Abstract
Malaria is a threat to human mankind and kills about half a million people every year. On the other hand, COVID‐19 resulted in several hundred thousand deaths since December 2019 and remains without an efficient and safe treatment. The antimalarials chloroquine (CQ) and its analog, hydroxychloroquine (HCQ), have been tested for COVID‐19 treatment, and several conflicting evidence has been obtained. Therefore, the aim of this review was to summarize the evidence regarding action mechanisms of these compounds against Plasmodium and SARS‐CoV‐2 infection, together with cytometry applications. CQ and HCQ act on the renin angiotensin system, with possible implications on the cardiorespiratory system. In this context, flow and image cytometry emerge as powerful technologies to investigate the mechanism of therapeutic candidates, as well as for the identification of the immune response and prognostics of disease severity. Data from the large randomized trials support the conclusion that CQ and HCQ do not provide any clinical improvements in disease severity and progression of SARS‐CoV‐2 patients, as well as they do not present any solid evidence of increased serious side effects. These drugs are safe and effective antimalarials agents, but in SARS‐CoV‐2 patients, they need further studies in the context of clinical trials. © 2020 International Society for Advancement of Cytometry
Keywords: Plasmodium, SARS‐CoV‐2, clinical trials, renin angiotensin system, viral invasion, autophagy, side effect
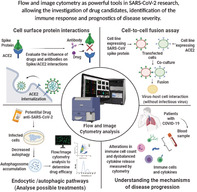
Flow and image cytometry as powerful tools in SARS‐CoV2 research allowing the investigation of drug candidates, identification of the immune response and prognostics of disease severity.
Chloroquine as an Antimalarial Drug
History of Chloroquine
The first antimalarial, quinine, was isolated from the bark of the Cinchona tree indigenous in South and Central America, an alkaloid compound categorized as quinoline methanol (for a comprehensive review, see Achan and co‐workers (1)), (2). Strategic and health‐related efforts during World War II led to the commercial production of the 4‐aminoquinoline chloroquine (CQ), in 1947 (3). CQ is among the safest and cheapest drugs of all time (4, 5). Further, chemical introduction of a hydroxyl group at position two of one of the N‐ethyl groups resulted in hydroxychloroquine (HCQ) (6, 7). Since the 1950s, CQ was used to eradicate malaria and its most devastating agent Plasmodium falciparum. However, that became officially impossible due to emerging resistance (5).
Uptake and Mode of Action of Chloroquine in Plasmodium
CQ is a weak diprotic base (pKa = 10.1; (8)), meaning it can be protonated in the acidic environments of the low pH organelles within the cell, where it accumulates as CQ2+ remaining entrapped (3, 9). CQ and its derivatives exhibit their main antimalarial activity in the asexual stages, that is, when the parasite infects the red blood cells (RBCs) and feeds on hemoglobin to generate amino acids (3, 10). The most accepted, but simplified hypothesis, is that the CQ accumulation inside of the food vacuole (FV) interferes with the detoxification of heme, the product of hemoglobin catabolism (3, 11). When Plasmodium catabolizes hemoglobin, toxic monomeric α‐hematin (ferriprotoporphyrin IX) is released as by‐product. α‐Hematin is an agent that catalyzes reactive oxygen species (ROS) production and can deposit on and damage cell membranes (10). Since the parasites lack the heme oxygenase pathway, they rely on a unique α‐hematin sequestration mechanism, to form inert hemozoin (β‐hematin), alias malaria pigment (2, 11, 12). This process is essential for Plasmodium survival, thus considered the parasites' Achilles heel (Fig. 1). The exact mechanisms by which Plasmodium manages hemozoin formation are still under discussion and point to involvement of lipids, proteins, and biocrystallization. Therefore, pinpointing the definite mode of action of CQ and derivatives in Plasmodium still is challenging. It is well established, though, that hematin crystals are formed by β‐hematin dimers, which then complex to bigger structures resulting in hemozoin crystals. Dimer formation is achieved by coordinate bonds between the prosthetic iron and the carboxylate side chain of β‐hematin. Dimers then interact via hydrogen bonding to form inert hemozoin crystals (12).
Figure 1.
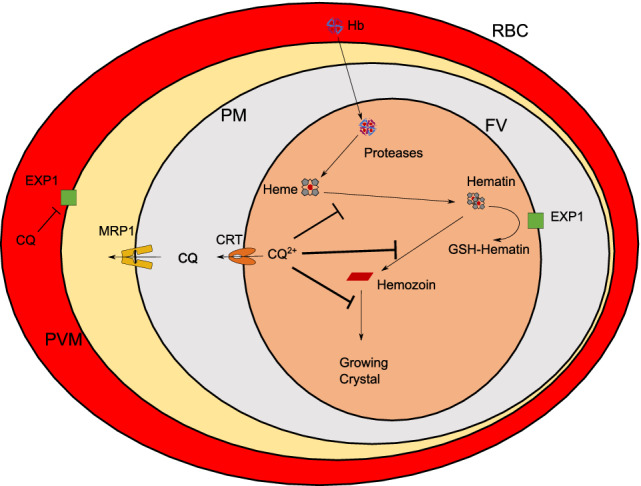
Suggested modes of action of chloroquine against Plasmodium falciparum parasites. Chloroquine can (1) prevent the formation of hemozoin by masking functional groups of hematin and the growing hemozoin crystal resulting in accumulation in the food vacuole (FV) as CQ2+ and (2) inhibit the activity of PfEXP1 which is involved in reduced glutathione (GSH)‐mediated detoxification of heme. CQ resistance in P. falciparum parasites is believed to be inferred by mutations in PfCRT and PfMRP1 transporters that promote efflux of CQ out of the FV and the parasite, respectively. RBC, red blood cell; PVM, parasitophorous vacuole membrane; PM, parasite membrane; FV, food vacuole; CQ, chloroquine; CQ2+, protonated chloroquine as present in the FV; Hb, hemoglobin; EXP1, Plasmodium falciparum exported protein 1 (a glutathione‐S‐transferase); CRT, Plasmodium falciparum CQ resistance transporter; MRP1, Plasmodium falciparum multidrug resistance‐associated protein 1. [Color figure can be viewed at wileyonlinelibrary.com]
In fact, one of the primary modes of action of different quinoline drugs, including CQ, is binding to heme and hematin, which then inhibits hemozoin crystal formation. Accumulation of the drug on the heme or hemozoin molecule masks any functional groups preventing formation or growth of the hemozoin crystal. Nonetheless, each drug exhibits specific binding modes that differ from each other. For instance, while CQ forms a Fe—N bond with a heme monomer, QN builds a Fe–O interaction (2).
In addition to hemozoin formation, the Plasmodium parasite can detoxify free heme by P. falciparum exported protein 1 (PfEXP1) enzyme catalysis. PfEXP1 is a glutathione‐S‐transferase located at the parasitophouros membrane that binds the thiol group of reduced glutathione (GSH) to the iron center of heme (12, 13). This mechanism was proposed as explanation to the substantial portion of free heme that escapes the FV, hence also biocrystallization into hemozoin. At the same time, it is known that minor concentrations in the micromolar range already effectively kill the parasite, suggesting additional mechanisms to be involved in heme detoxification (14).
In 2018, Lisewski and colleagues reported a direct inhibition of PfEXP1 by CQ at nanomolar levels, which may indicate a heme/hematin‐unrelated complementary effect of the drug (14) (Fig. 1). It is important to note that CQ and quinoline‐based derivatives are used to treat a broad range of conditions, including infectious and autoimmune diseases (15). Showing effects unrelated to heme‐detoxification underpins the pleiotropic character of the drugs.
CQ Derivatives
The emergence of CQ‐resistant strains urged the synthesis of CQ derivatives to overcome the health threat posed by malaria (5). While quinine (QN) was originally identified as a natural compound, it was soon replaced by the cost‐effective and safe 4‐aminoquinolines CQ and HCQ. QN is still used, though, as it is effective against CQ‐resistant Plasmodium strains (2). Amodiaquine (ADQ), also a 4‐aminoquinoline and QN derivative, was synthesized in the early 1940s and is still effective against CQ‐resistant strains. It is used as a partner drug in the front‐line malaria treatment artemisinin combination therapies (ACTs) (2, 4). Among the CQ derivatives, primaquine is the only 8‐aminoquinoline and was synthesized in the 1950s. Apart from the others, it potentially attacks the liver stages of P. vivax and P. ovale (5).
