Abstract
Objective
The large number of clinical variables associated with coronavirus disease 2019 (COVID‐19) infection makes it challenging for frontline physicians to effectively triage COVID‐19 patients during the pandemic. This study aimed to develop an efficient deep‐learning artificial intelligence algorithm to identify top clinical variable predictors and derive a risk stratification score system to help clinicians triage COVID‐19 patients.
Methods
This retrospective study consisted of 181 hospitalized patients with confirmed COVID‐19 infection from January 29, 2020 to March 21, 2020 from a major hospital in Wuhan, China. The primary outcome was mortality. Demographics, comorbidities, vital signs, symptoms, and laboratory tests were collected at initial presentation, totaling 78 clinical variables. A deep‐learning algorithm and a risk stratification score system were developed to predict mortality. Data were split into 85% training and 15% testing. Prediction performance was compared with those using COVID‐19 severity score, CURB‐65 score, and pneumonia severity index (PSI).
Results
Of the 181 COVID‐19 patients, 39 expired and 142 survived. Five top predictors of mortality were D‐dimer, O2 Index, neutrophil:lymphocyte ratio, C‐reactive protein, and lactate dehydrogenase. The top 5 predictors and the resultant risk score yielded, respectively, an area under curve (AUC) of 0.968 (95% CI = 0.87–1.0) and 0.954 (95% CI = 0.80–0.99) for the testing dataset. Our models outperformed COVID‐19 severity score (AUC = 0.756), CURB‐65 score (AUC = 0.671), and PSI (AUC = 0.838). The mortality rates for our risk stratification scores (0–5) were 0%, 0%, 6.7%, 18.2%, 67.7%, and 83.3%, respectively.
Conclusions
Deep‐learning prediction model and the resultant risk stratification score may prove useful in clinical decisionmaking under time‐sensitive and resource‐constrained environment.
Keywords: artificial intelligence, coronavirus, machine learning, pneumonia, prediction model
1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19) that causes severe respiratory illness was first reported in Wuhan, China in December 2019 1 – 3 and was declared a pandemic by the World Health Organization on March 11, 2020. 4 More than 4.2 million people have been infected and >290,000 have died of COVID‐19 worldwide (May 11, 2020). 5 The actual numbers are likely much higher due to testing shortages and under reporting. There are likely to be second waves and recurrence. 6 The current COVID‐19 pandemic has overwhelmed many hospitals around the world.
There is an urgent need by frontline physicians to effectively triage patients under time‐sensitive, stressful, and potentially resource‐constrained circumstances in this COVID‐19 pandemic. This is particularly challenging in light of the large array of clinical variables that have been identified to be associated with COVID‐19 infection, while the disease course remains incompletely understood and effective treatments are not yet available. The most commonly used reverse‐transcriptase polymerase chain reaction (RT‐PCR) has poor sensitivity (high false–negative rate 7 ) and long turnaround time (a few days to a week 8 ) during which the patients are assumed COVID‐19‐positive, potentially holding up valuable resources. Moreover, there are heterogeneous symptom presentations, and many patients are asymptomatic but may deteriorate rapidly. 1 , 2 , 3 Given these challenges, establishing a simplified risk stratification score system from studying the large array of clinical variables from large cohorts of patients could be helpful in COVID‐19 disease management. Similar risk stratification scores, such as Sequential Organ Failure Assessment (SOFA) and Modified Early Warning Score (MEWS), have been established for general emergency department triage. Unfortunately, SOFA and MEWS are not very helpful, because they failed to accurately stratify COVID‐19 patients. 9 , 10 , 11
Machine learning methods are increasingly being used in medicine. 12 , 13 , 14 Machine learning uses computer algorithms to learn relationships among different data elements to inform outcomes. In contrast to conventional analysis methods (ie, linear or logistic regression), the exact relationship among different data elements with respect to outcome variables does not need to be explicitly specified. The neural network method in machine learning, for example, is made up of a collection of connected nodes, which models the neurons in a biological brain. Each connection, like the synapses in a brain, can transmit signals to, and receive signals from, other nodes. Nodes and connections are initialized with weights that are adjusted during learning. In radiology, machine learning has been shown to be able to accurately detect lung nodules on chest X‐rays after it learns from a separate training dataset constructed with expert radiologist labels of nodules. 15 In cardiology, machine learning has been shown to be able to detect abnormal EKG patterns after it learns from a separate training dataset constructed with expert cardiologist labels of normal and abnormal EKGs. 16 Machine learning has also been used to estimate risk, such as in the Framingham Risk Score for coronary heart disease, 17 and to guide antithrombotic therapy in atrial fibrillation 18 and defibrillator implantation in hypertrophic cardiomyopathy. 19 In addition to approximating physician skills, machine learning algorithms can also find novel relationships not readily apparent to humans. Many studies have shown that machine learning outperforms logistic regression and classification tree models 20 as well as humans in many tasks in medicine. 21 Machine learning is particularly useful in dealing with large and complex datasets. With increasing computing power and growing relevance of big data in medicine, machine learning is expected to play an important role in clinical practice.
The goal of this study was to develop a deep‐learning artificial intelligence algorithm to identify the top predictors amongst the large array of clinical variables at admission to predict the likelihood of mortality in COVID‐19 patients. Based on these deep‐learning findings, we developed a simplified risk stratification score system to predict the likelihood of mortality. Prediction performance was compared with those obtained using standard CURB‐65 score for pneumonia infection, pneumonia severity index (PSI), and COVID‐19 severity score.
