Abstract
The development of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) serological tests is massive. The external validation of their performance is needed before use in clinical routine practice. Our study aims at assessing the analytical and clinical performance of two enzyme‐linked immunosorbent assay tests detecting antibodies directed against the virus nucleocapsid protein: The NovaLisa SARS‐CoV‐2 immunoglobulin G (IgG), immunoglobulin A (IgA), and immunoglobulin M (IgM) test (NovaTec) allowing a separate detection of each antibody and the Platelia SARS‐CoV‐2 Total Ab test (Bio‐Rad) detecting total antibodies (IgM, IgA, and IgG). Two‐hundred and eight coronavirus disease 2019 samples from 48 quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) confirmed patients were used to perform the sensitivity analysis. Non‐SARS‐CoV‐2 sera (n = 79) with a potential cross‐reaction to SARS‐CoV‐2 immunoassays were included in the specificity analysis. In addition, using receiver operator characteristic curves, adapted cut‐off for improvement of the performances were proposed. The kinetics of these antibodies was also assessed over 8 weeks. Two weeks after the RT‐qPCR positive detection, the NovaLisa test shows a sensitivity and specificity of 94.9% (95% confidence interval [CI]: 83.1%‐98.6%) and 96.2% (95% CI: 89.4%‐98.7%) for IgG, of 89.7% (95% CI: 76.4%‐95.9%) and 98.7% (95% CI: 93.2%‐98.8%) for IgA, and of 48.7% (95% CI: 33.9%‐63.8%) and 98.7% (95% CI: 93.2%‐99.8%) for IgM. With the Platelia system, the specificity and sensitivity were 97.4% (95% CI: 92.1%‐99.7%) and 94.9% (95% CI: 87.7%‐98.0%) for total antibodies using the adapted cut‐offs. The NovaLisa and the Platelia tests have satisfactory analytical performances. The clinical performances are excellent for IgG, IgA, and total antibodies especially if the cut‐off is optimized.
Keywords: antibodies; COVID‐19, ELISA; IgA; IgG; IgM; kinetics; SARS‐CoV‐2
Highlights
The sensitivity and the specificity of the NovaLisa (for immunoglobulin G [IgG] and immunoglobulin A [IgA]) and the Platelia were excellent.
Cut‐off optimizations improve the clinical performances of both assays.
Over a period of 8 weeks, IgM and IgA gradually disappeared while IgG remain persistent.
1. INTRODUCTION
End of 2019, a novel respiratory disease emerged in the city of Wuhan, Hubei Province of the People's Republic of China. On 7 January 2020, Chinese authorities determined that these severe cases of pneumonia were caused by a new coronavirus, temporarily named “2019‐nCoV.” 1 This virus, genetically related to the coronavirus responsible for the 2003 severe acute respiratory syndrome (SARS) outbreak, was renamed SARS coronavirus 2 (“SARS‐CoV‐2”) by the International Committee on Taxonomy of Viruses on 11 February 2020. 2
Since then an unprecedented health, economic and human crisis has quickly struck the world. By mid‐March 2020, the World Health Organization European Region had become the epicenter of the epidemic, reporting more than 40% of confirmed cases worldwide. As of 28 April 2020, the region was contributing 63% of the global mortality due to the virus. 3 On 8 June, the John Hopkins' University assessment showed that the virus has spread to 188 countries and territories, the number of confirmed cases exceeds 6 913 608 million and the number of deaths worldwide stands at 400 121 deaths. 4
In this context of continuous progression of knowledge of coronavirus disease 2019 (COVID‐19) and its evolution, several SARS‐CoV‐2 immunoassays have been developed. To date, more than 224 different CE marked tests have been identified, including 72 manual or automated immunoassays. 5 Various techniques are available, enzyme‐linked immunosorbent assay (ELISA), chemiluminescence enzyme immunoassays, fluorescence immunoassays, lateral flow immunoassays to detect immunoglobulin G (IgG), immunoglobulin A (IgA), immunoglobulin M (IgM) (separately or in combination) as well as different antibody targets (Spike [S], RBD and/or, nucleocapsid proteins).
It is essential for laboratories to independently validate these methods before broad introduction into routine clinical practice. In this context, more and more independent validations of serological tests are published by analyzing the same sample of sera against different techniques but also very often with different antibody targets. 6 , 7 , 8 , 9 , 10 , 11 , 12
The main objective of our study is to assess and compare the analytical and clinical performance of two ELISA tests detecting antibodies directed against the nucleocapsid protein of the virus: The NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgA, and IgM test (NovaTec) allowing a separate detection of each antibody and the Platelia SARS‐CoV‐2 Total Ab test (Bio‐Rad) detecting total antibodies (IgM, IgA, and IgG).
