Abstract
Testing is one of the commendable measures for curbing the spread of coronavirus disease (COVID‐19). But, it should be done using the most appropriate specimen and an accurate diagnostic test such as real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR). Therefore, a systematic review was conducted to determine the positive detection rate of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in different clinical specimens using qRT‐PCR. A total of 8136 pooled clinical specimens were analyzed to detect SARS‐CoV‐2, the majority were nasopharyngeal swabs (69.6%). A lower respiratory tract (LRT) specimens had a positive rate (PR) of 71.3% (95% confidence interval [CI]: 60.3%‐82.3%) while no virus was detected in the urinogenital specimens. Bronchoalveolar lavage fluid (BLF) specimen had the PR of 91.8% (95% CI: 79.9%‐103.7%), followed by rectal swabs; 87.8% (95% CI: 78.6%‐96.9%) then sputum; 68.1% (95% CI: 56.9%‐79.4%). A low PR was observed in oropharyngeal swabs; 7.6% (95% CI: 5.7%‐9.6%) and blood samples; 1.0% (95% CI: −0.1%‐2.1%) whereas no SARS‐CoV‐2 was detected in urine samples. Feces had a PR of 32.8% (95% CI:1 5.8%‐49.8%). Nasopharyngeal swab, a widely used specimen had a PR of 45.5% (95% CI: 31.2%‐59.7%). In this study, SARS‐CoV‐2 was highly detected in LRT specimens while no virus was detected in urinogenital specimens. BLF had the highest PR followed by rectal swab then sputum. Nasopharyngeal swab which is widely used had moderate PR. Low PR was recorded in oropharyngeal swab and blood samples while no virus was found in urine samples. Last, the virus was detected in feces, suggesting SARS‐CoV‐2 transmission by the fecal route.
Keywords: clinical sample, clinical specimen, COVID‐19, polymerase chain reaction, SARS‐CoV‐2
Abbreviations
- BLF
bronchoalveolar lavage fluid
- COVID‐19
coronavirus disease 2019
- GIT
gastrointestinal tract
- LRT
lower respiratory tract
- NOS
Newcastle Ottawa Scale
- qTR‐PCR
real‐time reverse transcription‐polymerase chain reaction
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- UGT
urinogenital tract
- URT
upper respiratory tract
1. INTRODUCTION
Coronavirus diseases 2019 (COVID‐19) is a highly infectious and an emerging respiratory disease caused by a severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). 1 Although its pathogenesis is still unclear but the current evidence associated SAR‐CoV‐2 infection with angiotensin‐converting 2 receptors. 2 , 4 On the other hand, real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR) of upper respiratory specimens, mainly nasopharyngeal swabs have been widely used to confirm the clinical diagnosis of COVID‐19. 5
However, there are reports of SARS‐CoV‐2 detections from other sites including feces, 6 , 7 and, therefore, extending the spectrum of specimens other than those from respiratory tract to be considered for clinical diagnosis of SARS‐CoV‐2. From the beginning, health authorities, that is, World Health Organization (WHO) 8 and Centre for Disease Control and Prevention (CDC) advocated massive and rapid testing of COVID‐19. Testing as one of commendable approaches in the fight against COVID‐19 pandemic needs to be done using both most appropriate specimen 6 and an accurate diagnostic test like PCR. 9 If testing is properly done especially during this time when the reopening is on its way in most of the countries, the risk of SARS‐CoV‐2 transmission will be minimized.
Currently, the detection profile of SARS‐CoV‐2 RNA from different clinical specimens using qRT‐PCR after onset of symptoms is not yet well established. A recent study by Wang et al 6 using 1070 clinical specimens such as bronchoalveolar lavage fluid (BLF), fibrobronchoscope brush biopsy (FBB), sputum, nasal and pharyngeal swabs, urine, feces, and blood collected from 205 patients revealed a dynamic profile with a high detection rate of virus from lower respiratory tract (LRT) specimens, that is, BLF and zero detection from urogenital tract specimen, that is, urine.