The Bottom Line
Researchers investigated if AI could predict death for COVID‐19 by extracting and analyzing 56 of 78 clinical variables on 181 hospitalized COVID‐19 patients in China. Their AI algorithm determined that top predictors of mortality were D‐dimer, O2 Index, neutrophil:lymphocyte ratio, C‐reactive protein, and lactate dehydrogenase. These top 5 variables had an AUC of 0.95 for predicting mortality. The AI model outperformed COVID‐19 severity score.
2. METHODS
2.1. Study population and data collection
The study was approved by the Ethics Committee of West Branch of Union Hospital affiliated Tongji Medical College of Huazhong University of Science and Technology (2020‐0197). This reporting adhered to the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) 22 and STROBE guideline. 23 This retrospective study consisted of 220 laboratory‐confirmed COVID‐19 patients from January 29, 2020 to February 19, 2020 in the West Branch of Union Hospital (Affiliated Tongji Medical College, Huazhong University of Science and Technology) in Wuhan, China, that treated moderate to severe COVID‐19 patients. Demographics, comorbidities, vital signs, symptoms, and laboratory tests at the initial presentation were collected, totaling 78 clinical variables. The primary outcome variable was mortality with March 31, 2020 being the final date of follow‐up.
2.2. Preprocessing
A few patients had some missing clinical variables. The clinical variable (brain natriuretic peptide) that was missing from >15% of the patients was removed from the dataset. For others where needed, missing data were imputed with predictive mean modeling using the Multivariate Imputation by Chained Equations in R (statistical analysis software 4.0). 24 The majority of the parameters had <5% missing data. Collinearity analysis in feature selection was used to remove correlated variables, resulting in 56 independent clinical variables being used in the neural network predictive model.
2.3. Deep neural network for feature selection
A deep neural network architecture consisting of 6 fully connected dense layers was constructed for feature selection with 56 clinical variables as inputs. Rectified linear units were chosen as activation functions and the output layer used a sigmoid function, because binary classification was used to classify patients into the expired and survived groups. Five‐fold cross validation was used to optimize model performance. Model prediction performance after fitting was used to rank the 56 features by importance from the neural network, using permutation importance methodology. 25 By shuffling single features, the error of the model was analyzed to select the features with the highest relevance. The optimal number of features necessary for classification was derived through comparing model accuracy of testing with different numbers of features. 26 Once the features were ranked and the optimal number of features was determined, the neural network architecture for classification was reduced to a simple model with 2 fully connected dense layers, rather than 6, to prevent overfitting for prediction using top (5) clinical variables or risk stratification scores. A sigmoid function for activation in the output layer was used.
2.4. Risk stratification score
The top 5 clinical variables derived from the neural networks were chosen to compute the risk score for mortality. Locally weighted smoothing (LOESS) 27 was used to plot the probability of mortality for each clinical variable. Different probabilities were evaluated and thresholds derived from 20% probability of mortality were found to achieve a balanced sample distribution across risk scores. Each of the 5 clinical variables had equal weight. The risk score ranged from 0–5.
2.5. Comparison with other indices
Performance of our deep neural network model and risk score model to predict mortality was compared with those of the PSI, 28 , 29 CURB‐65 pneumonia severity score, and COVID‐19 severity score. PSI (ranging from −10 to 285) is used to calculate the probability of morbidity and mortality. PSI requires a large set of input parameters. CURB‐65 (ranging from 0–5) composed of a point system based on an exhibition of confusion, urea nitrogen level in the blood, respiratory rate, blood pressure, and age over 65 (CURB‐65). 30 , 31 COVID‐19 severity score (ranging from 0–3 as mild, moderate, severe, critical rating) was computed according to the fifth edition of the diagnosis and treatment plan issued by the Chinese National Health Commission as: (1) mild: minimal clinical symptoms, no radiological pneumonia signs; (2) moderate: fever, respiratory tract symptoms, signs of radiological pneumonia findings; (3) severe: respiratory distress (respiratory rate ≥30 bpm), SaO2 ≤93% at resting state, or PaO2/FiO2 ≤300 mm Hg; and (4) critical: respiratory failure requiring mechanical ventilation, shock, or other organ failure requiring ICU treatment.
2.6. Statistical analysis and performance evaluation
Categorical variables described as frequencies and percentages were compared using χ2 test. Continuous variables presented as medians and interquartile ranges (IQR) were compared using Mann‐Whitney U tests. Statistical analyses were performed in SPSS v26. For performance evaluation, data were split 85% for training and 15% for testing. All prediction performance was evaluated by the area under the curve (AUC) of the receiver operating characteristic (ROC) curve for the test dataset. The average ROC curve and AUC were obtained with 5 runs.
3. RESULTS
3.1. Patient characteristics
This cohort consisted of 220 patients with confirmed COVID‐19 diagnosis in the West Branch of Union Hospital from January 29, 2020 to February 19, 2020. Thirty‐nine patients were excluded due to missing discharge or mortality information. Of the remaining 181 COVID‐19 patients, 142 survived and 39 expired. The primary outcome variable was mortality with March 31, 2020 being the final date of follow‐up.
