Abstract
The coronavirus disease 2019 (COVID‐19) pandemic has revealed major shortcomings in our ability to mitigate transmission of infectious viral disease and provide treatment to patients, resulting in a public health crisis. Within months of the first reported case in China, the virus has spread worldwide at an unprecedented rate. COVID‐19 illustrates that the biomaterials community was engaged in significant research efforts against bacteria and fungi with relatively little effort devoted to viruses. Accordingly, biomaterials scientists and engineers will have to participate in multidisciplinary antiviral research over the coming years. Although tissue engineering and regenerative medicine have historically dominated the field of biomaterials, current research holds promise for providing transformative solutions to viral outbreaks. To facilitate collaboration, it is imperative to establish a mutual language and adequate understanding between clinicians, industry partners, and research scientists. In this article, clinical perspectives are shared to clearly define emerging healthcare needs that can be met by biomaterials solutions. Strategies and opportunities for novel biomaterials intervention spanning diagnostics, treatment strategies, vaccines, and virus‐deactivating surface coatings are discussed. Ultimately this review serves as a call for the biomaterials community to become a leading contributor to the prevention and management of the current and future viral outbreaks.
Keywords: antivirals, biomaterials, COVID‐19, diagnostics, filtration
1. INTRODUCTION
Due to increased public hygiene, vaccination and antibacterial advancements, humanity is less likely than ever to succumb to infectious diseases of any kind. Despite this, in the past several months coronavirus disease 2019 (COVID‐19), caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has escalated to a global pandemic at an unprecedented rate. Identified as a novel beta‐coronavirus and closely related to other zoonotic SARS‐coronaviruses, it is believed that SARS‐CoV‐2 originally existed in a bat host and entered human populations via a wet‐market in Wuhan City, China. 1 To put this most recent outbreak into perspective, the waves of novel zoonotic viruses began largely with severe acute respiratory syndrome (SARS) in 2002, and include Middle East respiratory syndrome (MERS), various animal influenza strains, and now COVID‐19. Regardless of this growing trend, much of the biomaterial community has focused their research on antibacterial work; a cursory search on Pubmed, as of May 2020, shows 90 articles including the keywords “biomaterial and antiviral” in the last 5 years, but over 3,500 results for the same search with “biomaterial and antibacterial.” To facilitate rapid problem solving and scientific advancement during this viral pandemic, it is our goal to review the current clinical practices against COVID‐19 and how current and future biomaterials work can contribute.
1.1. Etiology, epidemiology, and morphology
The SARS‐CoV‐2 virus was first identified in December 2019 in Wuhan Province, China as a cluster of pneumonias. 2 Since then, the virus has spread globally at an astonishing rate and is expected to cause millions of infections and hundreds of thousands of deaths in the next 2 years. 3 A brief comparison of SARS‐CoV‐2 to the related viruses SARS coronavirus (SARS‐CoV) and MERS coronavirus (MERS‐CoV) provides some interesting insights.
The first SARS outbreak began in November 2002 in Guangdong Province, China and by July fifth, 2003 the World Health Organization (WHO) declared the SARS outbreak contained. Approximately 8,000 SARS cases and 774 deaths (9.2% mortality) were reported in total across 29 countries. The majority of these cases were people in close contact with animals and healthcare workers who were associated with aerosol generating procedures while the overall human‐to‐human transmission was limited. 4 The MERS outbreak began in September 2012 in Saudi Arabia and by June 2019, ~2,500 cases and 845 deaths (34.5% mortality) were reported in total across 27 countries. A similar transmission pattern to SARS is documented, with overall limited human‐to‐human transmission. 5
Although all three are zoonotically transmitted novel coronaviruses, COVID‐19 differentiates itself significantly in how widely and quickly it has spread, and how low the mortality rate is compared to the other two viruses. 4 , 5 To illustrate this point, consider that SARS and MERS were found to have a basic reproduction number (R0), a measure of how contagious an infectious disease is, between 2.5 and 3.5, whereas the most recent estimates for COVID‐19 are around 4–9 making it at least twice as contagious. 6 Not only is this number important to project rate of spread, but R0 is directly related to the concept of herd immunity – that in a given population, a threshold of immunized individuals confers protection from pathogen transmission to the unimmunized individuals. The basic relationship between R0 and herd immunity is the “herd immunity threshold” and is calculated as 1‐1/R0. 7 Given this equation, diseases with an R0 of 3 would require 67% of the population be immunized to achieve the herd immunity threshold, whereas an R0 of 5.5, such as COVID‐19, would require 82% to be immunized. 8
Taxonomically, SARS‐CoV‐2 is an enveloped positive sense RNA virus and part of the beta‐coronavirus family (Figure 1). The viral genome is around 30 kb and is divided into several open reading frames which encode both structural and nonstructural proteins. 9 Following successful entry into cells, viral RNA is released into the cell cytoplasm, where positive‐sense RNA is translated by host cells into proteins. The structural proteins include spike (S) glycoprotein, envelope (E) glycoprotein, membrane (M) glycoprotein, and nucleocapsid (N) protein. 10 Briefly, the S protein interfaces with angiotensin‐converting enzyme 2 (ACE2) to fuse with host cells and is of great interest for ongoing research. The E protein assists with assembly and budding of new virions in the endoplasmic reticulum. The M protein is the main structural protein. The N protein binds to viral RNA and is involved in processing and stabilizing the RNA. 10 , 11 Nonstructural proteins (NSPs) include enzymes that assemble to form replication‐transcription complexes involved in viral replication. 12 , 13 Replication is believed to take place within a network of modified endoplasmic reticulum membranes. Following assembly, virions are released via exocytosis into the surrounding extracellular space, where they go on to infect other host cells or are expelled or excreted from the body.
FIGURE 1.
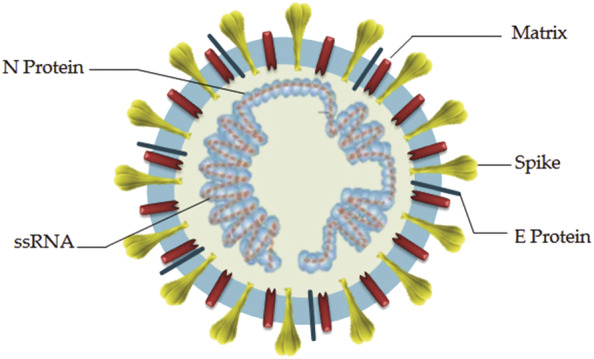
Structure of SARS‐CoV‐2 virus particle. Nucleocapsid (N), envelope (E), and spike (S) proteins along with matrix form a shell surrounding single‐stranded (ss) RNA. Reproduced with permission from Astuti et al. 10
1.2. Mechanism of transmission and infection
While originally a zoonotic virus, the primary method of transmission has transitioned to human‐to‐human via close contact, fomites, and droplet. 14 Of great concern is the “asymptomatic carrier” – a patient who sheds the virus for days to weeks before symptoms occur, if at all. 15 Additionally, there is growing evidence that SARS‐CoV‐2 is highly contagious partly due to its ability to travel in aerosols, which are widely defined as small respirable droplets <5 to 10 μm in diameter. Unlike large (>20 μm) droplets, these small aerosols do not immediately settle due to gravity but are instead capable of long‐range airborne transport. 16 , 17 Once the particles are inhaled, pathogenesis begins in the lungs due to direct invasion of host epithelial cells. This creates a localized inflammatory response driven by several cytokine‐mediated pathways. It has been shown that S protein interaction with ACE2 activates a signaling pathway which significantly upregulates chemokine (C‐C motif) ligand 2 production (CCL2), a cytokine which attracts T cells, monocytes and basophils. 18 Another important pathway involves intracellular antigenic peptide presentation via major histocompatibility complex (MHC) to cytotoxic T lymphocytes. These immune triggers are also responsible for a systemic inflammatory response which can range from a normal immune response to a cytokine storm—an uncontrolled production and circulation of pro‐inflammatory cytokines which can damage other organ systems and lead to superinfection, acute respiratory disease syndrome (ARDS), cardiac injury, stroke, and multi‐organ dysfunction. 18 , 19 , 20
The largest clinical report on COVID‐19 (72,314 cases) was published by the Chinese Center for Disease Control (CDC) and was summarized in the Journal of the American Medical Association in February 2020. It categorized patients by clinical manifestation into mild (81%), severe (14%), and critical infection (5%) groups. 21 Symptoms ranged from headache, gastroenteritis, and cough for mild infection, to ventilator‐dependent respiratory failure, sepsis and multi‐organ dysfunction (MOD) for those critically infected. 2 , 19 No fatalities were reported for mild and severe cases, but in critical infections the case fatality rate approached 50%. These numbers are similar to smaller studies in the United States. 22 , 23 Treatment of these patients revolves largely around supportive respiratory care and targeted intervention of secondary injuries. Successively more severe stages of acute respiratory failure require successively more invasive treatments. In practice respiratory status is monitored by using a pulse oximeter to assess blood oxygen saturation (SpO2), a measure of the percentage of hemoglobin which is oxygenated, with a target saturation of >90%. The most basic oxygen treatment includes the use of low flow nasal cannula. The next step up includes high flow nasal oxygen (HFNO) and noninvasive ventilation (NIV) treatments. These are newer therapies that show promise in decreasing intubation rates and intensive care unit (ICU) mortality and length of stay. HFNO utilizes a specialized nasal cannula to deliver high volumes of oxygen and due to such high flow also improves the physiology of breathing. NIV uses an oro‐nasal mask to deliver mechanical ventilation and oxygenation without the need for intubation. 24 , 25 However, if a patient continues to be hypoxic on these therapies then intubation with invasive mechanical ventilation becomes necessary. The details of ventilator management is outside the scope of this paper, but if a patient continues to be hypoxic despite maximum ventilatory support, then extracorporeal membrane blood oxygenation (ECMO) is a final rescue therapy. Such critically ill patients almost uniformly suffer from ARDS which severely inhibits oxygen diffusion across their lung alveolar membranes. ECMO bypasses the lungs and affords the patient time to recover by using a device to drain, perform gas exchange on, and then recirculate oxygenated blood back to the patient. 26 Despite their necessity, all these oxygen treatments carry risk to healthcare workers with viral particle aerosolization from the intubation procedure carrying the highest risk.