Mefloquine was screened after the emergence of CQ‐resistant Plasmodium strains as antimalarial drug of choice by the Walter Reed Army Institute of Research (WRAIR) in the 1980s. It belongs to the class of amino alcohols and is, like ADQ, still in use, as a partner drug for ACTs. Further, mefloquine is still effective against CQ‐resistant strains, in which an increased expression of PfEXP1 upon mefloquine treatment was observed (2, 5, 14).
Lumefantrine and Halofantrine also belong to the amino alcohol class of CQ analogs. Lumefantrine is used in combination with artemether in ACTs, while Halofantrine is only used in rare cases, due to its high toxicity (5, 16).
Piperaquine was first synthesized in the 1980s as another bis‐4‐aminoquinoline analog of CQ, linking together two quinoline molecules by their piperazine rings. This was thought to increase the positive charge and generate a bulky molecule that gets entrapped in the FV more efficiently (5).
It is noteworthy that some of the described derivatives show additional antimalarial effects in stages other than the asexual ones, despite their unknown mode of action (16). Except for halofantrine, all CQ analogs are on the World Health Organization (WHO) model list of essential medicines 2019 (4). Almost all antimalarials reviewed above were extensively studied for their potential antiviral effect and reviewed recently (17).
Resistance Mechanisms to CQ in Plasmodium falciparum
CQ resistance emerged quickly after the approval of the drug independently in several distinct regions of the world in the late 1950s. Soon, CQ‐resistant parasite strains spread continuously from Colombia, and the Mekong Subregion until sub‐Saharan Africa was entirely covered in the 1980s (18).
When talking about CQ resistance, it is important to distinguish between the in vitro determined increase in IC50 and the actual clinical outcome in vivo as the latter is also dependent on each individual's host factors, for example, metabolism or innate immunity (18). In fact, although infected with CQ‐resistant strains, individuals from a study in Mali cleared P. falciparum infection after CQ treatment, which could be shown to be age‐related (19). According to CQ's suggested modes of action, corresponding hypotheses exist to explain resistance. Among them are the import and export of CQ into the FV and the enhanced detoxification of CQ‐hematin complexes by GSH (20).
Resistance to CQ could be linked to several different markers, such as prevalent mutations in conserved genes encoding for transporter proteins (3). Parasites of the genus Plasmodium possess two main transporter types/families that exert xenobiotic trafficking in and out of the food vacuole: while P‐glycoprotein‐related transporters direct xenobiotics into the FV, members of the drug metabolite transporter family facilitate the export (Fig. 1). In Plasmodium, they are represented by PfMDR1 (Plasmodium falciparum multidrug resistance protein1), PfMDR‐2 (Plasmodium falciparum multidrug resistance protein2), and PfMRP‐1 (Plasmodium falciparum multidrug resistance‐associated protein1), and PfCRT (Plasmodium falciparum chloroquine resistance transporter), respectively (21).
PfCRT was identified in 2000 to be one of the main driving forces of CQ resistance (20). Its underlying gene pfcrt is highly polymorphic, with up to 20 codon variations known leading to altered amino acid sequences (22). The most important mutation conferring CQ resistance is K76T, despite its inability to confer resistance alone. According to the charged drug leak hypothesis, though, the K76T mutation introduces with threonine an uncharged amino acid, removing a positive charge (carried by lysine), which allows the double positively charged CQ to exit down its concentration gradient (23). Apart from its role in resistance mechanism, field studies corroborate the significance of K76T in determining clinical outcomes (3). Although not solely responsible for CQ resistance, PfCRT plays a predominant role, along with other proteins such as the aforementioned PfMDR‐1, the Na+/H+ exchanger1 (PfNHE1) and PfMRP1 (3, 24, 25).
The membrane‐associated transporter PfMDR1 imports nutrients into the FV but also transports hydrophobic compounds in the opposite direction. Mutations and copy number variations of PfMDR1 are mainly connected to MQ, HF, LMF, and QN resistance. In this context, the most abundant amino acid change is N86Y (3). Located at the parasite plasma membrane, PfMRP1 promotes efflux of CQ and QN, among other molecules such as glutathione, from the parasite. Woodland and colleagues recently showed binding of CQ and QN to PfMRP1 correlating resistance to mutations in the transporter (15). Some studies suggest a role for the putative Na+/H+ exchanger PfNHE1, which might be localized to the FV membrane. Potentially involved in maintaining the physiology of the FV it can affect QN susceptibility (3, 20).
As already described, resistance patterns and mechanisms are quite complex since different polymorphisms of several genes/proteins, especially PfCRT and PfMDR, interact. In addition, even identical haplotypes can exhibit fluctuating levels of CQ resistance, which were linked to further genes (26). To complicate the molecular interplay even more, clinical studies revealed that mutations in PfCRT also influence the expression of up to 45 unrelated genes, whose roles in the overall resistance could not be determined yet (27). This might be the parasite's response to cover the fitness loss accompanied by PfCRT mutations (20).
CQ as an Anti‐Sars‐Cov‐2 Drug
Potential Mechanism of Action in Mammalian Cells
Malaria is a threat to human mankind and kills about half a million people every year. On the other hand, COVID‐19 resulted in several hundred thousand deaths since December 2019 and remains without any efficient and safe treatment. The antimalarials CQ and its analog HCQ have been tested for COVID‐19 treatment. The first evidence that they might present anti‐SARS‐CoV‐2 effects came from an in vitro assay (6). Since then, several mechanistic studies and clinical trials have been performed around the world.
In mechanistic studies with mammalian cells infected with different viruses, CQ has presented several effects, including prevention of autophagy (28), neutralization of acidic compartments, such as lysosomes and endosomes, diminished endocytosis [by reducing phosphatidylinositol binding clathrin assembly protein (PICALM) expression] (29), and by acting as zinc ionophore facilitating extracellular zinc influx, which inhibits RNA polymerase (30). Another mechanism may involve inhibition of virion assembly in endoplasmic reticulum‐Golgi intermediate compartment (ERGIC)‐like structures (29). In fact, in the past, CQ was tested against several viruses, including the coronaviruses that cause severe acute respiratory syndrome (SARS) and Middle Eastern respiratory syndrome (MERS), and demonstrated important antiviral effects in vitro (31, 32, 33). However, until today, no therapeutic effects have been observed in humans (31).
The global pandemics caused by the coronavirus SARS‐CoV‐2 has led to an urgent search for strategies of inhibiting invasion, replication, and dissemination of the virus within the human organism. During cellular invasion, the SARS‐CoV‐2‐spike protein has been in the focus, since it enables the virus to invade cells through various mechanisms. Experimental evidence has been collected and demonstrated that SARS‐CoV‐2 invades host cells via two main receptors: CD147 (reviewed by Ulrich and Pillat (34)) and the angiotensin converting enzyme 2 (ACE2) (35). Fantini and co‐workers also identified a ganglioside‐binding domain at the N‐terminal site of the SARS‐CoV‐2 spike protein, which would bind to host cell surface gangliosides based on electrostatic and other noncovalent interactions (36). Within the compounds suggested for SARS‐CoV‐2, are CQ and HCQ (37), which had been already extensively studied for prevention and therapy of malaria. We discuss here common and different invasion methods of the two pathogens, and which tools cytometry provides for studying such mechanisms, based on an updated evidence collected on June 25, 2020 from data sources: PubMed (via MEDLINE), Scopus, bioRxiv, Preprints, ClinicalTrials.gov and World Health Organization. The work by Fantini and co‐workers (36) suggests that CQ and HCQ would have domains, which compete with SARS‐CoV‐2 for host cell ganglioside binding and thereby prevent host cell infection. Devaux and co‐workers reviewed that CQ interferes with several processes, including posttranslational modifications and biosynthesis of carbohydrates, such as sialic acid (38). Sialic acid biosynthesis involves action of quinone reductase 2 (39), which possibly might be inhibited by CQ (38, 40). Further, ACE2 glycosylation might be impaired. Due to changes in its glycosylation status, ACE2 subsequently is not anymore recognized as cellular SARS‐CoV‐2 receptor (41). Mechanisms depending on endosome alkalization were also described, in which the weak base CQ prevents acidification of the endosome. Under this condition, cleavage of the viral envelope and liberation of the viral gene into the cell cytoplasm would not occur (reviewed in reference (42)). Further, infection by SARS‐CoV‐2 induces high levels of pro‐inflammatory cytokines TNFα, IL‐1β, IL‐6, IL‐8 and IL‐17A in patients with COVID‐19 (43, 44), providing possible targets for CQ treatment. This drug affects the immune response by turning pro‐inflammatory features toward an anti‐inflammatory action by reducing the overproduction of TNFα and expression of TNFα receptors, as shown for SARS‐CoV infection of the human monocytes (9), IL‐6 detection in autopsy tissues of SARS‐CoV patients (45) and in the plasma of SARS‐CoV‐2 patients (46). Half maximal effective concentrations (EC50) of CQ and HQ against SARS‐CoV‐2, observed in studies in vitro, match possibly achievable tissue concentrations (6, 32, 41, 47, 48). Taken together, these results turned CQ and HCQ into attractive treatment options for SARS‐CoV‐2 infections. However, in the next sections, we will discuss possible toxic mechanisms of CQ and HCQ in SARS‐CoV‐2 infection and the latest clinical evidences for potential harms and benefits of these drugs.
