Abstract
The coronavirus disease 2019 (COVID‐19) pandemic has created a precipitous increase in the need for molecular diagnostics. Unfortunately, access to RNA extraction reagents can represent a bottleneck for quantitative real‐time reverse transcriptase‐polymerase chain reaction (qRT‐PCR)‐based methodologies, stemming from both extraordinary supply‐chain stresses and the global reach of the virus into resource‐limited settings. To provide flexible diagnostic options for such environments, we report here an “unextracted modification” for qRT‐PCR using the Centers for Disease Control's (CDC's) widely utilized primers/probe sets for severe acute respiratory syndrome coronavirus 2 (N1/N2/N3 targeting viral nucleocapsid and RP‐control targeting human RNase P). This approach replaces RNA extraction/purification with a heat‐inactivation step of viral transport media (VTM), followed by direct inoculation—with or without VTM spin concentration—into PCR master mixes. Using derivatives of care from our clinical workflow, we compared traditional and unextracted CDC methodologies. Although some decrease in analytic sensitivity was evident (by higher C t values) without extraction, in particular for the N2 primer/probe‐set, we observed high categorical positive agreement between extracted and unextracted results for N1 (unconcentrated VTM‐38/40; concentrated VTM‐39/41), N3 (unconcentrated VTM‐38/40; concentrated VTM‐41/41), and RP (unconcentrated and concentrated VTM‐81/81). The negative categorical agreement for N1/N2/N3 was likewise high. Overall, these results suggest that laboratories could adapt and validate unextracted qRT‐PCR protocols as a contingency to overcome supply limitations, with minimal impact on categorical results.
Keywords: COVID‐19, diagnostics, RNA, RT‐PCR, SARS‐CoV‐2, unextracted
1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) coronavirus, has created an unprecedented upsurge in the need for molecular diagnostic testing. 1 Many stakeholders—including government organizations, commercial entities, and individual diagnostic laboratories—have been forced to develop molecular assays rapidly and en masse. 2 This reality has created analytic, regulatory, and logistical issues, including the need to balance rigorous methodologies with real‐time dissemination of testing. 3 One notable challenge involves the ability of laboratories to obtain the necessary reagents, either premanufactured diagnostic kits or individual components to formulate their own assays. Nationally and internationally, diagnostic demand has outstripped supply and contributed to variable availability of local testing. 4
A number of emergent COVID‐19 diagnostics are based upon real‐time reverse transcriptase‐polymerase chain reaction (qRT‐PCR). 5 While potentially quantitative, these assays have been deployed predominantly in a qualitative/categorical manner, with a cycle‐threshold value (C t) indicating the presence of the virus. 6 qRT‐PCR allows laboratories to flexibly customize primers and probes with other reagents and instruments, validating their local combination for acceptable clinical performance and to meet regulatory requirements. 7 One common approach for SARS‐CoV‐2 has involved TaqMan‐based assays, with nucleocapsid‐targeting oligonucleotides developed by the Centers for Disease Control (CDC). 6 These primers/probes have been implemented with various reagents/instruments, including a number of combinations with FDA Emergency Use Authorization in the United States. 8 Initially promulgated with three locus‐targets (N1, N2, N3), this “CDC‐approach” has also been adapted by many labs to exclude N3, due to potential cross‐reactivity with other betacoronaviruses. 3
To implement this (or any) qRT‐PCR assay for SARS‐CoV‐2, viral RNA is first typically extracted from respiratory samples, often swab‐derived specimens in liquid viral transport media (VTM). 9 This extraction step serves two overarching purposes: (a) lysing viral particles to free genomic RNA and (b) removing potential PCR inhibitors. Various commercial products are available for this purpose, including automated and manual methods, although these reagents have experienced a massive surge in demand with widespread shortfalls in recent availability. 4 A potential solution to this bottleneck could direct inoculation of respiratory specimens into the qRT‐PCR master mix, as has been recently proposed for COVID‐19. 10 In theory, heat denaturation (before reverse transcription) could provide the necessary degree of virion denaturation and RNA access, while thoroughly inactivating the viable virus. 11 While this strategy does not attempt to remove inhibitors, similar methods have proven successful for other pathogens and specimen types. 12 , 13 , 14 In this context, we sought to evaluate this approach for the CDC primers/probes for SARS‐CoV‐2—both with and without additional viral preconcentration—to provide a contingency against supply‐chain insecurities. We describe these results here, which may prove valuable to laboratories with resource limitations, either at baseline or due to the pandemic.
