Abstract
Background
Severe acute respiratory infection (SARI) accounts for a large burden of illness in Indonesia. However, epidemiology of SARI in tertiary hospitals in Indonesia is unknown. This study sought to assess the burden, clinical characteristics, and etiologies of SARI and concordance of clinical diagnosis with confirmed etiology.
Methods
Data and samples were collected from subjects presenting with SARI as part of the acute febrile Illness requiring hospitalization study (AFIRE). In tertiary hospitals, clinical diagnosis was ascertained from chart review. Samples were analyzed to determine the “true” etiology of SARI at hospitals and Indonesia Research Partnership on Infectious Diseases (INA‐RESPOND) laboratory. Distribution and characteristics of SARI by true etiology and accuracy of clinical diagnosis were assessed.
Results
Four hundred and twenty of 1464 AFIRE subjects presented with SARI; etiology was identified in 242 (57.6%), including 121 (28.8%) viruses and bacteria associated with systemic infections, 70 (16.7%) respiratory bacteria and viruses other than influenza virus, and 51 (12.1%) influenza virus cases. None of these influenza patients were accurately diagnosed as having influenza during hospitalization.
Conclusions
Influenza was misdiagnosed among all patients presenting with SARI to Indonesian tertiary hospitals in the AFIRE study. Diagnostic approaches and empiric management should be guided by known epidemiology. Public health strategies to address the high burden of influenza should include broad implementation of SARI screening, vaccination programs, clinician education and awareness campaigns, improved diagnostic capacity, and support for effective point‐of‐care tests.
Keywords: diagnostic accuracy, etiology, Indonesia, influenza, severe acute respiratory infection
1. INTRODUCTION
Severe acute respiratory infection (SARI) was defined in 2011 for purposes of global surveillance. The 2011 definition harmonized heterogeneous definitions used by three WHO regions, thus facilitating comparisons. The revisions include one definition for all age groups to simplify implementation, dropping “shortness of breath” and “breathing difficulty,” adding “history of fever” and increasing the onset of symptoms to 10 days. SARI is now defined as an acute respiratory illness with a history of fever or measured fever of ≥38°C and cough, with onset within the past 10 days and requiring hospitalization. 1 This case definition enables monitoring of severe influenza‐related diseases and assessment of burden.
Severe acute respiratory infection criteria are not specific for influenza. A number of respiratory viruses other than influenza and bacterial etiologies in patients that met SARI criteria have been reported from previous studies. 2 , 3 , 4 Several viruses and bacteria that cause systemic diseases such as dengue virus, 5 , 6 chikungunya virus, 7 , 8 Salmonella spp., 9 Leptospira spp., 10 and Rickettsia typhi 11 may present with a respiratory illness and fulfill the SARI criteria. A few studies have evaluated the sensitivity and specificity of SARI criteria for influenza detection. Sensitivity and specificity range from 37% to 84% and 23% to 78%, respectively. 12 , 13 , 14 SARI criteria may also be used for pneumonia surveillance. 13
Severe acute respiratory infection and influenza‐like illness (ILI) surveillance have been conducted in Indonesia since 1999 at several hospitals and primary health centers. These studies reported that the proportion of influenza cases varied from 14% to 20% of all enrolled subjects. 15 , 16 , 17 , 18 As the aims of these studies were to confirm influenza infections, other causes of respiratory infections or “systemic” viral or bacterial infections were not analyzed.
Since bacterial, influenza, and non‐influenza viral respiratory infections are prevalent in Indonesia, there is a need to evaluate strategies for respiratory pathogen surveillance in the region. Our study aimed to identify pathogens associated with SARI in Indonesia, to compare these with hospital assessed etiology, and to describe their demographic and clinical characteristics. In addition, we determined performance characteristics of SARI criteria for identification of influenza virus.
2. METHODS
2.1. Participants
Patients fulfilling the criteria for SARI 1 were identified from an observational cohort study of patients hospitalized with acute febrile illness requiring hospitalization (AFIRE) conducted in Indonesia from 2013 to 2016. 19 The AFIRE study recruited patients presenting to 8 tertiary hospitals for evaluation of acute fever, at least 1 year old, hospitalized within the past 24 hours, and not hospitalized within the past 3 months. These hospitals were top referral provincial hospitals representing 7 major cities (Bandung, Denpasar, Jakarta, Makassar, Semarang, Surabaya, and Yogyakarta) in three populous islands. Patients were eligible for participation if they or a legal guardian/representative provided written consent following an explanation of the study objectives and procedures in Indonesian. Assent was obtained from children ≥13 years old or who were old enough to understand the proposed research.
Informed consent or assent was obtained prior to collection of clinical data, laboratory data, and specimens. Blood was collected from all subjects at enrollment, 14‐28 days, and 3 months after enrollment. Other biological specimens such as nasopharyngeal swabs, sputum, urine, or feces were collected per the attending physician. Records were reviewed for demographic and clinical information, including clinical diagnoses made during hospitalization. The “true” diagnosis was determined based on hospital and/or Indonesia Research Partnership on Infectious Disease (INA‐RESPOND) laboratory testing in conjunction with available clinical information.
Severe acute respiratory infection was assessed per the WHO definition requiring (a) an acute respiratory illness (ARI), (b) history of fever or measured fever of ≥38°C, (c) cough, (d) onset within the past 10 days, and (e) requiring hospitalization. 20 As no standard definition of ARI was available, we defined ARI as any sign or symptom related to the respiratory tract, including coryza, nasal congestion, sore throat, hemoptysis, and dyspnea. Presence of cough was determined by chart review. For patients who had pneumonia, it was assumed that cough was present even if not recorded. The requirements of fever, onset <10 days, and requiring hospitalization were already met by participants through AFIRE inclusion criteria. Ethical clearance was obtained from the institutional review board of the National Institute of Health Research and Development, Ministry of Health of Indonesia (number: KE 01.05/EC/407/ 2012).
2.2. Diagnostic testing
Diagnostic microbiology tests were conducted at hospitals immediately after specimens were collected and at the INA‐RESPOND laboratory retrospectively. The diagnostic laboratory evaluations and identified pathogens are shown in Figure 1. Details of the laboratory assays listed below and interpretation for each pathogen are listed in Table S1.
Figure 1.