Therefore, the aim of this systematic review was to establish the profile of detecting SARS‐CoV‐2 from different types clinical specimens using a standard diagnostic test (qRT‐PCR).
2. MATERIALS AND METHODS
2.1. Protocol development
A systematic review protocol was developed based on the question “What is the positivity rate for detecting SARS‐CoV‐2 using qRT‐PCR in different types of clinical specimens?”. The review was developed in accordance with Preferred Reporting Items for Systematic Reviews and Meta‐Analyses Protocols (PRISMA‐P) guidelines. 10 The protocol was registered in the International Prospective Register of Systematic Reviews (PROSPERO database: https://www.crd.york.ac.uk/PROSPERO with a registration number CRD42020189107).
2.2. Search strategy
A rigorous systematic search strategy was developed with the help from librarian using published guidelines of the Cochrane Collaboration. 11 A systematic search from PubMed/MEDLINE, Science Direct, and Google scholar 12 was conducted. We also searched the websites of key healthcare organizations such as WHO and CDC. With the help of Google, a supplementary search was done from grey literature sources, for example, preprints and journal's website (JAMA, Lancet, Nature Research, and NEMJ). Data from 31 December 2019 onward conducted in human beings and published in the English language qualified for inclusion. The strategy was primarily developed for PubMed using keywords (Supporting Information Additional File S1). The search terms were combined using Boolean logic “OR” for synonymous terms and “AND” across elements of PCO (population, comparability, and outcome). Filters were set to exclude nonhuman studies, limit the publication period exclude review, and case report articles, among others. Keywords such as “laboratory diagnosis,” “polymerase chain reaction,” “clinical sample,” “clinical specimen,” “novel coronavirus 2019”, “SARS‐CoV‐2,” COVID‐19 were used. This search strategy was adapted to the other databases' search. All searched articles from the different databases were exported into EndNote software version X7 (Thomson Reuters, 2015) where duplicates were identified and removed. The articles were grouped into relevant categories as indicated in the PRISMA flow diagram (Figure 1).
Figure 1.
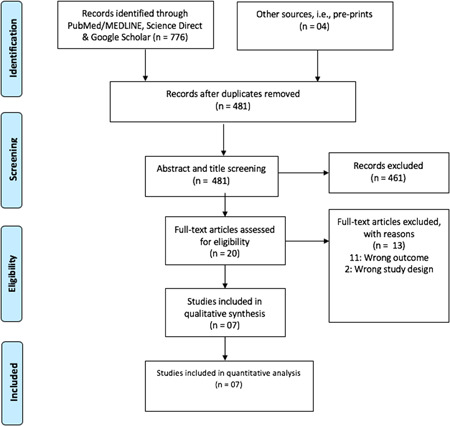
Prism flow chart showing study screening
2.3. Eligibility criteria
Both clinical trials and observational studies (including cross‐sectional, retrospective, and prospective studies) were eligible. Additionally, case series were included, however, considering at least one patient per study, most of the case reports were excluded. This review focused but not limited to studies which reported RNA extracted from clinical specimens and determined by qRT‐PCR targeting the open reading frame lab gene of SARS‐CoV‐2. A cycle threshold value of less than 40 was regarded as positive for SARS‐CoV‐2 RNA. In this case, the less the cycle threshold value the higher the viral load. Studies which were conducted to determine the diagnostic accuracy, reviews, and nonhuman articles were excluded. For inclusion in the final analysis, at least two simultaneously tested samples were required to be reported per study.