2. METHODS
For routine clinical care, our CLIA‐accredited laboratory performs COVID‐19 testing using a variation of the CDC assay. Swab specimens are submitted in VTM: Hanks balanced salt solution (HBSS), pH 6.8, 11 μg/mL phenol red, without Ca/Mg; 0.5% gelatin; 100 μg/mL gentamicin; 200 U/mL penicillin; 2 μg/mL amphotericin. RNA is extracted on the Biomerieux easyMAG NUCLISENS platform (200 μL VTM input, 60 μL H2O eluate). Five microlitres extract is added to each of three master mixes (15 μL each), as described in the EUA product insert 15 : N1, N2, and RP (an internal control targeting human RNase P). Primers/probes are provided by Integrated DNA Technologies, together with TaqPath 1‐Step qRT‐PCR enzyme mixture (Life Technologies). Note that while the N3 primer/probe‐set is excluded from our routine clinical testing, it was incorporated into the study activities here. Thermocycling was performed on a 7500 Fast DX real‐time platform (Applied Biosystems), with FAM‐signal normalized against ROX: 25°C for 2 minutes, 50°C for 15 minutes, 95°C for 2 minutes, (95°C for 3 seconds, 55°C for 30 seconds) × 45. For all analyses conducted here, positivity was considered individually for each primer/probe‐set (0.1 ΔR n within 45 cycles). This differs from our clinical‐use protocols for the CDC assay, in which only a composite detection is reported, requiring positivity of all tested targets at C t < 40 (extremely rare diagnostic specimens with repeatable single‐component detections, but not at EUA‐defined standards, are considered indeterminate, none of which arose during the performance of this study).
For the concentration of VTM, 500 μL specimen was centrifuged to approximately 20 μL using an Amicon Ultra 0.5‐mL filter and a standard Thermo benchtop microcentrifuge (50 000 Da MWCO, 30 minutes at 14 000g) and reconstituted with 30 μL H2O. For all experiments (concentrated and unconcentrated), VTM specimens were first heat‐inactivated by placing the specimen in a preheated heat‐block (95°C, 10 minutes) before further manipulation within a biological safety cabinet. Specimens/data were permanently deidentified of patient information and blinded to the performing technologist, as approved by the Vanderbilt Human Subjects Protection Program. In evaluating percent agreement between different methodologies, proportional confidence intervals were calculated by Wilson's method.
Two sets of experiments were conducted to evaluate the performance of CDC's primers/probes for detecting SARS‐CoV‐2 when omitting a dedicated extraction step. Fresh extracts from nasopharyngeal swab specimens—residuals from clinical testing—were analyzed, paired with two variations of unextracted VTM: (a) 5 μL of VTM added directly to each master mix; and (b) 5 μL of spin‐concentrated VTM added to each master mix. We conducted both concentrated and unconcentrated experiments to determine the impact of VTM concentration on analytic sensitivity (with a potentially greater concentration of both virus and PCR inhibitors). In both experiments, extracted VTM comparators were not preconcentrated, ensuring a direct comparison with clinical‐use protocols. To assess positive agreement for each experiment, we utilized all specimens from our clinical workflow with a “detected” result over a ∼24 hour period. At the time, COVID‐19 testing at our institution covered exclusively symptomatic individuals from both outpatient and hospital/emergency department settings (ie, no asymptomatic screens or tests‐of‐cure). To assess negative agreement, we employed a commensurate number of “not detected” specimens from clinical testing, randomly selected from the same times. Different specimen cohorts (positive and negative) were employed for experiments with unconcentrated and concentrated VTM.
