has been demonstrated to be a successful intervention strategy. 94 The first report of successful passive antibody therapy (in combination with ribavirin and methylprednisolone) for coronavirus was published in March 2003 during the outbreak of SARS in Hong Kong. 95 Later, a larger study involving 80 patients was done to test the efficacy of convalescent plasma of patients with SARS. 96 The study highlighted that earlier administration of convalescent plasma (before day 14) was more effective and more likely to give a good outcome than administration after day 14, consistent with the concept of viremia peaks in the first week of infection. 96 During the current COVID‐19 outbreak in China, administration of convalescent plasma has reported a reduction in viral load and morbidity of patients. 93 , 97 The mechanism of how passive antibody therapy works is given in brief in Figure 1.
According to the latest data, approximately 20% of patients have recovered from COVID‐19 worldwide (COVID‐19 Global Cases). 7 Neutralizing antibodies can be prepared from pooled convalescent plasma or monoclonal antibodies can be developed by immortalizing B‐cell repertoires of convalescent plasma. 98 To increase efficacy, several factors, including the timing of plasma administration, antibody titer of the administered plasma, and screening of convalescent plasma for blood borne pathogens, need to be considered. 93 To provide a timely response to the COVID‐19 pandemic, the US Food and Drug Administration (FDA) has approved convalescent sera to be delivered to patients seriously ill with COVID‐19, under the emergency investigational new drug protocol. 99 The eligibility of the donor and the recipient has been extensively described. 99 The FDA has stipulated that convalescent sera must only be collected from individuals who have recovered, with complete resolution of symptoms at least 14 days prior to donation and with a SARS‐CoV‐2 neutralizing antibody titer greater than 1:320. 99 In addition, not only can convalescent plasma serve as a safe therapy for patients critically ill with COVID‐19, such neutralizing antibodies also have the potential to be used as short prophylactic molecules for those at a high risk for COVID‐19, including health‐care providers and vulnerable individuals with comorbidities.
5.2. Interferon α/β
IFNs limit the spread of viral infection through induction of interferon stimulated genes (ISGs) that encode cytokines and antiviral proteins (Figure 1). 100 These antiviral proteins exhibit antiviral effects both directly, by inhibiting viral replication, and indirectly, by stimulating the adaptive immune system. 100 IFNα and IFNβ form one of the first lines of host defence against invading viruses. 101 Strong antiviral activities of IFNα and IFNβ support the use of IFNs as antiviral therapy. High‐dose IFNα or IFNβ monotherapy or in combination with ribavirin restricts SARS‐CoV and MERS‐CoV replication in cell cultures. 35 , 102 , 103 , 104 Patients with SARS who received IFN alfacon‐1 with corticosteroids were reported to exhibit better disease outcome than those who received corticosteroids alone. 105 Similarly, early combination therapy of pegylated IFNα‐2a and ribavirin was reported to improve survival in patients with MERS‐CoV. 106 Although the treatment group exhibited anemia, no other adverse effect was present. 106 Given the inhibitory effects of IFN on MERS‐CoV and SARS‐CoV, IFN therapy in SARS‐CoV‐2 has the potential to be used for treatment of patients with COVID‐19. Currently, several clinical trials involving IFNs either as monotherapy 99 , 107 or in combination 108 , 109 , 110 are currently under way to evaluate the efficacy and safety of IFN in treating patients with COVID‐19. Interestingly, standard care of patients with chronic viral infections has minimized the use of IFNs due to emerging evidence of IFN‐associated immunopathology. 111 Although IFN may be useful as adjunctive treatment, it can also exacerbate the clinical condition of critically ill patients, and therefore more results from controlled clinical trials are required before IFN can be used to treat COVID‐19.