Study flow, specimen tested, and pathogen by diagnostic method. Urine and CSF are not included because no pathogens were identified from urine samples (n = 23) and CSF specimens were unavailable. The number of cases is shown in parenthesis when more than 1 case was identified
2.3. Hospital laboratory
At each hospital, all blood specimens, and other biological specimens, when available were cultured. Bacterial culture and identification were performed via VITEK® 2 (bioMérieux) and BD Phoenix™ (Becton‐Dickinson) automated systems, according to manufacturer instructions. Based on the standard of care, the two most common “systemic” pathogens, dengue virus and Salmonella typhi, were tested using rapid NS1 antigen and IgM/IgG antibody test; IgM Salmonella typhi was evaluated by Tubex TF. In a subset of cases, microscopic examination was performed on sputum or feces.
2.4. INA‐RESPOND laboratory testing
2.4.1. “Systemic infection” pathogens
We used the term “systemic infections” to cover infections by several pathogens that are usually circulating in the blood and not commonly found in respiratory specimens, including dengue virus, Salmonella spp., Rickettsia typhi, Leptospira spp., and chikungunya virus. Given that dengue is endemic in Indonesia, all blood specimens, regardless of the clinical presentation, were tested for dengue virus by real‐time polymerase chain reaction (rRT‐PCR), NS1 antigen ELISA, dengue IgM, and IgG ELISA assays. As other “systemic infections” are prevalent in Indonesia, they were tested when dengue testing was negative, using molecular and serological assays.
2.4.2. Real‐time polymerase chain reaction for respiratory pathogens
Bacterial DNA was extracted from sputum or viral transport media (VTM) containing a respiratory swab using the QIAamp Bacterial DNA Mini Kit (Qiagen, Hilden) according to the manufacturer's protocol. Bacterial DNA was eluted in 100 µL of AE buffer and used as template for real‐time PCR assay or stored at −80°C. RNA was extracted from VTM containing a respiratory swab or sputum using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden) according to the manufacturer's protocol. Viral RNA was eluted in 60 µL of AVE buffer and stored at −80°C.
2.4.3. Detection of pathogens using real‐time PCR
Pathogen detection was performed with the QuantiTect Probe RT‐PCR Kit (QIAGEN; Cat#: 204443) in an Applied Biosystems 7500 Fast Real‐time PCR System (Thermo Fisher Scientific). The positive control was a synthetic plasmid carrying the nucleotide sequence of the detection target. Primers were synthesized and used to amplify the corresponding genome segments of influenza A and B, respiratory syncytial virus A and B, adenovirus, human metapneumovirus, parainfluenza virus 1, 2, 3, and 4, parechovirus, enterovirus, bocavirus, coronavirus, Legionella pneumophila, Mycoplasma pneumoniae, Chlamydia pneumoniae, Chlamydophila psittaci, Haemophilus influenzae, Bordetella pertussis, and Streptococcus pneumoniae.
2.5. Serology/ ELISA
Serologic testing was performed for influenza A, influenza B, respiratory syncytial virus (RSV), measles, and rubella. Influenza A (Cat# ESR1231M) and B (Cat# ESR1232M), virus IgM kits and influenza A (Cat# ESR1231G) and B (Cat# ESR1232G), and virus IgG kits (SERION ELISA classic kit Institut Virion/Serion GMBH‐Germany) were used according to manufacturer instructions. ELISA kits (Serion) were used to test IgM and IgG antibodies of RSV, measles, and rubella viruses. Since we used a semi‐quantitative ELISA method, we considered influenza/RSV/measles and rubella infection present when sero‐conversion or at least twofold increase of IgM and/or IgG titers was observed, consistent with manufacturer specifications and “Quality Standards in Microbiological/Infectiological Diagnostics.” 21
2.6. Data analysis
Patient characteristics, clinical, hematology and chemistry data, and outcomes were collected via paper case report form, entered into the Open Clinica database, and tabulated according to SARI etiology. Pathogens were evaluated according to clinical diagnoses of respiratory and non‐respiratory conditions at discharge. Group comparisons were performed using chi‐square. Comparisons between continuous variables were performed by t test using STATA 17. Logistic regression was used to explore univariate relationships between the variables and influenza status. Regarding a multivariate model, we were faced with the issue of having a limited number of influenza cases relative to the number of variables under consideration. Thus, following guidance from Harrell (2015, Sections 4.3 and 4.7) 22 for scenarios when there are a limited number of events (influenza cases) relative to the number of variables, we fit a multivariate logistic regression with LASSO penalty as an exploratory variable selection exercise. Statistical analyses were performed using SPSS version 22 (IBM Corporation) and R version 3.6.0.; the level of significance was set at P < .05. SARI criteria for diagnosing influenza were evaluated using randomly selected SARI and non‐SARI cases. Influenza PCR and/or serology assays were considered the gold standard.
3. RESULTS
Of 1464 subjects enrolled in the parent AFIRE study, 420 met the SARI criteria. Eighty‐nine had blood and respiratory specimens, and 331 had only blood specimens. The first screening for dengue and 4 other “systemic infections” (Salmonella spp., Rickettsia spp., chikungunya virus, and Leptospira spp.) contributed to 26% (109/420) of the total SARI cases. Influenza diagnostic tests were performed in all 420 SARI cases. Influenza was identified in 12.1% (51/420) cases, consisting of influenza A untypeable (32), influenza B (13), influenza A H3N2 (5), and influenza A H1N1 pdm (1). A total 43 out of 51 (83.7%) influenza cases were confirmed by serology only, including two cases that also had evidence of chikungunya virus and Rickettsia typhi infections, and 8 cases by RT‐PCR and serology including one mixed infection with Moraxella catarrhalis. Among 200 randomly selected SARI cases, 24 influenza cases were identified and among 200 randomly selected non‐SARI subjects, influenza was confirmed in 10 subjects, suggesting the sensitivity of SARI criteria to identify influenza cases was 70.6% (24/34), but the specificity was only 50.5% (190/366).
