Abstract
COVID‐19 is highly contagious pathogenic viral infection initiated from Wuhan seafood wholesale market of China on December 2019 and spread rapidly around the whole world due to onward transmission. This recent outbreak of novel coronavirus (CoV) was believed to be originated from bats and causing respiratory infections such as common cold, dry cough, fever, headache, dyspnea, pneumonia, and finally Severe Acute Respiratory Syndrome (SARS) in humans. For this widespread zoonotic virus, human‐to‐human transmission has resulted in nearly 83 lakh cases in 213 countries and territories with 4,50,686 deaths as on 19 June 2020. This review presents a report on the origin, transmission, symptoms, diagnosis, possible vaccines, animal models, and immunotherapy for this novel virus and will provide ample references for the researchers toward the ongoing development of therapeutic agents and vaccines and also preventing the spread of this disease.
Keywords: COVID‐19 pneumonia, diagnosis, entry mechanism, possible vaccines, animal models and immunotherapy, symptoms, transmission
This review presents a report on the origin, transmission, symptoms, diagnosis, possible vaccines, animal models and immunotherapy for this novel virus and will provide sample references for the researchers towards the ongoing development of therapeutic agents and vaccines and also preventing the spread of this disease.
Abbreviations
- aAPC
artificial antigen‐presenting cells
- ACE2
angiotensin‐converting enzyme 2
- ARDS
acute respiratory distress syndrome
- CoV
coronavirus
- COVID‐19
coronavirus disease 19
- CQDs
carbon quantum dots
- CSSE
Center for Systems Science and Engineering
- DPP4
dipeptidyl peptidase 4
- ELISA
enzyme‐linked immunosorbent assay
- GGO
ground‐glass opacity
- HAT
human airway trypsin‐like protease
- HIV
human immunodeficiency virus
- HPV
human papilloma virus
- mAb
monoclonal antibody
- MERS
Middle East respiratory syndrome
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- MHV
mouse hepatitis virus
- MTB
Mycobacterium tuberculosis
- NCIP
Novel COVID‐19‐infected pneumonia
- POCT
Point‐of‐care testing
- RBD
receptor‐binding domain
- RT‐PCR
reverse transcription–polymerase chain reaction
- SARS
severe acute respiratory syndrome
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- TMPRSS2
transmembrane protease serine 2
1. INTRODUCTION
Coronavirus (CoV) is a positive single‐stranded RNA virus that causes respiratory infections such as common cold, flu, rarely pneumonia, and finally Severe Acute Respiratory Syndrome (SARS) (Tyrrell & Bynoe, 1966). It belongs to Coronaviridae family in the Nidovirales order. The external surface creates crown‐like spikes of 9–12 nm long (Wei et al., 2020) and hence, termed as coronavirus (Latin: corona means crown). The genome (nucleic material) size varies from 26 to 32 kbs in length (Shereen, Khan, Kazmi, Bashir, & Siddique, 2020). There are four subfamilies, namely alpha‐, beta‐, gamma‐, and deltacoronaviruses, exist till date. The alpha‐ and betacoronaviruses are originated from mammals (bats), whereas gamma‐ and deltacoronaviruses are originated from birds and pigs. There are seven human coronaviruses identified till date (Table 1; Chan et al., 2020; Franquet, 2011; Hansell et al., 2008; Lee, 2020; Malainou & Herold, 2019). Among these seven subtypes, betacoronaviruses are causing severe diseases and fatalities, while alphacoronaviruses cause mildly symptomatic infections. As per the genomic analysis, SARS‐CoV‐2 is identical with bat CoV (96%) and pangolin CoV (86%–92%), so bats could be the possible primary reservoir (GISAID, 2020; Shereen et al., 2020; Zhou, Yang, et al., 2020). The major four structural genes encode the spike protein (S), nucleocapsid protein (N), membrane glycoprotein (M), and small membrane protein (SM) with an additional hemagglutinin esterase (HE) if present (HE is only present in some betacoronaviruses). The genome RNA is complexed with N protein to form a helical capsid structure and found within the viral membrane. Trimers of S protein give the virion a crown‐like shape. In some cases, HE protein also forms smaller spikes (Figure 1).
Table 1.
Comparative analysis for seven human coronaviruses
| Name | Type | Year of finding | Key hosts | Cellular receptor | Symptoms | CT imaging |
|---|---|---|---|---|---|---|
| 229E | Alpha | 1966 | Bats | Human aminopeptidase N (CD13) | Common cold, running nose, fever, headache, malaise, bronchiolitis, pneumonia (in neonates) | Bibasilar pleural effusions and diffuse consolidations plus GGOs |
| NL63 | Alpha | 2004 | Bats, Palm Civets | ACE2 | Common cold, bronchiolitis/croup in children, fever, malaise, sore throat, cough and rhinitis | Acute upper respiratory tract infections |
| OC43 | Beta | 1967 | Cattle | 9‐O‐Acetylated sialic acid | Common cold, running nose, fever, headache, malaise, bronchiolitis, pneumonia (in neonates) | Acute upper respiratory tract infections |
| HKU1 | Beta | 2005 | Mice | 9‐O‐Acetylated sialic acid | Common cold, dyspnea, chronic respiratory disease | Acute upper respiratory tract infections |
| SARS‐CoV ( Severe Acute Respiratory Syndrome ) (Franquet, 2011; Lee, 2020) | Beta | 2002 | Bat, palm civets, raccoon dogs | ACE2 | Fever, headache, diarrhea, shivering, cough, dyspnea, pneumonia | Subpleural GGO and consolidation, interlobular septal and intralobular septal thickening |
| MERS‐CoV (Middle East Respiratory Syndrome) (Hansell et al., 2008; Malainou & Herold, 2019 ) | Beta | 2012 | Bat, camel | DPP4 | Fever, dry cough, sore throat, dyspnea, pneumonia | Extensive GGO and occasional septal thickening and pleural effusions, basilar, bilateral, and subpleural airspace |
| SARS‐CoV−2 ( Severe Acute Respiratory Syndrome ) (Chan et al., 2020) | Beta | 2019 | Bat, Pangolin (anteater) | ACE2 | Fever, dry cough, sore throat, short of breath, headache, myalgia, anosmia, rarely pneumonia | Multifocal patchy GGOs with subpleural distribution, Diffuse heterogeneous consolidation with GGO |
FIGURE 1.
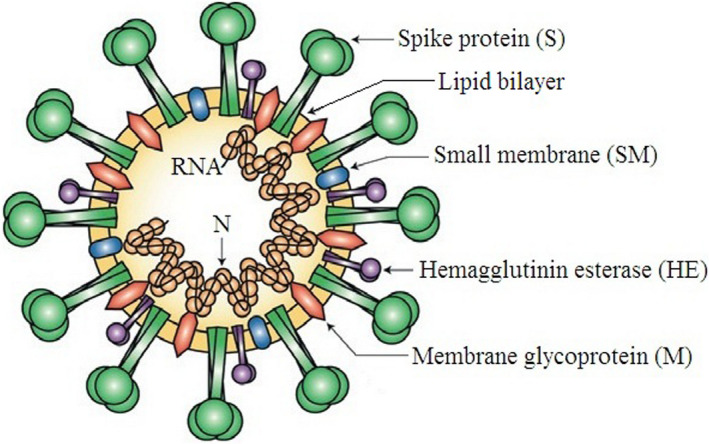
Structure of coronavirus virion. The genome RNA is complexed with nucleocapsid protein (N) within the viral membrane; (Reproduced from Finlay & Hancock, 2004, Nature reviews: Microbiology ©Springer Nature)
SARS‐CoV‐2 is a new strain of CoV for humans making the pandemic coronavirus disease (COVID‐19). It is a pathogenic viral infection initiated from Huanan Seafood wholesale market of Wuhan (Hubei province, central China) on 31 December 2019, where seafood and live animals were sold (Huang, Wang, et al., 2020; Li, Guan, et al., 2020). This recent outbreak of novel CoV has expanded throughout China and spread rapidly around the whole world due to onward transmission (Lipsitch, Swerdlow, & Finelli, 2020). The virus was confirmed by Chinese authorities on 7 January 2020 (Lu, Stratton, & Tang, 2020; WHO, 2020a) and the transmission appeared to be from animals into human (Chan et al., 2020; Hui et al., 2020; Nishiura et al., 2020). The exact route of transmission is not clear yet. But the spike protein (S) may binds to ACE2 as an entry receptor for viral entry into the host cell and infects lung alveolar epithelial cells (Franquet, 2011; Kirchdoerfer et al., 2016; Xu, Chen, et al., 2020). After penetration into the body, all RNA‐related machinery will be hacked. These viruses are responsible for diarrhea in pigs and cows and respiratory diseases in chickens. The animal‐specific viruses are not normally infecting humans. But, may cause life‐threatening pneumonia due to novelty of the virus, if they pass to humans. This is really very dangerous for the immune‐suppressed individuals having diabetes, cardiovascular diseases and leads to pulmonary failure.
The important consequence of COVID‐19 is pneumonia means this virus infects mainly respiratory system (Zhu, Zhang, et al., 2020). It may be transmitted through respiratory droplets and even via fecal‐oral transmission (Chan et al., 2020; Wang, Cao, et al., 2020). That means the human coronaviruses are transmitted from one infected person to another mainly by direct contact through saliva, coughing or sneezing (within a range of about 1–1.5 m). Presently, there are no specific drugs or vaccines available for the treatment of human CoV (Han, Lin, Jin, & You, 2020). Therefore, it is very essential to understand the nature of this virus and its clinical characteristics. For this widespread zoonotic virus (Table 2), human‐to‐human transmission has been confirmed in more than 83 lakh cases of nearly 213 countries and territories as on 19 June 2020 (WHO, 2020). The common clinical symptoms for current COVID‐19 are fever, dry cough, dyspnea, muscle soreness, headache, anosmia, and fatigue (Wang, Tang, & Wei, 2020). Initially, about 20% of cases were severe, and mortality rate was approximately 3% (Wang, Horby, Hayden, & Gao, 2020). For which, WHO has declared a global health emergency on 30 January 2020 (Mahase, 2020). It was too early to conclude the mortality rate. From the closed cases, the recent mortality rate is reached at ~9%, which may vary depending upon the current cases. Furthermore, the outbreak has destroyed the global economy and stock market. This is the seventh CoV which infect humans (Zhu, Zhang, et al., 2020) and other two are SARS and MERS. According to Jung et al., the current epidemic has a substantial potential for causing global pandemic (R0 > 1) (Jung et al., 2020). This study will make a brief report on 2019‐nCoV, which may be helpful for the researchers as well as society.
Table 2.
Comparison between SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2
| Name | Origin | Number of infected | Number of deaths | Number of countries | R0 a | Mortality (%) |
|---|---|---|---|---|---|---|
| SARS‐CoV ( Severe Acute Respiratory Syndrome ) | Guangdong, southern China | 8,098 | 778 | 29 | 2–4 | 10 |
| MERS‐CoV ( Middle East Respiratory Syndrome ) | Jeddah, Saudi Arabia | 2,434 | 876 | 27 | 1 | 35.9 |
| SARS‐CoV‐2 ( Severe Acute Respiratory Syndrome ) | Hubei, central China | 8,385,440 | 4,50,686 | 213 | 1.4–5.5 | ~9 (from the closed cases, till date) |
R0, Basic Reproduction Number.
1.1. Three important types of coronavirus which infect humans
1.1.1. Severe Acute Respiratory Syndrome coronavirus (SARS‐CoV)
SARS‐CoV was the first infectious disease originated from Guangdong province of southern China in November 2002 and subsequently spread worldwide (Booth et al., 2003; Dwosh, Hong, Austgarden, Herman, & Schabas, 2003; Lee et al., 2003; Tsang et al., 2003; Weiss & Leibowitz, 2011; WHO, 2003). This acute and often severe CoV was spread via airborne droplets and through fomites (Donnelly et al., 2003), which have infected nearly 8,000 peoples with 778 deaths (WHO, 2003) before the episode was shutdown. The coronavirus virion structure (Finlay & Hancock, 2004) is shown in Figure 1. As per the phylogenetic analyses, it was placed in the early branch of betacoronaviruses category and the genus includes MHV (Snijder et al., 2003; Zhu & Chen, 2004). The patients infected by SARS‐CoV, showed fever, sore throat, nonproductive cough, and myalgia in the first phase of the illness. In the second phase, dyspnea, hypoxia, and diarrhea with continued fever became more prominent followed by severe lung damage. Deaths occurred from day 4 and severity of the disease increased with age, with mortality 50% for patients over 60 age (Booth et al., 2003; Lee et al., 2003; Tsui, Kwok, Yuen, & Lai, 2003).
Initially, it was suggested that Himalayan palm civets (Paguma larvata) and raccoon dogs (Nyctereutes procyonoides) as possible sources for human infections (Guan et al., 2003). But civets in the wild did not have evidence of infection with SARS‐CoV and the discovery of SARS‐like bat coronaviruses with approximately 90% sequence identity with SARS‐CoV in Chinese horseshoe bats (Rhinolophus sinicus) suggests that this or a related species of bat is likely origin of SARS‐CoV (Lau et al., 2005; Li et al., 2005). At early phases, the lungs of infected humans contained hyaline membranes, small vessel thrombi, edema, fibrin exudates, loss and sloughing of pneumocytes, and a mixed cellular infiltrate of lymphocytes. Whereas, at later phases, pneumonitis and consolidation with type II pneumocyte hyperplasia, squamous metaplasia and bronchiolitis obilterans was found (Beijing Group, 2003; Cameron et al., 2007; Wong et al., 2004).
1.1.2. Middle East respiratory syndrome coronavirus (MERS‐CoV)
The novel lethal MERS coronavirus infection was first identified from Jeddah, Saudi Arabia in June 2012 (Bauerfeind et al., 2016; Zaki, van Boheemen, Bestebroer, Osterhaus, & Fouchier, 2012). The case was noticed in a man who was admitted to the hospital with pneumonia and acute kidney injury (McIntosh, Hirsch, & Thorner, 2017). A similar virus with 99.5% identity was reported from Qatar in September 2012 who had traveled to Saudi Arabia before the disease progression exacerbated (Wise, 2012). Since then, nearly 2,500 confirmed human infections have been reported in 27 countries with mortality rate more than 30% (WHO, 2020b). The virologic research revealed that this infection was originated by dromedary camels (Camelus dromedarius), which infects human respiratory tract (Al‐Tawfiq, 2013; Azhar, El‐Kafrawy, et al., 2014). All related cases have been noticed in Middle East, East Asia, North Africa, Europe, and United States (WHO, 2018).