5.3. IL‐6R inhibitor
Emerging evidence shows that patients with COVID‐19 exhibit the cytokine storm syndrome, characterized by the release of a high level of proinflammatory cytokines, including a marked elevation of IL‐6. 85 , 112 , 113 The increased level of IL‐6 was reported to be a reliable indicator of poor outcome in COVID‐19 patients with severe forms of the disease. 45 A high level of inflammatory cytokines is associated with pulmonary inflammation and severe lung damage. 114 Treatment of hyperinflammation using existing, approved therapies that are readily available would benefit patients with severe COVID‐19. Blocking the IL‐6 receptor, using the relatively safe tocilizumab, has shown therapeutic benefit against inflammatory disease and rheumatoid arthritis. 115 Tocilizumab, a humanized monoclonal antibody, competitively binds both membrane‐bound and soluble IL‐6 receptors, thus preventing IL‐6 from binding to its receptor (Figure 1). 116 Inhibition of the IL‐6 signaling cascade leads to a decrease in circulating immune cells. 116 Moreover, a clinical trial (ChiCTR2000029765) using tocilizumab was reported to be effective in quick control of fever along with improvement in respiratory function in patients with severe form of COVID‐19. 45 A number of clinical trials are currently being conducted to evaluate the safety and efficacy of tocilizumab for patients with COVID‐19. 117 A retrospective study reported that the use of tocilizumab effectively improved clinical symptoms and inhibited disease progression in patients with severe COVID‐19, in China. 118 The targeting of IL‐6 and its receptor by tocilizumab and siltuximab could mitigate the cytokine storm‐related clinical symptoms in severe form of COVID‐19. 119 However, IL‐6 antagonism may weaken the immune system of the patient; severely ill patients have been found to be coinfected with bacteria and fungi 69 and the risk of patients becoming more susceptible to secondary bacterial infection may outweigh the benefits of the treatment. Furthermore, the optimal timing of tocilizumab administration has not yet been determined. Although IL‐6R inhibitors have been used to treat COVID‐19 during the current pandemic, the results from ongoing controlled clinical trials are critical for further evaluation of this treatment.
6. CONCLUSION
The current COVID‐19 pandemic continues to be a significant threat to global health. During SARS‐CoV infection, lymphocytes become exhausted, which is characterized by the increased expression of inhibitory receptors such as PD‐1, CTLA‐4, and TIGIT on T cells and NKG2A on NK cells, resulting in the inhibition of their antiviral activities. Although the production of antibodies in patients with COVID‐19 is evident, the effectiveness of these antibodies in controlling viral replication remains to be elucidated. Uncontrolled release of proinflammatory cytokines may explain the severe clinical manifestation in some patients as it induces unnecessary influx of immune cells to the infection site, enabling these cells to release toxic granules, which are harmful to the lung cells. Therefore, strategies involving use of agents, such as the IL‐6R blocker, to control the hyperinflammatory response could be used as a potential approach to treat patients with COVID‐19. In addition, strategies to enhance viral clearance, such as the use of passive antibodies and IFN‐I therapy, could have be promising in treating patients with severe COVID‐19.
CONFLICT OF INTEREST
The authors declare no competing interests.
ACKNOWLEDGEMENTS
S.K. would to acknowledge the support from Australian Government and the University of Western Australia (UWA) through Research Training Program (RTP) and UWA University Postgraduate Awards (UPA). D.M. acknowledges the Indonesia Endowment Fund for Education (LPDP) of Republic of Indonesia for providing support for her study. K.D., S.K.P., and R.T. acknowledge their respective institute/university.