2. OPPORTUNITIES FOR BIOMATERIALS RESEARCH
Following the outbreak of the COVID‐19 pandemic, SARS‐CoV‐2 rapidly became one of the most important subjects in biomedical research. In the following sections, we outline a series of focus areas for biomaterials researchers by first providing a brief description of current clinical standards and then opportunities for biomaterials intervention. In many cases, multidisciplinary approaches involving biomaterials are expected to play a major role in disease detection and management.
2.1. Diagnostics
2.1.1. Nucleic acid testing
Nucleic acid testing via reverse transcription polymerase chain reaction (RT‐PCR) has emerged as the most common means of screening patient samples for current SARS‐CoV‐2 infection globally. 19 , 27 Polymerase chain reaction (PCR)‐based assays are useful clinical tools as they offer exceptional detection limits and excellent pathogen specificity. Following the COVID‐19 outbreak, international collaboration facilitated the rapid development of these RT‐PCR‐based SARS‐CoV‐2 detection kits, a major accomplishment for global medicine. Most of these tests employ the same three steps: first, biological samples undergo chemical treatment to extract and purify RNA. This is typically achieved by lysing viral membranes to release nucleic acids, which are then chemically precipitated and collected via centrifugation. Next, reverse transcriptase enzymes are used to transcribe RNA to complimentary DNA (cDNA), which is subsequently completed by the same reverse transcriptase enzyme to form highly stable double‐stranded cDNA. Through iterative cycles of heating and cooling (thermocycling), target sequences of cDNA corresponding to custom‐designed primers can be specifically amplified. Thermocycling consists of sequential denaturation, annealing, and extension phases. During denaturation, cDNA is heated to induce separation of double‐stranded cDNA into single strands. During annealing, the reaction mixture is cooled to induce the formation of double‐stranded primer/cDNA structures. During extension, the reaction mixture is again heated to an intermediate temperature at which the polymerase enzyme binds to primer‐containing regions of DNA and replicates the target DNA strand. When fluorescent reporter probes are included in the reaction mixture, accumulation of target cDNA sequences can be monitored in real‐time. Notably, current probes are designed to increase fluorescence in direct proportion to target nucleic acid sequence accumulation. In this way, low detection limits are achieved by amplifying target cDNA prior to detection.
POC nucleic acid tests
Current SARS‐CoV‐2 RT‐PCR testing has several limitations at sample collection and processing stages, introducing bottlenecks in testing workflows and severely limited access to testing in regions that lack PCR instrumentation infrastructure. Given these limitations, point‐of‐care (POC) strategies are an attractive alternative and accordingly are a subject of current research. As the name implies, POC tests can rapidly provide diagnoses without requiring off‐site laboratory processing of samples. Ideally, POC solutions eliminate the need for specialized equipment, allowing tests to be conducted in the field or even self‐administered at home. This dramatically reduces the risk of disease spread during centralized testing and processing, increases testing throughput and capacity, and expands testing access for less developed or lower income areas. Biomaterials research can provide key solutions to the challenges and limitations faced by current diagnostic approaches. At the most rudimentary level, biomaterials laboratories can help manufacture key disposables, including test swabs used to collect specimens. For example, the company Formlabs has developed and validated 3D printed nasal swabs for collection of respiratory samples. 28 Beyond offsetting disposables shortages, biomaterials can also play a supporting role in the development of POC tests. Emerging POC nucleic acid tests combine state‐of‐the‐art molecular and synthetic biology discoveries with biomaterials advances to facilitate low technology disease detection. 27 To bypass instrumentation needs, multidisciplinary research is now underway to develop novel methods of purifying genetic material from complex biological samples, amplifying genetic material of interest, detecting target sequences, and conveying those results in simple and intuitive ways. 29 , 30 , 31 , 32
Isothermal RT‐PCR tests
In conjunction with emerging biology‐based advances, biomaterials scientists can contribute important technologies to facilitate next‐generation diagnostic test development. To aid in nucleic acid purification for example, Ali et al. developed a magnetic nanoparticle‐based scheme that allowed for rapid isolation of RNA and DNA from biological samples via application of a magnetic field. 33 In the same study, the authors also developed a magnetic nanoparticle‐based chemiluminescent reporter system that allowed for detection of target sequences with relatively simple instrumentation. Biomaterials also form the basis of colorimetric readout options that allow for interpretation of assay results without the use of any specialized optics, making them ideal tests for POC detection. Biomaterials‐based surface plasmon resonance biosensors such as gold nanoparticles or quantum dots have been demonstrated to change color according to size, shape, surface chemistry, and aggregation state thus making them excellent optical probes. 34 , 35 This can be exploited to produce simple colorimetric readouts from complex molecular reactions. 10 For example, Kellner et al. leveraged CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR‐associated) technology to develop a novel nucleic acid test called SHERLOCK (specific high‐sensitivity enzymatic reporter unlocking), with a colorimetric readout option. 36 This strategy capitalizes on nonspecific nuclease activity exhibited by some Cas proteins—upon exposure to target RNA, the Cas proteins become activated and cleave reporter molecules in the immediate vicinity (Figure 2a). By incorporating antigen‐RNA‐biotin reporter molecules, detection was made possible with commercially available lateral flow dipsticks containing antibody‐conjugated gold nanoparticles. For the end user, making a diagnosis from a complex assay is incredibly simple: positive and negative tests can be determined by the location of a dark purple line on a paper strip. Efforts to apply this technology (and other similar ones) to develop COVID‐19 tests are underway. 2 , 22 One SHERLOCK test has received US Food and Drug Administration Emergency Use Authorization (FDA EUA) for COVID‐19 detection in clinical samples.
FIGURE 2.