Mechanism of Action of CQ or HCQ on the RAS: Implications for the Cardiorespiratory System
Actions of CQ and HCQ on the renin angiotensin system (RAS) may explain beneficial effects in vitro and possible undesired side effects in humans during treatment of COVID‐19. RAS is a crucial component in the regulation of several tissues and organ functionality, playing a central role in blood pressure and fluid‐electrolyte homeostasis, and also in processes of inflammation and fibrosis (49, 50). RAS is controlled by three major enzymes: (I) renin that cleaves angiotensinogen to originate angiotensin I (Ang I); (II) angiotensin‐converting enzyme (ACE) converting Ang I into angiotensin II (Ang II), whose actions are mediated by Ang II receptor type 1 (AT1R) and Ang II receptor type 2 (AT2R); (III) angiotensin‐converting enzyme 2 (ACE2), which hydrolyzes Ang II into Angiotensin‐(1‐7) (Ang 1–7) that exerts its biological function through the Mas receptor (Mas R) (49, 51). Then, the activity of ACE elevates Ang II concentration, whereas ACE2 catalyzes the cleavage of Ang II into Ang 1–7, characterizing the pressor axis (ACE/Ang II/AT1R) and the depressor axis (ACE2/Ang 1‐7/Mas R) (52), respectively. Alterations of activity and/or expression of one of these components cause an imbalance of RAS, hence, inducing cardiorespiratory problems.
ACE2, one of the main components of RAS, is also the invasion receptor for SARS‐CoV‐2, which bound to this enzyme enters the cell mainly through endocytosis, promoting loss of ACE2 function (53, 54). Low ACE2 activity increases the ACE/Ang II/AT1R pressor axis at the expense of the depressor ACE2/Ang1‐7/Mas R axis, rising the concentration of Ang II and reducing the Ang 1–7 concentration. Ang II binding to the AT1R stimulates blood pressure increase, vascular permeability, inflammatory cells into tissues and cytokine production (55). Furthermore, activation of NAD(P)H oxidase, stimulated by AT1R activation, produces ROS, mitochondrial dysfunction, and cellular injury (56, 57). These pathophysiological changes could alter lung parenchyma, favoring acute respiratory lung distress (ARDS) observed in patients with COVID‐19 (58, 59, 60). On the other hand, Ang 1–7 effects are opposite to those attributed to Ang II. Ang 1–7 induces nitric oxide production and decreases oxidative stress, which drives cardioprotective effects, improving heart function, preventing heart and vasculature remodeling, and protects against cardiac arrhythmias (61, 62, 63). In addition, Ang 1–7 downregulates leucocyte infiltration, proinflammatory cytokine production (TNFα, IL‐1β and IL‐6) and fibrosis, besides upregulating production of the anti‐inflammatory cytokine IL‐10. Taken together, Ang 1–7 attenuates inflammatory status (49, 54, 64).
Hence, ACE2 exhibits a controversial scenario in COVID‐19. While it enables viral entry, ACE2 can also protect the lung from SARS‐CoV‐2‐induced injury (64). ACE2 may reduce inflammation by decreasing of the Ang II/Ang 1–7 ratio because of its enzyme activity (50, 65). Many efforts, including HCQ studies, are being made to find a way to prevent viral invasion or replication. In this context, it is known that sialic acid biosynthesis involves action of quinone reductase 2 (39), which possibly might be inhibited by HCQ (38, 40). Further, ACE2 glycosylation might be impaired. Variations in ACE2 glycosylation status might prevent ACE2‐SARS‐CoV‐2 interaction, inhibiting viral invasion (41, 66).
However, up to now, there is no favorable scientific evidence to support the use of any dose of CQ and HCQ in patients with COVID‐19 (67, 68). In contrast, there are studies that suggest potential harm of patients infected with SARS‐CoV‐2 by HCQ treatment, which may be associated with a significant occurrence of ventricular arrhythmias (69) and increased risk of QT prolongation (69, 70). ACE2 is expressed in similar quantities at the cell surface in absence or presence of treatment with CQ; however, impaired glycosylation might reduce ACE2 activity (71). Although enzyme activity is not relevant for virus infection, it is extremely important for Ang II conversion into Ang 1–7 that is a physiological antiarrhythmic agent (63). COVID‐19 patients present downregulation of ACE2 expression in the plasmatic membrane interaction with SARS‐CoV2. We hypothesized that the loss of ACE2 functions corroborates to CQ‐impaired ACE2 glycosylation, leading to arrhythmia, increase of oxidative stress, vascular permeability and fibrosis, as well as to proinflammatory cytokine production (Fig. 2). All these consequences may be attributed to the imbalance of RAS, due to the increase ofthe pressor axis, aggravating ARDS and elevating death risk of COVID‐19 patients.
Figure 2.
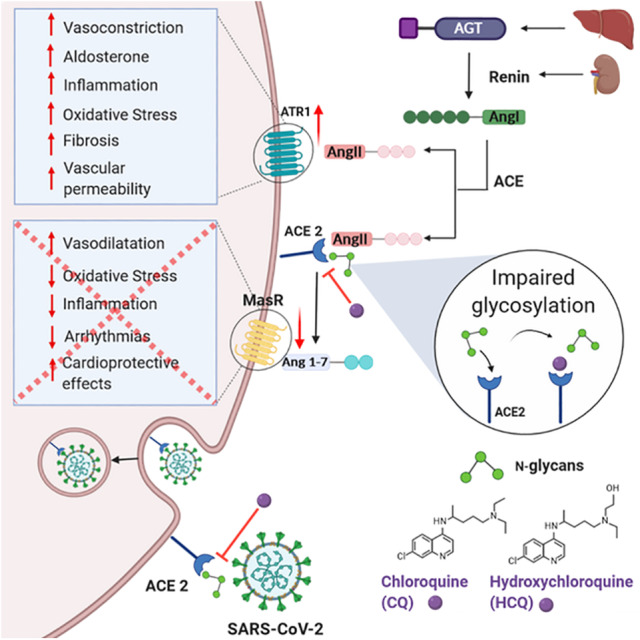
Interference of chloroquine and hydroxychloroquine in the renin‐angiotensin system (RAS). Angiotensinogen, produced in the liver, is cleaved by the renin protease produced in the kidney. Cleavage of Ang I by ACE produces the active octapeptide Ang II that acts via the AT1R, inducing vasoconstriction, production of aldosterone, increased inflammation, oxidative stress, fibrosis, and vascular permeability. Ang II levels are regulated by ACE2 that cleaves Ang II and produces Ang 1–7, a heptapeptide that acts via the Mas receptor, inducing vasodilatation and cardioprotective effects, while decreasing oxidative stress, inflammation and arrhythmias. Expression of ACE2, the SARS‐CoV‐2 cell receptor, is decreased by the endocytosis process that allows viral entry. CQ and HCQ inhibit viral entry by impairing terminal glycosylation of ACE2, which may reduce enzyme activity, elevating Ang II concentration and favoring the pressor axis. CQ, chloroquine; HCQ, hydroxychloroquine; AGT, angiotensinogen; Ang I, angiotensin I; Ang II, angiotensin II; ACE, angiotensin‐converting enzyme; ACE2, angiotensin‐converting enzyme 2; AT1R, Ang II receptor type 1; MasR, Mas receptor. [Color figure can be viewed at wileyonlinelibrary.com]
CQ or HCQ for the Prophylaxis or Treatment of COVID‐19?