3. RESULTS
For unextracted/unconcentrated VTM, the categorical agreement with extracted VTM (detected/undetected) is summarized in Table 1A for each primer/probe‐set. The positive agreement was high for N1 (95%), N3 (95%), and RP (100%), although increased pairwise C t values were still observed, unextracted versus extracted (ΔC t = 5.2 ± 4.0, 5.2 ± 3.5, and 6.9 ± 3.3, respectively). C t discrepancies were even more pronounced for N2 (>15 ΔC t for 37/40 specimens), with a low resultant positive agreement (33%, 13/40). False positives in the unextracted/unconcentrated samples occurred at a low frequency (2/41 for N1, 0/41 for N2, and 1/41 for N3). Figure 1A summarizes, for each primer/probe, the range of C t values for extracted specimens that were also detected when unextracted, along with all discordant examples. Not surprisingly, discordant results for N1/N3 reflect specimens with higher extracted C t values, while discordant results for N2 occurred broadly. On a per‐specimen basis, 40 of 41 positive extracted specimens were detected by at least one nucleocapsid primer/probe when unextracted/unconcentrated. The observed negative agreement was likewise ≥95% for each target.
Table 1.
Categorical and C t value agreement between extracted/unextracted specimens
| Primer/probe | Positive percent agreement | ΔC t | Negative percent agreement | |
|---|---|---|---|---|
| (A) Extracted versus unextracted/unconcentrated | N1 |
95.0% (38/40) 95% CI: 83.5%‐98.6% |
5.2 ± 4.0 |
95.1% (39/41) 95% CI: 83.4%‐98.7% |
| N2 |
32.5% (13/40) 95% CI: 20.0%‐48.0% |
>15 C t—37/40 specimensa |
100% (41/41) 95% CI: 91.4%‐100% |
|
| N3 |
95.0% (38/40) 95% CI: 83.5%‐98.6% |
5.2 ± 3.5 |
97.6% (40/41) 95% CI: 87.4%‐99.6% |
|
| RP |
100% (81/81) 95% CI: 95.5%‐100% |
6.9 ± 3.3 | NA* | |
| (B) Extracted versus unextracted/concentrated | N1 |
95.1% (39/41) 95% CI: 83.4%‐98.7% |
2.6 ± 2.5 |
97.5% (39/40) 95% CI: 87.1%‐99.6% |
| N2 |
85.4% (35/41) 95% CI: 85.4%‐93.1% |
8.7 ± 2.5 |
100% (40/40) 95% CI: 91.2%‐100% |
|
| N3 |
100% (41/41) 95% CI: 91.4%‐100% |
2.4 ± 1.9 |
95.0% (38/40) 95% CI: 83.5%‐98.6% |
|
| RP |
100% (81/81) 95% CI: 95.5%‐100% |
4.2 ± 2.7 | NA* |
Note: Summarized here are the observed categorical positive and negative agreement (detected/nondetected) for each CDC primer/probe‐set, between extracted and unextracted specimens (percentage agreement with 95% CI). Data are summarized for comparisons between (a) extracted VTM versus unextracted/unconcentrated VTM and (b) extracted VTM versus unextracted/concentrated VTM. For each evaluation of positive agreement, the table also includes the pairwise C t‐value difference between extracted and unextracted specimens (ΔC t = C t‐unextracted − C t‐extracted, mean ± standard deviation).
Abbreviations: CDC, Centers for Disease Control; CI, confidence interval; C t, cycle‐threshold value; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; VTM, viral transport media.
A standard deviation could not be meaningfully calculated for N2‐unextracted/unconcentrated, given the preponderance of specimens generated an undetected (>45) C t value.
Negative percent agreement for the RP primer/probe‐set was not applicable (NA), as these SARS‐CoV‐2‐negative specimens still appropriately demonstrate positivity for the RP internal control.