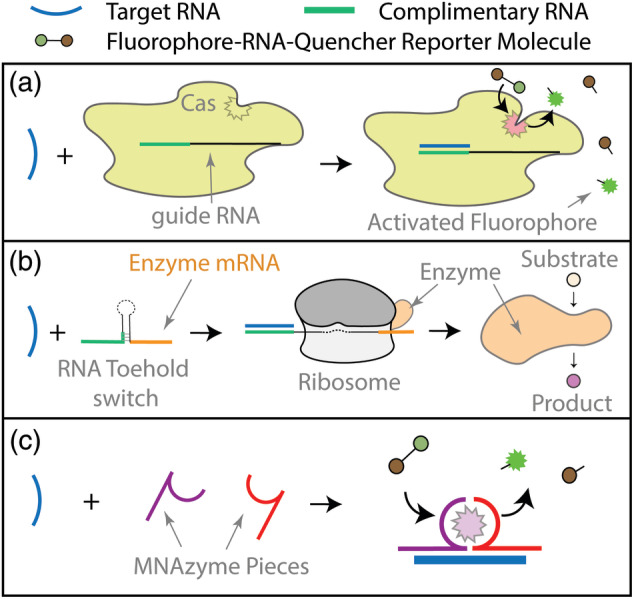
Diagnostic strategies for SARS‐CoV‐2 nucleic acid detection. (a) CRISPR/Cas‐based detection; in the presence of target RNA sequences (blue), Cas proteins become activated and cleave fluorophore‐RNA‐quencher reporter molecules, resulting in an increase in fluorescence. (b) RNA toehold sensors; in the presence of target RNA, toehold sensors unfold, allowing ribosomes to bind and synthesize enzymes encoded by a messenger RNA (mRNA) sequence located downstream from the loop region. Newly synthesized enzymes then convert substrate into colored product. (c) MNAzymes; multi‐part nucleic acid (MNA)‐based enzymes assemble in the presence of target RNA and subsequently cleave reporter molecules to generate signal
An alternative colorimetric method involves the use of RNA toehold switches coupled with paper‐based, cell‐free protein expression platforms. Although traditionally RNA has not been considered a biomaterial, a recent definition expands the field to include any material “designed to take a form that can direct, through interaction with biological systems, the course of any therapeutic or diagnostic procedure.” 37 This includes materials built from biomolecules, such as RNA, using synthetic biology principles, as reflected by the increasing number of synthetic biology‐focused sessions that are now held at major biomaterials conferences. RNA toehold switches are rationally designed, synthetic RNA molecules comprised of toehold and coding regions separated by a hairpin structure that blocks translation by sequestering the ribosome‐binding site and start codon. Upon exposure to a specific trigger RNA, the target sequence binds to the toehold region to induce a conformational shift, unfolding the hairpin and allowing for ribosome binding and subsequent gene expression (Figure 2b). Pardee et al. designed toehold switches that, in combination with ribosomes, generated LacZ enzyme upon exposure to Zika‐derived trigger RNA. 38 When substrate was added to the mix, colored product was generated by the enzyme, resulting in easy detection by the naked eye. To improve detection limits, an isothermal amplification step was added, allowing viral RNA to be converted into cDNA, amplified, and then converted back into RNA to trigger toehold switches. With amplification, target sequences could be detected in clinically relevant low femtomolar concentrations. In addition, inclusion of CRISPR/Cas9 editing technology allowed differentiation between viral RNA sequences differing in just a single nucleotide. Most importantly, all test reagents (including buffers, enzymes, ribosomes, and substrates) could be lyophilized onto a single paper disk, allowing for one‐step testing of clinical samples. In addition to requiring minimal instrumentation this platform was shown to be highly economical, expanding access to testing. Work is currently underway to extend this technology to develop tests for COVID‐19. 39 Both the CRISPR and RNA‐toehold based detection strategies make use of enzymatic reporter systems that not only transduce but also amplify signal. Conventional nucleic acid probes allow for signal growth in direct proportion to target sequence accumulation, but enzymatic reporters allow for exponential relationships between target sequence concentration and signal intensity. When combined with excellent specificity, this can theoretically allow for detection of much lower concentrations of target nucleic acids. From a COVID‐19 perspective, lower detection limits could allow for detection of exceptionally low viral loads associated with asymptomatic but infectious patients.
Nucleic acid biosensors
Emerging biosensors with ultra‐low limits of detection may one day allow for complete elimination of reverse transcription and/or cDNA amplification steps, providing faster and simpler one‐step detection. Biomaterials can play key roles in the development of such sensors. An enormous range of nucleic acid biosensors and transducers have been reported, including surface‐functionalized nanostructures and programmable DNA‐responsive smart materials. 40 , 41 Qiu et al. for example, built a gold nanoisland‐based plasmonic biosensors to aid in the detection of SARS‐CoV‐2 RNA. 42 Nanoislands were functionalized with complimentary DNA, allowing for selective capture of target RNA. Changes in localized surface plasmon resonance could be detected in response to RNA binding. In addition, biomaterials strategies can be utilized to enhance existing sensor technology. For example, synthetic multi‐component nucleic acid enzymes, or MNAzymes, can be designed to self‐assemble in the presence of defined target sequences (Figure 2c). Upon assembly, MNAzymes become catalytically active and process substrates to yield fluorescent or otherwise detectable products. To enhance this signal amplification and reduce the need for cDNA amplification, Gao et al. developed a cationic copolymer that assisted in the assembly of MNAzymes, dramatically increasing the catalytic ability of the enzyme to yield a 200 times faster rate of substrate conversion to detectable product. 43 In the presence of copolymer, nanomolar concentrations of MNAzymes could detect picomolar amounts of target sequence at physiological temperature.
2.1.2. Protein testing
Protein tests are employed to detect viral antigens or antibodies raised against them. While the presence of viral antigens indicates active infection, antibodies can take several days to appear in the blood. 2 , 44 Thus, antibody tests are more routinely used to demonstrate evidence of previous infection. Although it is currently unclear if blood antibodies raised against SARS‐CoV‐2 confer immunity, antibody tests are still expected to prove useful for disease tracking and surveillance as well as for monitoring vaccine efficacy. In addition, as the relationship between antibodies and immunity is further revealed, individuals testing positive for antibodies may be able to resume normal daily activities on a shorter timeline or even donate plasma to aid in the treatment of sick patients. To date, enzyme‐linked immunosorbent assays (ELISAs) and other immunoassays have been used to screen for the presence of antibodies raised against SARS‐CoV‐2N or S protein antigens. Significantly more FDA EUAs have been granted to immunoglobulin (Ig) G tests over IgM, most likely because IgM responses generally precede IgG responses and fade quickly, while serum levels of IgG antibodies are sustained and believed to be more closely related to conferred immunity. 45
Lateral flow immunoassays and ELISA
Biomaterials make the basis for emerging immunoassays for antibody detection. In many cases, colloidal gold‐antigen conjugates can be used as optical probes. 46 For example, Cellex™ offers a gold nanoparticle‐based lateral flow assay for detection of IgG and IgM antibodies. When sample is added to the test strip, antibodies in the sample bind to antigen immobilized onto the particle surface. In the event of a positive test, two strips of anti‐human IgG or IgM secondary antibody located further along the strip capture gold complexes, resulting in the development of a dark red line. Magnetic nanoparticles have also found use in antibody detection applications—the biotechnology company Abbott has been granted an EUA for an IgG test that utilizes viral antigen‐coated magnetic nanoparticles. Nanoparticles can be immobilized after capturing antibodies, washed extensively, and tagged with anti‐human IgG conjugates capable of producing chemiluminescent signals.
In the US, several ELISA‐based tests have also been granted EUA status. ELISAs typically exhibit lower limits of detection than colloidal gold lateral flow assays because they include a signal amplification step—when antibodies of interest are present, captured enzymes generate detectable product. In addition, ELISAs typically use 96‐well plate formats or similar, allowing many samples to be tested at once. Despite these advantages, processing of samples can be labor‐intensive and involves many wash steps that can introduce error. In addition, samples must be processed at centralized facilities, resulting in increased turnaround times. Microfluidics allow for the design of POC ELISA tests that require very little user input. 47 , 48 , 49 Microfluidic lab‐on‐a‐chip devices consist of micron‐scale channels and chambers and allow multi‐step diagnostic assays to be conducted rapidly and efficiently. 50 For broad surveillance of population antibody titers, these microfluidic ELISA tests are appealing—they can be conducted in the field, reduce user error, dramatically reduce required sample volume, and minimize reagent use. Tan et al. are currently developing a microfluidic ELISA for quantitative detection of IgG antibodies raised against SARS‐CoV‐2 S proteins. 51 The test requires only 10 μl of sample and returns results in 15–20 min.