Keam S, Megawati D, Patel SK, Tiwari R, Dhama K, Harapan H. Immunopathology and immunotherapeutic strategies in severe acute respiratory syndrome coronavirus 2 infection. Rev Med Virol. 2020;30:e2123. 10.1002/rmv.2123
Abbreviations: ACE2, angiotensin‐converting enzyme 2; ADCC, antibody dependent cellular cytotoxicity; ARDS, acute respiratory distress syndrome; ASC, antibody‐secreting cells; CCL‐2, chemokine ligand‐2; CoV, coronavirus; COVID‐19, coronavirus disease 2019; CRS, cytokine storm syndrome; CTLA‐4, cytotoxic T lymphocyte associated antigen‐4; CXCL‐10, CXC chemokine ligand‐10; DC, dendritic cell; eIF2α, eukaryotic initiation factor 2 subunit‐α; FDA, US Food and Drug Administration; GCSF, granulocyte colony stimulating factor; ICU, intensive care unit; IFN, interferon; Ig, immunoglobulin; IL, interleukin; IL‐6R, IL‐6 receptor; IP10, interferon gamma‐induced protein 10; ISG, interferon stimulated genes; MCP1, monocyte chemoattractant protein 1; MERS‐CoV, Middle East Respiratory Syndrome Coronavirus; MHC‐I, major histocompatibility complex I; MIP1A, macrophage inflammatory protein 1‐alpha; NK, natural killer; NKG2A, natural killer group 2 member A; NLR, nucleotide‐binding and oligomerization domain (NOD)‐like receptor; NP, internal nucleoprotein; PD‐1, programmed death receptor‐1; PD‐L1, programmed death ligand‐1; PHEIC, Public Health Emergency of International Concern; PKP, protein kinase R; PRR, pattern recognition receptor; R0, Reproductive number; RBD, receptor binding domain; RLR, retinoic acid‐inducible gene I (RIG‐I)‐like receptors; SARS‐CoV, Severe Acute Respiratory Syndrome Coronavirus; SARS‐CoV‐2, Severe Acute Respiratory Syndrome Coronavirus 2; ssRNA, single‐stranded RNA; TCR, T cell receptor; TFH, follicular helper T; TIGIT, T‐cell immunoglobulin and ITIM domain; TLR, Toll‐like receptor; TNF‐α, tumor necrotic factor‐α; WHO, World Health Organization.
REFERENCES
- 1. Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J Med Virol. 2020;92:401‐402. 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burki TK. Coronavirus in China. Lancet Respir Med. 2020;8:238. 10.1016/S2213-2600(20)30056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chatterjee P, Nagi N, Agarwal A, et al. The 2019 novel coronavirus disease (COVID‐19) pandemic: a review of the current evidence. Indian J Med Res. 2020;151(2):147‐159. 10.4103/ijmr.IJMR_519_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dhama K, Sharun K, Tiwari R, et al. Coronavirus disease 2019—COVID‐19. 2020. Preprints 2020030001
- 5. Zheng J. SARS‐CoV‐2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16(10):1678‐1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270‐273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis. 2020;20:533‐534. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callaway E, Cyranoski D. China coronavirus: six questions scientists are asking. Nature. 2020;577:605‐607. [DOI] [PubMed] [Google Scholar]
- 9. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019‐nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao S, Lin Q, Ran J, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019‐nCoV) in China, from 2019 to 2020: a data‐driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chappell B. Coronavirus: COVID‐19 Is Now Officially A Pandemic, WHO Says. Washington: NPR; 2020. https://www.npr.org/sections/goatsandsoda/2020/03/11/814474930/coronavirus‐covid‐19‐is‐now‐officially‐a‐pandemic‐who‐says. Published 2020. Accessed March 11, 2020. [Google Scholar]
- 12. Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses. 2015;1282:1‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burrell C, Howard C, Murphy F. Fenner and White’s Medical Virology. 5th ed. Cambridge: Academic Press; 2016. [Google Scholar]
- 15. Weiss SR, Navas‐Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635‐664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801‐8811. 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu X, Liu Q, Du L, et al. Receptor‐binding domain as a target for developing SARS vaccines. J Thorac Dis. 2013;5:S142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team . Vital surveillances: the epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID‐19)—China, 2020. China CDC Weekly. 2020;2(2):113‐122. [PMC free article] [PubMed] [Google Scholar]