Figure 1.
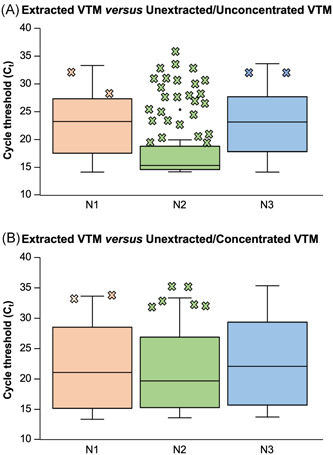
C t range of extracted VTM specimens with a positive unextracted agreement. These box‐and‐whisker plots summarize—for each primer/probe‐set (N1/N2/N3)—the range of extracted C t values (vertical axis) for SARS‐CoV‐2‐positive specimens that were also detected by qRT‐PCR when unextracted. Pairwise data are shown for (a) extracted VTM versus unextracted/unconcentrated VTM and (b) extracted VTM versus unextracted/concentrated VTM. Also depicted (X's) are the individual extracted C t values for SARS‐CoV‐2‐positive specimens where the corresponding unextracted specimen was not detected. C t, cycle‐threshold value; qRT‐PCR, quantitative real‐time reverse transcriptase‐polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; VTM, viral transport media
For unextracted/concentrated VTM versus extracted VTM, corresponding data are summarized in Table 1B and Figure 1B. Categorical positive agreement remained high for N1 (95.1%, 39/41), N3 (100%, 41/41), and RP (100%, 81/81). Preconcentration of VTM mitigated the pairwise C t differences, although the added benefit was modest (ΔC t = 2.6 ± 2.5, 2.4 ± 1.9, and 4.2 ± 2.7, respectively), potentially reflecting co‐concentration of both virus and PCR‐interfering substances. For N2, the C t‐differential was not as pronounced as with concentrated VTM (ΔC t = 8.7 ± 2.5), although categorical agreement (85%, 35/41) was still the lowest among primer/probe sets. False positives in the unextracted/concentrated samples occurred at a low frequency (1/41 for N1, 0/41 for N2, and 2/41 for N3). On a per‐specimen basis, all positive extracted specimens were detected by at least one nucleocapsid primer/probe when unextracted/concentrated; negative agreement remained ≥95% for each target.
4. DISCUSSION
In summary, the current study addresses a potential (and very practical) solution for a challenging workflow scenario—extraction reagent shortages—confronting many clinical laboratories during the COVID‐19 pandemic. Although we observed a decrease in analytic sensitivity for all CDC primer/probe sets with unextracted VTM, indicated by higher pairwise C t values, categorical agreement between unextracted and extracted specimens was high for N1, N3, and RP, both with and without VTM concentration. These findings reflect the assay's qualitative nature—with any amplification categorized as “detected”—along with the high viral burden within most positive specimens (ie, far above the assay's limit of detection). As a result, the assay tolerated a loss of analytic sensitivity without a commensurate loss of diagnostic sensitivity. The N2 primer/probe‐set represents a notable exception, as the prominent loss of sensitivity also compromised categorical results. In additional experiments (not shown), we determined that N2‐inhibition was due to the salt‐solution base of the VTM itself. The addition of either HBSS (without Mg/Ca/phenol red) or saline alone to master mix drastically inhibited N2‐amplification of defined positive specimens, but only marginally impacted N1/N3/RP. Accordingly, the reduced N2‐inhibition for the unextracted but concentrated VTM may be attributable (at least in part) to the aqueous dilution of VTM after spin concentration (see Section 2). It remains unclear why N2, among the CDC primer/probe sets, was particularly compromised by these salt conditions, although we hypothesize that it may due to its run of five consecutive cytosine residues. 4
Overall, this study suggests that laboratories might employ unextracted modifications of qRT‐PCR assays for symptomatic COVID‐19 testing, with only minimal diagnostic impact. Of note, however, we are not advocating for this approach when sufficient extraction reagents remain available to an institution (and we have not yet needed to implement it ourselves). Nevertheless, the pandemic highlights how the diagnostic supply‐chain can be stretched beyond its capacity, and the global scope of COVID‐19 threatens regions where molecular resources are already limited. Affected supplies include both reagents for laboratory‐developed assays, as well as kits for (more expensive) all‐in‐one molecular platforms. Our protocols might provide laboratories with a cost‐effective and rapid alternative when extraction supplies are scarce. Moreover, after heat denaturation and (if employed) spin concentration, all molecular steps of the procedure occur within a single vessel, simplifying the overall workflow.