2.4. Data extraction
Study selection was managed using EndNote software version X7 (Thomson Reuters, 2015) where two independent reviewers (GMB and BJN) evaluated articles for potential inclusion by screening titles and abstracts followed by full‐text screening to determine eligibility for final inclusion. Discrepancies were resolved by consensus, and/or consulting a third reviewer where necessary. Data extracted from study documents, included author's information, year of publication, study design, and positivity rate (positive tests/total specimens) (Table 1). Unavailable, unclear information and additional details were requested from the corresponding author. In some of the studies, 7 , 13 , 14 patient was regarded as positive, if one of the specimens tested positive. Additionally, at least two specimens reported per test qualified for analysis where a positive patient was considered when one of the specimens tested positive, and recovered patient was considered when at least two qRT‐PCR consecutive tests tested negative in all tested specimen. The number of laboratory tests was counted based on the specimen tested not the number of patients.
Table 1.
Characteristics of the reviewed studies
| Author | Design | Type of specimen | Positive test | Total test | Risk assessment (NOS); 0: high risk, 10: low risk |
|---|---|---|---|---|---|
| Wang et al 6 | Cross‐sectional | Bronchoalveolar lavage fluid | 14 | 15 | 6 |
| Fibrobronchoscope brush biopsy | 6 | 13 | |||
| Sputum | 75 | 104 | |||
| Nasal swabs | 5 | 8 | |||
| Pharyngeal swabs | 126 | 398 | |||
| Feces | 44 | 153 | |||
| Blood | 3 | 307 | |||
| Urine | 0 | 72 | |||
| Xu et al 13 | Prospective study | Nasopharyngeal swab | 22 | 49 | 6 |
| Rectal swab | 43 | 49 | |||
| Lo et al 15 | Prospective study | Nasopharyngeal swab | 57 | 84 | 4 |
| Sputum | 1 | 1 | |||
| Urine | 0 | 49 | |||
| Feces | 46 | 79 | |||
| Chan et al 14 | Case series | Nasopharyngeal swab | 4 | 5 | 6 |
| Throat swab | 2 | 3 | |||
| Sputum | 2 | 2 | |||
| Serum | 1 | 3 | |||
| Plasma | 0 | 4 | |||
| Urine | 0 | 5 | |||
| Feces | 0 | 4 | |||
| Chen et al 7 | Retrospective study | Pharyngeal swab | 65 | 167 | 8 |
| Sputum | 155 | 206 | |||
| Feces | 17 | 64 | |||
| Liu et al 5 | Cross‐sectional | Sputum | 28 | 57 | 4 |
| Bronchoalveolar lavage fluid | 4 | 5 | |||
| Nasopharyngeal swab | 1843 | 4818 | |||
| Wang et al 16 | Retrospective study | Nasopharyngeal swab | 134 | 706 | 4 |
| Oropharyngeal swab | 54 | 706 |
Abbreviation: NOS, Newcastle Ottawa Scale.
2.5. Data synthesis for included studies
DerSimonian–Laird (DL) random‐effects analysis was performed to establish a summary estimate (positivity rate/proportion) by a random‐effects model using Open Meta Analyst software 17 and expressed using by pooled effect estimates and their 95% confidence intervals (CIs). The narrative was written by the lead reviewer (GMB) and then checked independently by three other reviewers (BJN, MVM, and AM). Heterogeneity in the analyzed studies was determined using I 2 statistic while subgrouping analysis was performed based on the type of clinical specimen (Figure 2) and sampling site (Figure 3).
Figure 2.