Viral antigen testing
Although nucleic acid testing is the predominant means of diagnosing current COVID‐19 patients, biomaterials strategies can also be designed to detect viral antigens presented at the surface of intact virus. Such diagnostics would prove immensely beneficial, as they require little to no sample preprocessing and can directly sense the presence of virus in complex biological fluids. Because no amplification is involved, extremely low detection limits are required to confer clinical relevance. In addition, as detection limits decrease, excellent pathogen selectivity is paramount to maintaining good specificity. The development of such biosensors has been an active area of biomaterials research for years—promising technologies include piezoelectric, electrochemical, or optical biosensors that can detect incredibly minute changes in mass, electrical activity, or optical characteristics. 52 , 53 , 54 , 55 , 56 To replace current standards, emerging biomaterials sensors must demonstrate an ability approach or surpass the sensitivity and specificity of conventional nucleic acid tests. Applications of biosensors to SARS‐CoV‐2 detection should screen for S protein antigens, as these are presented on the outside of viral particles and readily accessible. For example, Seo et al. developed a field‐effect transistor biosensor that could detect SARS‐CoV‐2 in unprocessed clinical samples. 57 To build the device, graphene sheets of a field‐effect transistor were modified with antibody raised against the SARS‐CoV‐2 S protein. When samples were added, viral particles were sequestered by the antibody, resulting in measurable changes in the electrical behavior of the transistor. Notably, the biosensor platform had a far lower limit of detection in clinical samples than ELISA. Antigen tests can also probe for other viral antigens, but disruption of viral particle structure is required. For example, Quidel® received EUA for a fluorescent immunoassay‐based antigen test that detects viral N proteins. The approach requires virus to first be denatured and uses a cassette‐based lateral flow sandwich immunoassay that generates fluorescent outputs that must be read with an instrument also sold by the firm.
2.2. Therapeutics and vaccines
In addition to diagnostics development, biomaterials technologies can be employed to develop novel treatment strategies for effective management of COVID‐19 infection. In the following section, opportunities for biomaterials‐based strategies are discussed.
2.2.1. Antiviral therapies
Traditional antiviral therapies can be classified into non‐specific antiviral agents (e.g., ribavirin, remdesivir, chloroquine) and targeted inhibitors of a specific viral element (e.g., HIV protease inhibitors, RNAi). 58 , 59 , 60 , 61 In addition, immunomodulatory agents (e.g., interferons and corticosteroids) have been used to decrease the inflammatory state associated with viral infection. These therapies are compelling but also have major shortcomings. Nonspecific antivirals are generally more prophylactic, not effective against all viruses, and can have significant side effect profiles. 59 , 61 , 62 At the same time, targeted inhibitors are definitionally limited by their specificity to one target and offer no guarantee of efficacy in future viral outbreaks. Finally, immunomodulation to control cytokine storm carries risk of suppressing beneficial parts of the immune response to the point of harm. In the case of SARS‐CoV‐2, it is fortunate that there is a large degree of similarity to previous viral outbreaks, otherwise a considerable amount of time would be required to find new targets and develop therapies against them. Instead, the acute and pressing need for treatment options has resulted in attempts to repurpose existing drugs. 63 , 64
Several drugs have come to the forefront of the medical and research community, including hydroxychloroquine, remdesivir, and dexamethasone. Hydroxychloroquine is a drug with multiple mechanisms of action, illustrated by its use as an antimalarial and anti‐inflammatory in several rheumatological diseases. Three main proposed mechanisms in COVID‐19 include blocking viral entry into host cells, decreased viral replication, and anti‐inflammatory properties. 65 Despite initial promise, hydroxychloroquine serves as a reminder to the medical community that even during a pandemic robust clinical data are required to make informed recommendations for treatments with known, severe side effects. Another consideration is the sudden shortage of hydroxychloroquine and the effect it has had on rheumatological patients who rely on it for chronic treatment. 66 , 67 Remdesivir is a broad spectrum antiviral prodrug which is converted to a nucleoside analog in cells and has been shown to be effective against Ebola, SARS‐CoV and MERS in vitro. 68 It is currently being studied in randomized studies with preliminary data supporting use in hospitalized COVID‐19 patients requiring oxygen therapy, though once again further data are pending. 69 Dexamethasone is a strong synthetic corticosteroid with anti‐inflammatory and immunosuppressive mechanisms of action. 70 Based on the current data from the RECOVERY trial, dexamethasone is the most promising repurposed drug with significant data showing decreased mortality in patients requiring either invasive or noninvasive oxygen therapy. 71 Despite these heroic efforts, relatively little progress has been made toward the development of COVID‐19 therapeutics that directly target SARS‐CoV‐2. Instead, hospitalized patients are provided with symptomatic relief, repurposed compassionate‐use drugs and life‐supporting interventions while they fight off the viral infection, as previously described. Biomaterials offers several opportunities to address the limitations of existing therapeutic strategies (Figure 3).
FIGURE 3.
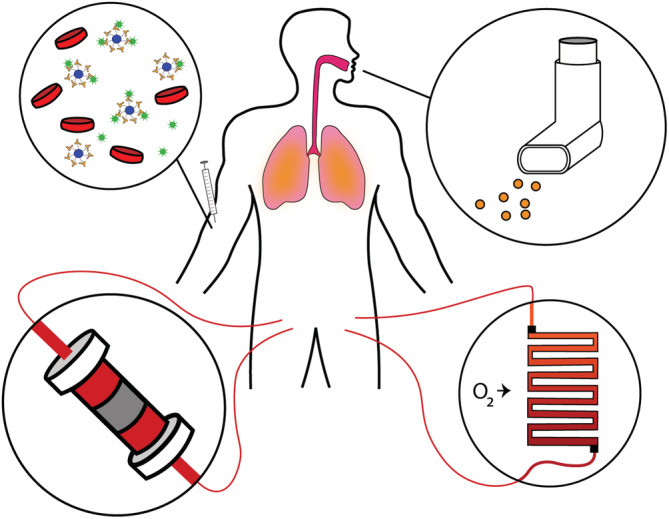
Biomaterials‐based therapeutic strategies for treatment of COVID‐19. Nanodecoys designed to trap and sequester virus can be directly injected into the blood (top left), while nanoparticles loaded with drugs can be formulated as inhalants to provide local delivery to lung tissue (top right). Extracorporeal blood treatments can replenish O2 (bottom right), modulate immune signaling via proinflammatory cytokine removal or anti‐inflammatory cytokine supplementation, or directly remove viral particles from the bloodstream (bottom left)
Drug delivery
Drug candidates may directly inhibit viral processes including infection or replication, modulate pro‐inflammatory cytokine signaling, or stimulate regenerative healing processes. Unfortunately, many of these drug candidates are not ideally suited for systemic administration using standard oral or intravenous routes. Antibodies, anti‐inflammatory cytokines, and other protein‐based drugs often exhibit short half‐lives, while sufficiently hydrophobic drugs suffer from low accumulation in target tissue. In addition, many existing antiviral medications demonstrate significant off‐target effects that hinder clinical use. 72 For example, some drug candidates that were found to robustly inhibit SARS‐CoV‐2 infection and replication in vitro did not yield clinical success in vivo due to adverse off‐target effects. 63 , 73 , 74 , 75 When therapeutic concentrations of a potential drug cannot be safely achieved in target tissue, use becomes infeasible, as lower doses confer no therapeutic benefit while higher doses cause safety concerns.