This novel zoonotic virus is responsible for acute human respiratory syndrome and the dromedary camels of East Africa and Arabian Peninsula are only the confirmed animal host with DPP4 receptor (Conzade et al., 2018; Falzarano et al., 2017; Miguel et al., 2017; Perera et al., 2013; Ramadan & Shaib, 2019; Reusken, Haagmans, et al., 2013). Humans are believed to acquire MERS‐CoV though contact with camels, camel products, and consumptions (Azhar, Hui, Memish, & Zumla, 2019). This virus is closely related with bats’ coronaviruses, suggesting bats may be the possible natural reservoir (Ramadan & Shaib, 2019), but not unconfirmed yet. Also, no vaccine is available for MERS‐CoV till date. Epidemiological and viral sequence data suggested that camels are the main culprit for virus transmission to humans (Azhar, El‐Kafrawy, et al., 2014; Azhar, Hashem, et al., 2014).
Camel meat is a common food and important source of nutrition for people in Middle East and African countries. Keeping in mind, El‐Duah et al., 2019 have reported the transmissibility of this virus through other domestic livestock species. No viral RNA or antibodies against MERS‐CoV (Ghanaian bats) were noticed in any of these tested livestock species. Hence, cattle, goat, donkey, sheep, and swine are not likely hosts of clade 2c coronaviruses. Sudan camels were rich with MERS‐CoV antibodies, but no infections were observed in camel workers, other livestock, or bats (Farag et al., 2019). Kandeil et al., 2019 have conducted the surveillance studies on Middle East and African camels by collecting 4,027 nasal swabs and 3,267 serum samples to elucidate MERS‐CoV infection and transmission. According to them, MERS‐CoV RNA and antibodies were noticed in nasal swab samples collected from Tunisia, Senegal, Egypt, and Saudi Arabia. Lau et al., 2020 have reported the presence of MERS‐CoV antibodies in Bactrian and hybrid camels in Dubai, UAE. In this study, the Bactrian camel serum samples collected from Xinjiang were tested negative for MERS‐CoV antibodies.
Camels and bats have been identified as the potential source of virus for MERS‐CoV. The role of spike protein (S) gene in virus attachment to host cells was examined (Sohrab & Azhar, 2019; Zumla, Chan, Azhar, Hui, & Yuen, 2016), which will be very valuable to design the therapeutic compounds and vaccines to prevent MERS‐CoV disease. The schematic representation for the genome organization of human betacoronaviruses is shown in Figure 2. The antibody‐based therapeutic modalities have received significant attention for the treatment of many diseases (Hu et al., 2019; Qiu et al., 2014; Zhu et al., 2006). Based on that, Wang et al., 2019 have reported the generation of antibody‐peptide bispecific fusion proteins for clinical use. The efficacy of a ChAdOx1 MERS vaccine in dromedary camels (seronegative and seropositive) was studied by Alharbi et al., 2019. From the study, it was cleared that antibody responses in seropositive camels were enhanced by using a single dose of this vaccine.
FIGURE 2.
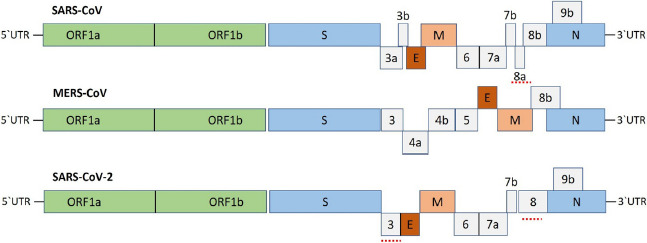
Genome organization for human betacoronaviruses (SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2), (Adapted with Permission from Shereen et al., 2020, Journal of Advanced Research, © Elsevier BV.)
1.1.3. Severe Acute Respiratory Syndrome SARS‐CoV‐2
The novel CoV (2019‐nCoV) infection was initiated from Wuhan of China, in December 2019 (Lu, Stratton, et al., 2020) and has been spread to almost all countries. On 11 February 2020, WHO announced the name “COVID‐19” for this epidemic disease caused by a new, viral, zoonotic pathogen 2019‐nCoV. This virus was again renamed as severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) by the International Committee on Taxonomy of Viruses (Gorbalenya et al., 2020). Also, WHO declared COVID‐19 as the sixth public health emergency of international concern (PHEIC). SARS‐CoV‐2 is closely related to bat‐SL‐CoVZXC21 and bat‐SL‐CoVZC45 and spread by human‐to‐human transmission via droplets or direct contacts (Carlos, Dela Cruz, Cao, Pasnick, & Jamil, 2020; Chang et al., 2020; Li, Guan, et al., 2020; Wang, Hu, et al., 2020). Moreover, the hospital‐related transmission was also suspected in 41% of patients (Wang, Hu, et al., 2020). The infection may have mean incubation period of 6.4 days and a basic reproduction number of 2.24–3.58. In this period, the patient may not display any symptoms (asymptomatic). The most common symptom for COVID‐19 is fever, pneumonia followed by cough. As per computed tomography images of chest, bilateral lung involvement with GGO is the most common finding (Lai, Shih, Ko, Tang, & Hsueh, 2020).
Emerging infections were shown among pregnant women and their fetuses (Rasmussen & Hayes, 2005) with the increased risk of complications with H1N1 influenza (Siston et al., 2010) and Zika virus (Moore et al., 2017; Rasmussen, Jamieson, Honein, & Petersen, 2016). Basing on that, Chen, Guo, et al., 2020 have reported the cesarean data for nine COVID‐19 patients in their third trimester. The patients had fever (seven), fever with cough (four), myalgia (three), sore throat (two), and malaise (two). Fetal distress was also observed in two cases. Furthermore, five patients had lymphopenia and three patients had increased aminotransferase concentrations. Nine live births were recorded. No neonatal asphyxia was observed in newborn babies. Amniotic fluid, neonatal throat swab, cord blood, and breast milk samples were tested and found negative for the virus. The clinical characteristics of pregnant women were similar with non‐pregnant patients. Since small number of cases analyzed, so more follow‐up studies should be done to evaluate the safety and health of pregnant women having COVID‐19 infection (Qiao, 2020). Zhu, Xu, et al. (2020)) have reported first time the clinical characteristics of a 52‐year‐old COVID‐19 patient, who had received kidney transplantation 12 years ago. The overall clinical features were similar with non‐transplanted COVID‐19 patients. The patient was successfully recovered with low‐dose methylprednisolone‐based therapy. These data may be helpful for the future treatment of such type of patients.
The epidemic has spread from China to 212 countries and territories. According to WHO as on 19 June 2020, the number of confirmed cases of COVID‐19 is 8,385,440 (WHO, 2020). The countries like USA, Brazil, Russia, India, UK, Italy, Spain, France, Germany, Turkey, Saudi Arabia, Peru, Chile, Pakistan, Mexico, Bangladesh, and Canada have reported more positive cases. The outbreak has killed 4,50,686 people to date. Moreover, Gilbert et al., 2020 have evaluated the preparedness and vulnerability of African countries against their risk toward COVID‐19. As per their study, Algeria, Egypt, and South Africa have highest importation risk, whereas, Sudan, Angola, Nigeria, Ethiopia, Tanzania, Kenya, and Ghana have moderate risk. Hence, it is most important to provide necessary epidemiological data so that appropriate decisions are made regarding Paralympic Games, Tokyo 2020 Olympic and similar type of mass gatherings (Gallego, Nishiura, Sah, & Rodriguez‐Morales, 2020).
2. ENTRY MECHANISM OF HUMAN CORONAVIRUSES
To examine the possibilities of domestic animals as reservoir, serological studies were performed on sheep, goats, cattle, and chickens in Saudi Arabia, UAE, Jordan, and Europe. All samples tested seronegative for MERS‐CoV (CDCP, 2016; Hemida et al., 2013; Meyer et al., 2015; Reusken, Ababneh, et al., 2013; Reusken, Haagmans, et al., 2013). But, dromedary camels are the primary animal host for MERS‐CoV (Al‐Tawfiq, 2013; Azhar, El‐Kafrawy, et al., 2014; Chan et al., 2014; Reusken et al., 2016). And human‐to‐human transmission occurs by close contacts and droplets or due to nosocomial infection (Bauerfeind et al., 2016; Sabir et al., 2016). It is uncertain how COVID‐19 transmission occurred from animals to humans. Initially, large numbers of infected people were linked to the wet animal market in Wuhan where live animals are routinely sold. So, it is suggested to be the zoonotic origin of the COVID‐19. At beginning, two species of snakes identified as possible reservoir of the COVID‐19. But, there is no consistent evidence of coronavirus reservoirs other than mammals and birds (Bassetti, Vena, & Giacobbe, 2020; Ji, Wang, Zhao, Zai, & Li, 2020). Genomic sequence analysis of COVID‐19 showed similarity with two bat‐derived SARS‐like coronaviruses (Lu, Stratton, et al., 2020; Wan, Shang, Graham, Baric, & Li, 2020), indicating that mammals are the most likely link between COVID‐19 and humans.
In the case of human‐to‐human transmission, the mechanism of transmission remains controversial (Chan et al., 2020; Li, Guan, et al., 2020; Phan et al., 2020) to some extent. The reports have suggested that human‐to‐human transmission is a likely route for spreading the novel CoV, which is supported by the cases that occurred within families or among the people who did not visit the wet animal market (Carlos et al., 2020; Wu, Hao, et al., 2020). Human‐to‐human transmission occurs via direct contact or through droplets spread by coughing or sneezing from an infected individual. In some cesarean cases of pregnant women having COVID‐19 positive, there was no evidence for transmission from mother to child. Hence, it is not clear whether transmission can occur during vaginal birth or not. Moreover, no evidence of in utero transmission was seen in SARS or MERS to date (Rasmussen, Smulian, Lednicky, Wen, & Jamieson, 2020).
SARS‐CoV‐2 possesses spike protein, nucleocapsid protein, membrane glycoprotein, small membrane protein, and hemagglutinin esterase (Wu, Hao, et al., 2020; Zhou, Yang, et al., 2020). The spike protein contains a 3‐D structure in the RBD region to maintain van der Waals forces (Xu, Chen, et al., 2020). The glycoprotein spikes present on the outer surface are mostly responsible for attachment and entry to the host cells (Figure 3). The RBD is loosely attached among viruses, so, the virus may infect multiple hosts (Perlman & Netland, 2009; Raj et al., 2013). SARS‐CoV and MERS‐CoV recognize exopeptidases as a key receptor for entry to human cells (Wang et al., 2013), while others mostly recognize aminopeptidases or carbohydrates. The entry mechanism of human coronavirus depends upon HAT and TMPRSS2, which splits spike protein and may establish further penetration changes (Bertram et al., 2011; Du et al., 2009). MERS coronavirus binds DPP4, while SARS‐coronavirus binds ACE2 as a key receptor (Raj et al., 2013; Wang et al., 2013) and both are found in lung cells. Also, it is not clear whether COVID‐19 is receiving ADE from other coronaviruses or not. We are at the beginning to understand the dynamics of COVID‐19 in humans and its impact (Tetro, 2020). COVID‐19 infection has caused pulmonary, renal, cardiac, and circulatory damage that proves fatal in patients. Keeping in mind, Baig, Khaleeq, Ali, & Syeda, 2020 have investigated the involvement and possible contribution of neurological tissue damage to the morbidity and mortality caused by COVID‐19.
FIGURE 3.
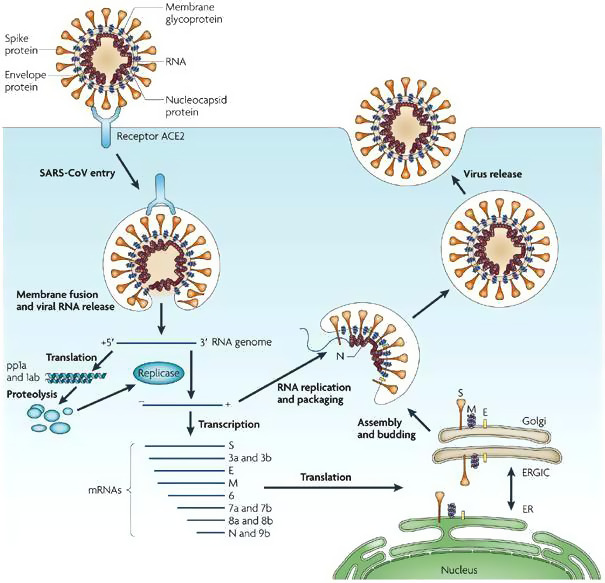
Entry mechanism of human coronaviruses (Reproduced from Du et al., 2009, Nature reviews: Microbiology ©Springer Nature)
3. DIAGNOSIS OF COVID‐19
A number of “unknown viral pneumonia” cases in China by a novel coronavirus (SARS‐CoV‐2) (Huang, Wang, et al., 2020; WHO, 2020c) were suddenly reported in December 2019. In just three months, this highly contagious virus has spread from Wuhan to almost all the continents. As the proper vaccination process for SARS‐CoV‐2 is not available, it is very important to identify it in early stage. By which immediate isolation of the infected person may be taken care from the healthy population. According to the latest guideline (trial sixth version) published by China government (General Office of National Health Committee., 2020), the diagnosis of COVID‐19 should be confirmed by RT‐PCR or gene sequencing for respiratory or blood specimens. Also, Chest CT is playing important role in the diagnosis of current COVID‐19 detection in epidemic areas. Keeping in mind, Ai et al., 2020 have reported the results of chest CT in comparison with RT‐PCR results in 1,014 patients with suspected COVID‐19.