CQ and HCQ pharmacokinetics shows large distribution together with slow elimination from the body, enabling toxic effects of this drug (72). HCQ has one hydroxyl group more than CQ and is associated with a lower incidence of adverse effects with chronic use (73). A randomized double‐masked clinical trial assessed, for the first time, low dosages of CQ (high dosage: 600 mg twice daily for10 days; low dosage: 450 mg twice on day 1 and once daily for 4 days) in patients with severe COVID‐19 (67). They did not observe any apparent benefit of CQ regarding lethality of enrolled patients, but they suggested that higher dosages of CQ should not be recommended for treatment of severe COVID‐19, because of safety concerns regarding QTc interval prolongation, favoring fatal arrhythmias (67).
Several HCQ trials for COVID‐19 treatment have been conducted around the world, evaluating the maximum dose of 600 mg. Some of these studies observed promising resultsin the therapy against SARS‐CoV‐2 (38, 47, 48, 74, 75), while others revealed no therapeutic effects in COVID‐19 (67, 76). A systematic review of these small studies concluded that results are conflicting and there is insufficient evidence about HCQ‐induced effects in COVID‐19 (68). Outcomes as mortality, progression of disease, symptoms, and viral load were evaluated (68).
In this context of conflicting and insufficient evidence, data from the largest trial, entitled “Randomised Evaluation of COVID‐19 Therapy” (RECOVERY), did not reveal any meaningful reduced mortality of hospitalized patients treated with HCQ (ClinicalTrials.gov Identifier: NCT04381936; (77, 78)). In this study, 25.7% of hospitalized patients treated with HCQ died compared to 23.5% of patients, who had usual care alone (endpoint of 28 days; 1,542 patients treated with HCQ vs. 3,132 control patients; hazard ratio 1.11 [95% confidence interval 0.98–1.26]). These preliminary results of the RECOVERY trial demonstrated that HQC did not evoke any beneficial effects in patients hospitalized with COVID‐19. Thus, hereupon, the RECOVERY, as well as, randomized worldwide clinical trial launched by the WHO, called “Treatments for COVID‐19: Canadian Arm of the SOLIDARITY Trial,” decided to stop enrolling participants to the HCQ and CQ arms ((79); ClinicalTrials.gov Identifier: NCT04330690). These data demonstrated the importance of large and randomized trials to provide accurate results about the efficacy and the safety of therapies.
Large randomized trials of HCQ have also been conducted to evaluated prophylaxis for COVID‐19. The study entitled “Treatment of Non‐severe Confirmed Cases of COVID‐19 and Chemoprophylaxis of Their Contacts as Prevention Strategy: a Cluster Randomized Clinical Trial (PEP CoV‐2 Study)” randomized more than 2,300 asymptomatic subjects, and no significant difference in progression of severe disease was observed between HCQ and control groups (ClinicalTrials.gov Identifier: NCT04304053; (77)). Similar results were also observed by Boulware and co‐workers with 821 asymptomatic participants receiving HCQ or placebo within 4 days after exposure ((80); ClinicalTrials.gov number, NCT04308668). The incidence of severe disease did not differ significantly between patients treated with HCQ (11.8%) and those treated with placebo (14.3%). On the other hand, the mild and medium side effects were more frequents in the HCQ group than in the placebo group (40.1% vs. 16.8%). Serious side effects were not identified ((80); ClinicalTrials.gov number, NCT04308668).
There are studies suggesting, based on preliminary evidence, that HCQ might increase the risk of adverse events in COVID‐19 patients. Some clinical trials suggest increased risk of QT prolongation (69, 70) and elevated frequency of arrhythmias in patients receiving HCQ compared to control subjects (16% vs. 10%) (69). However, large studies did not reveal any serious harm signals in patients treated or not with HCQ during SARS‐CoV‐2 infection (80, 81, 82). Finally, it is important to highlight that possible adverse effects of CQ accumulation including macular eye disease and cardiomyopathy should not be neglected (38, 83).
Cytometry Applications for Studying Molecular Interactions of Coronavirus Infection and Pathology
Cytometry applications focus at the investigation of virus–cell surface interactions as well as at determination of viral load to study efficiencies of drug and vaccine candidates. This is important for any functional study and screening of drug candidates, which might interfere with SARS‐CoV‐2 infection. For instance, culture media from Expi293F cells were collected, which secreted the SARS‐CoV‐2 recognition binding domain (RBD) fused to superfolder GFP (sfGFP), incubated with serial dilutions of Expi293F cells expressing myc‐tagged ACE2 and then analyzed by flow cytometry (84). This flow cytometry assay measuring ACE‐2 expression by detection with an anti‐myc Alexa 647‐coupled antibody versus RBD‐sfGFP allowed the screening of 30 single amino acid mutations of the RBD sequence. The T92Q substitution removing the N90 glycan increased the binding fluorescence signal, showing that this assay besides overall gross binding analysis would be able to detect small alterations in the ACE‐2 glycan surface coat as consequence of HCQ or CQ action (84).
Cell‐to‐cell fusion assays were established for studying virus–host cell interactions without the need of the infectious virus (85). For these assays, a cell line expressing the EGFP‐fused SARS‐CoV spike protein, recombinantly expressed by Vero E6 cells, and a second cell line, which expresses the virus entry receptor, for instance, ACE2, can be used. An advance of this technique was obtained by Sha et al., who developed a double fluorescence label assay, in which they transfected COS‐7 cells with a plasmid encoding the SARS‐CoV spike protein or ACE2 (86). Following selection of transfectants, recombinant SARS‐CoV spike protein expressing cells were transfected with a pDsRed2‐ER vector, while the ACE2 receptor expression was visualized by using an EGFP‐coding vector. Following co‐culture of SARS‐CoV spike protein‐red fluorescence and ACE2 green fluorescence labeled cells, cell fusion occurred and multinucleated syncytia with yellow fluorescence were detected by fluorescence microscopy. Efficiency of viral protein‐provoked cell fusion was quantified by flow cytometry analysis (86).
For studying cell surface protein interactions between the receptor‐binding domain (RBD) of the SARS‐CoV spike protein‐fused to human Fc domain (RBD‐Fc) and ACE2, image cytometry was also employed (87). The authors of this work also found that the spike protein RBD was internalized together with ACE2, and that removal of N‐linked glycosylation of the RBD did not have any effect on ACE2 internalization. Similarly, Wang and co‐worked evaluated antibody interference of spike binding to ACE2 receptor by flow cytometry (88) (Fig. 3). In order to evaluate, whether antibodies from immunized mouse bind to RBD of SARS‐CoV‐2 spike, RBD‐Fc molecules were preincubated with these immunoglobulins. After that, incubation mixtures were added to cells expressing ACE2‐GFP and Alexa Fluor 594 conjugated goat anti‐human IgG antibodies. The antibody entitled 47D11 interfered with spike/ACE2 interaction and only single‐positive cells were observed (88) (Fig. 3). The influence of other drugs, such as CQ and HCQ, in the spike/ACE2 interaction can also be evaluated by similar methods. Moreover, expression of the SARS‐CoV GFP‐fused 7a protein in HEK 293 cells led to inhibition of host cellular DNA synthesis and accumulation of cells in the G0/G1 phase, as studied by confocal scanning fluorescence and epifluorescence microscopy (89).
Figure 3.