- 20. Thevarajan I, Nguyen THO, Koutsakos M, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non‐severe COVID‐19. Nat Med. 2020;26(4):1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271‐280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus‐induced lung injury. Nat Med. 2005;11:875‐879. 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2:264‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cervantes‐Barragan L, Züst R, Weber F, et al. Control of coronavirus infection through plasmacytoid dendritic‐cell‐derived type I interferon. Blood. 2007;109:1131‐1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davidson S, Maini MK, Wack A. Disease‐promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res. 2015;35:252‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newton AH, Cardani A, Braciale TJ. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Sem Immunopathol. 2016;38:471‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vidal SM, Khakoo SI, Biron CA. Natural killer cell responses during viral infections: flexibility and conditioning of innate immunity by experience. Curr Opin Virol. 2011;1:497‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frieman M, Heise M, Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133:101‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Le Bon A, Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14:432‐436. [DOI] [PubMed] [Google Scholar]
- 30. Rabouw HH, Langereis MA, Knaap RC, et al. Middle East respiratory coronavirus accessory protein 4a inhibits PKR‐mediated antiviral stress responses. PLoS Pathog. 2016;12:e1005982. 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Comar CE, Goldstein SA, Li Y, et al. Antagonism of dsRNA‐induced innate immune pathways by NS4a and NS4b accessory proteins during mers coronavirus infection. mBio. 2019;10:e00319‐19. 10.1128/mBio.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Teijaro JR. Type I interferons in viral control and immune regulation. Curr Opin Virol. 2016;16:31‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen K‐L, Yang Y‐H. Diagnosis and Treatment of 2019 Novel Coronavirus Infection in Children: A Pressing Issue. World J Pediatr. 2020:1‐3. 10.1007/s12519-020-00344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuri T, Zhang X, Habjan M, et al. Interferon priming enables cells to partially overturn the SARS coronavirus‐induced block in innate immune activation. J Gen Virol. 2009;90:2686‐2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ströher U, DiCaro A, Li Y, et al. Severe acute respiratory syndrome‐related coronavirus is inhibited by interferon‐α. J Infect Dis. 2004;189:1164‐1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lokugamage K, Schindewolf C and Menachery V. SARS‐CoV‐2 sensitive to type I interferon pretreatment. bioRxiv 2020. In press. https://doi.org/101101/20200307982264. [Google Scholar]
- 37. Abbas AK, Lichtman AH, Pillai S. Basic immunology: Functions and disorders of the immune system. 6. Boston: Elsevier; 2019. [Google Scholar]
- 38. Dörner T, Radbruch A. Antibodies and B cell memory in viral immunity. Immunity. 2007;27:384‐392. [DOI] [PubMed] [Google Scholar]
- 39. Sörman A, Zhang L, Ding Z, Heyman B. How antibodies use complement to regulate antibody responses. Mol Immunol. 2014;61:79‐88. [DOI] [PubMed] [Google Scholar]
- 40. Zahavi D, AlDeghaither D, O’Connell A, Weiner LM. Enhancing antibody‐dependent cell‐mediated cytotoxicity: a strategy for improving antibody‐based immunotherapy. Antibody Therapeutics. 2018;1:7‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guo L, Ren L, Yang S, et al. Profiling early Humoral response to diagnose novel coronavirus disease (COVID‐19). Clin Infect Dis. 2020;1‐8. 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. To KK‐W, Tsang OT‐Y, Leung W‐S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet. 2020;20(5):565‐574. 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang B, Zhou X, Zhu C, et al. Immune phenotyping based on neutrophil‐to‐lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID‐19. medRxiv. 2020. In press. 10.1101/2020.03.12.20035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease. Clin Infect Dis; 2020. In press. 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cao X. COVID‐19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20:269‐270. 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang J, Zheng Y, Gou X, et al. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID‐19) infection: a systematic review and meta‐analysis. Int J Infect Dis. 2020;94:91‐95. 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708‐1720. 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan. China JAMA. 2020;323:1061. 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Centers for Disease Control and Prevention . Severe Outcomes Among Patients with Coronavirus Disease 2019 (COVID‐19)—United States, February 12–March 16, 2020; 2020. https://www.cdc.gov/mmwr/volumes/69/wr/mm6912e2.htm?s_cid=mm6912e2_w#F1_down. Accessed April 1, 2020).