Bernheim et al., 2020 have analyzed chest CT findings in 121 infected patients in relationship to the time between symptom onset and the initial CT scan. In this study, they observed that certain CT findings are more common depending on the time course of infection. The reported chest imaging features of COVID‐19 are similar with other types of CoV syndromes. Keeping in mind, Kooraki, Hosseiny, Myers, & Gholamrezanezhad, 2020 have reported the precautions and safety measures for medical staff members to manage patients with suspected NCIP. Clinical diagnosis of COVID‐19 is mainly based on epidemiological history, clinical manifestations, nucleic acid detection, CT scan, POCT of IgM/IgG, ELISA, and blood culture. CoV infections led to damage the lung and may cause pneumonia. In light of this, Li, Guan, et al., 2020 have discussed the current knowledge of diagnosis, treatment and molecular immune pathogenesis for COVID‐19. The combining assessment of laboratory and clinical findings could be helpful for early diagnosis of COVID‐19 pneumonia (Shi, Han, et al., 2020).
Radiologists are the first‐line health care person, exposed directly to COVID‐19 patients. The role of radiologists is given by Kim, 2020. According to him, with the detection of lung abnormality, they should suggest the disease severity and possible co‐infection in other hospitalized patients. Lei, Li, Li, & Qi, 2020 have discussed the CT images in a 33‐year‐old woman. According to their study, the bilateralism of the peripheral lung opacities, without subpleural sparing, is the common CT findings of COVID‐19 pneumonia. A low‐cost and disposable colorimetric PAD should be developed for simple, rapid screening and detecting in infectious diagnostics. Such alternative approach of detection for MERS‐CoV has also been developed (Teengam et al., 2017).
4. TRAVEL RESTRICTIONS AND TRANSMISSIBILITY REDUCTION
The ongoing epidemic of COVID‐19 is becoming unstoppable and has broken all previous records and declared pandemic by WHO (Callaway, 2020). Yang et al., 2020 have studied 149 confirmed patients in three tertiary hospitals of Wenzhou. 85 patients had Hubei residence/ travel history, 49 had contact with people of Hubei, and 15 had no travel/contact history. Most patients presented mild infection. Based on the rapid spread of this novel virus in Mainland China, Chinazzi et al., 2020 have reported a global metapopulation disease transmission model to understand the effect of travel limitations on transmission of this ongoing epidemic. Also, Dong, Du, & Gardner, 2020 have developed an interactive web‐based dashboard to track COVID‐19 in real time. This study was hosted by CSSE at Johns Hopkins University, USA, to visualize and track reported cases of COVID‐19 in real time.
The Diamond Princess Cruise ship having 3,711 people was infected by SARS‐CoV‐2 (Ministry of Health, Labour and Welfare of Japan, 2020). A significant number of positive cases have been identified, which may be due to human‐to‐human transmission as there were limited space and high population density (Zhang, Diao, et al., 2020). The novel COVID‐19 is highly contagious and is transmitted mostly through respiratory droplets. But, whether its transmission can be forwarded by touching a surface (ie, a fomite) is uncertain. Hence, a thorough understanding of the virus transmission should be essential as we know the transmission of MERS‐CoV occurs mainly through nonhuman, zoonotic sources (camels, bats; Mobaraki & Ahmadzadeh, 2019). The proposed transmission mechanism of human CoV is shown in Figure 4.
FIGURE 4.
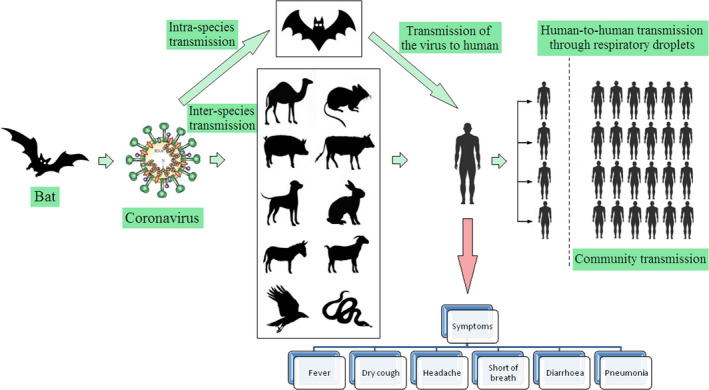
Transmission mechanism of human coronavirus
Isolation of positive cases and contact tracing is used to control the outbreak. Keeping in mind, Hellewell et al., 2020 have used a mathematical model to assess whether isolation and contact tracing are really able to control the outbreak. Moreover, Rothana & Byrareddy, 2020 have reported the pathogenesis, phylogenetic analysis, epidemiology, symptoms, transmission, and future directions to control the onward transmission. The containment measures in China are successfully reducing transmission, which decreased the epidemic growth (Roosa et al., 2020). Such reduction is not the case elsewhere, and USA, Brazil, Italy, Spain, Germany, France, Russia, and India have been affected a lot (Remuzzi & Remuzzi, 2020; WHO, 2020). In these countries, a large number of asymptomatic carriers and mild patients may be present in the community. It is very important to identify and isolate them from healthy population (Hu et al., 2020). As we all know that prevention is better than cure, preventing and controlling the outbreak must be focused on the early isolation of patients and quarantine from close contacts in families and healthy population (Tian, Hu, et al., 2020). Moreover, social distancing will play a positive effect to control the outbreak as it will decrease the community transmission.
5. SYMPTOMS OF COVID‐19 INFECTION
The symptoms of novel COVID‐19‐infected pneumonia appear after an incubation period of ~5.2 days (Li, Guan, et al., 2020). The common symptoms for COVID‐19 are identified by fever, dry cough, headache, fatigue, anosmia, and dyspnea. But in second phase, other symptoms such as sputum production, haemoptysis, diarrhea, and lymphopenia will be observed (Carlos et al., 2020; Huang, Wang, et al., 2020; Ren et al., 2020; Wang, Tang, et al., 2020). The clinical features of chest CT scan revealed pneumonia, RNAaemia, acute cardiac injury, and GGOs (Huang, Wang, et al., 2020).
Both SARS‐CoVs enter the cell via ACE2 receptor (Wan et al., 2020; Zhou, Yang, et al., 2020). The SARS‐Cov‐2 first infects lower airways and binds to ACE2 (found in lung) on alveolar epithelial cells. Both viruses activate immune cells and induce the secretion of inflammatory cytokines and chemokines. The “cytokine storm” is the proposed mechanistic theory for organ damage, which leads to death (Jiang et al., 2020). The period of death ranged from 6 to 41 days from the onset with a median of 14 days (Wang, Tang, et al., 2020). This depends on the age and immune system of the patient.
6. POSSIBLE VACCINES FOR COVID‐19 PNEUMONIA
No approved vaccines or specific treatments are yet available against SARS‐CoV, MERS‐CoV infection for potential therapy of humans. So many researchers are trying to develop a vaccine using a variety of platforms (Graham, Donaldson, & Baric, 2013; Kato et al., 2019; Schindewolf & Menachery, 2019). It may take months and years to develop particular vaccines for such a rapidly mutating SARS‐CoV‐2 as these studies are in progress and more laboratory and clinical trials still should be explored. Furthermore, antigen presentation may be affected by CoV (Menachery et al., 2018). According to WHO, minimum 18 months required for the development of COVID‐19 vaccines (Huaxia, 2020).
Since SARS‐CoV‐2 is a kind of RNA virus, so it possesses some similar functional proteins as of HIV. Hence, the HIV protease inhibitors may be effective for potential therapy of humans. Recently, lopinavir/ritonavir (LPV/r) combination has been recommended for the treatment of COVID‐19 pneumonia. This combination was previously confirmed for the effective treatment of MERS‐CoV and SARS‐CoV (Chu et al., 2004; Momattin, Al‐Ali, & Al‐Tawfiq, 2019). Baricitinib may be suggested as a potential drug for COVID‐19 treatment which might reduce the process of both virus invasion and inflammation (Richardson et al., 2020). As per molecular docking studies, another anti‐HIV drug nelfinavir was anticipated to be active against SARS‐CoV‐2 (Xu, Peng, et al., 2020). Remdesivir and chloroquine could inhibit COVID‐19 infection in vitro (Wang, Cao, et al., 2020). Moreover, Arbidol has a broad‐spectrum antiviral activity against respiratory viruses. The clinical trials of safety and efficacy of these drugs are going on. In addition, SARS‐CoV‐specific human mAb (CR3022) could bind potently with 2019‐nCoV receptor‐binding domain (Tian, Li, et al., 2020). So, CR3022 should be developed to prevent and treat new coronavirus infections.
It is also reported that 75 patients were treated by using 75 mg oseltamivir, 500 mg lopinavir, 500 mg ritonavir, and 0.25 g ganciclovir (intravenous) for 3–14 days (Chen, Guo, et al., 2020; Chen, Zhou, et al., 2020). Moreover, one patient treated by lopinavir/ritonavir showed improvements in his chest radiographs after four days (Huang, Tu, et al., 2020). This combination was also reported by Bhatnagar et al., 2020 with the protocol as per the guidelines of ICMR and WHO (WHO, 2020d). Also, a case of COVID‐19 pneumonia is responding well to remdesivir in USA, which is now undergoing a clinical trial in China (Cao, Deng, & Dai, 2020; Lai et al., 2020). Hence, it may be used as a possible therapeutic option for COVID‐19 pneumonia (Al‐Tawfiq, Al‐Homoud, & Memish, 2020; Ko et al., 2020). The safety and efficacy of chloroquine for the treatment of COVID‐19 are discussed by Cortegiani, Ingoglia, Ippolito, Giarratano, & Einav, 2020. Furthermore, hydroxychloroquine may be effective in inhibiting COVID‐19 infection due to less toxic nature (Liu, Cao, et al., 2020; Liu, Xiao, et al., 2020; Sivabakya & Srinivas, 2020). Caly, Druce, Catton, Jans, & Wagstaff, 2020 have also reported recently that the anti‐parasitic Ivermectin inhibits the replication of SARS‐CoV‐2 in vitro. The ongoing advances in designing vaccines and therapeutics to counter COVID‐19 pneumonia have been highlighted by Dhama et al., 2020. Moreover, Lin, Lu, Cao, & Li, 2020 have recommended high‐doses intravenous immunoglobulin and low‐molecular‐weight heparin anticoagulant therapy as an effective treatment for severe and critical type patients. In addition, some recent clinical trials on drug repositioning for COVID‐19 pneumonia treatment were also listed (Fisher & Heymann, 2020; Rosa & Santos, 2020). The novel virus may damage the central nervous system of the patient (Zhou, Zhang, Wang, & Gao, 2020). In that case, the health workers should focus on intensive care. The combined effect of hydroxychloroquine and ivermectin is also suggested by Patrì & Fabbrocini, 2020. But there is no in vitro evidence reported on the combined effect. Moreover, sitagliptin may be useful for the patients having pre‐existing cardiovascular and (or) diabetic issues (Bardaweel, Hajjo, & Sabbah, 2021). Due to the involvement of immune hyperactivation and ARDS in fatal cases of COVID‐19, anti‐rheumatic drugs such as hydroxychloroquine and tocilizumab have been proposed by Zhong, Tang, Ye, & Dong, 2020 as potential therapies for the treatment of COVID‐19. Triple antiviral therapy with lopinavir–ritonavir, interferon beta‐1b, and ribavirin were safe and superior to lopinavir–ritonavir alone for mild to moderate COVID‐19 patients (Hung et al., 2020). In addition, Teicoplanin, an antibiotic exhibited significant efficacy to inhibit the first stage of MERS‐CoV previously. Its activity may be beneficial against this novel virus (Baron, Devaux, Colson, Raoult, & Rolain, 2020). Some existing drugs (Figure 5) used against COVID‐19 infection are listed in Table 3. However, more research is urgently needed to identify novel chemotherapeutic drugs for treating and controlling COVID‐19 infections. Several clinical trials are going on to assess the adequacy of therapeutic agents (Menni, Sudre, Steves, Ourselin, & Spector, 2020). Such drugs will help to reduce the morbidity and mortality, whereas, prophylactics and vaccines will prevent further disease transmission (Park, Thwaites, & Openshaw, 2020).
FIGURE 5.
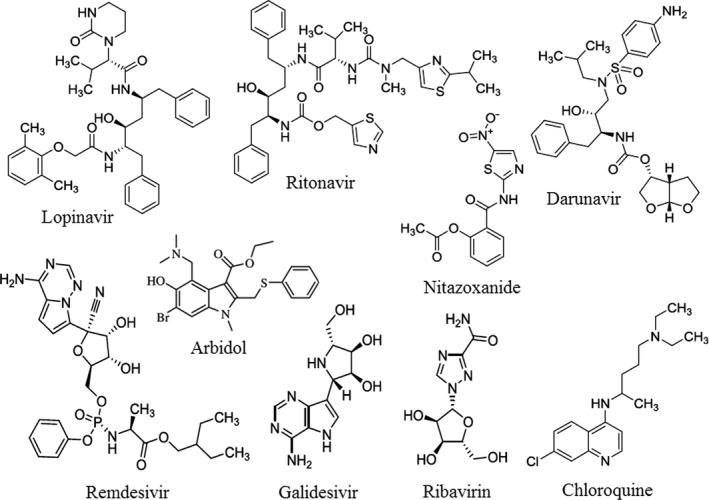
The possible drugs used against COVID‐19 pneumonia
Table 3.