The demographic, comorbidity, symptoms, vitals, and laboratory findings of patients who expired versus those who survived are shown in Table 1 . Patients who expired were significantly older compared to those survived (median age 65 [IQR, 68–78] vs 58 [IQR, 48–66] years old, P < 0.001). There were more male (66.7%) patients who died than female, and fewer male (47.9%) patients survived than female, but the sex variable as a whole was not significantly different between group in this cohort (P > 0.05).
TABLE 1.
Demographic, comorbidity, symptoms, imaging findings, vitals, and laboratory findings of the mortality group compared with the survival group
| Died | Survived | ||
|---|---|---|---|
| (n = 39) | (n = 142) | P | |
| Age | 65 (68, 78) | 58 (48, 66.25) | <0.001 |
| Sex | |||
| Female | 13 (33.3%) | 74 (52.1%) | 0.26 |
| Male | 26 (66.7%) | 68 (47.9%) | |
| Height | 163 (158, 170) | 168 (158, 170) | 0.345 |
| Weight | 65 (60, 70) | 65 (56, 70) | 0.966 |
| Symptoms | |||
| Dyspnea | 35 (89.7%) | 71 (50.0%) | <0.001 |
| Fatigue | 39 (100%) | 109 (76.8%) | 0.004 |
| Diarrhea | 12 (30.8%) | 23 (16.2%) | 0.017 |
| Sputum | 15 (38.5%) | 30 (21.1%) | 0.036 |
| Date since symptoms onset | 10 (7, 15) | 12 (8, 16) | 0.068 |
| Cough | 33 (84.6%) | 107 (75.4%) | 0.347 |
| Fever | 34 (87.2%) | 123 (86.6%) | 0.713 |
| Headache | 7 (17.9%) | 26 (18.3%) | 0.892 |
| Comorbidities | |||
| Heart failure | 1 (2.56%) | 0 | 0.045 |
| Other respiratory disease | 4 (10.3%) | 4 (2.82%) | 0.052 |
| Hypertension | 16 (41.0%) | 41 (28.9%) | 0.089 |
| Coronary artery disease | 6 (15.4%) | 10 (7.04%) | 0.12 |
| Smoking history | 2 (5.13%) | 2 (1.41%) | 0.174 |
| Stroke | 1 (2.56%) | 1 (0.70%) | 0.339 |
| Malignancy | 1 (2.56%) | 7 (4.93%) | 0.503 |
| Liver disease | 0 | 1 (0.70%) | 0.612 |
| Exposure history | 3 (7.69%) | 8 (5.63%) | 0.67 |
| Diabetes | 6 (15.4%) | 18 (12.7%) | 0.713 |
| Chronic kidney disease | 1 (2.56%) | 3 (2.11%) | 0.813 |
| Vitals | |||
| O2 concentration | 0.3 (0.2, 0.4) | 0.9 (0.9, 1) | <0.001 |
| Oxygen saturation (SaO2) | 87 (83, 92) | 97 (95, 98) | <0.001 |
| PaO2 | 56 (53, 65) | 98 (80, 100) | <0.001 |
| O2 index | 60 (56, 72) | 276 (212, 345) | <0.001 |
| Respiratory rate | 31 (30, 34) | 22 (20, 29) | <0.001 |
| Heart rate | 93 (80, 110) | 87 (80, 98) | 0.079 |
| Systolic blood pressure | 130 (120, 150) | 125 (120, 138) | 0.422 |
| Diastolic blood pressure | 79 (73, 86) | 80 (76, 86) | 0.33 |
| Highest temperature | 39 (38, 39) | 39 (38, 39) | 0.44 |
| Chemistry | |||
| Creatinine clearance rate (CCr) | 69.13 (52.0, 84.1) | 97.30 (73.6, 114.5) | <0.001 |
| Lactate dehydrogenase, U/L (LDH) | 549 (399, 714) | 223 (183, 309) | <0.001 |
| Ailirubin, direct, umol/L | 4.6 (3.3, 8.2) | 3 (2.2, 4.3) | <0.001 |
| Aspartate aminotransferase, U/L (AST) | 53 (34, 86) | 27.5 (19.75, 38) | <0.001 |
| Albumin, g/L | 28.4 (25.4, 30.5) | 32.4 (29.5, 36.2) | <0.001 |
| Glucose, mmol/L | 7.0 (6.1, 9.1) | 5.8 (5.3, 6.7) | 0.002 |
| Bilirubin, total, umol/L | 15 (9, 22) | 10 (8, 14) | 0.003 |
| Blood urea nitrogen, mmol/dL | 7.3 (4.7, 10.8) | 4.3 (3.3, 5.7) | 0.004 |
| Alanine aminotransferase, U/L (ALT) | 47 (25, 60) | 30 (21, 50) | 0.043 |
| Creatinine, umol/L | 75.5 (69.9, 94.2) | 63.650 (54.1, 75.4) | 0.214 |
| Sodium, mmol/L | 136.2 (135.5, 141.4) | 139.1 (137.6, 141.8) | 0.267 |
| Triglycerides mmol/L | 1.3 (1.2, 2.1) | 1.41 (1.0, 2.0) | 0.467 |
| Total protein, g/L | 62.9 (57.9, 69.8) | 63.4 (59.8, 67.5) | 0.707 |
| Potassium, mmol/L | 3.9 (3.5, 4.3) | 4.0 (3.6, 4.2) | 0.748 |
| Hematology | |||
| Neutrophile:lymphocyte ratio (NE/LY) | 8 (4, 13) | 2 (2, 3) | <0.001 |
| Neutrophil, % | 87 (85, 92) | 67 (58, 76) | <0.001 |