Drug delivery systems can be designed to administer less drug overall while selectively targeting affected tissue, dramatically reducing systemic exposure and expanding the candidate pool of potential drugs. In addition, delivery of drugs with poor stability or low bioavailability to targets of interest can be more readily achieved. In our lab, this concept has already been applied to challenging hydrophobic cancer therapeutics by using novel self‐assembling tyrosine‐derived polymeric nanospheres (Tyrospheres) to minimize toxic side effects and maintain bioactivity. 76 , 77 , 78 , 79 Tyrospheres and other polymeric particle systems built from commonly used polyesters have been shown to locally deliver immunosuppressants and antibacterial therapeutics. 80 , 81 , 82 Other methods of creating polymeric drug delivery devices include electrospun fibermats and drug eluting stents. 83 , 84 , 85 These techniques can be used to overcome the challenges of implementing current therapeutics for COVID‐19 that have failed due to unacceptably severe side effects. Because SARS‐CoV‐2 has been shown to target the lung epithelial tissue, formulation of drug delivery systems such as polymeric particles as inhalants may hold promise and have previously been demonstrated for treatment of other diseases like lung cancers. 86 , 87 Concerns have been raised regarding the potential for exhaled particles to aerosolize viruses and facilitate disease spread, but careful design of particle size may be able to ameliorate this risk. 88 Notably, inhalants could be administered at home, or even as a prophylactic measure. For intubated patients, drug‐eluting coatings of endotracheal tubes placed into the airway could similarly target affected tissue while sparing the rest of the body; such coatings have been previously reported. 89
Nanodecoys
In an alternate nanomaterials‐centered strategy, nanostructures can be designed to mimic living cells. These particles, called nanodecoys, are built from, or incorporate cell membrane‐derived materials to trap and sequester viruses. Rao et al. captured Zika virus with nanodecoys comprised of gelatin nanoparticles cloaked in mosquito‐derived cell membrane, attenuating infection and preventing passage of the virus into the fetal mouse brain. 75 Current knowledge of routes of entry used by SARS‐CoV‐2 could be exploited to create nanoscale cell‐mimicking decoys for viral trapping. In particular, SARS‐CoV‐2 fusion with host cell membranes is now known to proceed through interactions between viral S proteins, ACE2 and proteases such as the transmembrane protein TMPRSS2. 9 , 51 , 90 Administration of soluble ACE2 has been proposed for blocking S proteins from interacting with membrane‐bound ACE2, and efforts to identify short peptide fragments of the full length protein with equivalent or enhanced S protein‐binding capabilities have been reported. 51 , 91 Moreover, decorating cell‐mimetic nanoparticles with the ACE2 protein or related peptide fragments could provide an even more biomimetic presentation of the protein. At the same time, nanodecoys could present cocktails of surface proteins in defined spatial arrangements to more closely mimic host cells. Such strategies could actively engage S protein fusion machinery rather than simply block S protein receptor‐binding domains, resulting in deactivation of the virus.
Extracorporeal blood treatment
Another therapeutic strategy involves the use of extracorporeal blood treatments to mitigate the most damaging aspects of COVID‐19. ECMO devices are used clinically for the most critically ill patients, but these machines are typically in even shorter supply than ventilators, are costly to operate, and are typically only found at specialized centers. 92 , 93 Alternatively, emerging nanoparticle and microparticle oxygen carriers could lead to the development of more accessible extracorporeal blood oxygenation strategies. 94 For severely ill patients however, oxygenation alone may not be sufficient to reduce mortality rates. A recent retrospective analysis of Chinese patient data revealed that almost half of the COVID‐19 patients treated with ECMO still died from septic shock or multi‐organ failure. 63 To improve survival, mitigation of inflammatory signaling is crucial. Pro‐inflammatory cytokine removal can be achieved by flowing blood through polymeric sorbent cartridges, while administration of mesenchymal stem cell (MSC)‐derived factors can exert potent anti‐inflammatory immunomodulatory effects. 95 , 96 For example, CytoSorbents™ developed an extracorporeal blood purification device called CytoSorb®. The device, which looks like a large cylindrical cartridge, utilizes proprietary polymer beads to sequester pro‐inflammatory mediators from blood, is designed for use with standard dialysis equipment, and recently received EUA for the treatment of COVID‐19 patients. Alternatively, Sentien Biotech has developed a combination product called SBI‐101 that houses allogeneic MSCs in a hollow‐fiber hemofilter device. By flowing blood through the device, anti‐inflammatory MSC‐derived molecules can perfuse patient blood without necessitating cell transplantation, eliminating risks of immune rejection while improving cell survival and overcoming pharmacokinetic barriers associated with transplantation. This strategy has shown promise for treatment of kidney and liver failure in animal model, and a Phase I/II clinical trial evaluating use in human acute kidney injury patients is underway (NCT03015623). 97 , 98 , 99 In late June 2020, another Phase I/II trial was also announced to investigate the use of SBI‐101 for treatment of COVID‐19 patients suffering from acute kidney injury and already receiving renal replacement therapy (NCT04445220). Finally, a more direct approach to hemofiltration has been taken by ExThera Medical with the Seraph® 100 filter, a heparin‐functionalized ultra‐high molecular weight polyethylene bead‐based filter. This technology has been shown to sequester a variety of pathogens from methicillin‐resistant Staphylococcus aureus to cytomegalovirus, without filtering out anti‐infectious drugs, and has been granted an EUA for treatment of COVID‐19 patients. 100 , 101 , 102 , 103
2.2.2. Vaccines
While therapeutics are designed to treat actively infected patients, vaccines effectively reduce population susceptibility to infection. The development of a SARS‐CoV‐2 vaccine holds the most potential for attenuating COVID‐19 spread, motivating a flurry of research in vaccine development as recently reviewed by Liu et al. 2 , 104 Previous work on SARS vaccine development focused on live attenuated whole virus vaccines and S protein subunit vaccines; an inactivated virus MERS vaccine was also developed. During in vivo studies however, these vaccines were derailed by evidence of an immune hypersensitive‐type lung pathology on exposure of vaccinated subjects to a live virus challenge. 33 , 105 Another problem with the S protein subunit vaccines was antibody‐dependent enhancement—post‐immunization antibodies which actually enhance viral cell entry upon live virus challenge. A commentary by Hotez et al. attempts to address both these issues and suggests that for COVID‐19 a subunit vaccine of just the receptor binding domain of the S protein may confer immunity without inducing immunopathologies. 105
With that in mind, the ideal vaccine elicits a robust immune response characterized by sustained antibody production and conferred immunity across a large and diverse patient population. Biomaterials strategies to augment conventional vaccine design and development have been extensively reviewed. 47 , 106 The key advantage to biomaterials‐based approaches is enhanced control over the vaccination process. Conventional vaccine strategies are essentially comprised of one to several bolus injections of antigenic material, while engineered systems allow for fine manipulation of antigen delivery rate and presentation as well as host immune cell response. As our understanding of acquired immunity continues to evolve, biomaterials can provide the tools for precise immune modulation.
Multiple biomaterials‐based solutions such as nanoparticles, liposomes, scaffolds, and microneedles have been proposed. Nanoscale structures can be designed to protect antigen cargo, present antigens in biomimetic formats, and allow for specific immune cell targeting. 47 This can be achieved by leveraging modifications in nanoparticle size, shape, surface chemistry (particle charge, hydrophobicity), and material composition. 107 Microneedles are applied topically and slowly dissolve allowing for pain‐free sustained antigen delivery into the skin, a highly immune‐reactive organ. 108 , 109 Dr. Mark Prausnitz and colleagues have conducted extensive work to develop microneedle‐based polio, influenza, and hepatitis B vaccines. 110 , 111 , 112 Kim et al. developed a polymeric microneedle array‐based MERS vaccine using a MERS‐CoV S protein subunit trimer as an antigen, and demonstrated robust and sustained antigen‐specific antibody responses in mice. 113 Ongoing efforts to develop the MERS vaccine were then applied to rapidly design and produce a similar microneedle vaccine based on SARS‐CoV‐2 S protein subunit trimers that similarly showed promise in mice. Plans are underway to coordinate a first‐in‐man Phase I clinical trial. 45 In some cases, polymeric materials may also act as adjuvants. Adjuvants are broadly defined as a substance that enhances the antigenicity of an antigen. Babensee et al. have shown that certain biomaterials have the ability to selectively upregulate the differentiation and maturation of dendritic cells, which play an important role in antigen presentation, and therefore act as adjuvants. 114 Another example of this is when mice mounted robust antibody responses to bovine serum albumin when antigen was delivered from subcutaneously implanted biodegradable tyrosine‐derived polymer devices. 115 This was due to adjuvant activity of one of the polymer degradation byproducts. In addition, anti‐bovine serum albumin (BSA) antibody titers were significantly higher when BSA was delivered from tyrosine‐derived polymer devices as compared to poly(bisphenol A‐iminocarbonate) controls.