Some existing drugs for the possible use against COVID‐19
| Sl. No. | Name of the drug | Possible mechanism | Disease | Trials, clinical studies, outcome |
|---|---|---|---|---|
| 1 | Lopinavir | May inhibit the viral proteases | HIV infection | No benefit, In combination Lopinavir and Ritonavir not found to improve COVID‐19 patients |
| 2 | Ritonavir | May increase the levels of other protease inhibitors | ||
| 3 | Darunavir | Act as a antiretroviral protease inhibitor | HIV infection | In vitro study revealed no benefit |
| 4 | Remdesivir | May block viral nucleotide synthesis to stop viral replication | Ebola infection | Potential benefit found in preliminary results |
| 5 | Galidesivir | May leads to the structural change in the viral enzyme and results premature termination of the elongating RNA | Ebola, hepatitis C, Marburg virus | In vitro experiments suggest benefits |
| 6 | Ribavirin | May inhibits viral mRNA polymerase and leading to a decrease in viral replication (or production of defective virions) | Hepatitis C, RSV infection | Results awaited |
| 7 | Chloroquine | May elevate endosomal pH and interfere with ACE2 glycosylation | Malarial infection | Potential benefit |
| 8 | Nitazoxanide | May inhibit viral protein expression | Helminthic and protozoal infection | Potential benefit |
| 9 | Arbidol (Umifenovir) | May prevent viral entry to the target cell | influenza infection | No benefit |
| 10 | Ivermectin | Viral Protease | Anti‐parasitic agent, anti‐HIV | Results awaited |
| 11 | Hydroxychloroquine | Blocking Virus–Cell Membrane Fusion | Antimalarial and antiautoimmune Agent | Mixed, No benefit |
| 12 | Tocilizumab | IL‐6 inhibition | Inflammation, cytokine release syndrome (CRS) | Potential benefit |
| 13 | Glucocorticosteroids | Downregulation of inflammatory cytokines | Allergy, hypersensitivity, autoimmune diseases | No benefit |
| 14 | Intravenous immunoglobulin (IVIG) | Pleiotropic immunomodulation | anti‐inflammatory and cytoprotective | Potential benefit |
CoV is biologically diverse and rapidly mutating. So, anticoronavirus therapy is somewhat challenging. Zhang, Lin, et al. (2020)) have designed some broad‐spectrum antiviral compounds and suggested to exhibit excellent properties against MERS‐CoV, SARS‐CoV, and the novel BetaCoV/Wuhan/2019 as well. Łoczechin et al., 2019 have reported the antiviral activity of seven different CQDs for the treatment of human coronavirus infections. In addition, Suwannarach et al., 2020 have also studied the immunomodulatory activities of fungal bioactive compounds as potential therapeutic agents against CoV. Liu, Zhou, et al., 2020 have discussed some patents disclose methodologies for treating and preventing CoV infections, which may be applicable to COVID‐19. As respiratory failure is the most important findings of COVID‐19, attention should be given for critically ill patients during mechanical ventilation in hospitals (Linga, Joynta, Lipman, Constantine, & Joannes‐Boyau, 2020). Till date, more than 500 clinical trials (randomized/ non‐randomized) were registered with standard of care. Other trials are also enrolling over remdesivir, IL‐6 inhibitors (tocilizumab and sarilumab), stem‐cell therapy, convalescent plasma therapy, corticosteroids, vaccine candidates, direct‐acting antiviral drugs, and traditional Chinese medicines. Most of these trials are giving comparative efficacy (Table 3) (Thorlund et al., 2020; Tu et al., 2020). In addition, six vaccine candidates namely mRNA (NCT04283461), aAPC (NCT04299724), DNA (NCT04336410), a lentiviral vector (NCT04276896), chimpanzee adenoviral vector ChAdOx1 (NCT04324606), and adenoviral vector 5 (NCT04313127) have entered phase I clinical trials to assess their safety and immunogenicity (Kim, Dema, & Reyes‐Sandoval, 2020).
7. ZOONOTIC TRANSMISSION
Although it was believed that bats and pangolins were potential hosts for SARS‐CoV‐2 transmission, but the origin of SARS‐CoV‐2 remains unclear and the evidence of transmission from animals to human is not established with certainty (Zhai, Wei, Lv, Xu, et al., 2020). As reported earlier, the initial infected individuals had a common exposure spot; Wuhan Seafood wholesale market of China, where various domestic animals, wild animals and live animals including poultry, bats, rabbits, turtles, marmots, pangolins, and snakes were sold for human consumption (Hui et al., 2020; Ji et al., 2020; Lu et al., 2020). The Chinese people are commonly consumed wild animal meat and their products due to their believed medicinal value and health promoting effects (Harypursat & Chen, 2020). So, the host range now gets expanded and it is very essential to understand the risks to other animals and possible zoonotic transmission.
On 28 February 2020, a 17‐year‐old Pomeranian dog in Hong Kong had tested positive as stated by Agriculture, Fisheries and Conservation Department (AFCD) (AFCD, 2020). The dog had initially tested negative for antibodies specific to COVID‐19 as antibody formation may take 14 days or more (Almendros, 2020a). Since then, another case was reported in a two‐year‐old German shepherd dog on 19 March (AFCD, 2020a,2020b). In both the cases, the dogs were living in close contact with SARS‐CoV‐2 positive owners (American Veterinary Medical Association, 2020). After that, AFCD of Hong Kong had conducted tests on 17 dogs and 8 cats, out of which only two dogs had tested positive. Both dogs later tested negative on RT‐PCR, which is the most sensitive test for the diagnosis of COVID‐19 (Wang, Kang, Liu, & Tong, 2020). This study supported human‐to‐animal transmission. But it is still not confirmed that dogs can transmit this virus to human beings (Almendros, 2020b).
On 31 March, a cat had tested positive on RT‐PCR from oral, nasal, and fecal samples in Hong Kong by the GHKSAR (GHKSAR, 2020). Furthermore, a cat tested positive at the University of Liège in Belgium. After this issue, Shi, Wen, et al., 2020; Shi, Han, et al., 2020 have reported an experimental infection of a number of companion and domestic animals by using two SARS‐CoV‐2 viruses isolated from environment and human COVID‐19 patient. They confirmed that cats and ferrets are highly susceptible to COVID‐19 (Li, 2020). They also suggested that SARS‐CoV‐2 was transmitted in cats via respiratory droplets. Moreover, cats in Wuhan were infected with SARS‐CoV‐2 during the outbreak, which clearly suggested true infection in them (Zhang, Wang, Qi, Shen, & Li, 2020). The infected cats may transmit the virus to other animals living in close contact with them (Martina et al., 2003). The first human to nondomestic animal transmission was reported in New York City, where a tiger was surprisingly infected by a positive asymptomatic zookeeper (USDA, 2020). Hence, it is suggested that the owners should adopt appropriate precautions and keep a suitable distance from the animals to prevent the spread of this virus (Rodriguez‐Morales, Dhama, Sharun, Tiwari, & Bonilla‐Aldana, 2020; Zhai, Wei, Lv, Xu, et al., 2020; Zhai, Wei, Lv, Zhai, et al., 2020).
Due to the broad host range, the virus keeps on circulating between vertebrates, various animal species, and humans (Tiwari et al., 2020). The wide host range of this novel virus is believed to be due to instability of the replicase enzyme, RNA‐dependent RNA polymerase, O‐linked glycans, polybasic furin cleavage site, higher rate of mutations, and genetic recombination (Chen, 2020; Patel & Jernigan, 2020). Initially, it was believed that bats or pangolins are the probable sources of origin of this novel virus (Andersen, Rambaut, Lipkin, Holmes, & Garry, 2020; Wu, Zhao, et al., 2020; Zhang, Wu, & Zhang, 2020). But the actual intermediate host and the nature are yet to be explored.
8. ANIMAL MODELS
Basic research on SARS‐CoV‐2 is essential to support the process of development of the therapeutic agents. It is useful to realize some models that can authentically imitate the comportments of coronavirus and reproduce the pathology of COVID‐19 (Takayama, 2020). Here, we enumerated briefly the applicable models: cell lines, organoids, and animal models.
Cell lines and organoids for SARS ‐CoV‐2
The in vitro cell model can offer valuable information about the virus infection. The cell lines (Human airway epithelial, Vero E6, Caco‐2, Calu‐3, HEK 293T, Huh 7) and organoids (human bronchial, human lung, human kidney, human liver ductal, human intestinal, human blood vessel) are a method to study the virus and the interactions inside host cells (Takayama, 2020).
Animal models for SARS ‐CoV‐2
Animal models are providing significant contributions to evaluate the vaccines for a particular disease. It will help in understanding the disease process and developing prophylactics and therapeutics (Dhama et al., 2020). The pathology of COVID‐19 can be replicated and observed in a tissue‐specific by using animal models. Small animals (wild‐type mice, human ACE2 transgenic mice, Syrian hamsters) and large animals (ferrets, cats, Cynomolgus macaques, Rhesus macaques) are being used for evaluating vaccines and drugs against SARS‐CoV‐2 (Takayama, 2020; Tiwari et al., 2020). Rhesus macaques displayed high virus amount in oral‐naso and rectal swabs while working on SARS‐CoV‐2. In studying the pathogenesis of the disease, the apparent lesions and clinical signs contribute significantly in developing vaccines and antivirals (Munster et al., 2020).
Golden Syrian hamsters were suggested to be the promising animal model when investigated against SARS type strains for revealing CoV pathology and pathogenesis (Roberts et al., 2008). Furthermore, transgenic mice are now considered to be compatible model for SARS‐CoV‐2 but initially, mice were challenging due to the difference in receptor ACE2 usage pattern (Liu, Cao, et al., 2020; Liu, Cao, et al., 2020; Liu, Zheng, et al., 2020; Liu, Xiao, et al., 2020; Wang, Tang, et al., 2020; Zhou, Yang, et al., 2020). On modeling cats, ferrets, pigs, orangutans, monkeys, bats, and humans are akin based on the structural similarity of their ACE2 receptors (Jarvis, 2020). Hence, these may contribute significantly as animal models to investigate further studies.
9. IMMUNOTHERAPY
As there are no approved vaccines or antiviral therapeutic agents for the treatment of COVID‐19, the existing pieces of evidence are discussed herewith. It is suggested that immunotherapy is an efficient therapeutic option against the infected patients (Jafari & Ghasemi, 2020). Acute respiratory distress syndrome (ARDS) can induce the apparition of other complications, including secondary bacterial infection and lung fibrosis. Recent clinical data have led to the conclusion that severe COVID‐19 is determined by an over reactive immune response leading to a cytokine storm and the development of ARDS (Bonam, Kaveri, Sakuntabhai, Gilardin, & Bayry, 2020). For severe cases, host‐directed immunotherapy is an adjunct therapy to reduce inflammation and inflammation‐associated lung damage and to prevent ICU hospitalization. Several immunotherapeutic strategies targeting either inflammatory mediators, or passively neutralize SARS‐CoV‐2, or prevent them from viral entry are under evaluation (Bonam et al., 2020; Groß, Jahn, Cushman, Bär, & Thum, 2020; Jafari & Ghasemi, 2020; Shih, Wu, Tu, & Chi, 2020).
Till date, all immunotherapy attempts for this novel virus comprise ACE2 immunoadhesin, immunoglubolins, polypeptide hormone for maturation of T cells, a monoclonal antibody against interleukin‐6 (IL‐6) and polyclonal antibody by plasma therapy. Immunoglobulins and plasma therapy intervention for SARS‐CoV‐2 infected patients could improve clinical outcomes (Jafari & Ghasemi, 2020). Moreover, to block the virus entry and attachment, monoclonal antibodies are preferred due to their purity, specificity, safety, low risk of blood‐borne pathogen contamination (Shanmugaraj, Siriwattananon, Wangkanont, & Phoolcharoen, 2020). ACE2 immunoadhesin has not been tested yet. Some of these methods were discussed in a clinical trial study reported by Zhang, Zhang, et al. (2020)).
Most of the COVID‐19 patients were observed to have increased in IL 6, leading to lung tissue damage. The same elevated serum levels of IL‐6, TNF‐α, and IFN‐γ were observed in cytokine release syndrome (CRS). Interleukin‐6 (IL‐6) acts as both anti‐inflammatory myokine and pro‐inflammatory cytokine. A rapid resolution of CRS symptoms in COVID‐19 patients has to target either IL‐6 receptor (tocilizumab, a recombinant humanized monoclonal antibody) or IL‐6 (siltuximab, a chimeric monoclonal antibody) to avoid the pulmonary edema, hypotension, and cardiac dysfunction (Le et al., 2018). Human monoclonal antibody (sarilumab), Anti‐IL‐6 receptor antibodies (tocilizumab), and anti‐IL‐6 antibody (siltuximab) are under evaluation in different centers (Baruah & Bose, 2020; Kruse, 2020; Liu, Zhang, et al., 2020). ASCIA recommends that tocilizumab may be considered as an off‐label treatment option for the patients with ARDS, and its clinical trials.
Furthermore, other interventions such as nanoparticles, viral‐vectors, and corticosteroids were also suggested for SARS‐CoV‐2. In this contest, Liao et al., 2020 have discussed the bronchoalveolar immune microenvironment design underlying immunopathogenesis in COVID‐19. Reis & Lima, 2020 have proposed a modified version of the CHA2DS2‐VASc score to reduce thromboembolic complications before the condition becomes severe. WHO advised “Do not routinely give systemic corticosteroids for the treatment of viral pneumonia outside clinical trials” (WHO, 2020d). However, ASCIA supports its use because the available data were generally low quality with small patient numbers (ASCIA, 2020). In addition, Klimek et al., 2020 have suggested the allergen‐specific immunotherapy (AIT) that confers a long‐term clinical benefit (activates immune tolerance responses) for the diseases like allergic rhinoconjunctivitis, allergic bronchial asthma, etc. (Bousquet, Lockey, & Malling, 1998; Jutel et al., 2013). The immune mechanisms in AIT and COVID‐19 were also discussed (Klimek et al., 2020). This report may provide ample references for the researchers to develop vaccines and antivirals against this novel emerging virus.
10. CONCLUSIONS
This manuscript has discussed various elements of COVID‐19 infection and its onward transmissions. After originating from Wuhan, this zoonotic virus has spread rapidly to 213 countries and territories in the globe and causing SARS in humans, which forced to declare it “Pandemic” by WHO, as R0 > 1. As per the laboratory confirmed positive cases, respiratory failure is the most common findings of COVID‐19. But the common symptoms are dry cough, fever, headache, loss of smell or taste, muscle pain, dyspnea, diarrhea, and pneumonia. This recent outbreak believed to be originated from bats and has resulted 4,50,686 deaths as on 19 June 2020 due to human‐to‐human transmission. Due to this onward transmission, more than 83 lakh positive cases are found in the globe till date and still increasing rapidly. Some countries are trying to fight by preventing community transmission to control the outbreak. Due to unclear mechanism for COVID‐19 transmission, it is difficult to develop evidence‐based infection control protocols to prevent community transmissions. According to the latest guideline published by China government, the diagnosis of COVID‐19 should be confirmed by RT‐PCR or gene sequencing for respiratory or blood specimens. Furthermore, Chest CT is playing a vital role in early detection of current COVID‐19 pneumonia. The clinical features of chest CT scan revealed pneumonia, RNAaemia, acute cardiac injury, and GGOs. Some patients may present normal chest finding despite of testing COVID‐19 positive.