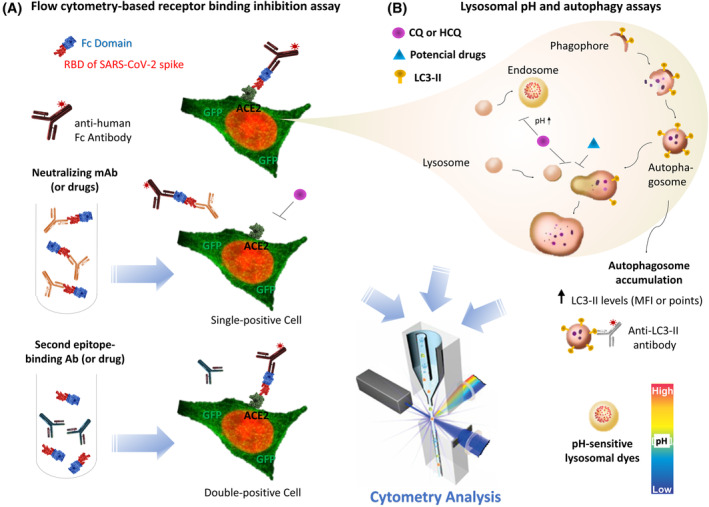
Cytometry applications for studying molecular interactions of coronavirus infection and pathology. (A) Antibody or drug interference of spike binding to ACE2 receptor can be evaluated by flow cytometry. The spike ectodomain is tagged to the human Fc domain (spike‐Fc), while ACE2 receptor expression was visualized by using an GFP‐coding vector. To study, whether drugs or monoclonal antibodies (mAbs) bind to the spike protein, these molecules can be preincubated with spike‐Fc to form complexes. After that, this mix can be incubated with cells expressing ACE2‐GFP and goat anti‐human IgG antibodies conjugated with fluorophore. If the tested antibody or drug interferes with spike/ACE2 interaction, only single‐positive cells will be observed in cytometry analysis. The influence of CQ and HCQ on spike/ACE2 interaction can also be evaluated by similar methods. (B) The influence of SARS‐CoV‐2 and possible treatments on lysosomal pH and autophagy can be investigated by cytometry analysis. CQ and HCQ, for example, may increase the endosomal and lysosomal pH, as evidenced by pH‐sensitive dyes. Moreover, CQ, HCQ and other potential anti‐SARS‐CoV‐2 drugs (corticosteroids, emtricitabine/tenofovir, interferon α‐2b, lopinavir/ritonavir and ruxolitinib) decrease autophagy by several mechanisms, including the inhibition of autophagosome fusion with lysosomes. This inhibition triggers autophagosome accumulation evidenced by high LC3‐II levels (detected as high MFI in flow cytometry and high number of LC3‐II+ points in image cytometry). Receptor binding domain, RBD; Ab, antibody; mAb, mouse antibody; MFI, median fluorescence intensity; CQ, chloroquine; HCQ, hydroxychloroquine; ACE2, angiotensin‐converting enzyme 2; LC3‐II, microtubule‐associated protein 1 light chain 3‐II. [Color figure can be viewed at wileyonlinelibrary.com]
The study of endocytic and autophagic pathways, to obtain data regarding possible treatments of SARS‐CoV‐2 by HCQ or CQ (90), is also a promising cytometry application. CQ, HCQ and other potential anti‐SARS‐CoV‐2 drugs, as corticosteroids, emtricitabine/tenofovir, interferon α‐2b, lopinavir/ritonavir and ruxolitinib, decrease autophagy by several mechanism, including the inhibition of autophagosome fusion with lysosomes (91, 92, 93, 94, 95, 96). This inhibition triggers autophagosome accumulation that can be evidenced by high LC3‐II levels (high MFI in flow cytometry and high number of LC3‐II+ points in image cytometry) (Fig. 3). In an in vitro study, Liu et al. infected Vero E6 cells with SARS CoV‐2 in the presence of increasing concentrations of CQ and CHQ and determined the effectivity of HCQ and CQ against SARS CoV‐2 infection using an immunofluorescence assay against the virus nucleoprotein (6).
Effects of CQ and derivatives on the endosomal pH possibly can also be evaluated using imaging with pH‐sensitive fluorescence dyes coupled to transferrin. Endocytosis of the complex formed by transferrin and its receptor and subsequent endocytic trafficking (97) will take the dye into the endosomes, enabling pH measurements in this organelle. In this context, endosome pH measurements, following conjugation of both rhodamine and fluorescein can be performed, by using flow or imaging cytometry, to determine the ratio of pH‐sensitive fluorescein over pH‐insensitive rhodamine fluorescence emissions (98). Another strategy of using nanoparticles for delivery of pH‐sensitive fluorophores into endosomes was described by Benjaminsen and co‐workers (99).
Cytometry applications, with focus on antibody‐enhancement (ADE) of SARS‐CoV2 infection are discussed in a recent paper of our group (100). Severe COVID‐19 is associated with a cytokine storm as well as depletion of CD8+ cells, increased numbers of neutrophils and lymphopenia as SARS‐CoV‐2 prognostics (101). Cossarizza and co‐workers (102) used flow cytometry for studying changes of lymphocyte subsets in patients with severe SARS‐CoV‐2, such as a decrease in T‐cell frequency together with an increase in the number of naive helper T‐cells and a reduction in the number of memory T‐cells, confirming previous results of Qin and co‐workers (103). ARDS as complication of SARS‐CoV‐2, possibly involving ACE‐2 dysfunction, can be also assessed by flow cytometry analysis of Treg cell phenotypes (104). CyTOF assays were used to determine signatures of the immune system in the COVID‐19 peripheral blood (105) showing immunological dysregulation with diminished T and NK cell numbers, while expression of CXCR3, CD28, and TGF‐β augmented. As shown above, alterations in the counts of immune cells and dysbalanced cytokine release, measured by cytometry, are important for the understanding of the mechanisms of disease progression. The inflammation marker NLR given as the neutrophil over lymphocyte count has gained importance for SARS‐CoV‐2 disease development (106), as shown before for cardiovascular disease prognostics (107).
Overall, cytometry is important for different fields of COVID‐19, from understanding the binding mechanism of SARS‐CoV‐2 to the definition of the immune status, vaccine development, and diagnostics for prognostics of disease severity. These parameters can be determined under conditions of HCQ or CQ treatment or treatment with another drug and provide a forecast of therapeutic efficiency. Therefore, future perspectives in cytometry applications in SARS‐CoV‐2 research will focus on multiplex immunophenotyping of infection rates and infected cell subtypes, analyses of cytokine production in single‐cells and in serum through the cytometric bead arrays and routine screening for drug and neutralizing antibody efficacies and undesired side effects, such as ADE. In this context, the involvement of innate lymphoid cells (ILCs), with ILC1, ILC2, and ILC3 profiles, in SARS‐CoV‐2 infection is unknown and requires futures cytometry experiments (107, 108). The ILC sorting from blood of infected subjects, enrichment of these populations, followed by several analyzes of function or sequencing single cells are required, since these lymphoid cells are closely involved in pulmonary disease. Moreover, the utilization of recombinant proteins of virus, such as spike RBD (Fc‐Tag), spike N‐terminal domains (NTD; Fc‐Tag), or cofactor of viral RNA polymerase (Fc‐Tag), can also be widely used in flow cytometry experiments, in other to test neutralizing‐antibodies, treatments or mechanisms for SARS‐CoV‐2 without requiring biosafety laboratories.
Taking together, data from the large randomized trials support the conclusion that CQ and HCQ do not provide any clinical improvement in disease severity and progression in SARS‐CoV‐2 patients, as well as, do not present any solid evidence of increased serious side effects (109). In this way, QH or HCQ administration in patients with SARS‐CoV‐2 is only recommended in the context of clinical trials. On the other hand, thus drugs are safe and effective antimalarial agents.