- 50. Hu Y, Li W, Gao T, et al. SARS coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25‐mediated RIG‐I ubiquitination. J Virol. 2017;91(8):e02143‐e02116. 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Siu K‐L, Kok K‐H, Ng M‐HJ, et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3· TANK· TBK1/IKKɛ complex. J Biol Chem. 2009;284:16202‐16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hu W, Yen Y‐T, Singh S, Kao CL, Wu‐Hsieh BA. SARS‐CoV regulates immune function‐related gene expression in human monocytic cells. Viral Immunol. 2012;25:277‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Spiegel M, Schneider K, Weber F, et al. Interaction of severe acute respiratory syndrome‐associated coronavirus with dendritic cells. J Gen Virol. 2006;87:1953‐1960. [DOI] [PubMed] [Google Scholar]
- 54. Yoshikawa T, Hill T, Li K, Peters CJ, Tseng CTK. Severe acute respiratory syndrome (SARS) coronavirus‐induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte‐derived macrophages and dendritic cells. J Virol. 2009;83:3039‐3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Winkler CW, Myers LM, Woods TA, Carmody AB, Taylor KG, Peterson KE. Lymphocytes have a role in protection, but not in pathogenesis, during La Crosse virus infection in mice. J Neuroinflammation. 2017;14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rauf A, Khatri M, Murgia MV, Saif YM. Fas/FasL and perforin–granzyme pathways mediated T cell cytotoxic responses in infectious bursal disease virus infected chickens. Results in Immunol. 2012;2:112‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shrestha B, Diamond MS. Fas ligand interactions contribute to CD8+ T‐cell‐mediated control of West Nile virus infection in the central nervous system. J Virol. 2007;81:11749‐11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vivier E, Raulet DH, Moretta A, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li Y, Sun R. Tumor immunotherapy: new aspects of natural killer cells. Chin J Cancer Res. 2018;30:173‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoffmann M, Pantazis N, Martin GE, et al. Exhaustion of activated CD8 T cells predicts disease progression in primary HIV‐1 infection. PLoS Pathog. 2016;12:e1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kahan SM, Wherry EJ, Zajac AJ. T cell exhaustion during persistent viral infections. Virology. 2015;479:180‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang C, Wang X‐m, S‐r L, et al. NKG2A is a NK cell exhaustion checkpoint for HCV persistence. Nat Commun. 2019;10:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID‐19 coronavirus (SARS‐CoV‐2) based on SARS‐CoV immunological studies. Viruses. 2020;12(3):254. 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dhama K, Patel S, Pathak M, et al. An update on SARS‐COV‐2/COVID‐19 with particular reference on its clinical pathology, pathogenesis, immunopathology and mitigation strategies—a review. 2020. Preprints 2020b, 2020030348. doi: 1020944/preprints2020030348v1. [DOI] [PMC free article] [PubMed]
- 65. Walls A, Park Y‐J, Tortorici MA, et al. Structure, function and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181(2):281‐292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ja T. Is COVID‐19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22(2):72‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu K, Chen Y, Lin R, Han K. Clinical features of COVID‐19 in elderly patients: A comparison with young and middle‐aged patients. J Infect. 2020;80:e14‐e18. 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507‐513. 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zheng M, Goa Y, Wang G, et al. Functional exhaustion of antiviral lymphocytes in COVID‐19 patients. Cell Mol Immunol. 2020;17:533‐535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Green AM, DiFazio R, Flynn JL. IFN‐γ from CD4 T cells is essential for host survival and enhances CD8 T cell function during Mycobacterium tuberculosis infection. J Immunol. 