As there are no specific vaccines available against COVID‐19 infection, the treatment may be carried out by using available broad‐spectrum antiviral drugs like nucleoside analogues and HIV protease inhibitors. Lopinavir/ritonavir (LPV/r) combination has been recommended for the treatment. The broad‐spectrum antiviral remdesivir and chloroquine are highly effective in the control of 2019‐nCoV infection. Also, the combined effect of hydroxychloroquine and ivermectin will play important rule in inhibiting 2019‐nCoV infection. It is also suggested that hydroxychloroquine and azithromycin may be useful for the early treatment of patients. Another most important thing is “isolation of positive cases, contact tracing, and social distancing will be very helpful to control the outbreak”. In this contest, Chu et al., 2020 have suggested that physical distancing, face masks, and eye protection to prevent person‐to‐person transmission. According to Hibberd, “If you can identify and quarantine most of the positive cases, then you do not have to lockdown everyone else” (Burki, 2020). Furthermore, we may boost our immunity with Tinospora cordifolia (Guduchi) as recommended by Ayurveda, India. In addition, preventives (Arsenic album) may be used as suggested by Department of Ayush, Government of India. This review presents a strong intellectual background on the origin, transmission, symptoms, diagnosis, possible vaccines, animal models, and immunotherapy for this novel COVID‐19 pneumonia, which gives an in‐depth insight into several aspects of this global pandemic and will provide ample references for the researchers as well as society. Moreover, the information's included in this review may be useful for the ongoing development of therapeutic agents and vaccines and also preventing the spread of this disease.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest in this work.
ACKNOWLEDGMENTS
The authors are highly thankful to the editor and reviewers for their valuable and constructive suggestions to modify the manuscript. RKM is also thankful to Principal, Government College of Engineering, Keonjhar, for his help and facilities.
Mohapatra RK, Pintilie L, Kandi V, et al. The recent challenges of highly contagious COVID‐19, causing respiratory infections: Symptoms, diagnosis, transmission, possible vaccines, animal models, and immunotherapy. Chem Biol Drug Des. 2020;96:1187–1208. 10.1111/cbdd.13761
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- AFCD . (2020). Blood test result of pet dog with low‐level infection of COVID‐19 released. Retrieved from www.afcd.gov.hk/english/publications/publications_press/pr2343.html
- AFCD . (2020a). Pet dog tests positive for COVID‐19 virus. Retrieved from www.afcd.gov.hk/english/publications/publications_press/pr2346.html
- AFCD . (2020b). Low‐level of infection with COVID‐19 in pet dog. Retrieved from www.afcd.gov.hk/english/publications/publications_press/pr2342.html
- Ai, T. , Yang, Z. , Hou, H. , Zhan, C. , Chen, C. , Lv, W. , … Xia, L. (2020). Correlation of chest CT and RT‐PCR testing in coronavirus disease 2019 (COVID‐19) in China: A Report of 1014 Cases. Radiology, In press. 10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alharbi, N. K. , Qasim, I. , Almasoud, A. , Aljami, H. A. , Alenazi, M. W. , Alhafufi, A. , … Balkhy, H. H. (2019). Humoral immunogenicity and efficacy of a single dose of ChAdOx1 MERS vaccine candidate in dromedary camels. Scientific Reports, 9, 16292. 10.1038/s41598-019-52730-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendros, A. (2020b). Can companion animals become infected with Covid‐19? The Veterinary Record, 186(12), 388–389. 10.1136/vr.m1194 [DOI] [PubMed] [Google Scholar]
- Al‐Tawfiq, A. (2013). Middle East Respiratory Syndrome‐coronavirus infection: An overview. Journal of Infection and Public Health, 6, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Tawfiq, J. A. , Al‐Homoud, A. H. , & Memish, Z. A. (2020). Remdesivir as a possible therapeutic option for the COVID‐19. Travel Medicine and Infectious Disease, 10.1016/j.tmaid.2020.101615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Veterinary Medical Association (2020). SARS‐CoV‐2 in animals, including pets. Retrieved from https://www.avma.org/resources-tools/animal-healthand-welfare/covid-19/sars-cov-2-animals-including-pets
- Andersen, K. G. , Rambaut, A. , Lipkin, W. I. , Holmes, E. C. , & Garry, R. F. (2020). The proximal origin of SARS‐CoV‐2. Nature Medicine, 26(4), 450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASCIA (2020). Position Statement ‐ Specific Treatments for COVID‐19, ASCIA Information for Health Professionals. Retrieved from www.allergy.org.au/members/covid-19
- Azhar, E. I. , El‐Kafrawy, S. A. , Farraj, S. A. , Hassan, A. M. , Al‐Saeed, M. S. , Hashem, A. M. , & Madani, T. A. (2014). Evidence for camel‐to‐human transmission of MERS coronavirus. New England Journal of Medicine, 370, 2499–2505. 10.1056/NEJMoa1401505 [DOI] [PubMed] [Google Scholar]
- Azhar, E. I. , Hashem, A. M. , El‐Kafrawy, S. A. , Sohrab, S. S. , Aburizaiza, A. S. , Farraj, S. A. , … Madani, T. A. (2014). Detection of the Middle East Respiratory Syndrome Coronavirus genome in an air sample originating from a camel barn owned by an infected patient. MBio, 5, e01450. 10.1128/mBio.01450-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar, E. I. , Hui, D. S. C. , Memish, Z. A. , Drosten, C. , & Zumla, A. (2019). The Middle East Respiratory Syndrome (MERS). Infectious Disease Clinics of North America, 33, 891–905. 10.1016/j.idc.2019.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig, A. M. , Khaleeq, A. , Ali, U. , & Syeda, H. (2020). Evidence of the COVID‐19 virus targeting the CNS: Tissue distribution, host–virus interaction, and proposed neurotropic mechanisms. ACS Chemical Neuroscience, 11(7), 995–998. 10.1021/acschemneuro.0c00122 [DOI] [PubMed] [Google Scholar]
- Bardaweel, S. K. , Hajjo, R. , & Sabbah, D. A. (2021). Sitagliptin: A potential drug for the treatment of COVID‐19? Acta Pharmaceutica, 71. 10.2478/acph-2021-0013 [DOI] [PubMed] [Google Scholar]
- Baron, S. A. , Devaux, C. , Colson, P. , Raoult, D. , & Rolain, J.‐M. (2020). Teicoplanin: An alternative drug for the treatment of coronavirus COVID‐19? International Journal of Antimicrobial Agents, 10.1016/j.ijantimicag.2020.105944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah, V. , & Bose, S. (2020. Immunoinformatics‐aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019‐nCoV. Journal of Medical Virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti, M. , Vena, A. , & Giacobbe, D. R. (2020). The Novel Chinese Coronavirus (2019‐nCoV) Infections: Challenges for fighting the storm. European Journal of Clinical Investigation, 50(3), e13209. 10.1111/eci.13209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerfeind, R. , von Graevenitz, A. , Kimmig, P. , Schiefer, H. G. , Schwarz, T. , Slenczka, W. , & Zahner, H. (2016). Zoonoses: Infectious diseases transmissible from animals to humans, 4th ed. Washington, DC: ASM Press. ISBN 9781555819255 [Google Scholar]
- Beijing Group of National Research Project for SARS . (2003). Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chinese Medical Journal (Engl), 116, 1283–1287. [PubMed] [Google Scholar]
- Bernheim, A. , Mei, X. , Huang, M. , Yang, Y. , Fayad, Z. A. , Zhang, N. , … Chung, M. (2020). Chest CT findings in coronavirus disease‐19 (COVID‐19): Relationship to duration of infection. Radiology, 10.1148/radiol.2020200463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, S. , Glowacka, I. , Müller, M. A. , Lavender, H. , Gnirss, K. , Nehlmeier, I. , … Pohlmann, S. (2011). Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin‐like protease. Journal of Virology, 85(24), 13363–13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar, T. , Murhekar, M. V. , Soneja, M. , Gupta, N. , Giri, S. , Wig, N. , & Gangakhedkar, R. (2020). Lopinavir/ritonavir combination therapy amongst symptomatic coronavirus disease 2019 patients in India: Protocol for restricted public health emergency use. Indian Journal of Medical Research, 151(2 & 3), 184–189. 10.4103/ijmr.IJMR_502_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonam, S. R. , Kaveri, S. V. , Sakuntabhai, A. , Gilardin, L. , & Bayry, J. (2020). Adjunct immunotherapies for the management of severely Ill COVID‐19 patients. Cell Reports Medicine, 1, 100016. 10.1016/j.xcrm.2020.100016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, C. M. , Matukas, L. M. , Tomlinson, G. A. , Rachlis, A. R. , Rose, D. B. , Dwosh, H. A. , … Detsky, A. S. (2003). Clinical features and short‐term outcomes of 144 patients with SARS in the greater Toronto area. JAMA, 289, 2801–2809. 10.1001/jama.289.21.JOC30885 [DOI] [PubMed] [Google Scholar]
- Bousquet, J. , Lockey, R. , & Malling, H. J. (1998). Allergen immunotherapy: Therapeutic vaccines for allergic diseases. A WHO position paper. The Journal of Allergy and Clinical Immunology, 102(4 Pt 1), 558–562. [DOI] [PubMed] [Google Scholar]
- Burki, T. K. (2020). Testing for COVID‐19. Lancet Respiratory Medicine, 8(7), e63–e64. 10.1016/S2213-2600(20)30247-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, E. (2020). Time to use the p‐word? Coronavirus enter dangerous new phase. Nature, 579, 12. [DOI] [PubMed] [Google Scholar]
- Caly, L. , Druce, J. D. , Catton, M. G. , Jans, D. A. , & Wagstaff, K. M. (2020). The FDA approved Drug Ivermectin inhibits the replication of SARS‐CoV‐2 in vitro . Antiviral Research, 178, 104787. 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, M. J. , Ran, L. , Xu, L. , Danesh, A. , Bermejo‐Martin, J. F. , Cameron, C. M. , … McGeer, A. J. (2007). Interferon‐mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. Journal of Virology, 81, 8692–8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y.‐C. , Deng, Q.‐X. , & Dai, S.‐X. (2020). Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID‐19: An evaluation of the evidence. Travel Medicine and Infectious Disease, 35, 101647. 10.1016/j.tmaid.2020.101647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos, W. G. , Dela Cruz, C. S. , Cao, B. , Pasnick, S. , & Jamil, S. (2020). Novel wuhan (2019‐nCoV) coronavirus. American Journal of Respiratory and Critical Care Medicine, 201(4), 7–8. 10.1164/rccm.2014P7 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2016). Middle East Respiratory Syndrome (MERS). Transmission.Retrieved from https://www.cdc.gov/coronavirus/mers/about/transmission.html
- Chan, J.‐F.‐W. , Yuan, S. , Kok, K. H. , To, K. K. , Chu, H. , Yang, J. , … Yuen, K.‐Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: A study of a family cluster. Lancet, 395(10223), 514–523. S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, R. W. Y. , Hemida, M. G. , Kayali, G. , Chu, D. K. W. , Poon, L. L. M. , Alnaeem, A. , … Peiris, J. S. M. (2014). Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: An in‐vitro and ex‐vivo study. Lancet Respir Med., 2, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D. , Lin, M. , Wei, L. , Xie, L. , Zhu, G. , Dela Cruz, C. S. , … Sharma, L. (2020). Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan. China. JAMA, 323(11), 1092–1093. 10.1001/jama.2020.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Guo, J. , Wang, C. , Luo, F. , Yu, X. , Zhang, W. , … Zhang, Y. (2020). Clinical characteristics and intrauterine vertical transmission potential of COVID‐19 infection in nine pregnant women: A retrospective review of medical records. Lancet, 395, 809–815. 10.1016/S0140-6736(20)30360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. (2020). Pathogenicity and transmissibility of 2019‐nCoV‐A quick overview and comparison with other emerging viruses. Microbes and Infection, 22(2), 69–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N. , Zhou, M. , Dong, X. , Qu, J. , Gong, F. , Han, Y. , … Zhang, L. I. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet, 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinazzi, M. , Davis, J. T. , Ajelli, M. , Gioannini, C. , Litvinova, M. , Merler, S. , … Vespignani, A. (2020). The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID‐19) outbreak. Science. 10.1126/science.aba9757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, C. M. , Cheng, V. C. C. , Hung, I. F. N. , Wong, M. M. L. , Chan, K. H. , Chan, K. S. , … Yuen, K. Y. (2004). Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax, 59, 252–256. 10.1136/thorax.2003.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, D. K. , Akl, E. A. , Duda, S. , Solo, K. , Yaacoub, S. , Schünemann, H. J. , … Schünemann, H. J. (2020). On behalf of the COVID‐19 Systematic Urgent Review Group Effort (SURGE) study authors, Physical distancing, face masks, and eye protection to prevent person‐to‐person transmission of SARS‐CoV‐2 and COVID‐19: A systematic review and meta‐analysis. The Lancet, 395(10242), 1973–1987. 10.1016/S0140-6736(20)31142-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzade, R. , Grant, R. , Malik, M. , Elkholy, A. , Elhakim, M. , Samhouri, D. , … Van Kerkhove, M. (2018). Reported direct and indirect contact with dromedary camels among laboratory‐confirmed MERS‐CoV cases. Viruses, 10(8), 425. 10.3390/v10080425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortegiani, A. , Ingoglia, G. , Ippolito, M. , Giarratano, A. , & Einav, S. (2020). A systematic review on the efficacy and safety of chloroquine for the treatment of COVID‐19. Journal of Critical Care, 57, 279–283. 10.1016/j.jcrc.2020.03.005. And the references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama, K. , Sharun, K. , Tiwari, R. , Dadar, M. , Malik, Y. S. , Singh, K. P. , & Chaicumpa, W. (2020). COVID‐19, an emerging coronavirus infection: Advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Human Vaccines & Immunotherapeutics, 16(6), 1232–1238. 10.1080/21645515.2020.1735227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, E. , Du, H. , & Gardner, L. (2020). An interactive web‐based dashboard to track COVID‐19 in real time. The Lancet Infectious Diseases, 20(5), 533–534. 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly, C. A. , Ghani, A. C. , Leung, G. M. , Hedley, A. J. , Fraser, C. , Riley, S. , … Anderson, R. M. (2003). Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet, 361, 1761–1766. 10.1016/S0140-6736(03)13410-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L. , He, Y. , Zhou, Y. , Liu, S. , Zheng, B.‐J. , & Jiang, S. (2009). The spike protein of SARS‐CoV ‐ a target for vaccine and therapeutic development. Nature Reviews Microbiology, 7(3), 226–236. 10.1038/nrmicro2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwosh, H. A. , Hong, H. H. , Austgarden, D. , Herman, S. , & Schabas, R. (2003). Identification and containment of an outbreak of SARS in a community hospital. CMAJ, 168, 1415–1420. [PMC free article] [PubMed] [Google Scholar]
- El‐Duah, P. , Sylverken, A. , Owusu, M. , Yeboah, R. , Lamptey, J. , Frimpong, Y. O. , … Adu‐Sarkodie, Y. (2019). Potential intermediate hosts for coronavirus transmission: No evidence of Clade 2c coronaviruses in domestic livestock from Ghana. Tropical Medicine and Infectious Disease, 4, 34. 10.3390/tropicalmed4010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano, D. , Kamissoko, B. , de Wit, E. , Maïga, O. , Cronin, J. , Samaké, K. , … Feldmann, H. (2017). Dromedary camels in northern Mali have high seropositivity to MERS‐CoV. One Health, 3, 41–43. 10.1016/j.onehlt.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag, E. , Sikkema, R. S. , Mohamedani, A. A. , de Bruin, E. , Munnink, B. B. O. , Chandler, F. , … Elrahman, S. H. A. (2019). MERS‐CoV in camels but not camel handlers, Sudan, 2015 and 2017. Emerging Infectious Diseases, 25(12), 2333–2335. 10.3201/eid2512.190882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay, B. B. , & Hancock, R. E. (2004). Can innate immunity be enhanced to treat microbial infections? Nature Reviews Microbiology, 2, 497–504. [DOI] [PubMed] [Google Scholar]
- Fisher, D. , & Heymann, D. (2020). Q&A: The novel coronavirus outbreak causing COVID‐19. BMC Medicine, 18, 57. 10.1186/s12916-020-01533-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franquet, T. (2011). Imaging of pulmonary viral pneumonia. Radiology, 260(1), 18–39. 10.1148/radiol.11092149 [DOI] [PubMed] [Google Scholar]
- Gallego, V. , Nishiura, H. , Sah, R. , & Rodriguez‐Morales, A. J. (2020). The COVID‐19 outbreak and implications for the Tokyo 2020 Summer Olympic Games. Travel Medicine and Infectious Disease, 34, 101604. 10.1016/j.tmaid.2020.101604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- General Office of National Health Committee . (2020). Notice on the issuance of a program for the diagnosis and treatment of novel coronavirus (2019‐nCoV) infected pneumonia (trial sixth edition) (2020–02‐18).