The secondary objectives are to describe the kinetics of these antibodies over a period of 8 weeks and to clarify the clinical interest of the independent detection of IgG, IgA, and IgM.
2. MATERIALS AND METHODS
2.1. Study design
This retrospective study was conducted from 8 May to 9 June 2020 at the clinical biology laboratory of the Iris Hospitals South (HIS‐IZZ, Brussels, Belgium). All the sera (n = 287) originate from blood samples taken during previous clinical requests for diagnostic purposes and were stored in the laboratory serum biobank at −20°C. Among these 287 samples, 79 samples were included in the specificity analysis and were collected before the COVID‐19 outbreak. The remaining 208 samples were included in the sensitivity analysis and were collected from patients hospitalized for COVID‐19 disease. This study has been approved by the ethical committee of the HIS‐IZZ (ethical agreement number: CEHIS/2020‐13).
2.2. Population
Blood samples positive for COVID‐19 were collected from symptomatic patients who came to the emergency room. Table 1 reports on the number and characteristics of subjects included in the study for gender, age, extend of disease based on computed tomography scan criteria, length of hospital stay, place of hospitalization and outcome. Patients were considered positive according to the results of the quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR). The delay between the first onset of symptoms and the RT‐qPCR is variable and has been estimated at 4 days (±1 day) in our cohort of 48 patients.
Table 1.
Demographic characteristics of patients included in the study
| Demography | |
| Age (median [min‐max; 95% CI]) | N = 48 (72.0 [21.5‐92.4; 4.4]) |
| Males | N = 28 |
| Females | N = 20 |
| Length of hospital stay [median (min‐max; 95% CI)] | N = 39a (21.0 [1.0‐69.0; 4.8]) |
| Delay between symptoms and PCR [median (min‐max; 95% CI)] | N = 36 (4.0 [0.0‐35.0; 2.1]) |
| Intubated in ICU | N = 11 |
| Not intubated in ICU | N = 3 |
| Hospitalized (non ICU) | N = 30 |
| Not hospitalized | N = 4 |
| Survivors | N = 42 |
| Non survivors | N = 6 |
| Extend of disease (CT scan criteria) | |
| Minimal | N = 1 |
| Moderate | N = 5 |
| Extended | N = 8 |
| Severe | N = 12 |
| Critical | N = 2 |
| Not categorized | N = 11 |
| Negative | N = 8 |
| No CT scan | N = 1 |
Abbreviations: CI, confidence interval; CT, computed tomography; ICU, intensive care unit; PCR, polymerase chain reaction.
1 patient referred to another hospital; 4 stays in progress.
2.3. Sample collection
Blood samples were collected in serum collection tubes (BD Vacutainer SST II advance, BD, Plymouth, UK) according to procedure previously described. 12
2.4. ELISA assays
The semi‐quantitative analysis of IgG, IgA, and IgM anti‐SARS‐CoV‐2 nucleocapsid antibodies was carried out by the NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgA, and IgM test (NovaTec Immundiagnostica GmbH, Dietzenbach, Germany) while the semi‐quantitative detection of total anti‐SARS‐CoV‐2 nucleocapsid antibodies (IgM/IgA/IgG) was carried out by the Platelia SARS‐CoV‐2 Total Ab method (Bio‐Rad, Marnes‐la‐Coquette, France). Both methods have been implemented on the ETI‐Max 3000 controller (DiaSorin) after specific programming according to the manufacturer's instructions. For each ELISA plate, a ratio between the extinction of the serum samples and the calibrator was calculated. The interpretation criteria provided by the manufacturers are provided in Table 2.
Table 2.
Interpretation criteria of the NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgA, and IgM test (NovaTec and of the Platelia SARS‐CoV‐2 Total Ab method (Bio‐Rad) on the ETI‐Max 3000 controller
| Test | Result | Interpretation |
|---|---|---|
| NovaLisa | Ratio <9 | Negative |
| Ratio ≥9 and ≤11 | Doubtfula | |
| Ratio >11 | Positive | |
| Platelia | Ratio <0.8 | Negative |
| Ratio ≥0.8 and <1.0 | Doubtfulb | |
| Ratio ≥1.0 | Positive |
Abbreviations: IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Procedure: Antibodies against the pathogen could not be detected clearly. It is recommended to repeat the test with a fresh sample in 2 to 4 weeks. If the result is equivocal again the sample is judged as negative.
Should be retested in duplicate before final interpretation. In case of repeated equivocal result, another specimen should be collected and tested few days later.
2.5. Evaluation and comparison of the clinical performances
2.5.1. Assessment of the clinical sensitivity
The clinical sensitivity was assessed at several time points since the confirmation of the diagnostic by RT‐qPCR (n = 208). Thirty‐nine, 35, 39, 34, 22, 17, 14, 6, and 2 sera from 48 positive patients with COVID‐19, collected respectively at 0 ± 2, 7 ± 2, 14 ± 2, 21 ± 2, 28 ± 2, 35 ± 2, 42 ± 2, 49 ± 2, and 56 ± 2 days since the date of respiratory sampling (t0), were analyzed to detect the appearance of the antibodies.