2013;190:270‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maloy KJ, Burkhart C, Junt TM, et al. CD4+ T cell subsets during virus infection: protective capacity depends on effector cytokine secretion and on migratory capability. J Exp Med. 2000;191:2159‐2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zheng H‐Y, Zhang M, Yang C‐X, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID‐19 patients. Cell Mol Immunol. 2020;17:541‐543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Buchbinder EI, Desai A. CTLA‐4 and PD‐1 pathways: similarities, differences, and implications of their inhibition. Am J Clin Oncol. 2016;39:98‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID‐19 in Wuhan, China. Clin Inf Dis. 2020;1‐7. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang H, Ma S. The cytokine storm and factors determining the sequence and severity of organ dysfunction in multiple organ dysfunction syndrome. Am J Emerg Med. 2008;26:711‐715. [DOI] [PubMed] [Google Scholar]
- 77. Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. D’Elia RV, Harrison K, Oyston PC, et al. Targeting the “cytokine storm” for therapeutic benefit. Clin Vac Immunol. 2013;20:319‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yiu HH, Graham AL, Stengel RF. Dynamics of a cytokine storm. PLoS ONE. 2012;7:e45027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hui DS, IA E, Madani TA, et al. The continuing 2019‐nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264‐266. 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bassetti M, Vena A, Giacobbe DR. The novel Chinese coronavirus (2019‐nCoV) infections: challenges for fighting the storm. Eur J Clin Invest. 2020;50:e13209. 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Harapan H, Itoh N, Yufika A, et al. Coronavirus Disease 2019 (COVID‐19): a Literature Review. J Infect Public Health. 2020;13(5):667‐673. 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. New Eng J Med. 2003;348:1986‐1994. [DOI] [PubMed] [Google Scholar]
- 84. Assiri A, Al‐Tawfiq JA, Al‐Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13:752‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mehta P, McAuley DF, Brown M, et al. COVID‐19: consider cytokine storm syndromes and immunosuppression. The Lancet. 2020;395:1033‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ruan Q, Yang K, Wang W, et al. Clinical predictors of mortality due to COVID‐19 based on an analysis of data of 150 patients from Wuhan, China. Intens Care Med. 2020;46(5):846‐848. 10.1007/s00134-020-06028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395:514‐523. 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun. 2020;12:4‐20. 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID‐19. J Pharmaceutical Anal. 2020;10:102‐108. 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dhama K, Sharun K, Tiwari R, et al. COVID‐19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020;2020:1‐7. 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rabaan A, Al‐Ahmed S, Sah R, et al. SARS‐CoV‐2/COVID‐19 and advances in developing potential therapeutics and vaccines to counter this emerging pandemic virus—a review. 2020, 2020040075 doi: 1020944/preprints2020040075v1 2020. [DOI] [PMC free article] [PubMed]
- 92. AminJafari A, Ghasemi S. The possible of immunotherapy for COVID‐19: a systematic review. Int Immunopharmacol. 2020;83:106455. 10.1016/j.intimp.2020.106455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Casadevall A, Pirofski L‐A. The convalescent sera option for containing COVID‐19. J Clin Invest. 2020;130:130‐1548. 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Marasco WA, Sui J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat Biotechnol. 2007;25:1421‐1434. 10.1038/nbt1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wong VW, Dai D, Wu AK, et al. Treatment of severe acute respiratory syndrome with convalescent plasma. Hong Kong Med J. 2003;9:199‐201. [PubMed] [Google Scholar]