Author Contributions
Micheli Pillat: Conceptualization; writing‐original draft; writing‐review and editing. Arne Krüger: Conceptualization; writing‐original draft; writing‐review and editing. Lara Guimarães: Writing‐original draft; writing‐review and editing. Claudiana Lameu: Conceptualization; funding acquisition; supervision; writing‐original draft; writing‐review and editing. Edmarcia de Souza: Writing‐original draft; writing‐review and editing. Carsten Wrenger: Conceptualization; funding acquisition; supervision; writing‐original draft; writing‐review and editing. Henning Ulrich: Conceptualization; funding acquisition; supervision; writing‐original draft; writing‐review and editing.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Acknowledgments
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo [São Paulo Research Foundation (FAPESP) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). M.M.P. acknowledges grant support by Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS); H.U., C.L., and C.W. acknowledge grant support by FAPESP Projects No. 2018/07366‐4, 2015/19128‐2, and 2015/26722‐8, respectively]; L.M.F.G. is recipient of a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) fellowship, and A.K. a recipient of a FAPESP fellowship [2018/08820‐0]. E.E.S. is a CAPES‐Interfarma best thesis award fellow..
Literature Cited
- 1. Achan J, Talisuna AO, Erhart A, Yeka A, Tibenderana JK, Baliraine FN, Rosenthal PJ, D'Alessandro U. Quinine, an old anti‐malarial drug in a modern world: Role in the treatment of malaria. Malar J 2011;144:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gorka AP, Dios A, Roepe PD. Quinoline drug‐Heme interactions and implications for antimalarial cytostatic versus cytocidal activities. J Med Chem 2013;56:5231–5246. [DOI] [PubMed] [Google Scholar]
- 3. Müller IB, Hyde JE. Antimalarial drugs: Modes of action and mechanisms of parasite resistance. Future Microbiol 2010;5:1857–1873. [DOI] [PubMed] [Google Scholar]
- 4. WHO, List 2019 . World Health Organization model list of essential medicines: 21st List 2019. 2019. https://apps.who.int/iris/handle/10665/325771.
- 5. Kaur K, Jain M, Reddy RP, Jain R. Quinolines and structurally related heterocycles as antimalarials. Eur J Med Chem 2010;45:3245–3264. [DOI] [PubMed] [Google Scholar]
- 6. Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, Li Y, Hu Z, Zhong W, Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS‐CoV‐2 infection in vitro. Cell Discov 2020;16:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McChesney EW. Animal toxicity and pharmacokinetics of hydroxychloroquine sulfate. Am J Med 1983;75:11–18. [DOI] [PubMed] [Google Scholar]
- 8.PubChem. n.d. ‘Chloroquine’. https://pubchem.ncbi.nlm.nih.gov/compound/2719 (accessed 8 April 2020).
- 9. Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: An old drug against today's diseases? Lancet Infect Dis 2003;3:722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fitch CD. Ferriprotoporphyrin IX, phospholipids, and the antimalarial actions of Quinoline drugs. Life Sci 2004;74:1957–1972. [DOI] [PubMed] [Google Scholar]
- 11. Hempelmann E. Hemozoin biocrystallization in plasmodium falciparum and the antimalarial activity of crystallization inhibitors. Parasitol Res 2007;100:671–676. [DOI] [PubMed] [Google Scholar]
- 12. Kumar S, Guha M, Choubey V, Maity P, Bandyopadhyay U. Antimalarial drugs inhibiting Hemozoin (beta‐hematin) formation: A mechanistic update. Life Sci 2007;80:813–828. [DOI] [PubMed] [Google Scholar]
- 13. Lisewski AM, Quiros JP, Ng CL, Adikesavan AK, Miura K, Putluri N, Eastman RT. Supergenomic network compression and the discovery of EXP1 as a glutathione transferase inhibited by artesunate. Cell 2014;158:916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lisewski AM, Quiros JP, Mittal M, Putluri N, Sreekumar A, Haeggström JZ, Lichtarge O. Potential role of Plasmodium falciparum exported protein 1 in the chloroquine mode of action. Int J Parasitol Drugs Drug Resist 2018;8:31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woodland JG, Hunter R, Smith PJ, Egan TJ. Chemical proteomics and super‐resolution imaging reveal that chloroquine interacts with Plasmodium falciparum multidrug resistance‐associated protein and lipids. ACS Chem Biol 2018;13:2939–2948. [DOI] [PubMed] [Google Scholar]
- 16. Delves M, Plouffe D, Scheurer C, Meister S, Wittlin S, Winzeler EA, Sinden RE, Leroy D. The activities of current antimalarial drugs on the life cycle stages of plasmodium: A comparative study with human and rodent parasites. PLoS Med 2012;9:e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. D'Alessandro S, Scaccabarozzi D, Signorini L, Perego F, Ilboudo DP, Ferrante P, Delbue S. The use of antimalarial drugs against viral infection. Microorganisms 2020;8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wellems TE, Plowe CV. Chloroquine‐resistant malaria. J Infect Dis 2001;184:770–776. [DOI] [PubMed] [Google Scholar]
- 19. Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, Coulibaly D. A molecular marker for chloroquine‐resistant falciparum malaria. N Engl J Med 2001;344:257–263. [DOI] [PubMed] [Google Scholar]
- 20. Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in cloroquine resistance. Mol Cell 2000;6:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ibraheem ZO, Majid RA, Noor SM, Sedik HM, Basir R. Role of different Pfcrt and Pfmdr‐1 mutations in conferring resistance to Antimalaria drugs in Plasmodium falciparum . Malaria Res Treatment 2014;2014:950424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cooper RA, Hartwig CL, Ferdig MT. Pfcrt is more than the Plasmodium falciparum Chloroquine resistance gene: A functional and evolutionary perspective. Acta Trop 2005;94:170–180. [DOI] [PubMed] [Google Scholar]
- 23. Martin RE, Marchetti RV, Cowan AR, Howitt SM, Bröer S, Kirk K. Chloroquine transport via the malaria parasite's chloroquine resistance transporter. Science 2009;325:1680–1682. [DOI] [PubMed] [Google Scholar]
- 24. Duraisingh MT, Cowman AF. Contribution of the Pfmdr1 gene to antimalarial drug‐resistance. Acta Trop 2005;94:181–190. [DOI] [PubMed] [Google Scholar]
- 25. Henry M, Briolant S, Zettor S, Pelleau S, Baragatti M, Baret E, Mosnier J, Amalvict R, Fusai T, Rogier C, et al. Plasmodium falciparum Na+/H+ exchanger 1 transporter is involved in reduced susceptibility to quinine. Antimicrob Agents Chemother 2009;53:1926–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen N, Russell B, Fowler E, Peters J, Cheng Q. Levels of chloroquine resistance in Plasmodium falciparum are determined by loci other than Pfcrt and Pfmdr1. J Infect Dis 2002;185:405–407. [DOI] [PubMed] [Google Scholar]
- 27. Jiang H, Patel JJ, Yi M, Mu J, Ding J, Stephens R, Cooper RA, Ferdig MT, Su X. Genome‐wide compensatory changes accompany drug‐ selected mutations in the Plasmodium falciparum Crt gene. PLoS One 2008;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17(9):528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu TY, Frieman M, Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID‐19. Nat Nanotechnol 2020;15:247–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Velthuis AJW, van den Worm SHE, Sims AC, Baric RS, Snijder EJ, van Hemert MJ. Zn(2+) inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog 2010;6(11):e1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Touret F, de Lamballerie X. Of chloroquine and COVID‐19. Antiviral Res 2020;177:104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keyaerts E, Vijgen L, Maes P, Neyts J, Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Biophys Res Commun 2004;323:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dyall J, Coleman CM, Hart BJ, Venkataraman T, Holbrook MR, Kindrachuk J, Johnson RF, Olinger GG Jr, Jahrling PB, Laidlaw M, et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother 2014;58:4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ulrich H, Pillat MM. CD147 as a target for COVID‐19 treatment: Suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep 2020;16:434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS‐CoV‐2 by full‐length human ACE2. Science 2020;367:1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fantini J, Chahinian H, Yahi N. Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS‐CoV‐2: What molecular dynamics studies of virus‐host interactions reveal. Int J Antimicrob Agents 2020;56(2):106020. 10.1016/j.ijantimicag.2020.106020. [DOI] [PubMed] [Google Scholar]
- 37. Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID‐19. Int J Antimicrob Agents 2020;55:105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID‐19? Int J Antimicrob Agents 2020;55:105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kwiek JJ, Haystead TA, Rudolph J. Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines. Biochemistry 2004;43:4538–4547. [DOI] [PubMed] [Google Scholar]