2.5.2. Evaluation of the kinetics appearance of antibodies
Assessment of the kinetics of IgG, IgA, and IgM appearance in COVID‐19 positive patients have been carried out by reporting the levels of antibodies every week for 8 weeks (ie, 7, 14, 21, 28, 35, 42, 49, and 56 days) from the time of the RT‐qPCR positive respiratory sample.
2.5.3. Assessment of the clinical specificity
Seventy‐nine samples were tested to assess the cross‐reactivity. Seventy‐three sera from COVID‐19 negative patients but who had other viral, bacterial, parasitic or autoimmune pathologies that could be considered as confounding factors were included in the study. Sera positive for the following viral, bacterial and infection from parasite origin were included to assess the possible cross‐reactivity: Hepatitis B surface antigen (n = 7), hepatitis A virus IgM (n = 3), adenovirus (n = 1), herpes simplex virus IgM and cytomegalovirus (CMV) IgM (n = 1), IgM CMV (n = 8), IgM parvovirus B19 (n = 5), human immunodefeciency virus (n = 1), antistreptolysin O (n = 4), anti‐Treponema pallidum antibody (n = 1), IgG Borrelia (n = 1), IgM Mycoplasma pneumoniae (n = 10), and Toxoplasma gondii IgM (n = 16). The cross‐reactivity of the following autoimmune pathologies was also assessed: rheumatoid factor (n = 1), anti‐thyroid peroxidase antibody (n = 7), search for irregular agglutinins (n = 4), direct coombs (n = 1). Finally, one serum with a high level of total IgM (9.01 g/L) (normal range, 0.40‐2.30 g/L), one serum with high total IgA (4.47 g/L) (normal range, 0.70‐4.00 g/L), and six sera from COVID‐19 negative healthy subjects, with no history of known autoimmune pathologies and without any acute infection of viral or bacterial origin were included in the study. In these six sera, residues from old viral infections were present: IgG parvovirus B19 (n = 1), viral capsid antigen and IgG CMV (n = 2), IgG herpes zoster virus and IgG Rubella (n = 2), and HBV antibody (n = 1). All these samples were collected in 2019 before the start of the COVID‐19 outbreak and were stored at −20°C.
2.6. Evaluation and comparison of the analytical performances
Evaluation of the performance was performed in accordance with the Clinical and Laboratory Standards Institute EP 15‐A3 document. 13 The acceptance criteria were defined according to the performance reported by the manufacturer and are summarized in Table 3.
Table 3.
Acceptance criteria for the evaluation of the analytical performances of the NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgA, and IgM test (NovaTec) and of the Platelia SARS‐CoV‐2 Total Ab method (Bio‐Rad)
| Validation step | NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgA, and IgM | Platelia SARS‐CoV‐2 Total Ab | ||
|---|---|---|---|---|
| Acceptance criteria according to manufacturer performances | Results | Acceptance criteria according to manufacturer performances | Results | |
| Trueness | Not reported by the manufacturer |
IgG: Low QC level: 15.72 ± 1.97 High QC level: 42.45 ± 5.54 IgA: Low QC level: 16.55 ± 0.93 High QC level: 106.00 ± 7.96 IgM: Low QC level: 18.20 ± 3.21 High QC level: 50.70 ± 5.64 |
Not reported by the manufacturer |
Total Ab: Low QC level: 1.34 ± 0.14 High QC level: 1.83 ± 0.24 |
| Precision |
Repeatability (CV): IgG: 4.06%‐8.71% IgA: Not reported by the manufacturer IgM: 2.75%‐10.30% Reproducibility (CV): IgG: 4.11%‐8.65% IgA: Not reported by the manufacturer IgM: 6.00%‐11.91% |
Repeatability (CV): IgG: 2.88%‐9.31% IgA: 2.48%‐4.21% IgM: 2.95%‐7.70% Reproducibility (CV): IgG: 10.64%‐11.71% IgA: 5.73%‐7.97% IgM: 9.57%‐14.06% |
Repeatability (CV): Total Ab: 3.3%‐4.0% Reproducibility (CV): Total Ab: 3.2%‐6.9% |
Repeatability (CV): Total Ab: 2.3%‐8.5% Reproducibility (CV): Total Ab: 10.5%‐10.6% |
| Limit of blank | Not reported by the manufacturer |
IgG: 0.05 IgA: 0.20 IgM: 0.01 |
Not reported by the manufacturer | Total Ab: 0.09 |
| Limit of detection | Not reported by the manufacturer |
IgG: 0.18 IgA: 0.44 IgM: 0.02 |
Not reported by the manufacturer | Total Ab: 0.25 |