| Lymphocyte, % | 7 (5, 11) | 23 (14, 30) | <0.001 |
| WBC count, G/L | 9 (7, 15) | 5 (4, 7) | <0.001 |
| Platelet count, G/L | 155 (109, 213) | 213 (178, 286) | <0.001 |
| Hemoglobin, g/L | 130 (114, 140) | 126 (112, 139) | 0.413 |
| Hematocrit, % | 39 (34, 43) | 38 (33, 41) | 0.466 |
| Coagulation | |||
| D‐dimer, μg/mL | 1.1 (0.6, 3.2) | 0.5 (0.2, 1.0) | <0.001 |
| Prothrombin time, s | 14 (14, 15) | 13 (12, 14) | <0.001 |
| Activated partial thromboplastin time, s | 37 (31, 43) | 36 (33, 39) | 0.403 |
| Immunology | |||
| C‐reactive protein, mg/L (CRP) | 98 (47, 128) | 11 (2, 40) | <0.001 |
| Procalcitonin, ng/mL | 0.3 (0.2, 0.5) | 0.06 (0.04, 0.1) | 0.028 |
| Other lab values | |||
| Creatine kinase‐MB, U/L | 19 (13, 36) | 10 (5, 13) | <0.001 |
| Apolipoprotein‐A, g/L (ApoA) | 0.7 (0.6, 0.8) | 0.9 (0.7, 1.0) | <0.001 |
| High density lipoprotein, mmol/L (HDL) | 0.8 (0.7, 1.1) | 0.9 (0.8, 1.2) | 0.004 |
| Troponin, ng/mL | 44 (8, 721) | 3 (2, 7) | 0.042 |
| Brain natriuretic peptide, ng/L (BNP) | 94 (49, 171) | 49 (27, 81) | 0.044 |
| Apolipoprotein‐B, g/L (ApoB) | 1.0 (0.9, 1.3) | 0.9 (0.8, 1.1) | 0.088 |
| Lipoprotein mg/Dl | 19 (9, 34) | 13 (7, 24) | 0.332 |
| Total cholesterol, mmol/L | 4.0 (3.6, 4.8) | 4.2 (3.7, 4.7) | 0.357 |
| Low density lipoprotein, mmol/L (LDL) | 2.3 (1.9, 3.1) | 2.4 (2.0, 2.9) | 0.654 |
| Treatments | |||
| Antibiotics | 39 (100%) | 93 (64.8%) | <0.001 |
| Steroid | 29 (74.4%) | 19 (13.4%) | <0.001 |
| Antiviral drug | 39 (100%) | 135 (95.1%) | 0.611 |
| Intravenous immunoglobulin | 6 (15.4%) | 23 (16.2%) | 0.842 |
| No O2 given (n = 37) | 0 | 37 (100%) | <0.001 |
| Non‐invasive ventilation (128) | 28 (21.9%) | 100 (78.1%) | <0.001 |
| Invasive ventilation (n = 15) | 11 (73.3%) | 4 (26.7%) | <0.001 |
| Clinical scores | |||
| CURB‐65 (range = 0–5) | 2 (1,3) | 0.5 (0, 1) | <0.001 |
| Pneumonia severity index (PSI) | 105 (88, 124) | 58 (0, 81) | <0.001 |
| COVID‐19 severity index (range = 0–3) | 3 (3, 3) | 2 (1, 2) | <0.001 |
| Respiratory failure | 39 (100%) | 75 (52.8%) | <0.001 |
| Length of hospitalization, day | 6 (3.5, 11) | 17 (10, 29) | <0.001 |
SI conversion factors: to convert alanine aminotransferase and lactate dehydrogenase to microkatal per liter, multiply by 0.0167; C‐reactive protein to milligram per liter, multiply by 10; D‐dimer to nmol/L, multiply by 0.0054; leukocytes to × 109 per liter, multiply by 0.001.
Ventilation information: no O2 given (n = 37), nasal cannula (n = 82), O2 mask (n = 36), high flow nasal cannula (n = 5), and noninvasive positive‐pressure ventilation (n = 6), and invasive ventilation (n = 15). One patient had missing data. Values are medians (interquartile ranges).
Sputum, dyspnea, and diarrhea symptoms were significantly different between groups (P < 0.05). There were no significant differences in comorbidities between groups (P > 0.05). For vitals, O2 saturation, PaO2, oxygen index, and respiratory rate were significantly different between the survived and expired groups (P < 0.05). Lab chemistry (creatinine clearance rate, lactate dehydrogenase, direct bilirubin, aspartate aminotransferase albumin, glucose, total bilirubin, blood urea nitrogen, and alanine aminotransferase) were significantly different (P < 0.05), and hematological variables (neutrophil:lymphocyte ratio, neutrophil, lymphocyte, WBC, and platelet counts) were significantly different between groups as well (P < 0.05). In addition, coagulation variables (D‐dimer and prothrombin), immunological variables (C‐reactive protein and procalcitonin), and other lab data (creatine kinase‐MB, ApoA, HDL, troponin, and brain natriuretic peptide) were significantly different between groups (P < 0.05).