2.3. Ex vivo antiviral strategies
2.3.1. Surface inactivation
As previously discussed, the main method of virus spread is through aerosolized particles. Large droplets (>20 μm) land on surfaces close to the point of emission while small droplets (<5–10 μm) begin to evaporate and become small enough to be transported by air several meters from the point of emission. 116 A major problem with viruses is their ability to remain active on surfaces. In this way, contaminated surfaces such as doorknobs, toilet handles, tables, and utensils used in restaurants, and touch screens can facilitate viral transfer from sick to healthy individuals. While surfaces can be sanitized with a variety of household cleaners, sterilizing after each individual use is difficult to maintain. This is especially problematic for areas with a high concentration of infected patients or a particularly susceptible population such as hospitals or senior living facilities. Surface stability varies by virus, but according to a recent study, SARS‐CoV‐2 can remain active on plastic, stainless steel, and cardboard for ~3.8, ~5.8, and ~ 6.5 hr, respectively at a temperature of 21–23°C with 40% humidity. 117 Strategies aimed at mitigating viral spread generally look to deactivate viruses upon landing on surfaces. 118 , 119 Existing antiviral surfaces see limited use in clinical practice even though ongoing research supporting different biomaterials coatings has been around for decades. 120 , 121 Biomaterials have been used for virus inactivation and have great potential to help combat the spread of SARS‐CoV‐2 and future viral outbreaks. 122 , 123 , 124 Indeed, reformulating antiviral materials as virucidal surface coatings can attenuate contamination of surfaces and minimize disease propagation. 125 , 126 In addition, antiviral biomaterials coatings can be applied to conventional personal protective equipment (PPE) to allow for multiple uses. 127
Metals
Biomaterials coatings with known antiviral properties can be made from metals, surfactants, and natural products. Metals, especially silver, gold, and copper have been used for decades as antiviral agents. 128 , 129 , 130 It is important to acknowledge the NIH and FDA's guidance on silver particles and that colloidal silver can cause serious side effects and are not meant for internal ingestion. 131 , 132 Silver nanoparticles have shown to inactivate HIV‐1, herpes simplex virus type 1, and influenza among other viruses. 133 , 134 , 135 , 136 The nanoparticle size plays a key role in HIV‐1 deactivation, where only particles within the range of 1–10 nm are able to bind to the sulfur‐bearing residues on the viral envelope. It is proposed that silver's antiviral activity is attributed to the silver nanoparticles physically inhibiting the binding between the virus and the host cell. 137 , 138 Gold nanoparticles behave in a similar manner by binding to virus particles thereby blocking the cell receptor and preventing the virus from starting the viral cycle. These nanoparticles suppress viral infection by selectively cleaving disulfide bonds, which blocks virus membrane fusion to the cell and prevents viral entry into the host cell (Figure 4a). 139 Gold nanoparticles have also shown to inhibit measles and have low cytotoxicity at viral inhibitory concentrations and may prove useful for viral inactivation for other enveloped viruses. 140 Copper oxide nanoparticles are used to inactivate both enveloped and nonenveloped RNA and DNA viruses. 141 Compared to silver nanoparticles which act as a physical barrier, copper oxide nanoparticles actively damage virus complexes. Copper oxide nanoparticles easily convert between Cu(I) and Cu(II) in vivo, and Cu(II) ions oxidatively damage biomolecules, including viruses. 141 Metal nanoparticles are toxic in vivo causing severe side effects, however by altering the concentration, particle size, or metal coating composition toxicity can be significantly reduced. 142 Mohammadyari, et al. reported that in doses <400 ppm, copper oxide nanoparticles 50 nm in size had no adverse side effects on rat kidney or liver; however, above 400 ppm the particles were toxic. 143 Silver and copper nanoparticles have been incorporated into clothing, wound dressings, and implant coatings due to their antiviral activity. 134 Although incorporation of metal nanoparticles into textiles has been extensively studied, their side effects and toxicity have been less intensively investigated. Due to their small size, nanoparticles can enter the human body through various routes and cause harmful side effects. 144 Further research into metal nanoparticle antiviral activity, toxicity, and cost‐effectiveness can further enhance the use of nanoparticles for both in vivo and ex vivo applications, which can be especially useful for incorporation into PPE. Copper's antiviral abilities have shown to be effective for the deactivation of influenza A virus through an increase in copper ion diffusion through the virus' lipid membrane. 145 Incorporation of Cu(II) held in place by zeolites into cotton textiles significantly increased inactivation of avian influenza H5 subtype virus. 146 Binding cationic copper ions into textiles, latex, and other polymer products during manufacture demonstrated virus inactivation properties for HIV‐1. 129 In addition to metals, treating textiles with a boron‐triclosan mixture showed efficacy at significantly reducing viral activity of human adenovirus and poliovirus type 1 strains. 147 Virus inactivating gloves are of particular interest in today's pandemic which can serve as a way to increase the safety of hospital workers. Incorporation of an anti‐infective agent, chlorhexidine, into the interior of examination gloves to deliver the anti‐viral agent within 10 minutes of exposure to a liquid has been patented in the 1990s. 148 However, a large amount of anti‐infective agent is required which does not provide a cost‐effective material.
FIGURE 4.
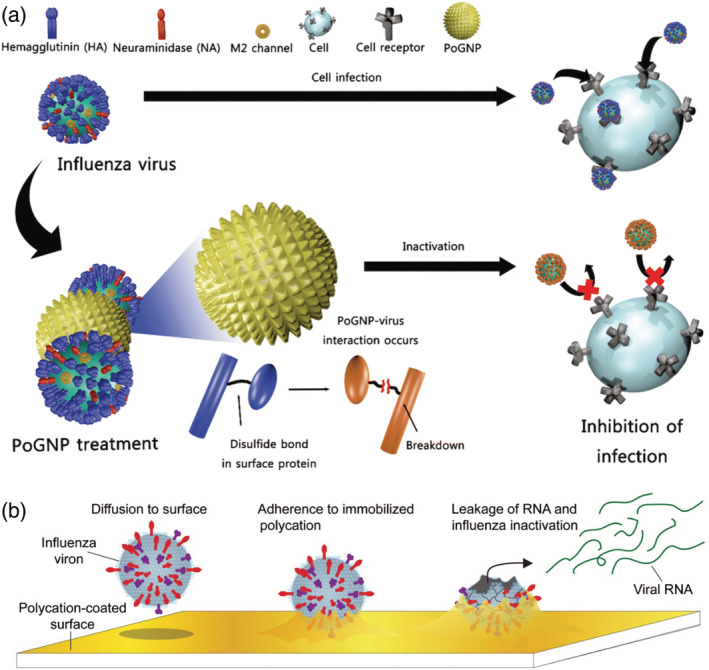
Mechanisms of materials to prevent virus spread and inactivation. (a) Use of porous gold nanoparticles (PoGNP) to prevent influenza virus attachment to cell surface; M2‐ matrix ion channel 2. Reproduced with permission from Kim et al 139 (b) Proposed mechanism of Influenza A virus inactivation by polymer N,N‐dodecyl methyl‐polyethylenimine (DM‐PEI) paint coated on surfaces. Reproduced with permission from Hsu et al. 160
Surfactants
Surfactants are chemical species that create self‐assembled micelles in solution and contain both a hydrophilic and a hydrophobic group. 149 They are often the common active ingredient in household disinfecting agents and have shown high antiviral activity. 150 The use of surfactants as sanitizing agents for virus inactivation has been recently reviewed by Lin et al. 151 Surfactants can be broken down into cationic—often found in fabric softeners, hair conditioners, antiseptic hand wash, and mouthwash; anionic—often found in detergent and personal‐care products; nonionic—often found in foaming and emulsifying agents; and zwitterionic species—often found in laundry and cosmetic products. All are capable of inactivating viruses by solvating and disrupting the lipid‐based envelope of the virus or by targeting the capsid proteins. 152 , 153 , 154 , 155 , 156 , 157 , 158 , 159 Although beneficial as sanitizing agents, surfactants are less commonly used to prepare virus deactivating surfaces. A significant research advancement lies in utilizing these antiviral agents in biomaterials applications. This approach has the potential to minimize the lifespan of viral particles on surfaces thereby minimizing transmission. One method uses surfactants as polymer paints, which can be applied to surfaces to halt the spread of viral infection. Cationic surfactant N,N‐dodecyl methyl‐polyethylenimine (DM‐PEI), a quaternary ammonium compound, has shown to be an antibacterial and enveloped antiviral polymer due to its ability to rupture the cell membranes by interaction with the polycationic chains. 160 The DM‐PEI coating acts as an adhesive for the virus particle, which immobilizes the virus, and causes damage from the hydrophobic polycation in DM‐PEI (Figure 4b). The virus' genetic material is released leaving residual inactivated virus particle adhered to the substrate surface. Biosurfactants are amphiphilic compounds, mostly produced at the microbial cell surface and are able to accumulate between fluid phases. 161 Similar to chemical surfactants, biosurfactants are used in industrial, agricultural, cosmetic, and pharmaceutical applications. 162 However, biosurfactants can be easily degradable, have a low toxicity, have high availability from raw materials, and often are not affected by environmental factors (pH, temperature, ionic strength). 163 Although mostly used for antimicrobial applications, biosurfactants have been used to inactivate retrovirus, herpes virus, and HIV, among others. 164 Biosurfactants are more commonly used against enveloped viruses due the physicochemical interactions between the virus lipid membrane and the membrane‐active surfactant causing viral disintegration. 165 Often biosurfactants are used in preventing biofilm formation and as anti‐adhesives in order to prevent bacterial and viral growth. 166 While underdeveloped, the use of surfactants as coatings to achieve antiviral activity has great potential to improve the safety of individuals in high trafficked areas such as airports or in places of high transmission rates such as hospitals.