Other viral and bacterial respiratory pathogens contributed to 16.7% (70/420) of SARI cases, whereas non‐respiratory pathogens other than the five pathogens denoted above contributed to 3% (13/420) of cases. The most common respiratory viruses after influenza virus were RSV and measles (2.6% (11/420) each), whereas the most common bacteria were Mycobacterium tuberculosis (2.9% (12/420)), Streptococcus pneumoniae, Pseudomonas aeruginosa, and Klebsiella pneumoniae(1.4% (6/420) each). Pathogens and the specimen type used for diagnostic tests are listed in Figure 1. A total 178 out of 420 (42.4%) cases had no pathogen identified. In addition, identified pathogens in SARI and non‐SARI groups are listed in Table S2.
3.1. Identified pathogens based on clinical diagnoses
Among the 420 subjects who met SARI criteria, 244 (58.1%) had a hospital discharge diagnosis of respiratory disease and 176 (41.9%) of non‐respiratory disease. No subject had a discharge diagnosis of influenza. The three most common discharge diagnoses among patients considered to have respiratory diseases were pneumonia (124 (50.8%)), upper respiratory tract infection (68 (28%)), and pulmonary TB (24 (9.8%)). Dengue and typhoid fever were equally the most frequent discharge diagnosis among SARI patients with non‐respiratory diseases (55 (31.3%)) (Figure 2).
Figure 2.
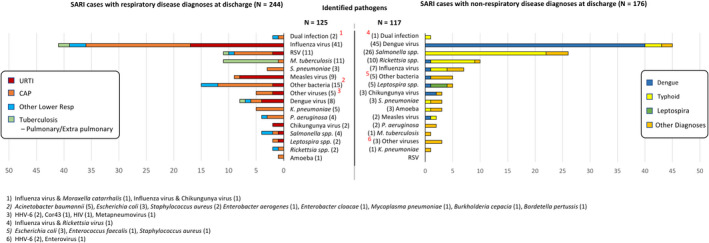
Identified pathogens based in clinical diagnoses
In this study, only 125 of the 244 subjects with a hospital discharge diagnosis of respiratory disease (51.2%) had identified causes, including influenza virus (41), other viruses (35), bacteria (46), mixed of influenza and bacteria (2), and parasite (1). Among 124 subjects diagnosed as having pneumonia, 59 (48%) had a confirmed etiology, consisting of 19 influenza cases, 13 other viral infections, 25 bacterial infections, 1 parasite, and 1 mixed influenza virus and bacteria; 65 remained unknown. In 68 clinically diagnosed URTI, influenza virus (17) and other viruses (10) were identified more frequently than bacteria (4) (Figure 2). Only 10/24 (41.7%) of the subjects with pulmonary TB as their discharge diagnosis were confirmed to have active M. tuberculosis infection with no other pathogens identified, while the rest of the causes were influenza (2) Salmonella typhi (2), dengue virus (1), RSV (1), and unidentified (8).
3.2. Demography, clinical manifestations, and hematological profiles
Unlike SARI cases associated with influenza that were distributed in all age groups, SARI associated with respiratory viruses was most common in children ≤5 years old, dengue in 5‐18 years old, and S. typhi and R. typhi in 5‐45 years old. The demography characteristics of SARI grouped by pathogens are listed in Table 1. Subjects with S. typhi and R. typhi were admitted to the hospitals significantly later compared to other groups. Among signs, symptoms, and hematological results, coryza was most common in respiratory viruses; headache, sore throat, vomiting, myalgia, arthralgia, leukopenia, thrombocytopenia, and lymphocytosis in dengue; chills, lethargy, abdominal pain, and diarrhea in rickettsiosis and typhoid fever; shortness of breath and leukocytosis in respiratory bacteria; and granulocytosis in non‐respiratory bacteria. Details of the proportion of these parameters in each pathogen group are listed in Table 1. and logistic regression in Table S3.
Table 1.
Demographic, clinical, and laboratory characteristics by illness category amongst Indonesian SARI patients (N = 420)
| Parameter | Dengue [N = 53] | Influenza [N = 48] | Salmonella and Rickettsia [N = 42] | Respiratory Bacteria a [N = 41] | Respiratory Viruses b [N = 29] | Other Bacteria c , [N = 16] | Unknown [N = 178] | P‐value d | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 1‐<5 y.o., N (%) | 8 (15.1) | 11 (22.9) | 4 (9.5) | 3 (7.3) | 20 (69.0) | 1 (6.3) | 54 (30.3) | <.001 | |||||||
| 5‐<18 y.o., N (%) | 26 (49.1) | 9 (18.8) | 16 (38.1) | 4 (9.8) | 3 (10.3) | 3 (18.8) | 38 (21.3) | <.001 | ||||||||
| 18‐<45 y.o., N (%) | 16 (30.2) | 14 (29.2) | 16 (38.1) | 15 (36.6) | 5 (17.2) | 3 (18.8) | 50 (28.1) | .462 | ||||||||
| 45‐<60 y.o., N (%) | 1 (1.9) | 8 (16.7) | 4 (9.5) | 8 (19.5) | 1 (3.4) | 2 (12.5) | 17 (9.6) | .066 | ||||||||
| ≥60 y.o., N (%) | 2 (3.8) | 6 (12.5) | 2 (4.8) | 11 (26.8) | 0 (0) | 7 (43.8) | 19 (10.7) | <.001 | ||||||||
| Male gender, N (%) | 27 (50.9) | 25 (52.1) | 20 (47.6) | 27 (65.9) | 18 (62.1) | 7 (43.8) | 90 (50.6) | .517 | ||||||||