- GHKSAR (2020). Pet cat tests positive for COVID‐19 virus. Retrieved from www.info.gov.hk/gia/general/202003/31/P2020033100717.html
- Gilbert, M. , Pullano, G. , Pinotti, F. , Valdano, E. , Poletto, C. , Boëlle, P.‐Y. , … Colizza, V. (2020). Preparedness and vulnerability of African countries against importations of COVID‐19: A modelling study. Lancet, 395, 871–877. 10.1016/S0140-6736(20)30411-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID: Global Initiative on Sharing All Influenza Data (2020). Phylogeny of SARS‐like beta coronaviruses including novel coronavirus (nCoV). Retrieved from https://nextstrain.org/groups/blab/sars‐like‐cov
- Gorbalenya, A. E. , Baker, S. C. , Baric, R. S. , de Groot, R. J. , Drosten, C. , Gulyaeva, A. A. , … Ziebuhr, J. (2020). Severe acute respiratory syndrome‐related coronavirus: the species and its viruses—a statement of the Coronavirus Study Group. bioRxiv. 10.1101/2020.02.07.937862 [DOI] [Google Scholar]
- Graham, R. L. , Donaldson, E. F. , & Baric, R. S. (2013). A decade after SARS: Strategies for controlling emerging coronaviruses. Nature Reviews Microbiology, 11, 836–848. 10.1038/nrmicro3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß, S. , Jahn, C. , Cushman, S. , Bär, C. , & Thum, T. (2020). SARS‐CoV‐2 receptor ACE2‐dependent implications on the cardiovascular system: From basic science to clinical implications. Journal of Molecular and Cellular Cardiology, 144, 47–53. 10.1016/j.yjmcc.2020.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y. , Zheng, B. J. , He, Y. Q. , Liu, X. L. , Zhuang, Z. X. , Cheung, C. L. , … Poon, L. L. M. (2003). Isolation and characterization of viruses related to the sars coronavirus from animals in Southern China. Science, 302, 276–278. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- Han, Q. , Lin, Q. , Jin, S. , & You, L. (2020). Coronavirus 2019‐nCoV: A brief perspective from the front line. Journal of Infection, 80, 373–377. 10.1016/j.jinf.2020.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansell, D. M. , Bankier, A. A. , MacMahon, H. , McLoud, T. C. , Müller, N. L. , & Fleischner, R. J. (2008). Society: Glossary of terms for thoracic imaging. Radiology, 246(3), 697–722. 10.1148/radiol.2462070712 [DOI] [PubMed] [Google Scholar]
- Harypursat, V. , & Chen, Y. K. (2020). Six weeks into the 2019 coronavirus disease (COVID‐19) outbreak‐ it is time to consider strategies to impede the emergence of new zoonotic infections. Chinese Medical Journal (Engl), 133(9), 1118–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellewell, J. , Abbott, S. , Gimma, A. , Bosse, N. I. , Jarvis, C. I. , Russell, T. W. , … van Zandvoort, K. (2020). Feasibility of controlling COVID‐19 outbreaks by isolation of cases and contacts. Lancet Glob Health, 8, e488–e496. 10.1016/S2214-109X(20)30074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida, M. , Perera, R. , Wang, P. , Alhammadi, M. , Siu, L. , Li, M. , … Peiris, M. (2013). Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Eurosurveillance Weekly, 18(50), 20659. 10.2807/1560-7917.es2013.18.50.20659 [DOI] [PubMed] [Google Scholar]
- Hu, D. , Zhu, Z. , Li, S. , Deng, Y. , Wu, Y. , Zhang, N. , … Ying, T. (2019). A broadly neutralizing germline‐like human monoclonal antibody against dengue virus envelope domain III. PLoS Path, 15, e1007836. 10.1371/journal.ppat.1007836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , Song, C. I. , Xu, C. , Jin, G. , Chen, Y. , Xu, X. , … Shen, H. (2020). Clinical characteristics of 24 asymptomatic infections with COVID‐19 screened among close contacts in Nanjing, China. Sci China Life Sci, 63, 706–711. 10.1007/s11427-020-1661-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Wang, Y. , Li, X. , Ren, L. , Zhao, J. , Hu, Y. I. , … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Tu, M. , Wang, S. , Chen, S. , Zhou, W. , Chen, D. , … Guo, L. (2020). Clinical characteristics of laboratory confirmed positive cases of SARS‐CoV‐2 infection in Wuhan, China: A retrospective single center analysis. Travel Medicine and Infectious Disease, 101606. 10.1016/j.tmaid.2020.101606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaxia . (2020) WHO says vaccines against novel coronavirus 18 months away, pushes global research. 2020. Xinhuanet. Retrieved from http://www.xinhuanet.com/english/202002/12/c_138777886.htm
- Hui, D. S. , Azhar, E. I. , Madani, T. A. , Ntoumi, F. , Kock, R. , Dar, O. , … Petersen, E. (2020). The continuing 2019‐nCoV epidemic threat of novel coronaviruses to global health ‐ the latest 2019 novel coronavirus outbreak in Wuhan, China. International Journal of Infectious Diseases, 91, 264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, I.‐N. , Lung, K.‐C. , Tso, E.‐K. , Liu, R. , Chung, T.‐H. , Chu, M.‐Y. , … Yuen, K.‐Y. (2020). Triple combination of interferon beta‐1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID‐19: An open‐label, randomised, phase 2 trial. The Lancet, 395(10238), 1695–1704. 10.1016/S0140-6736(20)31042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari, A. A. , & Ghasemi, S. (2020). The possible of immunotherapy for COVID‐19: A systematic review. International Immunopharmacology, 83, 106455. 10.1016/j.intimp.2020.106455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, C. (2020). Mar 16, 2020 which species transmit Covid‐19 to humans? we’re still not sure. [Accessed 2020 April 1). Retrieved from https://www.the-scientist.com/news-opinion/which-species-transmit-covid-19-to-humans-were-stillnot-sure-67272
- Ji, W. , Wang, W. , Zhao, X. , Zai, J. , & Li, X. (2020). Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross‐species transmission from snake to human. Journal of Medical Virology, 92(4), 433–440. 10.1002/jmv.25682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, F. , Deng, L. , Zhang, L. , Cai, Y. , Cheung, C. W. , & Xia, Z. (2020). Review of the clinical characteristics of coronavirus disease 2019 (COVID‐19). Journal of General Internal Medicine, 35(5), 1545–1549. 10.1007/s11606-020-05762-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S.‐M. , Akhmetzhanov, A. R. , Hayashi, K. , Linton, N. M. , Yang, Y. , Yuan, B. , … Nishiura, H. (2020). Real‐time estimation of the risk of death from novel coronavirus (COVID‐19) Infection: Inference using exported cases. Journal of Clinical Medicine, 9, 523. 10.3390/jcm9020523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutel, M. , Van de Veen, W. , Agache, I. , Azkur, K. A. , Akdis, M. , & Akdis, C. A. (2013). Mechanisms of allergen specific immunotherapy and novel ways for vaccine development. Allergology International, 62(4), 425–433. [DOI] [PubMed] [Google Scholar]
- Kandeil, A. , Gomaa, M. , Nageh, A. , Shehata, M. M. , Kayed, A. E. , Sabir, J. S. M. , … Kayali, G. (2019). Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) in Dromedary Camels in Africa and Middle East. Viruses, 11, 717. 10.3390/v11080717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, H. , Takayama‐Ito, M. , Iizuka‐Shiota, I. , Fukushi, S. , Posadas‐Herrera, G. , Horiya, M. , … Lim, C.‐K. (2019). Development of a recombinant replication deficient rabies virus‐based bivalent‐vaccine against MERS‐CoV and rabies virus and its humoral immunogenicity in mice. PLoS One, 14(10), e0223684. 10.1371/journal.pone.0223684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. (2020). Outbreak of novel coronavirus (COVID‐ 19): What is the role of radiologists? European Radiology, 30(6), 3266–3267. 10.1007/s00330-020-06748-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. C. , Dema, B. , & Reyes‐Sandoval, A. (2020). COVID‐19 vaccines: Breaking record times to first‐in‐human trials. Npj Vaccines, 5(1), 34. 10.1038/s41541-020-0188-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer, R. N. , Cottrell, C. A. , Wang, N. , Pallesen, J. , Yassine, H. M. , Turner, H. L. , … Ward, A. B. (2016). Pre‐fusion structure of a human coronavirus spike protein. Nature, 531, 118–121. 10.1038/nature17200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimek, L. , Jutel, M. , Akdis, C. , Bousquet, J. , Akdis, M. , Bachert, C. , … Zuberbier, T. (2020). Handling of allergen immunotherapy in the COVID‐19 pandemic: An ARIA‐EAACI statement. Allergy, 75(7), 1546–1554. 10.1111/ALL.14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, W.‐C. , Rolain, J.‐M. , Lee, N.‐Y. , Chen, P.‐L. , Huang, C.‐T. , Lee, P.‐I. , & Hsueh, P.‐R. (2020). Arguments in favor of remdesivir for treating SARS‐CoV‐2 infections. International Journal of Antimicrobial Agents, 55(4), 105933. 10.1016/j.ijantimicag.2020.105933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooraki, S. , Hosseiny, M. , Myers, L. , & Gholamrezanezhad, A. (2020). Coronavirus (COVID‐19) Outbreak: What the Department of Radiology should know. Journal of the American College of Radiology, 17(4), 447–451. 10.1016/j.jacr.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse, R. L. (2020). Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China, F1000. Philosophy and Phenomenological Research, 9, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C.‐C. , Shih, T.‐P. , Ko, W.‐C. , Tang, H.‐J. , & Hsueh, P.‐R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and coronavirus disease‐2019 (COVID‐19): The epidemic and the challenges. International Journal of Antimicrobial Agents, 55, 105924. 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S. K. P. , Li, K. S. M. , Luk, H. K. H. , He, Z. , Teng, J. L. L. , Yuen, K.‐Y. , Woo, P. C. Y. (2020). Middle East Respiratory Syndrome coronavirus antibodies in bactrian and hybrid camels from Dubai. mSphere, 5(1), e00898‐19. 10.1128/mSphere.00898-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S. K. , Woo, P. C. , Li, K. S. , Huang, Y. , Tsoi, H. W. , Wong, B. H. , … Yuen, K. Y. (2005). Severe acute respiratory syndrome coronavirus‐like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences of USA, 102, 14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, R. Q. , Li, L. , Yuan, W. , Shord, S. S. , Nie, L. , Habtemariam, B. A. , … Pazdur, R. (2018). FDA Approval Summary: Tocilizumab for Treatment of Chimeric Antigen Receptor T Cell‐Induced Severe or Life‐Threatening Cytokine Release Syndrome. The Oncologist, 23, 943–947. 10.1634/theoncologist.2018-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. S. (2020). Pneumonia associated with 2019 novel coronavirus: Can computed tomographic findings help predict the prognosis of the disease? Korean Journal of Radiology, 21(3), 257–258. 10.3348/kjr.2020.0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, N. , Hui, D. , Wu, A. , Chan, P. , Cameron, P. , Joynt, G. M. … Sung, J. J. (2003). A major outbreak of severe acute respiratory syndrome in Hong Kong. New England Journal of Medicine, 348, 1986–1994. [DOI] [PubMed] [Google Scholar]
- Lei, J. , Li, J. , Li, X. , & Qi, X. (2020). CT Imaging of the 2019 Novel Coronavirus (2019‐nCoV) Pneumonia. Radiology, 295, 18. 10.1148/radiol.2020200236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Guan, X. , Wu, P. , Wang, X. , Zhou, L. , Tong, Y. , … Feng, Z. (2020). Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. New England Journal of Medicine, 382, 1199–1207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Shi, Z. , Yu, M. , Ren, W. , Smith, C. , Epstein, J. H. , … Wang, L.‐F. (2005). Bats are natural reservoirs of SARS‐like coronaviruses. Science, 310, 676–679. [DOI] [PubMed] [Google Scholar]
- Li, X. (2020). Can cats become infected with Covid‐19? Veterinary Record, 186(14), 457.2–458. 10.1136/vr.m1455 [DOI] [PubMed] [Google Scholar]
- Liao, M. , Liu, Y. , Yuan, J. , Wen, Y. , Xu, G. , Zhao, J. , … Zhang, Z. (2020). Single‐cell landscape of bronchoalveolar immune cells in patients with COVID‐19. Nature Medicine, 26(6), 842–844. 10.1038/s41591-020-0901-9 [DOI] [PubMed] [Google Scholar]
- Lin, L. , Lu, L. , Cao, W. , & Li, T. (2020). Hypothesis for potential pathogenesis of SARS‐CoV‐2 infection–a review of immune changes in patients with viral pneumonia. Emerging Microbes & Infections, 9(1), 727–732. 10.1080/22221751.2020.1746199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linga, L. , Joynta, G. M. , Lipman, J. , Constantine, J.‐M. , & Joannes‐Boyau, O. (2020). COVID‐19: A critical care perspective informed by lessons learnt from other viral epidemics. Anaesthesia Critical Care & Pain Medicine, 39(2), 163–166. 10.1016/j.accpm.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch, M. , Swerdlow, D. L. , & Finelli, L. (2020). Defining the Epidemiology of Covid‐19 — Studies Needed. New England Journal of Medicine, 382, 1194–1196. 10.1056/NEJMp2002125 [DOI] [PubMed] [Google Scholar]