Antibiotic and steroid treatments were statistically different between groups (P < 0.05). Thirty‐seven patients had no O2 supplement, 82 patients had O2 via nasal cannula (n = 82), 36 patients had oxygen via O2 mask, 5 patients had oxygen via high flow nasal cannula, 5 patients had noninvasive positive‐pressure ventilation, and 15 patients had invasive ventilation. One patient had missing data. Modes of ventilation as a whole were statistically different between groups (P < 0.05).
The CURB‐65, PSI, COVID‐19 severity scores, and length of hospital stay were significantly different between groups (P < 0.05).
3.2. Top predictors of mortality
We first used collinearity analysis to remove highly correlated or irrelevant clinical variables, reducing the number of variables from 78 to 56 independent clinical variables. A deep neural network was used to rank the 56 independent clinical variables (Figure 1 ). The top 5 independent clinical variables associated with mortality were: D‐dimer, oxygen index, neutrophil to lymphocyte ratio (NE:LY), C‐reactive protein (CRP), and lactate dehydrogenase (LDH). We excluded diarrhea from the top 5 variables for subsequent calculation because it is a subjective symptom. The AUC for predicting mortality using the top 5 clinical variables was 0.968 (95% CI = 0.87–1.0) for the testing dataset. Note that treatment variables, the CURB‐65, PSI, and COVID‐19 severity scores were not part of the inputs in the neural network model.
FIGURE 1.
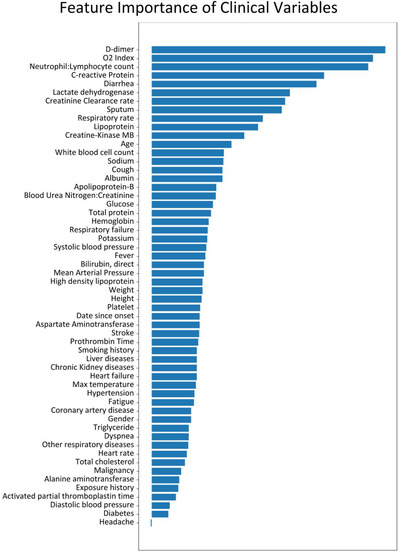
Neural network ranking of 56 independent clinical variables for predicting mortality
3.3. Risk stratification scores for mortality
Based on these top 5 variables, we also constructed a risk score stratification system to predict mortality (0 to 5, lowest to highest risk). The optimal thresholds that yielded a balanced sample distribution across risk scores were: >6.7 mg/L for D‐dimer, <94 for O2 index, >10 for NE:LY, >93 mg/L for CRP, and >450 U/L for LDH. To use the risk stratification score in practice, each of these variables that reached the respective thresholds would add 1 point to the total score. Figure 2 shows the results of risk score model for the testing dataset. The percentage of expired patients increased with increasing mortality risk score, and the percentage of patients who survived decreased with increasing risk score. Performance using the risk score yielded an AUC of 0.954 (95% CI = 0.80–0.99) for predicting mortality for the testing dataset. The mortality rates for the risk stratification scores of 0, 1, 2, 3, 4, and 5 were 0%, 0%, 6.7%, 18.2%, 67.7%, and 83.3%, respectively.
FIGURE 2.
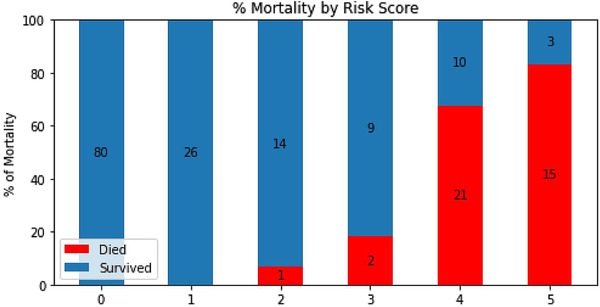
Risk scores predicting mortality (0 to 5, lowest to highest risk). Risk scores were constructed based on 5 top clinical variables
The ROC curves and AUC of the deep neural network of the top 5 clinical variables and the resultant risk score were compared with those of the COVID‐19 severity score, CURB‐65 score, and PSI (Figure 3 ). The prediction based on the 5 top clinical variables (AUC = 0.968) and the risk score (AUC = 0.954) outperformed in predicting mortality compared to COVID‐19 severity score (AUC = 0.756), CURB‐65 (AUC = 0.671), and PSI (AUC = 0.838).
FIGURE 3.