| Clinical symptoms | Shortness of breath, N (%) | 4 (7.5) | 21 (43.8) | 9 (21.4) | 33 (80.5) | 15 (51.7) | 8 (50.7) | 84 (47.2) | <.001 | |||||||
| Haemoptysis, N (%) | 0 (0) | 3 (6.3) | 0 (0) | 1 (2.4) | 0 (0) | 0 (0) | 5 (2.8) | .313 | ||||||||
| Coryza, N (%) | 19 (35.8) | 18 (37.5) | 11 (26.2) | 3 (7.3) | 20 (69.0) | 2 (12.5) | 67 (37.6) | <.001 | ||||||||
| Sore throat, N (%) | 21 (39.6) | 9 (18.8) | 6 (14.3) | 3 (7.3) | 2 (6.9) | 3 (18.8) | 25 (36.2) | <.001 | ||||||||
| Chills, N (%) | 4 (7.5) | 12 (25.0) | 15 (35.7) | 5 (12.2) | 2 (6.9) | 5 (31.3) | 24 (13.5) | .001 | ||||||||
| Lethargy, N (%) | 18 (34.0) | 12 (25.0) | 22 (52.4) | 16 (39.0) | 5 (17.2) | 8 (50.0) | 48 (27.0) | .007 | ||||||||
| Vomiting, N (%) | 19 (35.8) | 8 (16.7) | 9 (21.4) | 1 (2.4) | 1 (3.4) | 4 (25.0) | 11 (6.2) | <.001 | ||||||||
| Diarrheal, N (%) | 4 (7.5) | 1 (2.1) | 7 (16.7) | 0 (0) | 0 (0) | 2 (12.5) | 4 (2.2) | <.001 | ||||||||
| Myalgia, N (%) | 10 (18.9) | 3 (6.3) | 2 (4.8) | 1 (2.4) | 0 (0) | 3 (18.8) | 8 (4.5) | .002 | ||||||||
| Arthralgia, N (%) | 14 (26.4) | 3 (6.3) | 6 (14.3) | 0 (0) | 0 (0) | 2 (12.5) | 9 (5.1) | <.001 | ||||||||
| Abdominal pain, N (%) | 12 (22.6) | 4 (8.3) | 15 (35.7) | 3 (7.3) | 1 (3.4) | 3 (18.8) | 24 (13.5) | .001 | ||||||||
| Headache, N (%) | 29 (54.7) | 13 (27.1) | 17 (40.5) | 3 (7.3) | 2 (6.9) | 6 (37.5) | 29 (16.3) | <.001 | ||||||||
| Fatal cases, N (%) | 1 (1.9) | 2 (4.2) | 3 (7.1) | 8 (19.5) | 1 (3.4) | 1 (6.3) | 17 (9.6) | .054 | ||||||||
| Day of onset, (Mean ± SD) | 4.1 ± 1.5 | 4.7 ± 3.1 | 6.8 ± 3.5 | 5.5 ± 4.0 | 4.8 ± 2.7 | 4.6 ± 2.4 | 4.5 ± 2.8 | <.001 | ||||||||
| Haematology profile | Haemoglobin (g/dL), (Mean ± SD) | 12.92 ± 1.75 | 13.10 ± 2.00 | 12.69 ± 1.84 | 11.49 ± 2.40 | 11.57 ± 1.95 | 12.06 ± 1.61 | 11.92 ± 2.24 | <.001 | |||||||
| Haematocrit (%), (Mean ± SD) | 38.08 ± 5.47 | 38.37 ± 5.45 | 37.17 ± 5.12 | 34.41 ± 6.12 | 35.61 ± 5.06 | 35.75 ± 4.57 | 35.62 ± 6.43 | .005 | ||||||||
| Leukocyte(×103/mm3), (Mean ± SD) | 5.50 ± 3.99 | 12.19 ± 6.65 | 7.62 ± 4.07 | 15.20 ± 9.19 | 12.32 ± 7.98 | 12.31 ± 6.59 | 12.68 ± 7.18 | <.001 | ||||||||
| Platelet (×103/mm3), (Mean ± SD) | 139.89 ± 97.09 | 243.35 ± 107.30 | 171.62 ± 77.82 | 306.54 ± 169.14 | 294.61 ± 124.14 | 197.00 ± 138.44 | 262.18 ± 126.31 | <.001 | ||||||||
| Granulocyte (%), (Mean ± SD) | 59.31 ± 18.49 | 74.36 ± 14.80 | 66.18 ± 13.44 | 76.64 ± 17.32 | 63.00 ± 13.85 | 83.08 ± 8.12 | 71.96 ± 15.39 | <.001 | ||||||||
| Lymphocyte (%), (Mean ± SD) | 29.93 ± 18.23 | 16.98 ± 13.10 | 25.39 ± 11.95 | 12.79 ± 9.88 | 28.27 ± 12.96 | 11.75 ± 8.31 | 19.86 ± 13.33 | <.001 | ||||||||
Heat Map Index:
 >50%,
>50%,  40%‐50%,
40%‐50%,  30%‐40%,
30%‐40%,  20%‐30%,
20%‐30%,  10%‐20%,
10%‐20%,  1%‐10%.
1%‐10%.
Heat map of patient characteristics by etiology of illness.
Respiratory Bacteria; S pneumoniae (6), M tuberculosis (12), K pneumoniae (6), M pneumoniae (1), A baumannii (5), P cepacia (1), P aeruginosa (6), S aureus (3). B pertussis (1).
Respiratory Viruses; Cor‐43 (1), Enterovirus (1), HHV‐6 (4), Measles virus (11), hMPV (1), RSV (11).
Other Bacteria; Leptospiraspp. (7), E coli (6), E aerogenes, E cloacae, E faecalis (@1).
The chi‐square or ANOVA test were used to assess differences in frequency/mean amongst etiologies. Cells in bold show the highest or lowest percentage/value of parameters in each category when the mean difference was significant at the .05 level compared with two or more other categories by pairwise comparisons post‐hoc tests. Other Viruses (Chikungunya virus (5), HIV (1)), Amoeba (4), and co‐infection cases (3) were not included in the analysis since N < 10 in those each group.
3.3. Comorbidities and outcomes
The proportion of SARI in hospitalized patients with fever was higher in ≤5 years old and ≥60 years old (52.7% and 48.6%) than in the 5 to ≤18 years old and 18 to <60 years old groups (25.5% and 22.3%, respectively) (Table 2). However, the mortality rate among SARI cases was highest in ≥60 years and 45 to <60 years old groups (22.4% and 21.4%, respectively). Fatalities occurred more frequently in bacterial respiratory pathogens (19.5%) compared to influenza (6.3%) groups. Among these fatal cases, 27 were diagnosed as respiratory diseases and 8 were diagnosed as non‐respiratory diseases at discharge. Most fatal cases were associated with mono‐ or co‐infection of Mycobacterium tuberculosis and HIV. Only 3 of the 35 (8.6%) fatal cases did not have any underlying diseases.
Table 2.