| Carry‐over | Not reported by the manufacturer |
IgG: 0.18% IgA: 0.01% IgM: 0.11% |
Not reported by the manufacturer | Total Ab:0.8% |
| Specificity |
Cut‐off of the manufacturer (>11): IgG: 99.2% IgA: Not reported by the manufacturer IgM:100% |
Cut‐off of the manufacturer (>11): IgG: 98.7% IgA: 98.7% IgM: 100% Adapted cut‐off (≥7): IgG: 96.2% IgA: 98.7% IgM: 98.7% |
Cut‐off of the manufacturer ≥1: Total Ab: 99.6% |
Cut‐off of the manufacturer ≥1: Total Ab: 97.5% Adapted cut‐off ≥0.8: Total Ab: 94.9% |
| Sensitivity |
Cut‐off of the manufacturer after ≥12 d post symptoms: IgG: 100% IgA: Not reported by the manufacturer IgM: 57.1% |
Cut‐off of the manufacturer after ≥ 14 d post PCR: IgG: 89.7% IgA: 84.6% IgM: 30.8% Adapted cut‐off: IgG: 94.9% IgA: 89.7% IgM: 48.7% |
Cut‐off of the manufacturer between 11 and 20 d post symptoms: Total Ab: 97% |
Cut‐off of the manufacturer after ≥ 14 d post PCR: Total Ab: 94.7% Adapted cut‐off: Total Ab: 97.4% |
Abbreviations: Ab, antibody; COVID‐19, coronavirus disease 2019; CV, coefficient of variation; IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; PCR, polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
2.6.1. Trueness
For NovaLisa and Platelia tests, as there were no positive QC with two different levels indicated by the manufacturer, trueness has been estimated by comparing the average value obtained on 20 replicates of two levels of pool patients to target low and high values representative of our patient cohort.
2.6.2. Precision
Precision has been evaluated by analyzing the repeatability (expressed as intra‐run coefficient of variation [CV]) and the reproducibility (expressed as inter‐run CV) of the method. The two levels of pool patients were run in triplicate during 5 consecutive days.
2.6.3. Limit of blank and detection
The negative control for NovaLisa tests provided by the manufacturer was used as blank sample to determine the limit of blank (LoB) and limit of detection (LoD). As there were not enough negative control for Platelia tests, theses parameters were evaluated with the diluent provided by the manufacturer. The LoB has been determined by running the blank sample on three separate occasions. The LoD has been determined by running 30 analyses of the blank sample using the following equation according the SH GTA 04 document‐revision 1 of the COFRAC. 14
2.6.4. Evaluation of the carry‐over
A sample with a high level of antibodies was run in triplicate (A1, A2, and A3) followed by a negative sample also run in triplicate (B1, B2, and B3). The ratio is calculated using the following equation: (B1 − B3/A3− B3) × 100. Carry‐over below 1% is considered satisfactory and is not linked with significant interference.
2.7. Statistical analyses
Statistical analyses were carried out using MedCalc version 10.4.0.0 (MedCalc Software, Ostend, Belgium). Descriptive statistics were used to analyze the data. Sensitivity was defined as the proportion of correctly identified COVID‐19 positive patients who were initially positive by RT‐qPCR SARS‐CoV‐2 determination in respiratory samples. Specificity was defined as the proportion of naïve participants who classified as positive as analyzed by one of the two methods tested in this study. The adapted cut‐off of the NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgA, and IgM test (NovaTec) and the Platelia SARS‐CoV‐2 Total Ab method (Bio‐Rad) was determined using receiver operator characteristic (ROC) curves at 14 ± 2 days post RT‐qPCR. A Comparative ROC curves for the combined IgG, IgA, IgM (NovaTec) and total antibodies (Bio‐Rad) was performed. The highest value of one of the ratios (corresponding either to that of IgG, IgA, or IgM) was selected for the combined evaluation of the overall sensitivity of these three antibodies (NovaTec).
3. RESULTS
3.1. Evaluation and comparison of the clinical performances
For the 208 clinical samples, the ranges of calculated index values are as follow: IgG: (0.62‐44.34); IgA: (0.38‐113.99); IgM: (0.07‐43.90); total antibodies: (0.05‐3.84).