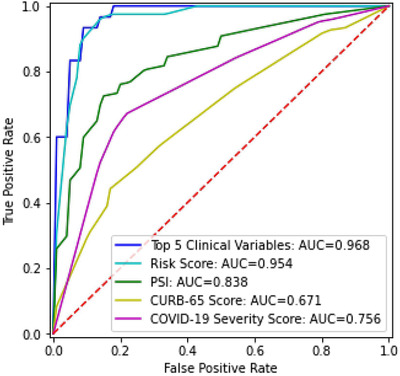
Receiver operating characteristic (ROC) curves and area under the curve (AUC) for top 5 clinical variables, risk score, pneumonia severity index (PSI), CURB‐65 score, and COVID‐19 severity score for the test dataset
4. DISCUSSION
We developed a deep neural network architecture to identify the top 5 predictive variables from 78 clinical variables to predict mortality in COVID‐19 patients early in their hospital course. The predictive model using the top 5 clinical variables as inputs yielded an AUC of 0.968 and the resultant risk stratification score yielded an AUC of 0.954 for predicting mortality for the testing dataset. This predictive model compared favorably with standard clinical predictive models for pneumonia (CURB‐65 and PSI) and a COVID‐19 severity score. Our deep neural network predictive model is flexible and can readily take on additional data to augment performance, and can be easily retrained with regional data. This predictive model and risk stratification score system have the potential to help direct patient flow and appropriate resources associated with COVID‐19 patient care.
The top predictors of mortality determined by deep neural network in this patient cohort were D‐dimer, O2 index, Ne:Ly, CRP, diarrhea, and LDH. Elevated D‐dimer, a small protein fragment present in the blood after a blood clot is degraded by fibrinolysis, suggests coagulation system dysfunction and inflammation, which may increase the likelihood of deep venous thrombosis. Reduced O2 index suggests a reduced ability of the lung to oxygenate blood, suggestive of disrupted lung function. Elevated Ne:Ly ratio could make it difficult for the body to eradicate infection, reflecting immunological disturbance due to COVID‐19 infection. Elevated CRP, a blood marker of inflammation, suggests inflammation and tissue damage in the body. Elevated LDH also indicates tissue damage in the body. These 5 top predictors reflect tissue inflammation, tissue damage, and compromised immunological defense. All of these parameters have been previously identified to be associated with COVID‐19 infection, 32 but the majority of published studies at the time of this writing did not rank these clinical features, or use quantitative prediction model and risk stratification scores to predict mortality.
Interestingly, unlike previous literature that generally cited age as an important predictor of mortality in COVID‐19, 1 , 2 , 3 , 32 our model did not find age to rank among the top predictors of mortality in our cohort, although age was significantly different between groups. None of the comorbidities and symptoms (except diarrhea) were among the top predictors in our cohort although some comorbidities and symptoms were significantly different between groups. These discrepancies might be because previous studies did not rank clinical features although other explanations (ie, different patient cohorts and hospital practice or environment) are possible. Although frontline physicians can readily identify patients to be at higher risk of mortality based on old age and physical distress, this and similar predictive models and advanced risk stratification tools based on large array of clinical variables from large patient cohorts may be helpful to objectively and robustly identify patients who ostensibly should do well but are at high risk of developing a severe disease course, complications, or poor prognosis in general. The value of such advanced risk stratification is, therefore, to identify patients for escalated care.
It is challenging for frontline physicians to effectively use such a large array of clinical variables in clinical decisionmaking, especially when the disease course is not well understood and clinical decisions need to be made in time‐sensitive, stressful, and potentially resource‐constrained environments. It is also challenging to determine the exact relationship amongst different clinical variables with respect to outcome by conventional methods. Therefore, we developed a practical risk stratification score system. To use the risk stratification score in practice, each of these variables that reached the respective thresholds would accumulate 1 point. There were 5 possible points. The mortality rates for our risk stratification scores (0–5) were 0%, 0%, 6.7%, 18.2%, 67.7%, and 83.3%, respectively. We envision such risk score to be used for risk stratification for triaging of patients with respect to ICU admission or mechanical ventilator use. These findings are significant and could have useful clinical applications.
Another novel finding is that prediction based on the 5 top clinical variables and the risk stratification score outperformed COVID‐19 severity score, CURB‐65, and PSI in predicting mortality in a head‐to‐head comparison. This finding is significant. A few studies have attempted to develop a predictive or risk score model to predict mortality and disease severity associated with COVID‐19 infection. General risk scores used in the emergency department, such as SOFA and MEWS, when applied to COVID‐19 infection unfortunately lack adequate sensitivity and specificity to predict mortality associated with COVID‐19 infection. 9 , 10 Lu et al created a 3‐tiered risk score based on only 2 variables, age and CRP thresholds, to determine mortality. 33 Ji et al used logistic regression and identified comorbidities, age, lymphocyte and LDH to be predictors of mortality but did not develop a risk score. 34 Xie et al reported age, lymphocyte count, LDH, and SpO2 to be independent predictors of mortality but did not develop a risk score. 35 Jiang et al used supervised learning (not deep learning) and found mildly elevated alanine aminotransferase, myalgia, and hemoglobin at presentation to be predictive of severe acute respiratory distress syndrome of COVID‐19 with 70%–80% accuracy. 36 This study had small and non‐uniform clinical variables from different hospitals. Ji et al predicted stable versus progressive COVID‐19 patients (n = 208) based on whether their conditions worsened during hospitalization. 37 They reported comorbidity, older age, lower lymphocyte, and higher lactate dehydrogenase at presentation to be independent high‐risk factors for COVID‐19 progression. A nomogram of these 4 factors yielded a concordance index of 0.86. Hu et al studied 105 patients to predict mortality using only demographic and vitals without laboratory tests. 38 They found an optimal cut‐off of Rapid Emergency Medicine Score (≥6) had a sensitivity of 89.5% and a specificity of 69.8%. None of these models used internal validation using independent datasets and none of these studies compared with other models. Although some of the predictors of mortality were shared among these and our studies, there were also differences. These differences could be due to different outcome measures being investigated (mortality, acute respiratory distress syndrome, and disease severity), patient cohorts, different disease severity at admission, hospital environment, and analysis methods used, among other factors. Therefore, a predictive model needs to be flexible to incorporate new and local data and new outcome variables.