- Liu, C. , Zhou, Q. , Li, Y. , Garner, L. V. , Watkins, S. P. , Carter, L. J. , … Albaiu, D. (2020). Research and development on therapeutic agents and vaccines for COVID‐19 and related human coronavirus diseases. ACS Central Science, 6, 315–331. 10.1021/acscentsci.0c00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Cao, R. , Xu, M. , Wang, X. I. , Zhang, H. , Hu, H. , … Wang, M. (2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS‐CoV‐2 infection in vitro. Cell Discovery, 6, 16. 10.1038/s41421-020-0156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Zheng, X. , Tong, Q. , Li, W. , Wang, B. , Sutter, K. , … Yang, D. (2020). Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS‐CoV, MERS‐CoV, and 2019‐nCoV. Journal of Medical Virology, 92(5), 491–494. 10.1002/jmv.25709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Zhang, J. , Yang, Y. , Zhang, L. , Ma, H. , Li, Z. , … Yi, J. (2020). The potential role of IL‐6 in monitoring coronavirus disease, 2019. SSRN Electronic Journal. 10.2139/ssrn.3548761 [DOI] [Google Scholar]
- Liu, Z. , Xiao, X. , Wei, X. , Li, J. , Yang, J. , Tan, H. , … Liu, L. (2020). Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. Journal of Medical Virology, 92(6), 595–601. 10.1002/jmv.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łoczechin, A. , Séron, K. , Barras, A. , Giovanelli, E. , Belouzard, S. , Chen, Y.‐T. , & Szunerits, S. (2019). Functional carbon quantum dots as medical countermeasures to human coronavirus. ACS Applied Materials & Interfaces, 11, 42964–42974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. , Stratton, C. W. , & Tang, Y. W. (2020). Outbreak of pneumonia of unknown etiology in Wuhan China: The mystery and the miracle. Journal of Medical Virology, 2020(92), 401–402. 10.1002/jmv.25678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. O. , Wu, H. , … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase, E. (2020). China coronavirus: WHO declares international emergency as death toll exceeds 200. BMJ, 368, m408. 10.1136/bmj.m408 [DOI] [PubMed] [Google Scholar]
- Malainou, C. , & Herold, S. (2019). Influenza. Internist (Berl), 60(11), 1127–1135. 10.1007/s00108-019-00670-6 [DOI] [PubMed] [Google Scholar]
- Martina, B. E. E. , Haagmans, B. L. , Kuiken, T. , Fouchier, R. A. M. , Rimmelzwaan, G. F. , van Amerongen, G. , … Osterhaus, A. D. M. E. (2003). SARS virus infection of cats and ferrets. Nature, 425, 915. 10.1038/425915a [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, K. , Hirsch, M. , & Thorner, A. (2017). Middle East respiratory syndrome coronavirus: Virology, pathogenesis, and epidemiology. In Post T. W. (Ed.), UpToDate. Waltham, MA: UpToDate Inc. [Google Scholar]
- Menachery, V. D. , Schäfer, A. , Burnum‐Johnson, K. E. , Mitchell, H. D. , Eisfeld, A. J. , Walters, K. B. , … Baric, R. S. (2018). MERS‐CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proceedings of the National Academy of Sciences USA, 115, E1012–E1021. 10.1073/pnas.1706928115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni, C. , Sudre, C. H. , Steves, C. J. , Ourselin, S. , & Spector, T. D. (2020). Quantifying additional COVID‐19 symptoms will save lives. The Lancet, 395(10241), e107–e108. 10.1016/S0140-6736(20)31281-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B. , García‐Bocanegra, I. , Wernery, U. , Wernery, U. , Wernery, R. , Sieberg, A. , … Eckerle, I. (2015). Serologic assessment of possibility for MERS‐CoV infection in equids. Emerging Infectious Diseases, 21, 181–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel, E. , Chevalier, V. , Ayelet, G. , Bencheikh, M. N. B. , Boussini, H. , Chu, D. K. , … Peiris, M. (2017). Risk factors for MERS coronavirus infection in dromedary camels in Burkina Faso, Ethiopia, and Morocco, 2015. Eurosurveillance Weekly, 22(13), 30498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobaraki, K. , & Ahmadzadeh, J. (2019). Current epidemiological status of Middle East respiratory syndrome coronavirus in the world from 1.1.2017 to 17.1.2018: A cross‐sectional study. BMC Infectious Diseases, 19, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momattin, H. , Al‐Ali, A. Y. , & Al‐Tawfiq, J. A. (2019). A systematic review of therapeutic agents for the treatment of the Middle East respiratory syndrome coronavirus (MERS–CoV). Travel Medicine and Infectious Disease, 30, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, C. A. , Staples, J. E. , Dobyns, W. B. , Pessoa, A. , Ventura, C. V. , da Fonseca, E. B. , … Rasmussen, S. A. (2017). Characterizing the pattern of anomalies in congenital Zika syndrome for pediatric clinicians. JAMA Pediatrics, 171, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster, V. J. , Feldmann, F. , Williamson, B. N. , van Doremalen, N. , Pérez‐Pérez, L. , Schulz, J. , … de Wit, E. (2020). Respiratory disease in rhesus macaques inoculated with SARS‐CoV‐2. Nature. 10.1038/s41586-020-2324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura, H. , Jung, S.‐M. , Linton, N. M. , Kinoshita, R. , Yang, Y. , Hayashi, K. , … Akhmetzhanov, A. R. (2020). The extent of transmission of novel coronavirus in Wuhan, China, 2020. Journal of Clinical Medicine, 9, 330. 10.3390/jcm9020330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M. , Thwaites, R. S. , & Openshaw, P. J. M. (2020). COVID‐19: Lessons from SARS and MERS. European Journal of Immunology, 50(3), 308–311. 10.1002/eji.202070035 [DOI] [Google Scholar]
- Patel, A. , & Jernigan, D. B. (2020). 2019‐nCoV CDC Response Team. 2020. Initial Public Health Response and Interim Clinical Guidance for the 2019 Novel Coronavirus Outbreak ‐ United States, December 31, 2019‐February 4, 2020. MMWR Morbidity and Mortality Weekly Report, 69(5), 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrì, A. , & Fabbrocini, G. (2020). Hydroxychloroquine and ivermectin: A synergistic combination for COVID‐19 chemoprophylaxis and/or treatment? Journal of the American Academy of Dermatology, 82(6), e221. 10.1016/j.jaad.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, R. A. , Wang, P. , Gomaa, M. R. , El‐Shesheny, R. , Kandeil, A. , Bagato, O. , Kayali, G. (2013). Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralization assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Eurosurveillance, 18(36). 10.2807/1560-7917.es2013.18.36.20574 [DOI] [PubMed] [Google Scholar]
- Perlman, S. , & Netland, J. (2009). Coronaviruses post‐SARS: Update on replication and pathogenesis. Nature Reviews: Microbiology, 7(6), 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, L. T. , Nguyen, T. V. , Luong, Q. C. , Nguyen, T. V. , Nguyen, H. T. , Le, H. Q. , … Pham, Q. D. (2020). Importation and human‐to‐human transmission of a novel Coronavirus in Vietnam. New England Journal of Medicine, 382, 872–874. 10.1056/NEJMc2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, J. (2020). What are the risks of COVID‐19 infection in pregnant women? The Lancet, 395, 760–762. 10.1016/S0140-6736(20)30365-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X. , Wong, G. , Audet, J. , Bello, A. , Fernando, L. , Alimonti, J. B. , … Kobinger, G. P. (2014). Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature, 514, 47–53. 10.1038/nature13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, V. S. , Mou, H. , Smits, S. L. , Dekkers, D. H. , Müller, M. A. , Dijkman, R. , … Haagmans, B. L. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus‐EMC. Nature, 495(7440), 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan, N. , & Shaib, H. (2019). Middle East respiratory syndrome coronavirus (MERS‐CoV): A review. GERMS, 9(1), 35–42. 10.18683/germs.2019.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, S. A. , & Hayes, E. B. (2005). Public health approach to emerging infections among pregnant women. American Journal of Public Health, 95, 1942–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, S. A. , Jamieson, D. J. , Honein, M. A. , & Petersen, L. R. (2016). Zika vrus and birth defects– Reviewing the evidence for causality. New England Journal of Medicine, 374, 1981–1987. [DOI] [PubMed] [Google Scholar]
- Rasmussen, S. A. , Smulian, J. C. , Lednicky, J. A. , Wen, T. S. , & Jamieson, D. J. (2020). Coronavirus Disease 2019 (COVID‐19) and Pregnancy: What obstetricians need to know. American Journal of Obstetrics and Gynecology, 222(5), 415–426. 10.1016/j.ajog.2020.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, P. E. O. , & Lima, M. C. B. (2020). Can we manage prophylactic therapy in COVID‐19 patients to prevent severe illness complications? Journal of Vascular Surgery, 19, e20200057. 10.1590/1677-5449.200057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remuzzi, A. , & Remuzzi, G. (2020). COVID‐19 and Italy: What next? The Lancet, 395(10231), 1225–1228. 10.1016/S0140-6736(20)30627-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, L.‐L. , Wang, Y.‐M. , Wu, Z.‐Q. , Xiang, Z.‐C. , Guo, L. I. , Xu, T. , … Wang, J.‐W. (2020). Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chinese Medical Journal, 133, 1015–1024. 10.1097/CM9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken, C. , Ababneh, M. , Raj, V. , Meyer, B. , Eljarah, A. , Abutarbush, S. , … Koopmans, M. (2013). Middle East Respiratory Syndrome coronavirus (MERS‐CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Eurosurveillance Weekly, 18(50), 20662. 10.2807/1560-7917.es2013.18.50.20662 [DOI] [PubMed] [Google Scholar]
- Reusken, C. B. E. M. , Haagmans, B. L. , Müller, M. A. , Gutierrez, C. , Godeke, G.‐J. , Meyer, B. , … Koopmans, M. P. G. (2013). Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. The Lancet Infectious Diseases, 13, 859–866. 10.1016/S1473-3099(13)70164-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken, C. B. E. M. , Schilp, C. , Raj, V. S. , Bruin, E. D. , Kohl, R. H. G. , Farag, E. A. B. A. , … Koopmans, M. P. G. (2016). MERS‐CoV infection of alpaca in a region where MERS‐CoV is endemic. Emerging Infectious Diseases, 22, 1129–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, P. , Griffin, I. , Tucker, C. , Smith, D. , Oechsle, O. , Phelan, A. , … Stebbing, J. (2020). Baricitinib as potential treatment for 2019‐nCoV acute respiratory disease. Lancet, 395, e30–e31. 10.1016/S0140-6736(20)30304-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, A. , Lamirande, E. W. , Vogel, L. , Jackson, J. P. , Paddock, C. D. , Guarner, J. , … Subbarao, K. (2008). Animal models and vaccines for SARS‐CoV infection. Virus Research, 133(1), 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Morales, A. J. , Dhama, K. , Sharun, K. , Tiwari, R. , & Bonilla‐Aldana, D. K. (2020). Susceptibility of felids to coronaviruses. Veterinary Record, 186(17), e21. 10.1136/vr.m1671 [DOI] [PubMed] [Google Scholar]
- Roosa, K. , Lee, Y. , Luo, R. , Kirpich, A. , Rothenberg, R. , Hyman, J. M. , … Chowell, G. (2020). Real‐time forecasts of the COVID‐19 epidemic in China from February 5th to February 24th, 2020. Infectious Disease Modelling, 5, 256–263. 10.1016/j.idm.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa, S. G. V. , & Santos, W. C. (2020). Clinical trials on drug repositioning for COVID‐19 treatment. Revista Panamericana De Salud Publica, 44, e40. 10.26633/RPSP.2020.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothana, H. A. , & Byrareddy, S. N. (2020). The epidemiology and pathogenesis of coronavirus disease (COVID‐19) outbreak. Journal of Autoimmunity, 109, 102433. 10.1016/j.jaut.2020.102433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir, J. S. M. , Lam, T.‐T.‐Y. , Ahmed, M. M. M. , Li, L. , Shen, Y. , Abo‐Aba, S. E. M. , … Guan, Y. (2016). Co‐circulation of three camel coronavirus species and recombination of MERS‐CoVs in Saudi Arabia. Science, 351, 81–84. [DOI] [PubMed] [Google Scholar]
- Schindewolf, C. , & Menachery, V. D. (2019). Middle East respiratory syndrome vaccine candidates: Cautious optimism. Viruses, 11(1), 74. 10.3390/v11010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugaraj, B. , Siriwattananon, K. , Wangkanont, K. , & Phoolcharoen, W. (2020). Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease‐19 (COVID‐19). Asian Pacific Journal of Allergy and Immunology, 38(1), 10–18. [DOI] [PubMed] [Google Scholar]