If extraction resources were available, but in limited quantities, one potential application of this strategy could involve a two‐step protocol in which unextracted VTM (concentrated or unconcentrated) is first screened with N1 and N3. Specimens demonstrating positivity with either primer/probe‐set could then be reflexed to full extraction and amplification with N1 and N2 for definitive analysis, greatly reducing the total need for extraction reagents. Such screening would have flagged 39 of 40 and 40 of 41 positive specimens, respectively, for our unconcentrated and concentrated analyses, with only minimal initial false positives. We surmise the latter due to carry‐over contamination between specimens, given that false positives occurred with high C t values—with a mean C t of 36.2 across all incidences—in wells neighboring true positives. Of note, this two‐step approach would exploit the N3 primer but still mitigate the need to rely upon it for the final adjudicating step, given its potential for cross‐reactivity. 3 In the event that extraction resources were completely unavailable, a single‐step unextracted protocol might instead be envisioned. In the current study, this approach would have again identified 39 of 40 unconcentrated positive specimens and 40 of 41 concentrated specimens. An intrinsic risk of this approach is that positive results would not be confirmed through the N2 primer/probe‐set, and given the reported cross‐reactivity of the N3 primer set, the inclusion of an additional primer/probe‐set (within the N‐gene or otherwise) might offset the theoretical risk.
With either strategy, false‐negative results are expected for occasional specimens with low viral burdens, near the limit‐of‐detection of the unmodified assay (the reason why our observed positive agreement was not uniformly 100%). The exact proportion of such low‐abundance specimens within a laboratory's workflow could vary with the clinical circumstances of local testing—that is, inpatient or outpatient, symptomatic evaluation, or asymptomatic screening, test‐of‐diagnosis or test‐of‐cure. In implementing an “unextracted protocol,” a laboratory would need to consider these factors as part of their preimplementation validation, as well as any other variations in VTM formulation, primers/probes (CDC‐developed or otherwise), reagents, and instrumentation. Again, we must note that the specimens evaluated here were in the context of initial diagnostic testing of symptomatic individuals. We could see the approach being particularly unsuitable for test‐of‐cure scenarios where molecular positivity can linger with high C t values, although this practice (in general) is increasingly coming under scrutiny due to the questionable clinical significance of such results vis‐à‐vis infectivity. 16 Overall, our data suggest that high categorical agreement can be achieved under real‐world scenarios where management is impacted.
In conclusion, the myriad challenges of the COVID‐19 pandemic include widespread limitations in laboratory supplies, including RNA extraction reagents and kits. Performing qRT‐PCR using unextracted VTM, concentrated or unconcentrated, may serve as a contingency for resource‐limited settings around the globe. Going forward, we could envision such places benefiting from unextracted PCR strategies (for COVID‐19 or more broadly), even after supply‐chain limitations return to baseline in resource‐abundant locations.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
NMA, ML, and FRH made the initial observations that lead to the study. NMA, ML, AB, CQ, FRH, and JES designed the study. NMA, ML, and AB conducted the study. CQ and JES oversaw the study. JES performed statistical analysis. NMA, ML, and JES wrote the manuscript.