Pathogen identified and outcome based on age group
| Age group | ||||||
|---|---|---|---|---|---|---|
| 1‐<5 y.o. | 5‐<18 y.o. | 18‐<45 y.o. | 45‐<60 y.o. | ≥60 y.o. | Total | |
| Enrolled subjects | 198 | 409 | 585 | 167 | 105 | 1464 |
| Subjects with SARI | 104 | 106 | 119 | 42 | 49 | 420 |
| Fatal cases, N(%) | 4 (3.8%) | 3 (2.8%) | 8 (6.7%) | 9 (21.4%) | 11 (22.4%) | 35 (8.3%) |
| Pathogens identified: | ||||||
| Non‐respiratory pathogens, N ( a ) | ||||||
| Dengue virus | 8 | 26 | 16 | 1 | 2 (1) | 53 (1) |
| Salmonellaspp. | 3 | 14 | 11 (1) | 2 (1) | 0 | 30 (2) |
| Rickettsia typhi | 1 | 2 | 5 | 2 | 2 (1) | 12 (1) |
| Leptospiraspp. | 0 | 2 | 2 | 2 | 1 | 7 |
| Chikungunya virus | 1 | 4 | 0 | 0 | 0 | 5 |
| Escherichia coli | 0 | 1 | 0 | 0 | 5 (1) | 6 (1) |
| Amoeba | 2 | 1 | 0 | 0 | 1 | 4 |
| Enterobacter aerogenes | 0 | 0 | 1 | 0 | 0 | 1 |
| Enterobacter cloacae | 0 | 0 | 0 | 0 | 1 | 1 |
| Enterococcus faecalis | 1 | 0 | 0 | 0 | 0 | 1 |
| HIV | 0 | 1 (1) | 0 | 0 | 0 | 1 (1) |
| Respiratory pathogens, N ( a ) | ||||||
| Influenza virus b | 11 | 10b | 14 | 9b (3) | 7b | 51 (3) |
| RSV | 9 | 1 | 0 | 1 (1) | 0 | 11 (1) |
| Measles virus | 4 | 2 | 5 | 0 | 0 | 11 |
| Mycobacterium tuberculosis | 0 | 0 | 6 (3) | 3 | 3 (2) | 12 (5) |
| Klebsiella pneumoniae | 1 | 0 | 2 | 1 | 2 | 6 |
| Streptococcus pneumoniae | 1 (1) | 1 | 1 | 2 | 1 | 6 (1) |
| HHV‐6 | 4 | 0 | 0 | 0 | 0 | 4 |
| Bordetella pertussis | 0 | 1 | 0 | 0 | 0 | 1 |
| Cor43 | 1 | 0 | 0 | 0 | 0 | 1 |
| Metapneumovirus | 1 | 0 | 0 | 0 | 0 | 1 |
| Mycoplasma pneumoniae | 0 | 0 | 0 | 1 | 0 | 1 |
| Enterovirus | 1 | 0 | 0 | 0 | 0 | 1 |
| Pseudomonas aeruginosa | 1 | 0 | 3 (1) | 1 | 1 | 6 (1) |
| Staphylococcus aureus | 0 | 1 | 2 | 0 | 0 | 3 |
| Acinetobacter baumannii | 0 | 1 | 1 | 0 | 3 (1) | 5 (1) |
| Burkholderia cepacia | 0 | 0 | 0 | 0 | 1 | 1 |
| Unknown | 54 (3) | 38 (2) | 50 (3) | 17 (4) | 19 (5) | 178 (17) |
Number of fatal case.
Three cases mixed infection with Moraxella catarrhalis, Rickettsia typhi, and chikungunya virus.
Three influenza cases were also fatal. The first was a 49‐year‐old woman with rheumatic heart disease, the second was a 49‐year‐old woman with bronchiectasis, and the third was 59‐year‐old woman with mediastinal tumor and heart disease.
Among the SARI subjects, 173 (41.2%) had one or more comorbidities. These were more common in subjects diagnosed with respiratory diseases (132/244 (54.1%)) compared to subjects diagnosed as having non‐respiratory diseases (39/176 (22.1%)).
4. DISCUSSION
The prevalence of influenza within SARI patients in our cohort (12.1%) was consistent with previous findings from Indonesia. A 2011 study across nine hospitals reported a prevalence of 6% 23 and a 2013‐2016 study across three hospitals reported a prevalence of 14%, suggesting the annual incidence of influenza‐associated SARI was 13‐19 per 100,000 population. 18 Similar proportions of influenza among SARI cases have been reported in other parts of the world, 11.8% in Middle Eastern countries in 2007‐2014, 24 and 12% in China in 2010‐2012. 25 Our study confirms that influenza is an important cause of hospitalization in Indonesia. Since influenza was never diagnosed during hospitalization, our findings highlight the need for improved diagnostic strategies, optimization of management, and a national influenza vaccination program in Indonesia, now only mandatory for pilgrims.
Sensitivity and specificity of SARI criteria to identify influenza in our study are comparable with other studies from Western Kenya and Canada (72.1% vs 65.3%‐84% for sensitivity and 29.5% vs. 22.5%‐24.7% for specificity). In contrast, a study from Northern India reported a sensitivity and specificity of 37% and 78%, respectively. 14 These disparate results may be due to use of the previous SARI criteria by the latter study. The low specificity of SARI criteria seems related to its broad clinical criteria, resulting in 176 (41.9%) cases of non‐respiratory illness meeting the definition of SARI.
“Systemic” viral and bacterial infections may manifest clinically with respiratory signs and/or may progress to complicated pneumonia. Dengue with respiratory manifestations has been reported in more than 60% of the schoolchildren in Colombia, 26 in 4 Taiwanese patients, and 1 returned traveler in the United States, whose nasopharyngeal swabs were also positive by RT‐PCR. 5 , 6 Dengue was also the third leading cause of acute respiratory distress syndrome in pediatric patients. 27 Similarly, leptospirosis may present with severe pulmonary manifestations 10 , 28 and the bacteria can be detected from throat swabs. 10 Other “systemic” diseases such as chikungunya, rickettsiosis, and typhoid fever may also present with respiratory manifestations. 8 , 9 , 11 Although SARI patients with discharge diagnoses of such “systemic” diseases constituted a large proportion of cases, missed diagnoses only occurred in 25% (27/109) because in the majority of cases, clinicians were able to make a diagnosis based on other clinical manifestations (eg, diarrhea, rash, and icterus) and hematology profiles (eg, leukopenia, thrombocytopenia, and granulocytosis).