- 40. Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis 2006;6:67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, Ksiazek TG, Seidah NG, Nichol ST. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology J 2005;2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Al‐Bari MAA. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol Res Perspect 2017;5:e00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schett G, Sticherling M, Neurath MF. COVID‐19: Risk for cytokine targeting in chronic inflammatory diseases. Nat Rev Immunol 2020;20:271–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vabret N, Britton GT, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD. Immunology of COVID‐19: Current state of the science. Immunity 2020;52:910–941. 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He L, Ding Y, Zhang Q, Che X, He Y, Shen H, Wang H, Li Z, Zhao L, Geng J, et al. Expression of elevated levels of pro‐inflammatory cytokines in SARS‐CoV‐infected ACE2+ cells in SARS patients: Relation to the acute lung injury and pathogenesis of SARS. J Pathol 2006;210:288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu B, Li C, Chen P, Zhou N, Wang L, Li J, Jiang H, Wang DW. Hydroxychloroquine application is associated with a decreased mortality in critically ill patients with COVID‐19. medRxiv 2020. 10.1101/2020.04.27.20073379. [DOI] [Google Scholar]
- 47. Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res 2020;30:269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, et al. In vitro antiviral activity and projection of optimized dosing Design of Hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Clin Infect Dis 2020;1:8. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simões E, Silva AC, Silveira KD, Ferreira AJ, Teixeira MM. ACE2, angiotensin‐(1‐7) and mas receptor axis in inflammation and fibrosis. Br J Pharmacol 2020;169:477–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Khan A, Benthin C, Zeno B, Albertson TE, Boyd J, Christie JD, Hall R, Poirier G, Ronco JJ, Tidswell M, et al. A pilot clinical trial of recombinant human angiotensin‐converting enzyme 2 in acute respiratory distress syndrome. Crit Care 2017;21:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jia H. Pulmonary angiotensin‐converting enzyme 2 (ACE2) and inflammatory lung disease. Shock 2016;46:239–248. [DOI] [PubMed] [Google Scholar]
- 52. Gironacci MM, Vicario A, Cerezo G, Silva MG. The depressor axis of the renin‐angiotensin system and brain disorders: A translational approach. Clin Sci 2018;132:1021–1038. [DOI] [PubMed] [Google Scholar]
- 53. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun 2020;11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, Raizada MK, Grant MB, Oudit GY. Angiotensin‐converting enzyme 2: SARS‐CoV‐2 receptor and regulator of the renin‐angiotensin system: Celebrating the 20th anniversary of the discovery of ACE2. Circ Res 2020;126:1456–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Benigni A, Cassis P, Remuzzi G. Angiotensin II revisited: New roles in inflammation, immunology and aging. EMBO Mol Med 2010;2:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. De Gasparo M. Angiotensin II and nitric oxide interaction. Heart Fail Rev 2002;7:347–358. [DOI] [PubMed] [Google Scholar]
- 57. de Cavanagh EMV, Inserra F, Ferder M, Ferder L. From mitochondria to disease: Role of the renin‐angiotensin system. Am J Nephrol 2007;27:545–553. [DOI] [PubMed] [Google Scholar]
- 58. Rodriguez‐Morales AJ, Cardona‐Ospina JA, Ocampo EG, Gutiérrez‐Ocampo E, Villamizar‐Peña R, Holguin‐Rivera Y, Escalera‐Antezana JP, Alvarado‐Arnez LE, Bonilla‐Aldana DK, Franco‐Paredes C, et al. Latin American network of coronavirus disease 2019‐COVID‐19 research (LANCOVID‐19) clinical, laboratory and imaging features of COVID‐19: A systematic review and meta‐analysis. Travel Med Infect Di 2020;34:101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C. Radiological findings from 81 patients with COVID‐19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect Dis 2020;20:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ranieri VM, Rubenfeld GD, Thompson BT. Acute respiratory distress syndrome: The Berlin definition. JAMA ‐ J Am Med Assoc 2012;307:2526–2533. [DOI] [PubMed] [Google Scholar]
- 61. Li Y, Wu J, He Q, Shou Z, Zhang P, Pen W, Zhu Y, Chen J. Angiotensin (1‐7) prevent heart dysfunction and left ventricular remodeling caused by renal dysfunction in 5/6 nephrectomy mice. Hypertens Res 2009;32:369–374. [DOI] [PubMed] [Google Scholar]
- 62. McKinney CA, Fattah C, Loughrey CM, Milligan G, Nicklin SA. Angiotensin‐(1‐7) and angiotensin‐(1‐9): Function in cardiac and vascular remodelling. Clin Sci 2014;126:815–827. [DOI] [PubMed] [Google Scholar]
- 63. Joviano‐Santos JV, Santos‐Miranda A, Joca HC, Cruz JS, Ferreira AJ. New insights into the elucidation of angiotensin‐(1–7) in vivo antiarrhythmic effects and its related cellular mechanisms. Exp Physiol 2016;101:1506–1516. [DOI] [PubMed] [Google Scholar]
- 64. South AM, Tomlinson L, Edmonston D, Hiremath S, Sparks MA. Controversies of renin‐angiotensin system inhibition during the COVID‐19 pandemic. Nat Rev Nephrol 2020;16:305–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hemnes AR, Rathinasabapathy A, Austin EA, Brittain EL, Carrier EJ, Chen X, Fessel JP, Fike CD, Fong P, Fortune N, et al. A potential therapeutic role for angiotensin‐converting enzyme 2 in human pulmonary arterial hypertension. Eur Respir J 2018;51:1702638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, Ksiazek TG, Seidah NG, Nichol ST. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J 2005;22:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Borba MGS, Val FFA, Sampaio VS, Alexandre MAA, Melo GC, Brito M, Mourão MPG, Brito‐Sousa JD, Baía‐da‐Silva D, Guerra MVF, et al. Effect of high vs low doses of Chloroquine Diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection: A randomized clinical trial. JAMA Netw Open 2020;3(4):e208857. [DOI] [PubMed] [Google Scholar]
- 68. Hernandez AV, Roman YM, Pasupuleti V, Barboza JJ, White M. Hydroxychloroquine or Chloroquine for treatment or prophylaxis of COVID‐19: A living systematic review. Ann Intern Med 2020. 10.7326/M20-2496. [DOI] [PubMed] [Google Scholar]
- 69. Rosenberg ES, Dufort EM, Udo T, Wilberschied LA, Kumar J, Tesoriero J, Weinberg P, Kirkwood J, Muse A, DeHovitz J, et al. Association of treatment with hydroxychloroquine or azithromycin with in‐hospital mortality in patients with COVID‐19 in New York state. JAMA 2020;323:2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mahevas M, Tran V‐T, Roumier M, Chabrol A, Paule R, Guillaud C, Gallien S, Lepeule R, Szwebel T‐A, Lescure X, et al. No evidence of clinical efficacy of hydroxychloroquine in patients hospitalized for COVID‐19 infection with oxygen requirement: results of a study using routinely collected data to emulate a target trial. BMJ 2020;369:m1844. 10.1101/2020.04.10.20060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Warner FJ, Smith AI, Hooper NM, Turner AJ. Angiotensin‐converting enzyme‐2: A molecular and cellular perspective. Cell Mol Life Sci 2004;61:2704–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smit C, Peeters MYM, van den Anker JN, Knibbe CAJ. Chloroquine for SARS‐CoV‐2: Implications of its unique pharmacokinetic and safety properties. Clin Pharmacokinet 2020;18:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ben‐Zvi I, Kivity S, Langevitz P, Shoenfeld Y. Hydroxychloroquine: From malaria to autoimmunity. Clin Rev Allergy Immunol 2012;42:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID‐19 associated pneumonia in clinical studies. Biosci Trends 2020;14:72–73. [DOI] [PubMed] [Google Scholar]
- 75. Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B. Hydroxychloroquine and azithromycin as a treatment of COVID‐19: Results of an open‐label non‐randomized clinical trial. Int J Antimicrob Agents 2020;56(1):105949. 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mahévas M, Tran VT, Roumier M, Chabrol A, Paule R, Guillaud C, Gallien S, Lepeule R, Szwebel T‐A, Lescure X, et al. No evidence of clinical efficacy of hydroxychloroquine in patients hospitalized for COVID‐19 infection with oxygen requirement: Results of a study using routinely collected data to emulate a target trial. MedRxiv preprint 2020. 10.1101/2020.04.10.20060699. [DOI] [Google Scholar]
- 77. Kupferschmidt K. Three big studies dim hopes that hydroxychloroquine can treat or prevent COVID‐19. Science 2020. 10.1126/science.abd2496. [DOI] [PubMed] [Google Scholar]
- 78.Statement from the Chief Investigators of the Randomised Evaluation of COVid‐19 thERapY (RECOVERY) Trial on hydroxychloroquine, 5 June 2020. No clinical benefit from use of hydroxychloroquine in hospitalised patients with COVID‐19. 2020. Available online at: https://www.recoverytrial.net/files/hcq-recovery-statement-050620-final-002.pdf
- 79.NIH halts clinical trial of hydroxychloroquine. Study shows treatment does no harm, but provides no benefit. 2020. Available online at: https://www.nih.gov/news-events/news-releases/nih-halts-clinical-trial-hydroxychloroquine
- 80. Boulware DR, Pullen MF, Bangdiwala AS, Pastick KA, Lofgren SM, Okafor EC, Skipper CP, Nascene AA, Nicol MR, Abassi M, et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid‐19. N Engl J Med 2020;383(6):517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Horby P, Landray M. Edinburgh, UK, 2020. Available at: https://www.recoverytrial.net/files/recovery-dmc-letter-24-may-2020.pdf
- 82. Joseph A. WHO Resumes Hydroxychloroquine Study for Covid‐19, after Reviewing Safety Concerns. Available at: https://www.statnews.com/2020/06/03/who-resuming-hydroxychloroquine-study-for-covid-19/.