ACKNOWLEDGMENTS
The authors would like to acknowledge the thoughtful input of Dr. Charles Stratton, Dr. Adam Seegmiller, and Susan Sefers, as well as the effort of the entire VUMC Molecular Infectious Diseases, Microbiology, and Virology Laboratories during the COVID‐19 pandemic. This study was supported in part by the NIH National Human Genome Research Institute (R42HG009470).
Adams NM, Leelawong M, Benton A, Quinn C, Haselton FR, Schmitz JE. COVID‐19 diagnostics for resource‐limited settings: Evaluation of “unextracted” qRT‐PCR. J Med Virol. 2021;93:559–563. 10.1002/jmv.26328
Nicholas M. Adams and Mindy Leelawong contributed equally to this study.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470‐473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Binnicker MJ. Emergence of a novel coronavirus disease (COVID‐19) and the importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin Chem. 2020;66:664‐666. 10.1093/clinchem/hvaa071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babiker A, Myers CW, Hill CE, Guarner J. SARS‐CoV‐2 testing. Am J Clin Pathol. 2020;153:706‐708. 10.1093/ajcp/aqaa052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marx V. Coronavirus jolts labs to warp speed. Nat Methods. 2020;17:465‐468. 10.1038/s41592-020-0827-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID‐19 International Summit, 23 March 2020: value of diagnostic testing for SARS‐CoV‐2/COVID‐19. mBio. 2020;11:e00722‐20. 10.1128/mBio.00722-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic testing for severe acute respiratory syndrome‐related coronavirus‐2: a narrative review. Ann Intern Med. 2020;172:726‐734. 10.7326/M20-1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharfstein JM, Becker SJ, Mello MM. Diagnostic testing for the novel coronavirus. JAMA. 2020;323(15):1437–1438. 10.1001/jama.2020.3864 [DOI] [PubMed] [Google Scholar]
- 8. US Food and Drug Administration . Policy for diagnostics testing in laboratories certified to perform high complexity testing under CLIA prior to emergency use authorization for coronavirus disease‐2019 during the public health emergency . https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-diagnostic-tests-coronavirus-disease-2019-during-public-health-emergency. Accessed April 22, 2020.
- 9. Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections—the state of the art. Emerg Microbes Infect. 2020;9:747‐756. 10.1080/22221751.2020.1745095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fomsgaard AS, Rosenstierne MW. An alternative workflow for molecular detection of SARS‐CoV‐2–escape from the NA extraction kit‐shortage, Copenhagen, Denmark, March 2020. Euro Surveill. 2020;25:2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chin AWH, Chu JTS, Perera MRA, et al. Stability of SARS‐CoV‐2 in different environmental conditions. Lancet Microbe. 2020;1(1):E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwab KJ, Estes MK, Neill FH, Atmar RL. Use of heat release and an internal RNA standard control in reverse transcription‐PCR detection of Norwalk virus from stool samples. J Clin Microbiol. 1997;35:511‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leelawong M, Adams NM, Gabella WE, Wright DW, Haselton FR. Detection of single‐nucleotide polymorphism markers of antimalarial drug resistance directly from whole blood. J Mol Diagn. 2019;21:623‐631. 10.1016/j.jmoldx.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pastorino B, Bessaud M, Grandadam M, Murri S, Tolou HJ, Peyrefitte CN. Development of a TaqMan RT‐PCR assay without RNA extraction step for the detection and quantification of African Chikungunya viruses. J Virol Methods. 2005;124(1‐2)::65‐71. 10.1016/j.jviromet.2004.11.002 [DOI] [PubMed] [Google Scholar]
- 15. Centers for Disease Control and Prevention . CDC 2019‐novel coronavirus (2019‐nCoV) Real‐Time RT‐PCR Diagnostic Panel. https://www.fda.gov/media/134922/download. Accessed April 15, 2020.
- 16. Bullard J, Dust K, Funk D, et al. Predicting infectious SARS‐CoV‐2 from diagnostic samples. Clin Infect Dis. 2020. 10.1093/cid/ciaa638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