Physicians should be aware that many diseases can mimic Influenza and engender diagnostic confusion as demonstrated in our study. In our study, none of clinical and hematological profiles can be used to distinguish influenza and infections by other pathogens.
Identification of dengue as the most frequent cause of SARI may simply reflect the high rates of dengue in the AFIRE cohort. The 53 dengue cases that met SARI criteria were only 11.3% of the total dengue cases in the AFIRE study. Identification of Influenza as the most common virus may also be biased as serological tests were only available and performed for influenza, RSV, and measles. However, the misdiagnosis of influenza in our cohort and the discharge diagnosis of a non‐respiratory illness in 7/51 (13.7%) confirmed influenza cases are concerning. Clinicians in Indonesia may need education about influenza infection to mitigate misdiagnoses. 29 Public health campaigns and continuing medical education will be important in this regard. Healthcare policymakers should consider implementing SARI criteria to screen for Influenza in Indonesia given the sensitivity of 70.6%.
Only 57.6% (242/420) of cases had an identified etiology, consistent with other studies and suggesting the need for improved diagnostic approaches. A study of ILI in the ICU setting identified at least one pathogen in 45% of ILI subjects, with 75.2% of those cases having only a single pathogen. 30 Laboratory capacity, specimen collection technique and handling, communication among multidisciplinary team members, and recognition of the appropriate differential diagnosis are critical for facilitating pathogen identification. Rapid influenza testing has low sensitivity (50%‐70%), while molecular diagnosis requires trained technicians and specialized instruments. 31 We herein confirmed that serologic testing as an adjunct to RT‐PCR was helpful for identification of viral pathogens in community‐acquired pneumonia. 32
By improving clinician awareness of influenza and SARI, diagnostic capacity, influenza vaccination coverage and targeted public health policy, etiology of illness among patients presenting to hospital with SARI is more likely to be elucidated. This will facilitate appropriate treatment, which could lead to shorter hospital stays and decrease treatment costs. 33 , 34 Identification of SARI etiology could also reduce inappropriate antibiotic use, thus minimizing the emergence of the antimicrobial resistance. 35 , 36 , 37 , 38 Etiologic diagnosis is also important for controlling transmission and preventing outbreaks. 39 , 40 For example, prevention strategies such as respiratory isolation or cohorting when individual rooms are unavailable could be implemented. One study reported that the RR of hospital‐acquired ILI was 5.4 for patients who had contact with at least one infectious health worker, 17.96 for patients with contact with at least one infectious patient, and 34.75 for those with contact with at least one infectious patient and one infectious health worker. 41
This study had several limitations. It was not designed specifically for identification of influenza or SARI. Thus, respiratory specimens were only collected from a subset of patients with diagnosis of respiratory diseases, suggesting that we may have missed some influenza cases although influenza serological test was performed in acute and convalescent plasma of all of these subjects. For serology, we also used a twofold increase in ELISA IgM/IgG titers as the criteria for acute influenza infection instead of the fourfold increase in traditional titer 21 assays that would increase the sensitivity at the expense of some false positives. However, it has been reported that increase in antibodies’ titers after infection ranges from 1.2‐ to 10.2‐fold and 39%–55% of infected persons would not have a fourfold or greater rise in antibody titer after infection. 42 As such, this group may be different from the group that had respiratory specimens, limiting generalizability of findings.
The high proportion of cases without an identified etiology also suggests that pathogen distribution might be different if etiologies were identified in every case. Although unavailability of specimens may have limited ability to identify an etiology, our rates of pathogen identification are consistent with those of other studies. In addition, this was an observational study conducted at 8 tertiary hospitals with varied clinical practices and patient populations, which can introduce bias.
A major strength of this study was testing for a complete panel of respiratory viral and bacterial pathogens by culture, molecular assays, and serological tests on acute and convalescent plasma. In addition, this study is the first to report the epidemiology of non‐respiratory pathogens associated with SARI and the first to evaluate SARI criteria for identifying influenza cases in Indonesia.
In conclusion, influenza is often overlooked as an etiology of febrile illness in Indonesia. Implementation of the SARI criteria in tertiary referral hospitals would help identify potential influenza infections. Public health strategies to address the high burden of influenza should include vaccination programs, clinician education and awareness campaigns, improved diagnostic capacity, and support for effective point‐of‐care tests.
CONFLICT OF INTEREST
The authors have declared that no competing interests exist.
AUTHOR CONTRIBUTIONS
Abu Tholib Aman: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Project administration (equal); Supervision (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Tri Wibawa: Data curation (equal); Investigation (equal); Methodology (equal); Resources (equal); Supervision (equal); Writing‐review & editing (equal). Herman Kosasih: Conceptualization (equal); Formal analysis (equal); Methodology (equal); Supervision (equal); Validation (equal); Writing‐original draft (lead); Writing‐review & editing (equal). Rizka Humardewayanti Asdie: Data curation (equal); Investigation (equal); Resources (equal). Ida Safitri: Data curation (equal); Investigation (equal); Resources (equal); Writing‐review & editing (equal). Umi Solekhah Intansari: Data curation (equal); Investigation (equal); Resources (equal); Writing‐review & editing (equal). Yuli Mawarti: Data curation (equal); Investigation (equal); Resources (equal); Writing‐review & editing (equal). Pratiwi Sudarmono: Conceptualization (equal); Data curation (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (equal); Writing‐review & editing (equal). Mansyur Arif: Conceptualization (equal); Data curation (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (equal); Writing‐review & editing (equal). Dwiyanti Puspitasari: Data curation (equal); Investigation (equal); Resources (equal); Writing‐review & editing (equal). Bachti Alisjahbana: Conceptualization (equal); Data curation (equal); Investigation (equal); Methodology (equal); Project administration (equal); Supervision (equal); Writing‐review & editing (equal). Ketut Tuti Merati Parwati: Conceptualization (equal); Data curation (equal); Investigation (equal); Methodology (equal); Project administration (equal); Supervision (equal); Writing‐review & editing (equal). Muhammad Hussein Gasem: Conceptualization (equal); Data curation (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (equal); Supervision (equal); Writing‐review & editing (equal). Dewi Lokida: Conceptualization (equal); Data curation (equal); Investigation (equal); Methodology (equal); Resources (equal); Writing‐review & editing (equal). Nurhayati Lukman: Data curation (equal); Investigation (equal); Supervision (equal); Writing‐review & editing (equal). Teguh Sarry Hartono: Data curation (equal); Investigation (equal); Supervision (equal); Writing‐review & editing (equal). Yan Mardian: Data curation (equal); Formal analysis (equal); Visualization (equal); Writing‐review & editing (equal). C Jason Liang: Formal analysis (equal); Methodology (equal); Software (equal); Validation (equal); Visualization (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Sophia Siddiqui: Conceptualization (equal); Methodology (equal); Writing‐review & editing (equal). Muhammad Karyana: Conceptualization (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (equal); Supervision (equal); Writing‐original draft (equal). Chuen‐Yen Lau: Supervision (equal); Validation (equal); Visualization (equal); Writing‐review & editing (equal).