Our predictive model differed from previous COVID‐19 predictive models in several ways. Our approach used a deep‐learning algorithm method that offers unique advantages over logistic regression or multivariate models. In typical regression models, clinical variables need to be dichotomized based on optimal cut‐off values using various methods typically to maximize the summation of sensitivity and specificity, 39 which could introduce errors. The neural network model, on the other hand, can model large number of clinical variables without the need to specify the exact relationship amongst different data elements. In addition, our model was validated internally using independent datasets. Our predictive model can be easily retrained with additional data, new local data, as well as additional clinical variables.
Finally, it is important to note that our machine learning predictive model and resultant risk stratification score system were established based on a single institutional data. This work is a first step toward clinical translation. Our predictive model and risk stratification score system need to be externally validated using multi‐institutional data and/or prospective studies before clinical application can be realized. Should our tool be externally validated in due course, we envision such predictive model and risk stratification score system can be used by frontline physicians to more effectively triage COVID‐19 patients.
4.1. Limitations
Our study had several limitations. This is a retrospective study based on a single institution as mentioned above. This study includes clinical variables only at presentation. Incorporating longitudinal clinical data should provide additional insights. Finally, it is important to note that the COVID‐19 pandemic circumstance is unusual and evolving. Flow of patients (ie, ICU) and mortality may depend on individual hospitals' patient load, practice, and available resources, which also differ among countries.
5. CONCLUSION
We implemented a deep‐learning neural network algorithm to identify top clinical variables and constructed a risk stratification score to predict mortality in COVID‐19 patients. Our predictive model compares favorably with standard CURB‐65, PSI, and COVID‐19 severity score. This approach has the potential to provide frontline physicians with a simple and objective tool to stratify patients based on risks so that they can triage COVID‐19 patients more effectively in time‐sensitive, stressful, and potentially resource‐constrained environments.
AUTHOR CONTRIBUTIONS
JSZ developed codes, analyzed data, and edited the paper. PG analyzed data and edited the paper. CJ, YZ, and LZ collected data and edited the paper. XL and ZZ contributed to data interpretation and processing and edited the paper. TQD designed the study, wrote and edited the paper, and supervised the study. TQD takes final responsibility of the article.
CONFLICTS OF INTEREST
None.
ETHICS APPROVAL
This study was approved by the Ethics Committee of West Branch of Union Hospital affiliated Tongji Medical College of Huazhong University of Science and Technology (2020‐0197).
ACKNOWLEDGMENTS
The authors thank all the patient participants for their contribution to this study.
Biography
Tim Duong, PhD, is Professor and Vice Chair for Research, Radiology Director for MRI Research, and Director of Preclinical MRI Center at Stony Brook University.

Zhu JS, Ge P, Jiang C, et al. Deep‐learning artificial intelligence analysis of clinical variables predicts mortality in COVID‐19 patients. JACEP Open. 2020;1:1364–1373. 10.1002/emp2.12205
Supervising Editor: Michael Blaivas, MD, MBA.
Funding and support: By JACEP Open policy, all authors are required to disclose any and all commercial, financial, and other relationships in any way related to the subject of this article as per ICMJE conflict of interest guidelines (see www.icmje.org). The authors have stated that no such relationships exist.
Contributor Information
Liming Zhang, Email: zhangliming@bjcyh.com.
Tim Q. Duong, Email: tim.duong@stonybrook.edu.
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. WHO . Coronavirus disease 2019 (COVID‐19) Situation Report. 2020; https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed April 26; 2020.
- 5. https://coronavirus.jhu.edu/map.html.