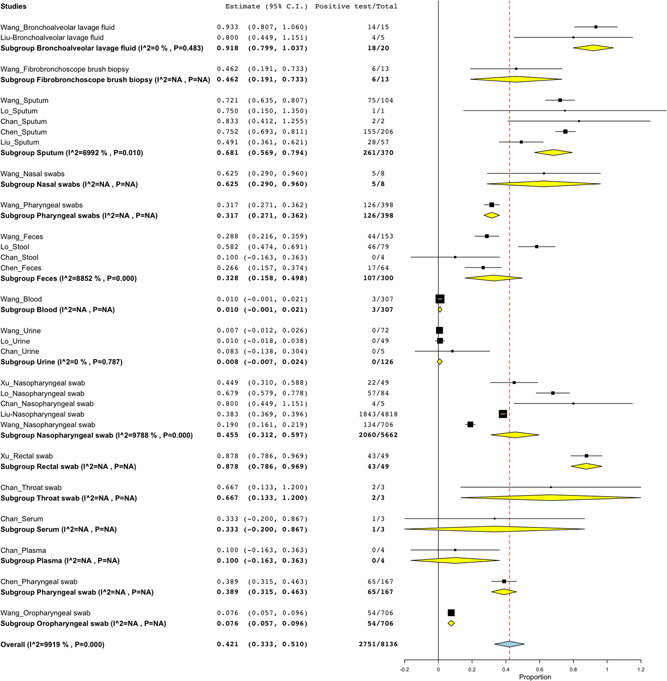
The rate of detection based on type of clinical specimen. BLF specimen had the positivity rate of 91.8% (18/20; 95% CI: 79.9%‐103.7%), 87.8% (43/49; 95% CI: 78.6%‐96.9%) for rectal swab, 68.1% (261/370; 95% CI: 56.9%‐79.4%) for sputum specimen, 62.5% (5/8; 95% CI: 29.0%‐96.0%) for nasal swabs, 46.2% (6/13; 95% CI: 19.1%‐73.3%) for FBB specimen, 31.7% (126/398; 95% CI: 27.1%‐34.1%) for pharyngeal swabs, 32.8% (107/300; 95% CI: 15.8%‐49.8%) for feces, 1.0% (3/3017; 95%CI: −0.1%‐2.1%) for blood sample, 0.8% (0/126; 95% CI: −0.7%‐2.4%) for urine sample, 45.5% (2060/5662; 95% CI: 31.2%‐59.7%) for nasopharyngeal swab, 87.8% (43/49; 95% CI: 78.6%‐96.9%), 66.7% (2/3; 95% CI: 11.3%‐120.0%) for throat swab, 33.3% (1/3; 95% CI: −20.0%‐82.8%) for serum, 10.0% (0/4; 95% CI: −16.3%‐36.3%) for plasma sample, 38.9% (65/167; 95% CI: 31.5%‐46.3%) and 7.6% (54/706; 95% CI: 5.7%‐9.6%) for oropharyngeal swab. BLF, bronchoalveolar lavage fluid; CI, confidence interval; FBB, fibrobronchoscope brush biopsy
Figure 3.
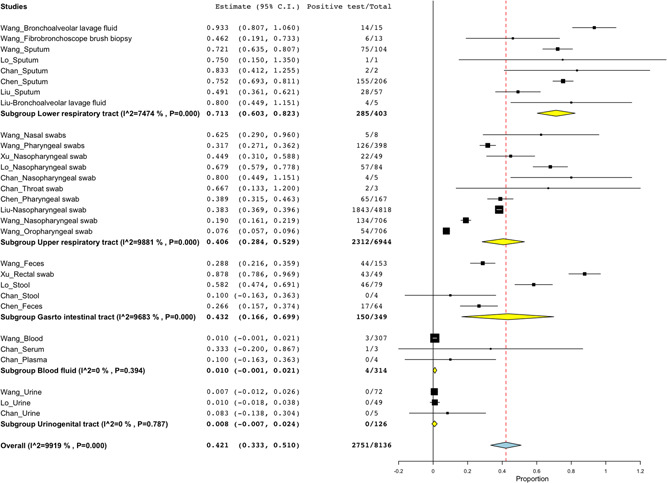
Positive detection rate based on sampling site. About 403 specimens were collected for detection of SARS‐CoV‐2 from LRT where 71.3% (285/403; 95% CI: 60.3%‐82.3%) tested positive. Specimens from URT had the positivity rate of 40.6% (2312/6944; 95% CI: 28.4%‐52.9%). Gastro intestinal tract (GIT) specimens recorded 43.2% positive SARS‐CoV‐2 (150/349; 95% CI: 16.6%‐69.9%). Blood fluids and UGT had the positivity rate of 1.0% (4/314; 95% CI: −0.1%‐2.1%) and 0.8% (0/126; 95% CI:−0.7%‐2.4%), respectively. CI, confidence interval; LRT, lower respiratory tract; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; UGT, urinogenital tract
2.6. Quality assessment
Output generated by Open Meta Analyst software through a cumulative forest plot was indirectly employed to assess the publication bias. 18 As such, the estimate of the proportions decreased with the increase in sample size. This decrease could be due to publication bias or it could be due to small‐study effects. However, when the analysis was limited to study with at least 100 sample size, the overall positive detection rate would have been 34.6% (95% CI: 17.9%‐51.3%) while that with all studies included regardless of the sample size was 42.7% (95% CI: 32.2%‐53.3%) (Supporting Information Additional File S2). On the other hand, the quality of the included studies (risk of bias) was assessed using the Newcastle Ottawa Scale adapted for cross‐sectional studies (Supporting Information Additional File S3) as previously described elsewhere. 19 The risk of bias was evaluated by two independent reviewers (GMB and BJN). Discrepancies were resolved by consensus, and/or consulting a third reviewer (MVM) where necessary.