- 96. Cheng Y, Wong R, Soo YOY, et al. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Inf Dis. 2005;24:44‐46. 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shen C, Wang Z, Zhao F, et al. Treatment of 5 critically ill patients with COVID‐19 with convalescent plasma. JAMA. 2020;323:1582. 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Traggiai E, Becker S, Subbarao K, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871‐875. 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. FDA . Investigational COVID‐19 Convalescent Plasma—Emergency INDs. Silver Spring, MD: Food and Drug Administration; 2020. [Google Scholar]
- 100. Fensterl V and Sen GC. Interferons and viral infections. BioFactors. 2009;35:14‐20. 10.1002/biof.6. [DOI] [PubMed] [Google Scholar]
- 101. Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778‐809, table of contents. 10.1128/cmr.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cinatl J, Morgenstern B, Bauer G, et al. Treatment of SARS with human interferons. Lancet; 2003;62:293‐294. 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Falzarano D, de Wit E, Martellaro C, Callison J, Munster VJ, Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon‐α2b and ribavirin. Sci Rep. 2013;3:1686. 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zielecki F, Weber M, Eickmann M, et al. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013;87:5300‐5304. 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Loutfy MR, Blatt LM, Siminovitch KA, et al. Interferon alfacon‐1 plus corticosteroids in severe acute respiratory syndrome: A preliminary study. JAMA. 2003;290:3222‐3228. 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- 106. Omrani AS, Saad MM, Baig K, et al. Ribavirin and interferon alfa‐2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Inf Dis. 2014;14:1090‐1095. 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Meng Z. Experimental TRIAL of rhIFNα Nasal Drops to Prevent 2019‐nCOV in Medical Staff. NCT04320238 2020; https://clinicaltrials.gov/ct2/show/NCT04320238. Accessed April 5, 2020.
- 108. Jiangxi Qingfeng Pharmaceutical Co. Ltd. Xiyanping Injection for the Treatment of New Coronavirus Infected Pneumonia. NCT04275388 2020; https://clinicaltrials.gov/ct2/show/NCT04275388 (April 5, 2020).
- 109. The Ninth Hospital of Nanchang . Evaluation of ganovo Danoprevir combined with ritonavir in the treatment of novel coronavirus infection. https://clinicaltrialsgov/ct2/show/NCT04291729. Accessed: April 5, 2020 2020.
- 110. Tongji Hospital . A prospective/retrospective randomized controlled clinical study of interferon atomization in the 2019‐nCoV pneumonia. NCT04254874. 2020; https://clinicaltrials.gov/ct2/show/NCT04254874. Accessed April 5, 2020.
- 111. Murira A, Lamarre A. Type‐I interferon responses: from friend to foe in the Battle against chronic viral infection. Front Immunol. 2016;7:609. 10.3389/fimmu.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Anders KL, Indriani C, Ahmad RA, et al. The AWED trial (Applying Wolbachia to Eliminate Dengue) to assess the efficacy of Wolbachia‐infected mosquito deployments to reduce dengue incidence in Yogyakarta, Indonesia: study protocol for a cluster randomised controlled trial. Trials. 2018;19:302. 10.1186/s13063-018-2670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Inf Dis. 2020;1‐9. 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wong CK, Lam CW, Wu AK, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95‐103. 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ogata A, Kato Y, Higa S, Yoshizaki K. IL‐6 inhibitor for the treatment of rheumatoid arthritis: a comprehensive review. Mod Rheumatol. 2019;29:258‐267. 10.1080/14397595.2018.1546357. [DOI] [PubMed] [Google Scholar]
- 116. Rubbert‐Roth A, Furst DE, Nebesky JM, et al. A review of recent advances using tocilizumab in the treatment of rheumatic diseases. Rheumatol Ther. 2018;;5:21‐42. 10.1007/s40744-018-0102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.ClinicalTrials. 2020. https://clinicaltrials.gov/ct2/results?cond=COVID-19&term=tocilizumab&cntry=&state=&city=&dist=. Acesssed April 14, 2020 .
- 118. Xu X, Han M Li T, et al. Effective Treatment of Severe COVID‐19 Patients with Tocilizumab. http://chinaxivorg/abs/20200300026 2020. [DOI] [PMC free article] [PubMed]
- 119. Liu T, Zhang J, Yang Y, et al. The potential role of IL‐6 in monitoring coronavirus disease 2019. Available at SSRN 3548761. 2020.