AUTHORS’ CONTRIBUTION
ATA, HK, PS, MA, BA, TP, MHG, DL, MK, and SS designed the study and prepared the protocol. MK, MHG, TW, RH, IS, USI, YM, DP, BA, ATA, DL, PS, TP, MA, HK, N, and TSH conducted and supervised the study. MK, DL, CJL, HK, YM, TW, RH, YM, ATA, SS, and CYL analyzed the data and prepared the draft of the manuscript. All reviewed and finalized the manuscript.
Supporting information
Tables S1‐S3
ACKNOWLEDGEMENTS
We are grateful for the support of the INA‐RESPOND network, especially the research assistants. We also thank the patients at each site for their participation in the study: 1. Site 510 RSUP dr Hasan Sadikin/ Faculty of Medicine Universitas Padjajaran, Bandung; 2. Site 520 RSUP Sanglah/ Faculty of Medicine Universitas Udayana, Bali; 3. Site 530 RSUPN dr Cipto Mangunkusumo/ Faculty of Medicine Universitas Indonesia, Jakarta; 4. Site 540 RSPI Soelianti Soeroso, Jakarta; 5. Site 550 RSUP dr Wahidin Sudirohusodo/ Faculty of Medicine Universitas Hasanuddin, Makassar; 6. Site 560 RSUP dr Kariadi/ Faculty of Medicine Universitas Diponegoro, Semarang; 7. Site 570 RSUD dr Soetomo/ Faculty of Medicine Universitas Airlangga, Surabaya; and 8. Site 580 RSUP dr Sardjito/ Faculty of Medicine Universitas Gadjah Mada, Yogyakarta.
Funding Information
This project has been funded in whole or in part with Federal funds from the NIAID, NIH, under contract Nos. HHSN261200800001E and HHSN261201500003I. NIAID collaborators contributed to design of the study; collection, analysis, and interpretation of data; and writing of the manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The peer review history for this article is available at https://publons.com/publon/10.1111/irv.12781.
REFERENCES
- 1. Fitzner J, Qasmieh S, Mounts AW, et al. Revision of clinical case definitions: influenza‐like illness and severe acute respiratory infection. Bull World Health Organ. 2018;96(2):122‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Razanajatovo NH, Guillebaud J, Harimanana A, et al. Epidemiology of severe acute respiratory infections from hospital‐based surveillance in Madagascar, November 2010 to July 2013. PLoS One. 2018;13(11):e0205124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Assane D, Makhtar C, Abdoulaye D, et al. Viral and Bacterial Etiologies of Acute Respiratory Infections Among Children Under 5 Years in Senegal. Microbiol Insights. 2018;11:1178636118758651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nguyen HKL, Nguyen SV, Nguyen AP, et al. Surveillance of Severe Acute Respiratory Infection (SARI) for Hospitalized Patients in Northern Vietnam, 2011–2014. Jpn J Infect Dis. 2017;70(5):522‐527. [DOI] [PubMed] [Google Scholar]
- 5. Tavakoli NP, Tobin EH, Wong SJ, et al. Identification of dengue virus in respiratory specimens from a patient who had recently traveled from a region where dengue virus infection is endemic. J Clin Microbiol. 2007;45(5):1523‐1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng NM, Sy CL, Chen BC, Huang TS, Lee SS, Chen YS. Isolation of dengue virus from the upper respiratory tract of four patients with dengue fever. PLoS Negl Trop Dis. 2017;11(4):e0005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rios L, Rodriguez‐Morales AJ, Castrillon‐Spitia JD, et al. Chikungunya virus infection, immunosuppression and respiratory tract infections: are they associated? Int Marit Health. 2018;69(2):149‐150. [DOI] [PubMed] [Google Scholar]
- 8. Hsu CH, Cruz‐Lopez F, Vargas Torres D, et al. Risk factors for hospitalization of patients with chikungunya virus infection at sentinel hospitals in Puerto Rico. PLoS Negl Trop Dis. 2019;13(1):e0007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breiman RF, Cosmas L, Njenga M, et al. Severe acute respiratory infection in children in a densely populated urban slum in Kenya, 2007–2011. BMC Infect Dis. 2015;15:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gasem MH, Farida H, Ahmed A, et al. Are Pathogenic Leptospira Species Agents of Community‐Acquired Pneumonia? Case Reports of Leptospirosis Presenting as Pneumonia. J Clin Microbiol. 2016;54(1):197‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aung AK, Spelman DW, Murray RJ, Graves S. Rickettsial infections in Southeast Asia: implications for local populace and febrile returned travelers. Am J Trop Med Hyg. 2014;91(3):451‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Amini R, Gilca R, Douville‐Fradet M, Boulianne N, De Serres G. Evaluation of the New World Health Organization Case Definition of Severe Acute Respiratory Infection for Influenza Surveillance During the Peak Weeks of Two Influenza Seasons in Quebec. Canada. J Pediatric Infect Dis Soc. 2017;6(3):297‐300. [DOI] [PubMed] [Google Scholar]
- 13. Makokha C, Mott J, Njuguna HN, et al. Comparison of severe acute respiratory illness (sari) and clinical pneumonia case definitions for the detection of influenza virus infections among hospitalized patients, western Kenya, 2009–2013. Influenza Other Respir Viruses. 2016;10(4):333‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta V, Dawood FS, Rai SK, et al. Validity of clinical case definitions for influenza surveillance among hospitalized patients: results from a rural community in North India. Influenza Other Respir Viruses. 2013;7(3):321‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beckett CG, Kosasih H, Ma'roef C, et al. Influenza surveillance in Indonesia: 1999–2003. Clin Infect Dis. 2004;39(4):443‐449. [DOI] [PubMed] [Google Scholar]
- 16. Kosasih H, Roselinda N, et al. Surveillance of influenza in Indonesia, 2003–2007. Influenza Other Respir Viruses. 2013;7(3):312‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adam K, Pangesti KN, Setiawaty V. Multiple Viral Infection Detected from Influenza‐Like Illness Cases in Indonesia. Biomed Res Int. 2017;2017:9541619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Susilarini NK, Haryanto E, Praptiningsih CY, et al. Estimated incidence of influenza‐associated severe acute respiratory infections in Indonesia, 2013–2016. Influenza Other Respir Viruses. 2018;12(1):81‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gasem MH, Kosasih H, Tjitra E, et al. An observational prospective cohort study of the epidemiology of hospitalized patients with acute febrile illness in Indonesia. PLoS Negl Trop Dis. 2020;14(1):e0007927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. WHO . WHO surveillance case definitions for ILI and SARI. 2014. https://www.who.int/influenza/surveillance_monitoring/ili_sari_surveillance_case_definition/en/2019.