3. RESULTS
3.1. Characteristics of the included studies
Of 780 pooled studies, seven studies qualified for final analysis (Table 1). Among the seven articles, information reported on positive detection rate was extracted from four studies 5 , 6 , 15 , 16 for suspected cases, while in three studies 7 , 13 , 14 information was recorded if at least one specimen tested positive (confirmed cases) from the simultaneously tested specimens and the recording of positive tests were stopped when at least two consecutive qTR‐PCR tested negative in all specimens. 7 , 13 Different types of clinical specimens were tested for SARS‐CoV‐2, BLF, 5 , 6 FBB, 6 sputum. 5 , 7 , 14 , 15 , 16 Some of the studies reported pharyngeal specimen without specifying whether the route was nasal of oral. 6 , 7 One study reported nasal and pharyngeal swabs combined, but for the purpose of this review, it was categorized as nasopharyngeal swabs. 5 Nasal swabs were reported in one study, 6 feces, 6 , 7 blood, 6 urine, 6 , 14 , 15 nasopharyngeal, 5 , 13 , 16 sputum, 5 , 7 , 14 rectal swab, 13 oropharyngeal, 16 serum, and plasma. 14
3.2. Positive detection rates for different types of clinical specimens
A total of 8136 specimens of the suspected cases were tested for SARS‐CoV‐2, where the majority of the specimens were nasopharyngeal swabs (69.6%; 5662/8136), with only three specimens for throat swabs and serum sample (Figure 2). Regarding the sampling site (Figure 3), most of the specimens were collected from the upper respiratory tract (URT) (85.3%; 6944/8136) whereas few specimens were reported from the urinogenital tract (UGT) (1.5%; 126/8126). There was high positive rate of 91.8% (18/20; 95% CI: 79.9%‐103.7%) and 71.3% (285/403; 95% CI: 60.3%‐82.3%) for BLF clinical specimens and LRT site, respectively. While low detection rate of 0.8% (0/126; 95% CI: −0.7%‐2.4%) for urine sample and 0.8% (0/126; 95% CI:−0.7%‐2.4%) for specimens collected from UGT.
4. DISCUSSION
This review aimed at profiling the detection rate of SARS‐CoV‐2 in different clinical specimens was conducted to guide the selection of samples for clinical diagnosis of COVID‐19. In this study, SARS‐CoV‐2 was detected in specimens from 8136 specimens of patients suspected and/or confirmed with COVID‐19, with LRT specimens being the most effective in detecting the virus by 71.3%. There was no evidence of detecting virus from the urinogenital specimens.