3.1.1. Assessment of clinical sensitivity
The sensitivity of the NovaLisa test was 89.7% (95% CI: 76.4%‐95.9%) for IgG, 84.6% (95% CI: 70.3%‐92.8%) for IgA and 30.8% (95% CI: 18.6%‐46.4%) for IgM according to manufacturer's cut‐offs in samples collected 2 weeks after RT‐qPCR positive detection. Best performances are observed 3 weeks after the RT‐qPCR with values of 91.2% (95% CI: 77.0%‐97.0%) for IgG and IgA and 38.2% (95% CI: 23.9%‐55.0%) for IgM. For the Platelia Total Ab test, the sensitivity 94.7% (95% CI: 83.1%‐98.6%) according to manufacturer's cut‐offs in samples collected 2 weeks after RT‐qPCR positive detection. Results were summarized in Figure 1. Among the 118 samples evaluated (39 results expected positive and 79 expected negative) 2 weeks after the RT‐qPCR positive detection, and according to manufacturer's cut‐off, the NovaLisa SARS‐CoV‐2 IgG, IgA, and IgM kit respectively identified 35, 33, 12 true positive and 78, 78, 79 true negative. Respectively, 4, 6, and 27 samples were classified as false negative and 1, 1, 0 as false positive with the IgG, IgA, and IgM kit. On the same cohort, the Platelia SARS‐CoV‐2 Total Ab method identified 36 true positive and 77 true negative. Two samples were false positive, and two samples were false negative.
Figure 1.
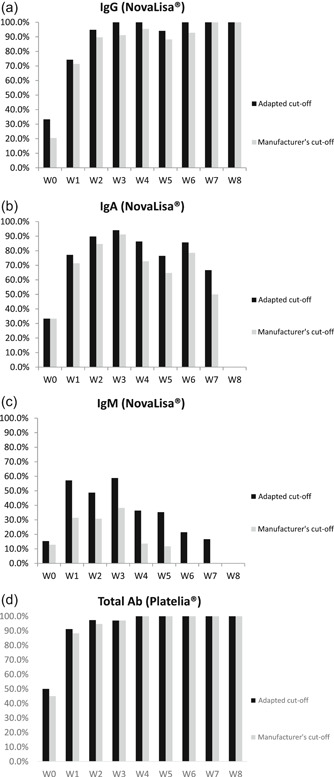
Evolution of the clinical sensitivity over 8 weeks according to the manufacturer's cut‐off and the adapted cut‐off for the NovaLisa SARS‐CoV‐2 (COVID‐19) IgG (A), IgA (B), and IgM (C) tests and (D) for the Platelia SARS‐CoV‐2 Total Ab method (Bio‐Rad). IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
The cut‐offs provided by the ROC curve analyses (ie, ≥7.0 and ≥0.8 for the NovaLisa kit and the Platelia system, respectively) improve the sensitivity up to 94.9% (95% CI: 83.1%‐98.6%) for IgG, 89.7% (95% CI: 76.4%‐95.9%) for IgA, 48.7% (95% CI: 33.9%‐63.8%) for IgM and 97.4% (95% CI: 92.1%‐99.7%) for total antibodies. Among the 118 samples tested, the use of these adapted cut‐offs permits the correct reclassification respectively of the 2, 2, and 7 false negative with the NovaLisa IgG, IgA, and IgM kit to the detriment of only one false positive case for IgG. Specificity was not changed for IgA and IgM. For the Platelia test, the use of the adapted cut‐offs permitted the correct reclassification of the one false negative to the detriment of two positive case. The sensitivity and specificity were 94.9% (95% CI: 83.1%‐98.6%) and 96.2% (95% CI: 89.4%‐98.7%) for IgG, and 89.7% (95% CI: 76.4%‐95.9%) and 98.7% (95% CI: 93.2%‐98.8%) for IgA, and 48.7% (95% CI: 33.9%‐63.8%) and 98.7% (95% CI: 93.2%‐99.8%) for IgM, and 97.4% (95% CI: 92.1%‐99.7%) and 94.9% (95% CI: 87.7%‐98.0%) for total antibodies (Table 3) and the kappa index were 0.90 (IgG), 0.90 (IgA), 0.54 (IgM), and 0.87 (IgG, IgA, IgM) for the NovaLisa and 0.90 (total antibodies) for the Platelia system, respectively using the adapted cut‐offs.
3.1.2. Assessment of clinical specificity
From the results obtained above, interference with certain antibodies or antigens produced following viral, bacterial or parasitic infections or following autoimmune pathologies reveals to be relatively low with a specificity of 98.7% (95% CI: 93.2%‐99.8%) for IgG, 98.7% (95% CI: 93.2%‐99.8%) for IgA, 100% (95% CI: 95.4%‐100%) for IgM with the NovaLisa tests and 97.5% (95% CI: 91.2%‐99.3%) for total antibodies with the Platelia system, respectively using the cut‐offs provided by the manufacturers. Using the adapted cut‐offs, the specificity was 96.2% (95% CI: 89.4%‐98.7%) for IgG and 98.7%(95% CI: 93.2%‐98.8%) for IgA, and 98.7% (95% CI: 93.2%‐99.8%) for IgM and 94.9% (95% CI: 87.7%‐98.0%) for total antibodies.