- 83. Ratliff NB, Estes ML, Myles JL, Shirey EK, McMahon JT. Diagnosis of chloroquine cardiomyopathy by endomyocardial biopsy. N Engl J Med 1987;316:191–193. [DOI] [PubMed] [Google Scholar]
- 84. Procko E. The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus. BioRxiv 2020. 10.1101/2020.03.16.994236. [DOI] [Google Scholar]
- 85. Petit CM, Chouljenko VN, Iyer A, Colgrove R, Farzan M, Knipe DM, Kousoulas KG. Palmitoylation of the cysteine‐rich endodomain of the SARS‐coronavirus spike glycoprotein is important for spike‐mediated cell fusion. Virology 2007;360:264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sha Y, Wu Y, Cao Z, Xu X, Wu W, Jiang D, Mao X, Liu H, Zhu Y, Gong R, et al. A convenient cell fusion assay for the study of SARS‐CoV entry and inhibition. IUBMB Life 2006;58:480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang S, Guo F, Liu K, Wang H, Rao S, Yang P, Jiang C. Endocytosis of the receptor‐binding domain of SARS‐CoV spike protein together with virus receptor ACE2. Virus Res 2008;136:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang C, Li W, Drabek D, Okba NMA, Haperen RV, Osterhaus ADME, Kuppeveld FJMV, Haagmans BL, Grosveld F, Bosch B‐J. A human monoclonal antibody blocking SARS‐CoV‐2 infection. Nat Commun 2020;11(1):2251. 10.1101/2020.03.11.987958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yuan X, Wu J, Shan Y, Yao Z, Dong B, Chen B, Zhao Z, Wang S, Chen J, Cong Y. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology 2006;346:74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yang N, Shen HM. Targeting the Endocytic pathway and autophagy process as a novel therapeutic strategy in COVID‐19. Int J Biol Sci 2020;16:1724–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome‐lysosome fusion. Autophagy 2018;14:1435–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kyrmizi I, Gresnigt MS, Akoumianaki T, Samonis G, Sidiropoulos P, Boumpas D, Netea MG, van de Veerdonk FL, Kontoyiannis DP, Chamilos G. Corticosteroids block autophagy protein recruitment in Aspergillus fumigatus phagosomes via targeting dectin‐1/Syk kinase signaling. J Immunol 2013;191:1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tripathi A, Thangaraj A, Chivero ET, Periyasamy P, Callen S, Burkovetskaya ME, Guo ML, Buch S. Antiretroviral‐mediated microglial activation involves dysregulated autophagy and lysosomal dysfunction. Cell 2019;8:1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhao J, Wang ML, Li Z, Gao DM, Cai Y, Chang J, Wang SP. Interferon‐alpha‐2b induces autophagy in hepatocellular carcinoma cells through Beclin1 pathway. Cancer Biol Med 2014;11:64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zha BS, Wan X, Zhang X, Zha W, Zhou J, Wabitsch M, Wang G, Lyall V, Hylemon PB, Zhou H. HIV protease inhibitors disrupt lipid metabolism by activating endoplasmic reticulum stress and inhibiting autophagy activity in adipocytes. PLoS One 2013;8:e59514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kusoglu A, Bagca BG, Ozates Ay NP, Saydam G, Avci CB. Ruxolitinib regulates the autophagy machinery in multiple myeloma cells. Anticancer Agents Med Chem 2020. Preprint. 10.2174/1871520620666200218105159. [DOI] [PubMed] [Google Scholar]
- 97. Mayle KM, Le AM, Kamei DT. The intracellular trafficking pathway of transferrin. Biochim Biophys Acta 2012;1820:264–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dunn KW, Park J, Semrad CE, Gelman DL, Shevell T, McGraw TE. Regulation of endocytic trafficking and acidification are independent of the cystic fibrosis transmembrane regulator. J Biol Chem 1994;269:5336–5345. [PubMed] [Google Scholar]
- 99. Benjaminsen RV, Sun H, Henriksen JR, Christensen NM, Almdal K, Andresen TL. Evaluating nanoparticle sensor design for intracellular pH measurements. ACS Nano 2011;5:5864–5873. [DOI] [PubMed] [Google Scholar]
- 100. Ulrich H, Pillat MM, Tárnok A. Dengue Fever, COVID‐19 (SARS‐CoV‐2) and antibody‐dependent Enhancement (ADE) – A perspective. Cytometry A 2020;97:662–667. 10.1002/cyto.a.24047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kuppalli K, Rasmussen AL. A glimpse into the eye of the COVID‐19 cytokine storm. EBioMedicine 2020;55:102789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cossarizza A, De Biasi S, Guaraldi G, Girardis M, Mussini C. SARS‐CoV‐2, the virus that causes COVID‐19: Cytometry and the new challenge for Global Health. Cytometry Part A 2020;97A:340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, et al. Dysregulation of immune response in patients with COVID‐19 in Wuhan, China. Clin Infect Dis 2020;71(15):762–768. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Halter S, Aimade L, Barbié M, Brisson H, Rouby JJ, Langeron O, Klatzmann D, Rosenzwajg M, Monsel A. T regulatory cells activation and distribution are modified in critically ill patients with acute respiratory distress syndrome: A prospective single‐Centre observational study. Anaesth Crit Care Pain Med 2020;39:35–44. [DOI] [PubMed] [Google Scholar]
- 105. Wang W, Su B, Pang L, Qiao L, Feng Y, Ouyang Y, Guo X, Shi H, Wei F, Su X, et al. High‐dimensional immune profiling by mass cytometry revealed immunosuppression and dysfunction of immunity in COVID‐19 patients. Cell Mol Immunol 2020;17:650–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu Y, Du X, Chen J, Jin Y, Peng L, Wang HHX, Luo M, Chen L, Zhao Y. Neutrophil‐to‐lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID‐19. J Infect 2020;81:6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bhat T, Teli S, Rijal J, Bhat H, Raza M, Khoueiry G, Meghani M, Akhtar M, Costantino T. Neutrophil to lymphocyte ratio and cardiovascular diseases: A review. Expert Rev Cardiovasc Ther 2013;11:55–59. [DOI] [PubMed] [Google Scholar]
- 108. Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, et al. Immunology of COVID‐19: Current state of the science. Immunity 2020;53:910–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.WHO discontinues hydroxychloroquine and lopinavir/ritonavir treatment arms for COVID‐19 4 July 2020. Available at: https://www.who.int/news-room/detail/04-07-2020-who-discontinues-hydroxychloroquine-and-lopinavir-ritonavir-treatment-arms-for-covid-19