Regarding the type of clinical specimens, BLF had a positivity rate of 91.8% followed by rectal swab (87.8%) then sputum specimens (68.1%). Nasopharyngeal swab which is commonly and widely used 1 had a positive detection rate of 45.5%. A low detection rate was observed in oropharyngeal swab (7.6%) with zero detection from urine samples. There was low (1.0%) detection of SARS‐CoV‐2 in the blood sample, but moderate in serum (33.3%) and plasma (10.0%). More importantly, the virus was detected in 32.8% of feces, suggesting that SARS‐CoV‐2 resist the acidic medium of the human gut and can be transmitted by the fecal route. In support of fecal route transmission, Wang et al 6 reported live virus from the stool specimens.
The majority of the specimens were nasopharyngeal swabs (69.6%). This is in‐line with the study which reported the clinical characteristics of patients with COVID‐19 where nasopharyngeal swab was typical. 20 Sampling of the nasopharyngeal swab is less invasive when compared to other specimens such as BLF 21 which makes it more preferable sample. Sputum which is another LRT sample was found to have a good detection rate but dry cough as the common clinical presentation for COVID‐19 patients limit its availability. 22 On the other hand, the rectal swab was the second in recording higher positive detection rate above URT specimens such as sputum. In this regard, a rectal swab can be considered as a representative of the gastrointestinal specimen.
This study was limited from the symptoms suggestive for COVID‐19 in four studies, 5 , 6 , 15 , 16 hence limiting the determination of true negative which had an impact on the denominator (total samples tested). However, in three studies 7 , 13 , 14 information was recorded if at least one specimen tested positive (confirmed cases) from the simultaneously tested specimens and the recording of positive tests was stopped when at least two consecutive qTR‐PCR tested negative in all specimens. 7 , 13 In addition, this review was limited to poor quality of the study design and small sample size for some of the investigated specimens, that is, BLF and rectal swabs. Most of the studies did not report on the stage of the disease which may affect the virus detection
5. CONCLUSION
In this study, SARS‐CoV‐2 was highly detected in LRT, with zero detection of virus from the urogenital specimens. Regarding the type of clinical specimens, BLF had the highest positive rate followed by rectal swab then sputum specimens. Nasopharyngeal swab which is commonly and widely used had a moderate detection rate. A low positive rate was recorded in oropharyngeal swab and blood samples while no virus was found in urine samples. More importantly, the virus was detected in feces, suggesting SARS‐CoV‐2 transmission by the fecal route. The use of specimens such as BLF, sputum, and the rectal swab is recommended for clinical diagnosis of COVID‐19.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHORS CONTRIBUTIONS
GMB: conceptualization, data curation, formal analysis, methodology, manuscript drafting. BJN: data curation, formal analysis, methodology, manuscript review, and editing. MVM: formal analysis and manuscript review and editing. AM: methodology and manuscript review and editing. All authors have read and approved the final version of this manuscript.
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
Authors, thank Deodatus Sabas, a Librarian at Muhimbili University of Health and Allied Sciences, Dar es Salaam (MUHAS), Tanzania for his support during the literature search. The authors acknowledge the training support received from MUHAS through Systematic Review and Meta‐analysis workshop funded by the Swedish International Development Cooperation Agency (Sida), Sweden.
Bwire GM, Majigo MV, Njiro BJ, Mawazo A. Detection profile of SARS‐CoV‐2 using RT‐PCR in different types of clinical specimens: A systematic review and meta‐analysis. J Med Virol. 2021;93:719–725. 10.1002/jmv.26349
DATA AVAILABILITY STATEMENT
All relevant data are within the manuscript and its Supporting Information files (Additional Files S1‐S3).