Naturally derived products
Many naturally derived products have been used to prevent virus transmission; cellulose has been used in meat products and as the wax coating on fruits and vegetables. 167 Antiviral coatings have been approved for use on food items and packaging to prevent the spread of human noroviruses and hepatitis A virus, among others. 168 These coatings are often made from polysaccharides, cellulose, starch, pectin, alginate, lipids, or other natural polymers and are generally termed safe for ingestion. 169 Active compounds derived or extracted from grape seed, green tea, Aloe vera, Eriobotryae folium, and cinnamaldehyde are a few naturally derived compounds used in active packaging against marine norovirus. 170 , 171 , 172 Catechins present in green tea extract contain antioxidant, anticarcinogenic, anti‐inflammatory and antimicrobial properties and have recently been shown to exhibit antiviral activity against the food‐borne pathogens hepatitis A and murine norovirus. 173 , 174 Cyclodextrins are another group of naturally occurring macrocyclic molecules that have been used for their virucidal activity. 175 These naturally occurring products are most commonly used in food packaging applications and have not been translated to applications outside of the food industry.
2.3.2. Viral filtration
To prevent the spread of human‐to‐human transmission, face masks have become mandated in many countries when traveling, shopping, or when simply leaving the house. Face masks have received significant review since the beginning of the pandemic; which fabrics are the best for particle filtration, can masks be reused, what are their utility lifespans, and what happens with the extra waste produced when they are discarded. One commonality most often agreed upon is that they are necessary to help filter aerosolized droplets from the air to minimize viral transmission. Existing filtration strategies see more regular clinical use, especially in the context of negative pressure isolation rooms and face masks. Currently they strive to capture viral particles from the air, but do not deactivate the particles. As a result, air filtration devices such as respirators and masks must be changed frequently when exposure to virus is suspected, a practice that contributes to ongoing PPE shortages and contributes to the generation of nondegradable waste. Biomaterials strategies can be employed to develop novel filtration devices capable of capturing and deactivating SARS‐CoV‐2 and other viruses.
Face masks
The most commonly used viral filtration devices are face masks. Masks are a necessary component for the health and safety of individuals who are exposed to aerosolized virus particles; however, not all masks are designed for virus filtration. 176 The most common masks used are (a) surgical masks which are capable of filtering 98% of 3 μm particles making them effective in blocking large‐particle droplets, but not able to filter aerosolized particles transmitted by coughs or sneezes; and (b) performance level face masks such as N95 masks which are capable of filtering 95% of 0.1–0.3 μm particles. 177 , 178 , 179 To reduce the spread and transmission of viruses, it is important to design masks that have a minimum porosity threshold of an individual virus particle—20–300 nm. 180 , 181 There is an abundance of current research focusing on chemically modifying masks to provide viral deactivation properties, which would allow for multiple uses, reducing PPE shortages, and nondegradable waste generation. Most masks are made from non‐biodegradable, non‐renewable petroleum‐based polymers that are already contributing to environmental pollution worldwide. 182 , 183 To reduce this and future pandemics' environmental impact, it is important to design masks that are made from biodegradable materials or can be multi‐use. Ideally, these masks would also have antiviral activity and be more effective at filtering virus particles. One example of a virus‐functionalized mask is by Tiliket et al. who chemically modified the mask layers by incorporating DM‐PEI functionalized Kimwipes® Lite (non‐woven cellulosic fiber filter) layers into a low‐cost commercial medical mask which improved the affinity for trapping airborne influenza virus A (H5N2). 180 Other approaches to modify masks for viral filtration efficacy include the incorporation of natural extracts from green tea (QR‐435, catechin, theaflavin) into the masks layers, which successfully block the passage of virus or inactivate viruses by inhibiting viral replication; impregnating copper oxide into N95 respiratory masks to mitigate viral titer recovery of influenza virus; and incorporating sialic acid, which mimics human cell receptor sites, to create an affordable, easy‐to‐produce filter capable of removing viruses such as influenza. 184 , 185 , 186 , 187 , 188 Due to more strict requirements to wear a face covering, Konda et al. studied the efficacy of different commonly available fabrics for preventing the inhalation of aerosol particles. 189 They found that tighter weaves with low porosity (i.e., high thread count cotton sheets) are ideal for mechanical filtration whereas natural silk, chiffon, and flannel provide high electrostatic filtering capabilities. A hybrid mask incorporating both cotton with silk or chiffon had increased filtration efficiency. Designing and implementing a mask that is able to filter or deactivate virus particles, is cost effective, is environmentally degradable, and has the potential to change the rate of virus spread worldwide.
Air filtration
A similar avenue of research is based on the use of ventilators which typically are fitted with an inhalant and an expiratory filter. However, the chemical make‐up of these filters is of extreme importance. When placed on mechanical ventilation, infected patients can contribute to the spread of the virus through the expiratory valve aerosolizing droplets into ambient air. Several filters, PALL Ultipor® 25, Ultipor® 100, and BB50T among others, are available for use on the expiratory limb of ventilators that have shown to filter >99.999% of H1N1 virus. 190 , 191 Most filters fitted on respirators are high efficiency particulate air (HEPA) filters which are adequate for bacterial, dust, pollen, mold filtration, and other particles larger than 0.2 μm in size; however they are not sufficient for virus particle filtration. 32 , 191 It is important to design a filtration device that can be attached to ventilators to mitigate the spread of aerosolized virus particles. Simultaneously, it is important to filter building air, especially in a hospital setting where aerosolized virus particles can be at a high concentration. Buildings are often fitted with HVAC systems that have a filter for airborne pathogens. 192 Robust filters also exist for virus filtration that involve carbon‐fiber ionizers designed to generate air ions and charge aerosolized virus particles which allows for the filter to capture the virus particles. 193 Home filter devices have been designed to inactivate common airborne viruses through the use of palladium‐titanium dioxide catalysts with vacuum ultraviolet irradiation. 194 It is important to implement these virus filtration systems in high traffic areas or areas with a high virus concentration to create a constant airflow and deactivate virus particles. It has been shown that when virus particles become electrostatically charged, they are attracted to an oppositely charged surface. 195 , 196 Hagbom et al. developed a small‐scale device using charged surface technology to both inactivate and filter Influenza A and H1N1 virus particles by placing the device between an infected and healthy individual. 197 It was proposed that reduced virus infectivity is caused by damaging the viral lipid envelope through peroxidation reactions between the reactive oxygen species and the lipid and protein. When the ionizing device was placed between two guinea pig cages, one infected with Influenza A and one healthy, none of the exposed guinea pigs tested positive for influenza‐specific immune response serum compared to 75% in exposed guinea pigs where an inactive ionizing device was present. Such a device, if scaled up appropriately, has the potential to be either used in hospital settings where hospital personnel are closely interacting with infected patients or to create a highly sensitive device to capture virus particles to help in the detection and identification of novel viruses. Similar virus filtration and inactivation systems (filtration, microfiltration, hydrophobic or ionic surface modifications) have been studied for their ability to trap viruses during water filtration. 198 Of particular interest is the use of previously mentioned DM‐PEI as a polyelectrolyte coating on commercially available microfiltration membranes which not only remove but also inactivate hepatitis E surrogates. 199
3. CONCLUSIONS
Much of our current knowledge stems from the previous viral outbreaks, SARS and MERS, but COVID‐19 has transcended those in overall mortality, prevalence, and social and economic impact. This review has described several specific opportunities for the biomaterials community to contribute significantly in the fight against the COVID‐19 pandemic. Importantly, this is not an exhaustive list but serves to illustrate a variety of recent advances and future directions for biomaterial research. Given how universal this outbreak has been, global readiness and scientific collaboration are imperative.