The comparative ROC curves for the combination of IgG, IgA, IgM (NovaTec) and total antibodies (Bio‐Rad) (Figure 3) showed an area under curve (AUC) at 0.939 for IgG, IgA, IgM (NovaTec) versus an AUC at 0.933 for total antibodies (Bio‐Rad). No statistically significant difference was observed between the two tests (P = 0.749).
Figure 3.
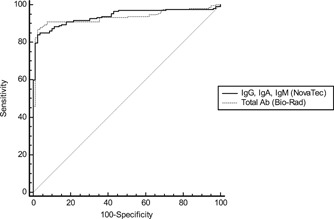
Comparative ROC curves for IgG, IgA, IgM (NovaTec) and total antibodies (Bio‐Rad) n = 287. Ab, antibody; IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; ROC, receiver operator characteristic
3.1.3. Evaluation of the kinetics appearance of antibodies
A peak in antibody production is observed for IgA, IgG, and IgM 3 weeks after PCR.
The values reached for IgA are significantly higher than those for IgG. Then the IgA values gradually decrease and are no longer detectable at week 8. IgM follow the same decreasing kinetics also from the third week with an earlier total disappearance of antibodies from the fourth week. Conversely, IgG reaches a plateau at the third week and the antibodies maintain this level until the eighth week (Figure 2). The appearance rate of total antibodies is much faster than each antibody measured separately and is observed from the first week post PCR.
Figure 2.
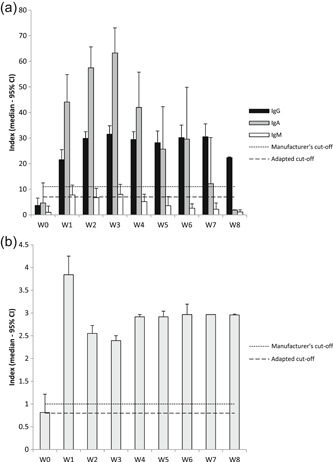
A, Anti‐SARS‐CoV‐2 IgG, IgA, IgM and (B) total antibody kinetics at different weeks after the RT‐qPCR positive detection in 48 patients on a total of 208 samples. IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; RT‐qPCR, quantitative reverse transcription‐polymerase chain reaction
Total antibodies quickly reach a plateau and remain detectable until week 8.
3.2. Evaluation and comparison of the analytical performances
Table 3 summarizes the analytical performances of the NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgA, and IgM test and the Platelia SARS‐CoV‐2 Total Ab method. As internal QC were not provided by the manufacturer, we only report the mean and the standard deviation of the two levels of patients' pool for each method, as instructed by the manufacturer. The precision shows appropriate repeatability and reproducibility and are in line with the CVs provided by the manufacturer. The intra‐ and inter‐run CVs were close to the range reported by the manufacturer for the two levels of QC. The most extreme CV value observed was 14.06% for the low QC reproducibility of IgM. The LoB and detection are below the adapted cut‐offs with the two tests and the carry‐over is negligible.
4. DISCUSSION
The interest of serological tests is well recognized today by many national regulatory authorities in combination with RT‐PCR (which remains the first‐line test for the diagnosis of the acute phase of COVID‐19) for serosurvelliance or seroepidemiology to identify people being or having been in contact with the virus. Their external validation of their performance is however needed before use in clinical routine practice and some improvement of their performance might be recommended. This study is the first to describe and compare the analytical and clinical performances of the NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgM, and IgA kit from NovaLisa and Platelia SARS‐CoV‐2 Total Ab Assay from Bio‐Rad. We found that the sensitivity increased with time from the first day until the second week post PCR.
For the two techniques, adaptation of the cut‐offs permits to achieve a sensitivity of 91.2% for total antibodies for samples collected 1 week after PCR until 97.4% in the second week. In these samples, the analyses of different immunoglobulins reveal sensitivities of 94.9% for IgG, 89.7% for IgA, and 48.7% for IgM. In lights of these results and the data provided by the manufacturer, assessment of IgM remained limited to be used in clinical practice, as already reported for other kits assessing IgM suggesting that technical improvements should be investigated with the firm.
Using the cut‐off provided by the manufacturer, only two false positives were observed with NovaLisa. These false positives samples resulted probably from a cross‐reaction with the IgG testing and were observed in a sample positive for IgM CMV. The second false positive result originates from serum also positive for IgM CMV and was reported falsely positive for IgA. For the Platelia, two false positives were observed, one with IgM Mycoplasma pneumoniae and one with IgM Toxoplasma gondii. However, as it is the case for most validations currently published, and given the scarcity of these samples, we were not able to assess the specificity towards other coronavirus like the strains 229E (alpha), NL63 (alpha), OC43 (beta), HKU1 (beta), SARS or Middle East respiratory syndrome.