REFERENCES
- 1. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet [Internet]. 2020;395(10229):1054‐1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet [Internet]. 2020;395(10224):565‐574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single‐cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019‐nCov. bioRxiv [Internet]. 2020. 10.1101/2020.01.26.919985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brake SJ, Barnsley K, Lu W, McAlinden KD, Eapen MS, Sohal SS. Smoking upregulates angiotensin‐converting enzyme‐2 Receptor: a potential adhesion site for novel coronavirus SARS‐CoV‐2 (Covid‐19). J Clin Med. 2020;9(3):841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu R, Han H, Liu F, et al. Positive rate of RT‐PCR detection of SARS‐CoV‐2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta [Internet]. 2020;505:172‐175. 10.1016/j.cca.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang W, Xu Y, Gao R, et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA—J Am Med Assoc. 2020;323(18):1843‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Chen, Guiju G, Xu Y, et al. Sputum and feces after conversion of pharyngeal samples in patients with COVID‐19. Ann Intern Med. 2020;172:832‐834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. East M, Committee I, Who E, Surveillance G Laboratory testing for coronavirus disease 2019 (COVID‐19) in suspected human cases. 2019. 2020. [Google Scholar]
- 9. Wu J, Liu J, Zhao X, et al. Clinical characteristics of imported cases of coronavirus disease 2019 (COVID‐19) in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020. 10.1093/cid/ciaa199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shamseer L, Moher D, Clarke M, et al. Preferred reporting items for systematic review and meta‐analysis protocols (prisma‐p) 2015: elaboration and explanation. BMJ [Internet]. 2015;349(January):1‐25. 10.1136/bmj.g7647 [DOI] [PubMed] [Google Scholar]
- 11. Sampson M, Mcgowan J, Cogo E, Grimshaw J, Moher D, Lefebvre C. An evidence‐based practice guideline for the peer review of electronic search strategies. J Clin Epidemiol. 2009;62(9):944‐952. [DOI] [PubMed] [Google Scholar]
- 12. Tober M. PubMed, ScienceDirect, Scopus or Google Scholar—which is the best search engine for an effective literature research in laser medicine? Med Laser Appl [Internet]. 2011;26(3):139‐144. 10.1016/j.mla.2011.05.006 [DOI] [Google Scholar]
- 13. Xu Y, Li X, Zhu B, et al. Characteristics of pediatric SARS‐CoV‐2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26(April):502‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chan JF, Yuan S, Kok K, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lo IL, Lio CF, Cheong HH, Lei CI, Cheong TH, Zhong X. Evaluation of SARS‐CoV‐2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID‐19 in Macau. Int J Biol Sci. 2020;16:16‐1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Tan L, Wang X, Liu W, Lu Y. Comparison of nasopharyngeal and oropharyngeal swabs for SARS‐CoV‐2 detection in 353 patients received tests with both specimens simultaneously Xiong. Int J Infect Dis [Internet]. 2020;94:107‐109. 10.1016/j.ijid.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wallace BC, Schmid CH, Lau J, Trikalinos TA. Meta‐Analyst: software for meta‐analysis of binary, continuous, and diagnostic data. BMC Med Res Methodol. 2009;9(1):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ocan M, Akena D, Nsobya S, et al. Persistence of chloroquine resistance alleles in malaria endemic countries: a systematic review of burden and risk factors. Malar J [Internet]. 2019;18(1):1‐15. 10.1186/s12936-019-2716-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herzog R, Álvarez‐pasquin MJ, Díaz C, et al. Are healthcare workers’ intentions to vaccinate related to their knowledge, beliefs and attitudes? a systematic review. 2013. [DOI] [PMC free article] [PubMed]
- 20. Wang D, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. 2020;1–9. [DOI] [PMC free article] [PubMed]
- 21. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID‐19 patients. 2020;1751. [DOI] [PMC free article] [PubMed]
- 22. Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID‐19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis [Internet]. 2020;2019(20):1‐8. 10.1016/S1473-3099(20)30198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files (Additional Files S1‐S3).