- Shereen, M. A. , Khan, S. , Kazmi, A. , Bashir, N. , & Siddique, R. (2020). COVID‐19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91–98. 10.1016/j.jare.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Han, X. , Jiang, N. , Cao, Y. , Alwalid, O. , Gu, J. , … Zheng, C. (2020). Radiological findings from 81 patients with COVID‐19 pneumonia in Wuhan, China: A descriptive study. The Lancet Infectious Diseases, 20(4), 425–434. 10.1016/S1473-3099(20)30086-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. Z. , Wen, Z. Y. , Zhong, G. , Yang, H. , Wang, C. , Liu, R. , … Bu, Z. (2020). Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS‐coronavirus‐2. BioRxiv. 10.1101/2020.03.30.015347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, H.‐I. , Wu, C.‐J. , Tu, Y.‐F. , & Chi, C.‐Y. (2020). Fighting COVID‐19: A quick review of diagnoses, therapies, and vaccines. Biomedical Journal. 10.1016/j.bj.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siston, A. M. , Rasmussen, S. A. , Honein, M. A. , Fry, A. M. , Seib, K. , Callaghan, W. M. , … Jamieson, D. J. (2010). Pandemic 2009 influenza A(H1N1) virus illness among pregnant women in the United States. JAMA, 303, 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivabakya, T. K. , & Srinivas, G. (2020). Will the antimalarial drug take over to combat COVID‐19? Journal of Public Health: from Theory to Practice. 10.1007/s10389-020-01293-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J. , Bredenbeek, P. J. , Dobbe, J. C. , Thiel, V. , Ziebuhr, J. , Poon, L. L. , … Gorbalenya, A. E. (2003). Unique and conserved features of genome and proteome of SARS‐coronavirus, an early split‐off from the coronavirus group 2 lineage. Journal of Molecular Biology, 331, 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrab, S. S. , & Azhar, E. I. (2019). Genetic diversity of MERS‐CoV spike protein gene in Saudi Arabia. J. Infect Public Health, 13(5), 709–717. 10.1016/j.jiph.2019.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwannarach, N. , Kumla, J. , Sujarit, K. , Pattananandecha, T. , Saenjum, C. , & Lumyong, S. (2020). Natural bioactive compounds from fungi as potential candidates for protease inhibitors and immunomodulators to apply for coronaviruses. Molecules, 25, 1800. 10.3390/molecules25081800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, K. (2020). In vitro and animal models for SARS‐CoV‐2 research. Trends in Pharmaceutical Sciences, 10.1016/j.tips.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teengam, P. , Siangproh, W. , Tuantranont, A. , Vilaivan, T. , Chailapakul, O. , & Henry, C. S. (2017). Multiplex paper‐based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid‐induced AgNPs aggregation for detecting MERS‐CoV, MTB, and HPV oligonucleotides. Analytical Chemistry, 89, 5428–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetro, J. A. (2020). Is COVID‐19 receiving ADE from other coronaviruses? Microbes and Infection, 22, 72–73. 10.1016/j.micinf.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlund, K. , Dron, L. , Park, J. , Hsu, G. , Forrest, J. I. , & Mills, E. J. (2020). A real‐time dashboard of clinical trials for COVID‐19. The Lancet: Digital‐Health, 2, e286–e287. 10.1016/S2589-7500(20)30086-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, S. , Hu, N. , Lou, J. , Chen, K. , Kang, X. , Xiang, Z. , … Zhang, J. (2020). Characteristics of COVID‐19 infection in Beijing. Journal of Infection, 80, 401–406. 10.1016/j.jinf.2020.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X. , Li, C. , Huang, A. , Xia, S. , Lu, S. , Shi, Z. , … Ying, T. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerging Microbes & Infections, 9, 382–385. 10.1080/22221751.2020.1729069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, R. , Dhama, K. , Sharun, K. , Iqbal Yatoo, M. , Malik, Y. S. , Singh, R. , … Rodriguez‐Morales, A. J. (2020). COVID‐19: Animals, veterinary and zoonotic links. Veterinary Quarterly, 40(1), 169–182. 10.1080/01652176.2020.1766725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, K. W. , Ho, P. L. , Ooi, G. C. , Yee, W. K. , Wang, T. , Chan‐Yeung, M. , … Lai, K. N. (2003). A cluster of cases of severe acute respiratory syndrome in Hong Kong. New England Journal of Medicine, 348, 1977–1985. [DOI] [PubMed] [Google Scholar]
- Tsui, P. T. , Kwok, M. L. , Yuen, H. , & Lai, S. T. (2003). Severe acute respiratory syndrome: Clinical outcome and prognostic correlates. Emerging Infectious Diseases, 9, 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y.‐F. , Chien, C.‐S. , Yarmishyn, A. A. , Lin, Y.‐Y. , Luo, Y.‐H. , Lin, Y.‐T. , … Chiou, S.‐H. (2020). A review of SARS‐CoV‐2 and the ongoing clinical trials. International Journal of Molecular Sciences, 21, 2657. 10.3390/ijms21072657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell, D. A. , & Bynoe, M. L. (1966). Cultivation of viruses from a high proportion of patients with colds. Lancet, 1, 76–77. [DOI] [PubMed] [Google Scholar]
- USDA (2020). USDA statement on the confirmation of COVID‐19 in a tiger in New York. Retrieved from www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/ny-zoo-covid-19 [Google Scholar]
- Wan, Y. , Shang, J. , Graham, R. , Baric, R. S. , & Li, F. (2020). Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade‐long structural studies of SARS. Journal of Virology, 94(7), e00127‐20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Horby, P. W. , Hayden, F. G. , & Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. Lancet, 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Hu, B. O. , Hu, C. , Zhu, F. , Liu, X. , Zhang, J. , … Peng, Z. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA, 323(11), 1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Xu, J. , Kong, Y. U. , Liang, R. , Li, W. , Li, J. , … Ying, T. (2019). Engineering a Novel Antibody‐Peptide Bispecific Fusion Protein Against MERS‐CoV. Antibodies, 8, 53. 10.3390/antib8040053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Cao, R. , Zhang, L. , Yang, X. , Liu, J. , Xu, M. , … Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro . Cell Research, 30, 269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Shi, X. , Jiang, L. , Zhang, S. , Wang, D. , Tong, P. , … Wang, X. (2013). Structure of MERS‐CoV spike receptor‐binding domain complexed with human receptor DPP4. Cell Research, 23(8), 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Tang, J. , & Wei, F. (2020). Updated understanding of the outbreak of 2019 novel coronavirus (2019‐nCoV) in Wuhan, China. Journal of Medical Virology, 92(4), 441–447. 10.1002/jmv.25689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Kang, H. , Liu, X. , & Tong, Z. (2020). Combination of RT‐qPCR testing and clinical features for diagnosis of COVID‐19 facilitates management of SARS‐CoV‐2 outbreak. Journal of Medical Virology, 92, 538–539. 10.1002/jmv.25721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Q. , Wang, Y. , Ma, J. , Han, J. , Jiang, M. , Zhao, L. , Liu, J. (2020). Description of the first strain of 2019‐nCoV, C‐Tan‐nCoV Wuhan Strain‐National Pathogen Resource Center, China, 2020. Retrieved from http://weekly.chinacdc.cn/en/article/id/e3a460f1-661b-4180-b562-ecd8e9502082 [PMC free article] [PubMed] [Google Scholar]
- Weiss, S. R. , & Leibowitz, J. L. (2011). Coronavirus pathogenesis. Advances in Virus Research, 81, Chapter‐4, 85–164. Elsevier. 10.1016/B978-0-12-385885-6.00009-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, J. (2012). Patient with new strain of coronavirus is treated in intensive care at London hospital. BMJ, 345, e6455. [DOI] [PubMed] [Google Scholar]
- Wong, C. K. , Lam, C. W. , Wu, A. K. , Ip, W. K. , Lee, N. L. , Chan, I. H. , & Sung, J. J. (2004). Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clinical and Experimental Immunology, 136, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2003). Severe Acute Respiratory Syndrome. (SARS): Multi‐country outbreak. [Google Scholar]
- World Health Organization (2018). Middle East respiratory syndrome coronavirus (MERS‐CoV). Retrieved from https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov) [Google Scholar]
- World Health Organization . (2020). Coronavirus disease 2019 (COVID‐19), Situation Report – 151.
- World Health Organization (2020a). Novel coronavirus–China. Retrieved from http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ [Google Scholar]
- World Health Organization . (2020b). Middle East respiratory syndrome coronavirus (MERS‐CoV). Retrieved from https://www.who.int/emergencies/mers-cov/en/
- World Health Organization . (2020c). Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected: interim guidance.
- World Health Organization (2020d). Clinical management of severe acute respiratory infection (SARI) when Covid‐19 disease is suspected. Interim Guidance. Retrieved from https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected
- Wu, F. , Zhao, S. U. , Yu, B. , Chen, Y.‐M. , Wang, W. , Song, Z.‐G. , … Zhang, Y.‐Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P. , Hao, X. , Lau, E. H. Y. , Wong, J. Y. , Leung, K. S. M. , Wu, J. T. , … Leung, G. M. (2020). Real‐time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Eurosurveillance Weekly, 25(3), 2000044. 10.2807/1560-7917.ES.2020.25.3.2000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Chen, P. , Wang, J. , Feng, J. , Zhou, H. , Li, X. , … Hao, P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, 63, 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Peng, C. , Shi, Y. , Zhu, Z. , Mu, K. , Wang, X. , Zhu, W. (2020). Nelfinavir was predicted to be a potential inhibitor of 2019‐nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. bioRxiv, 2027, 921627. 10.1101/2020.01.27.921627 [DOI] [Google Scholar]
- Yang, W. , Cao, Q. , Qin, L. E. , Wang, X. , Cheng, Z. , Pan, A. , … Yan, F. (2020). Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID‐19): A multi‐center study in Wenzhou city, Zhejiang, China. Journal of Infection, 80, 388–393. 10.1016/j.jinf.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki, A. M. , van Boheemen, S. , Bestebroer, T. M. , Osterhaus, A. D. , & Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine, 367, 1814–1820. [DOI] [PubMed] [Google Scholar]
- Zhai, S. L. , Wei, W. K. , Lv, D. H. , Xu, Z.‐H. , Chen, Q.‐L. , Sun, M.‐F. , … Wang, D. (2020). Where did SARS‐CoV‐2 come from? The Veterinary Record, 186, 254. [DOI] [PubMed] [Google Scholar]
- Zhai, S.‐L. , Wei, W.‐K. , Lv, D.‐H. , Zhai, Q. I. , Wen, X.‐H. , Xu, Z.‐H. , … Chen, Q.‐L. (2020). Can cats become infected with Covid‐19? Veterinary Record, 186(17), e20. 10.1136/vr.m1581 [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Lin, D. , Kusov, Y. , Nian, Y. , Ma, Q. , Wang, J. , … Hilgenfeld, R. (2020). α‐Ketoamides as Broad‐Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure‐Based Design, Synthesis, and Activity Assessment. Journal of Medicinal Chemistry, 63(9), 4562–4578. 10.1021/acs.jmedchem.9b01828 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Wang, Y. , Qi, C. , Shen, L. , & Li, J. (2020). Clinical trial analysis of 2019‐nCoV therapy registered in China. Journal of Medical Virology, 92, 540–545. 10.1002/jmv.25733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Zhang, H. J. , Huang, K. , Yang, Y. , Hui, X. , Gao, J. , … Jin, M. (2020). SARSCoV‐2 neutralizing serum antibodies in cats: A serological investigation. BioRxiv. 10.1101/2020.04.01.021196 [DOI] [Google Scholar]
- Zhang, S. , Diao, M. Y. , Yu, W. , Pei, L. , Lin, Z. , & Chen, D. (2020). Estimation of the reproductive number of novel coronavirus (COVID‐19) and the probable outbreak size on the Diamond Princess cruise ship: A data‐driven analysis. International Journal of Infectious Diseases, 93, 201–204. 10.1016/j.ijid.2020.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Wu, Q. , & Zhang, Z. (2020). Pangolin homology associated with 2019‐nCoV. bioRxiv. 10.1101/2020.02.19.950253 [DOI] [Google Scholar]
- Zhong, J. , Tang, J. , Ye, C. , & Dong, L. (2020). The immunology of COVID‐19: Is immune modulation an option for treatment? The Lancet Rheumatology, 2(7), e428–e436. 10.1016/S2665-9913(20)30120-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Zhang, M. , Wang, J. , & Gao, J. (2020). Sars‐Cov‐2: Underestimated damage to nervous system. Travel Medicine and Infectious Disease, 101642. 10.1016/j.tmaid.2020.101642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X.‐L. , Wang, X.‐G. , Hu, B. , Zhang, L. , Zhang, W. , … Shi, Z.‐L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, G. , & Chen, H. W. (2004). Monophyletic relationship between severe acute respiratory syndrome coronavirus and group 2 coronaviruses. Journal of Infectious Diseases, 189, 1676–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. , Xu, X. , Ma, K. , Yang, J. , Guan, H. , Chen, S. , … Chen, G. (2020). Successful recovery of COVID‐19 pneumonia in a renal transplant recipient with long‐term Immunosuppression. American Journal of Transplantation, 20, 1859–1863. 10.1111/ajt.15869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. A. , Zhang, D. , Wang, W. , Li, X. , Yang, B. O. , Song, J. , … Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382, 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z. , Dimitrov, A. S. , Bossart, K. N. , Crameri, G. , Bishop, K. A. , Choudhry, V. , … Dimitrov, D. S. (2006). Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. Journal of Virology, 80, 891–899. 10.1128/JVI.80.2.891-899.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A. , Chan, J. F. W. , Azhar, E. I. , Hui, D. S. C. , & Yuen, K.‐Y. (2016). Coronaviruses‐drug discovery and therapeutic options. Nature Reviews: Drug Discovery, 15, 327–347. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