- 21. Wellinghausen NA‐H M, Enders M, Fingerle V et al. MiQ Immunological Methods for the Detection of Infectious Diseases. Munich: Dustri‐Verlag Dr. Karl Feistle; 2016. [Google Scholar]
- 22. Harrell FE Jr. Regression modeling strategies: with applications to linear models, logistic and ordinal regression, and survival analysis. Berlin, Germany: Springer; 2015. [Google Scholar]
- 23. Pangesti KN, Susilarini NK, Pawestri HA, Setiawaty V. Influenza cases from Surveillance Acute Respiratory Infection in Indonesia, 2011. Health Sci Indonesia. 2014;5(1):7‐11. [Google Scholar]
- 24. Horton KC, Dueger EL, Kandeel A, et al. Viral etiology, seasonality and severity of hospitalized patients with severe acute respiratory infections in the Eastern Mediterranean Region, 2007–2014. PLoS One. 2017;12(7):e0180954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huai Y, Guan X, Liu S, et al. Clinical characteristics and factors associated with severe acute respiratory infection and influenza among children in Jingzhou. China. Influenza Other Respir Viruses. 2017;11(2):148‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Restrepo BN, Piedrahita LD, Agudelo IY, Parra‐Henao G, Osorio JE. Frequency and clinical features of dengue infection in a schoolchildren cohort from medellin. Colombia. J Trop Med. 2012;2012:120496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thong MK. Dengue shock syndrome and acute respiratory distress syndrome. Lancet. 1998;352(9141):1712. [DOI] [PubMed] [Google Scholar]
- 28. Helmerhorst HJ, van Tol EN, Tuinman PR, et al. Severe pulmonary manifestation of leptospirosis. Neth J Med. 2012;70(5):215‐221. [PubMed] [Google Scholar]
- 29. Kreslake JM, Wahyuningrum Y, Iuliano AD, et al. The intersection of care seeking and clinical capacity for patients with highly pathogenic Avian Influenza A (H5N1) Virus in Indonesia: knowledge and treatment practices of the public and physicians. Disaster Med Public Health Prep. 2016;10(6):838‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tramuto F, Maida CM, Napoli G, et al. Burden and viral aetiology of influenza‐like illness and acute respiratory infection in intensive care units. Microbes Infect. 2016;18(4):270‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ito Y. Clinical Diagnosis of Influenza. Methods Mol Biol. 2018;1836:23‐31. [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y, Sakthivel SK, Bramley A, et al. Serology enhances molecular diagnosis of respiratory virus infections other than influenza in children and adults hospitalized with community‐acquired pneumonia. J Clin Microbiol. 2017;55(1):79‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. von der Beck D, Seeger W, Herold S, Günther A, Löh B. Characteristics and outcomes of a cohort hospitalized for pandemic and seasonal influenza in Germany based on nationwide inpatient data. PLoS One. 2017;12(7):e0180920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Putri W, Muscatello DJ, Stockwell MS, Newall AT. Economic burden of seasonal influenza in the United States. Vaccine. 2018;36(27):3960‐3966. [DOI] [PubMed] [Google Scholar]
- 35. Choi SH, Chung JW, Kim T, Park KH, Lee MS, Kwak YG. Late diagnosis of influenza in adult patients during a seasonal outbreak. Korean J Intern Med. 2018;33(2):391‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Houten CB, Naaktgeboren C, Buiteman BJM, et al. Antibiotic overuse in children with respiratory syncytial virus lower respiratory tract infection. Pediatr Infect Dis J. 2018;37(11):1077‐1081. [DOI] [PubMed] [Google Scholar]
- 37. Tillekeratne LG, Bodinayake CK, Dabrera T, et al. Antibiotic overuse for acute respiratory tract infections in Sri Lanka: a qualitative study of outpatients and their physicians. BMC Fam Pract. 2017;18(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shallcross LJ, Davies DS. Antibiotic overuse: a key driver of antimicrobial resistance. Br J Gen Pract. 2014;64(629):604‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Besney J, Moreau D, Jacobs A, et al. Influenza outbreak in a Canadian correctional facility. J Infect Prev. 2017;18(4):193‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eibach D, Casalegno JS, Bouscambert M, et al. Routes of transmission during a nosocomial influenza A(H3N2) outbreak among geriatric patients and healthcare workers. J Hosp Infect. 2014;86(3):188‐193. [DOI] [PubMed] [Google Scholar]
- 41. Vanhems P, Voirin N, Roche S, et al. Risk of influenza‐like illness in an acute health care setting during community influenza epidemics in 2004–2005, 2005–2006, and 2006–2007: a prospective study. Arch Intern Med. 2011;171(2):151‐157. [DOI] [PubMed] [Google Scholar]
- 42. Freeman G, Perera RA, Ngan E, et al. Quantifying homologous and heterologous antibody titre rises after influenza virus infection. Epidemiol Infect. 2016;144(11):2306‐2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1‐S3