4.1. Target antigen and antibody isotype comparison
IgG tests perform better compared with IgA or IgM ones and show better sensitivity when the samples were taken minimum 2 weeks after the RT‐qPCR positive detection. Moreover, a combined IgG/IgA/IgM test seems to be a better choice in terms of sensitivity than measuring either antibody alone.
Insofar as serological tests have no indication in the acute diagnosis of COVID‐19, the advantage of detecting each antibody separately seems very limited in clinical practice. At best IgA appears earlier than IgG and are detectable from the 1st week.
Regarding the antigenic target, It is essential to compare tests that target the same antibody detection. 15 In this study, we compared for the first time the analytical and clinical performance of two ELISA tests detecting antibodies directed against the nucleocapsid protein. The clinical significance of these differences remains unknown. Do they only witness an infection, or will they witness a protective and lasting immunity over time?
However, there is a general consensus that SARS‐CoV‐2 neutralizing antibody responses are targeting the S protein. 16 , 17 Of note, in the longer term, and in the event that a vaccine would become available, measuring antibody responses to the nucleocapsid (N) antigen would be informative because most vaccine candidates are targeting the S protein. 18 Measuring the antibody response may help discriminate between vaccinated (responding to S only) and SARS‐CoV‐2 exposed individuals (responding to both S and N).
4.2. Kinetics over 8 weeks
To ensure protective and long‐lasting immunity, it should be known whether the antibodies are protective and persistent over months. The majority of studies have focused on the clinical performance of serological tests during the first 3 weeks post‐symptoms or post‐PCR. 19 , 20 , 21 , 22 This study, by assessing the antibody kinetics over a period of 8 weeks provide important data on the persistence of antibodies in infected patients. The delay between the first onset of symptoms and the RT‐qPCR is not so variable and has been estimated at 4 days (±1 day) in our cohort of 48 patients. Previous studies have assessed the presence of antibodies in serum collected from 0 up to 49 days post symptoms or post PCR. 23 , 24 The kinetics observed in this study is in line with litterature 25 showing a gradual appearance of antibodies between the first and second week post‐symptom. A production peak was then observed appearing at the third week followed by a decrease in IgM and IgA from week 4 and 8, respectively (Figure 2). Based on the demographic characteristics of patients included in the study and described in Table 1, we did not observe any clear association between the IgG, IgM, IgA or total antibodies results and disease severity, although this was already reported in some studies.
Finally, the present study has some limitations. First, monitoring of antibody dynamics was extended to a maximum of 56 days post PCR, kinetic analysis over several months will be necessary to confirm the persistence of antibodies. Second, the number of patients included in week 8 is too low to confirm a total disappearance of IgA (n = 2). In addition, the adaptation of the cut‐off must consider the population studied. In our study, the cohort studied only focused on symptomatic patients who came to the emergency room. Further investigations are needed to verify whether the appropriate cut‐offs are also applicable to pauci‐symptomatic and asymptomatic patients.
5. CONCLUSION
This study is the first to report the external validation of a new NovaLisa SARS‐CoV‐2 (COVID‐19) IgG, IgA, and IgM test (NovaTec) and Platelia SARS‐CoV‐2 Total Ab method (Bio‐Rad) directed against SARS‐CoV‐2 nucleocapsid. The clinical performances are excellent for IgG, IgA and total antibodies especially if the cut‐off is optimized.
CONFLICT OF INTERESTS
Among the authors, Jonathan Douxfils is chief executive officer and founder of QUALIblood sa and reports personal fees from Diagnostica Stago, Roche, Roche Diagnostics, Daiichi‐Sankyo, and Portola, outside the submitted work.
ETHICS STATEMENT
The study was approved by the ethical committee of the Iris Hospitals South.
ACKNOWLEDGMENTS
The authors thank also all the members of the clinical laboratory staff for technical assistance.
Tré‐Hardy M, Wilmet A, Beukinga I, et al. Analytical and clinical validation of an ELISA for specific SARS‐CoV‐2 IgG, IgA and IgM antibodies. J Med Virol. 2021;93:803–811. 10.1002/jmv.26303
REFERENCES
- 1. Chan JF‐W, Yuan S, Kok K‐H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet Lond Engl. 2020;395(10223):514‐523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organisation . Naming the coronavirus disease (COVID‐19) and the virus that causes it; 2020. Technical guidance. Published online 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
- 3. World Health Organisation . Coronavirus disease (COVID‐19) pandemic. Published online 2020. http://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/novel-coronavirus-2019-ncov
- 4. John Hopkins University . Coronavirus resource center. https://coronavirus.jhu.edu/map.html. Published 2020. https://coronavirus.jhu.edu/map.html. Accessed June 7, 2020.