- 6. Leung K, Wu JT, Liu D, Leung GM. First‐wave COVID‐19 transmissibility and severity in China outside Hubei after control measures, and second‐wave scenario planning: a modelling impact assessment. Lancet. 2020;395(10233):1382‐1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim HA, Hong HE, Yoon SH. Diagnostic performance of CT and RT‐PCR for COVID‐19: a meta‐analysis. Radiology. 2020, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yelin AN I, Tamer ES, Argoetti, Messer E , et al. Evaluation of COVID‐19 RT‐qPCR test in multi‐sample pools. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang X, Du R, Wang R, et al. Comparison of hospitalized patients with acute respiratory distress syndrome caused by COVID‐19 and H1N1. Chest. 2020;158(1):195‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zuo Y, Hua W, Luo Y, Li L. Skin Reactions of N95 masks and medial masks among health care personnel: a self‐report questionnaire survey in China. Contact Dermatitis. 2020;n/a(n/a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deo RC. Machine learning in medicine. Circulation. 2015;132(20):1920‐1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santos MK, Ferreira Junior JR, Wada DT, Tenorio APM, Barbosa MHN, Marques PMA. Artificial intelligence, machine learning, computer‐aided diagnosis, and radiomics: advances in imaging towards to precision medicine. Radiol Bras. 2019;52(6):387‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hwang TJ, Kesselheim AS, Vokinger KN. Lifecycle regulation of artificial intelligence‐ and machine learning‐based software devices in medicine. JAMA. 2019. [DOI] [PubMed] [Google Scholar]
- 15. Harris M, Qi A, Jeagal L, et al. A systematic review of the diagnostic accuracy of artificial intelligence‐based computer programs to analyze chest x‐rays for pulmonary tuberculosis. PLoS One. 2019;14(9):e0221339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson KW, Torres Soto J, Glicksberg BS, et al. Artificial intelligence in cardiology. J Am Coll Cardiol. 2018;71(23):2668‐2679. [DOI] [PubMed] [Google Scholar]
- 17. Alaa AM, Bolton T, Di Angelantonio E, Rudd JHF, van der Schaar M. Cardiovascular disease risk prediction using automated machine learning: A prospective study of 423,604 UK Biobank participants. PLoS One. 2019;14(5):e0213653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lip GY, Nieuwlaat R, Pisters R, Lane DA, Crijns HJ. Refining clinical risk stratification for predicting stroke and thromboembolism in atrial fibrillation using a novel risk factor‐based approach: the euro heart survey on atrial fibrillation. Chest. 2010;137(2):263‐272. [DOI] [PubMed] [Google Scholar]
- 19. O'Mahony C, Jichi F, Pavlou M, et al. A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM risk‐SCD). Eur Heart J. 2014;35(30):2010‐2020. [DOI] [PubMed] [Google Scholar]
- 20. Lette J, Colletti BW, Cerino M, et al. Artificial intelligence versus logistic regression statistical modelling to predict cardiac complications after noncardiac surgery. Clin Cardiol. 1994;17(11):609‐614. [DOI] [PubMed] [Google Scholar]
- 21. Killock D. AI outperforms radiologists in mammographic screening. Nat Rev Clin Oncol. 2020;17(3):134. [DOI] [PubMed] [Google Scholar]
- 22. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD). Ann Intern Med. 2015;162(10):735‐736. [DOI] [PubMed] [Google Scholar]
- 23. von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147(8):573‐577. [DOI] [PubMed] [Google Scholar]
- 24. van Buuren S, Groothuis‐Oudshoorn K. Mice: multivariate imputation by chained equations in R. J Stat Software. 2011;45(3):1‐67. [Google Scholar]
- 25. Altmann A, Tolosi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26(10):1340‐1347. [DOI] [PubMed] [Google Scholar]
- 26. Chen Z, Pang M, Zhao Z, et al. Feature selection may improve deep neural networks for the bioinformatics problems. Bioinformatics. 2020;36(5):1542‐1552. [DOI] [PubMed] [Google Scholar]
- 27. Zhang Z, Zhang H, Khanal MK. Development of scoring system for risk stratification in clinical medicine: a step‐by‐step tutorial. Ann Transl Med. 2017;5(21):436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marras TK, Gutierrez C, Chan CK. Applying a prediction rule to identify low‐risk patients with community‐acquired pneumonia. Chest. 2000;118(5):1339‐1343. [DOI] [PubMed] [Google Scholar]
- 29. Fine MJ, Auble TE, Yealy DM, et al. A prediction rule to identify low‐risk patients with community‐acquired pneumonia. N Engl J Med. 1997;336(4):243‐250. [DOI] [PubMed] [Google Scholar]
- 30. Lim WS, Macfarlane JT, Boswell TC, et al. Study of community acquired pneumonia aetiology (SCAPA) in adults admitted to hospital: implications for management guidelines. Thorax. 2001;56(4):296‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim WS, van der Eerden MM, Laing R, et al. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58(5):377‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng Z, Peng F, Xu B, et al. Risk factors of critical & mortal COVID‐19 cases: a systematic literature review and meta‐analysis. J Infect. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu J, Hu S, Fan R, et al. ACP risk grade: a simple mortality index for patients with confirmed or suspected severe acute respiratory syndrome coronavirus 2 disease (COVID‐19) during the early stage of outbreak in Wuhan, China. medRxiv. 2020. [Google Scholar]
- 34. Zhang P, Zhu L, Cai J, et al. Association of inpatient use of angiotensin converting enzyme inhibitors and angiotensin ii receptor blockers with mortality among patients with hypertension hospitalized With COVID‐19. Circ Res. 2020;126(12):1671‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xie J, Hungerford D, Chen H, et al. Development and external validation of a prognostic multivariable model on admission for hospitalized patients with COVID‐19. medRxiv. 2020, in press. [Google Scholar]
- 36. Jiang X, Coffee M, Bari A, et al. Towards an Artificial Intelligence Framework for Data‐Driven Prediction of Coronavirus Clinical Severity. Computers Materials Continua. 2020;63:537‐551. [Google Scholar]
- 37. Ji D, Zhang D, Xu J, et al. Prediction for Progression Risk in Patients with COVID‐19 Pneumonia: the CALL Score. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu H, Yao N, Qiu Y. Comparing rapid scoring systems in mortality prediction of critically ill patients with novel coronavirus disease. Acad Emerg Med. 2020;27(6):461‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fluss R, Faraggi D, Reiser B. Estimation of the youden index and its associated cutoff point. Biometrical J. 2005;47(4):458‐472. [DOI] [PubMed] [Google Scholar]