- 5. FindDx.org. SARS‐CoV‐2 diagnostic pipeline. FindDx. Published 2020. https://www.finddx.org/covid-19/pipeline/?avance=Commercialized&type=all&test_target=all&status=CE-IVD§ion=immunoassays&action=default#diag_tab. Accessed June 7, 2020.
- 6. GeurtsvanKessel CH, Okba NMA, Igloi Z, et al. Towards the next phase: valuation of serological assays for diagnostics and exposure assessment. Infect Dis (except HIV/AIDS). 2020. 10.1101/2020.04.23.20077156 [DOI] [Google Scholar]
- 7. Jääskeläinen AJ, Kekäläinen E, Kallio‐Kokko H, et al. Evaluation of commercial and automated SARS‐CoV‐2 IgG and IgA ELISAs using coronavirus disease (COVID‐19) patient samples. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull. 2020;25(18), 10.2807/1560-7917.ES.2020.25.18.2000603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kontou PI, Braliou GG, Dimou NL, Nikolopoulos G, Bagos PG. Antibody tests in detecting SARS‐CoV‐2 infection: a meta‐analysis. Diagn Basel Switz. 2020;10(5), 10.3390/diagnostics10050319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Montesinos I, Gruson D, Kabamba B, et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti‐SARS‐CoV‐2 antibodies. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2020;128:104413. 10.1016/j.jcv.2020.104413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Plebani M, Padoan A, Negrini D, Carpinteri B, Sciacovelli L. Diagnostic performances and thresholds: the key to harmonization in serological SARS‐CoV‐2 assays? Clin Chim Acta Int J Clin Chem. 2020;509:1‐7. 10.1016/j.cca.2020.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Traugott M, Aberle SW, Aberle JH, et al. Performance of SARS‐CoV‐2 antibody assays in different stages of the infection: comparison of commercial ELISA and rapid tests. J Infect Dis. 2020;222:362‐366. 10.1093/infdis/jiaa305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tré‐Hardy M, Wilmet A, Beukinga I, Dogné J‐M, Douxfils J, Blairon L. Validation of a chemiluminescent assay for specific SARS‐CoV‐2 antibody. Clin Chem Lab Med CCLM. 2020;0(0), 10.1515/cclm-2020-0594 [DOI] [PubMed] [Google Scholar]
- 13. Carey RN, Clinical and Laboratory Standards Institute . User Verification of Precision and Estimation of Bias: Approved Guideline. Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- 14. COFRAC . Guide technique d'Accréditation de vérification (Portée A)/Validation (Portée B) des méthodes en biologie médicale. Published online 2015. https://tools.cofrac.fr/documentation/SH-GTA-04
- 15. Kohmer N, Westhaus S, Rühl C, Ciesek S, Rabenau HF. Brief clinical evaluation of six high‐throughput SARS‐CoV‐2 IgG antibody assays. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2020;129:104480. 10.1016/j.jcv.2020.104480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS‐CoV‐2 and other human coronaviruses. Trends Immunol. 2020;41(5):355‐359. 10.1016/j.it.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. To KK, Tsang OT, Leung W‐S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565‐574. 10.1016/S1473-3099(20)30196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poh CM, Carissimo G, Wang B, et al. Two linear epitopes on the SARS‐CoV‐2 spike protein that elicit neutralising antibodies in COVID‐19 patients. Nat Commun. 2020;11(1):2806. 10.1038/s41467-020-16638-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu L, Liu W, Zheng Y, et al. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in 238 admitted hospital patients. Microbes Infect. 2020;22:206‐211. 10.1016/j.micinf.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Long Q‐X, Liu B‐Z, Deng H‐J, et al. Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med. 2020;26:845‐848. 10.1038/s41591-020-0897-1 [DOI] [PubMed] [Google Scholar]
- 21. Lou B, Li T‐D, Zheng S‐F, et al. Serology characteristics of SARS‐CoV‐2 infection since exposure and post symptom onset. Eur Respir J. 2020. 10.1183/13993003.00763-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suhandynata RT, Hoffman MA, Kelner MJ, McLawhon RW, Reed SL, Fitzgerald RL. Longitudinal monitoring of SARS‐CoV‐2 IgM and IgG seropositivity to detect COVID‐19. J Appl Lab Med. 2020. 10.1093/jalm/jfaa079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beavis KG, Matushek SM, Abeleda APF, et al. Evaluation of the EUROIMMUN Anti‐SARS‐CoV‐2 ELISA assay for detection of IgA and IgG antibodies. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2020;129:104468. 10.1016/j.jcv.2020.104468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Favresse J, Eucher C, Elsen M, Marie T‐H, Dogné J‐M, Douxfils J. Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS‐CoV‐2 total antibodies. Clin Chem. 2020. 10.1093/clinchem/hvaa131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests for SARS‐CoV‐2. JAMA. 2020. 10.1001/jama.2020.8259 [DOI] [PubMed] [Google Scholar]