ACKNOWLEDGMENTS
RBS and DC were funded by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number T32EB005583.
Chakhalian D, Shultz RB, Miles CE, Kohn J. Opportunities for biomaterials to address the challenges of COVID‐19. J Biomed Mater Res. 2020;108:1974–1990. 10.1002/jbm.a.37059
Funding information National Institute of Biomedical Imaging and Bioengineering, Grant/Award Number: T32EB005583
The copyright line for this article was changed on 3 November 2020 after original online publication.
REFERENCES
- 1. Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID‐19) outbreak. J Autoimmun. 2020;109:102433. 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, et al. And others. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis. 2020;20(5):533‐534. 10.1016/s1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. WHO. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS): World Health Organization; 2003. Retrieved from: https://www.who.int/csr/sars/WHOconsensus.pdf?ua=1 [Google Scholar]
- 5. WHO . WHO MERS‐CoV Global Summary and Assessment of Risk, July 2019 (WHO/MERS/RA/19.1). Geneva, Switzerland: World Health Organization; 2019. Retrieved from: https://apps.who.int/iris/bitstream/handle/10665/326126/WHO-MERS-RA-19 [Google Scholar]
- 6. Sanche S, Lin YT, Xu C, Romero‐Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(7):1470‐1477. 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fine P, Eames K, Heymann DL. "Herd immunity": a rough guide. Clin Infect Dis. 2011;52(7):911‐916. 10.1093/cid/cir007. [DOI] [PubMed] [Google Scholar]
- 8. Metcalf CJE, Ferrari M, Graham AL, Grenfell BT. Understanding herd immunity. Trends Immunol. 2015;36(12):753‐755. 10.1016/j.it.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 9. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260‐1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Astuti I, Ysrafil Y. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2): an overview of viral structure and host response. Diabetes Metab Syndr. 2020;14(4):407‐412. 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nieto‐Torres JL, Dediego ML, Alvarez E, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69‐82. 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses ‐ drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327‐347. 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knoops K, Kikkert M, Worm SH, et al. SARS‐coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang H, Fan C, Li M, et al. And others. COVID‐19: a call for physical scientists and engineers. ACS Nano. 2020;14(4):3747‐3754. 10.1021/acsnano.0c02618. [DOI] [PubMed] [Google Scholar]
- 15. Huff HV, Singh A. Asymptomatic transmission during the COVID‐19 pandemic and implications for public health strategies. Clin Infect Dis. 2020. 10.1093/cid/ciaa654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson EL, Turnham P, Griffin JR, Clarke CC. Consideration of the aerosol transmission for COVID‐19 and public health. Risk Anal. 2020;40(5):902‐907. 10.1111/risa.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tellier R, Li Y, Cowling BJ, Tang JW. Recognition of aerosol transmission of infectious agents: a commentary. BMC Infect Dis. 2019;19(1):101. 10.1186/s12879-019-3707-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kang S, Peng W, Zhu Y, et al. Others. Recent progress in understanding 2019 novel coronavirus (SARS‐CoV‐2) associated with human respiratory disease: detection, mechanisms and treatment. Int J Antimicrob Agents. 2020;55(5):105950. 10.1016/j.ijantimicag.2020.105950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lupia T, Scabini S, Mornese Pinna S, Di Perri G, De Rosa FG, Corcione S. 2019 novel coronavirus (2019‐nCoV) outbreak: a new challenge. J Glob Antimicrob Resist. 2020;21:22‐27. 10.1016/j.jgar.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Markus HS, Brainin M. COVID‐19 and stroke‐a global world stroke organization perspective. Int J Stroke. 2020;15(4):361‐364. 10.1177/1747493020923472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239‐1242. 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 22. Aggarwal S, Garcia‐Telles N, Aggarwal G, Lavie C, Lippi G, Henry BM. Clinical features, laboratory characteristics, and outcomes of patients hospitalized with coronavirus disease 2019 (COVID‐19): early report from the United States. Diagnosi. 2020;7(2):91‐96. 10.1515/dx-2020-0046. [DOI] [PubMed] [Google Scholar]
- 23. Bhatraju PK, Ghassemieh BJ, Nichols M, et al. Evans L and others. Covid‐19 in critically ill patients in the Seattle region ‐ case series. N Engl J Med. 2020;382(21):2012‐2022. 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price S, Singh S, Ledot S, et al. Respiratory management in severe acute respiratory syndrome coronavirus 2 infection. Eur Heart J Acute Cardiovasc Care. 2020;9(3):229‐238. 10.1177/2048872620924613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frat JP, Coudroy R, Thille AW. Non‐invasive ventilation or high‐flow oxygen therapy: when to choose one over the other? Respirology. 2019;24(8):724‐731. 10.1111/resp.13435. [DOI] [PubMed] [Google Scholar]
- 26. Ramanathan K, Antognini D, Combes A, et al. Planning and provision of ECMO services for severe ARDS during the COVID‐19 pandemic and other outbreaks of emerging infectious diseases. Lancet Respir Med. 2020;8(5):518‐526. 10.1016/s2213-2600(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yeh HC, Ho YP, Wang TH. Quantum dot‐mediated biosensing assays for specific nucleic acid detection. Nanomedicine. 2005;1(2):115‐121. 10.1016/j.nano.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 28. Formlabs . 2020. 3D printed test swabs for COVID‐19 testing. Retrieved from https://formlabs.com/covid-19-response/covid-test-swabs/
- 29. Myhrvold C, Freije CA, Gootenberg JS, et al. Field‐deployable viral diagnostics using CRISPR‐Cas13. Science. 2018;360(6387):444‐448. 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleosides Nucleotides Nucleic Acids. 2008;27(3):224‐243. 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- 31. Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115(22):12491‐12545. 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 32. USEPA . 2019. Indoor air quality (IAQ). Retrieved from https://www.epa.gov/indoor-air-quality-iaq/what-hepa-filter-1
- 33. Agrawal AS, Tao X, Algaissi A, et al. Immunization with inactivated middle east respiratory syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum Vaccin Immunother. 2016;12(9):2351‐2356. 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hong Y, Huh Y‐M, Yoon DS, Yang J. Nanobiosensors based on localized surface plasmon resonance for biomarker detection. J Nanomater. 2012;2012:1‐13. 10.1155/2012/759830. [DOI] [Google Scholar]
- 35. Guo P, Wei C. Quantum dots for robust and simple assays using single particles in nanodevices. Nanomedicine. 2005;1(2):122‐124. 10.1016/j.nano.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 36. Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14(10):2986‐3012. 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biomaterials II. Biomedical materials. In: Williams D, Zhang X, eds. Definitions of Biomaterials for the Twenty‐First Century, Cambridge, MA: Elsevier; 2019:15‐23. [Google Scholar]
- 38. Pardee K, Green AA, Takahashi MK, et al. Fan M and others. Rapid, low‐cost detection of zika virus using programmable biomolecular components. Cell. 2016;165(5):1255‐1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 39. Fraumeni P. 2020. U of T researcher's ‘lab‐in‐a‐box’ promises to boost COVID‐19 testing capacity. Retrieved from https://www.utoronto.ca/news/u‐t‐researcher‐s‐lab‐box‐promises‐boost‐covid‐19‐testing‐capacity
- 40. English MA, Soenksen LR, Gayet RV, et al. Programmable CRISPR‐responsive smart materials. Science. 2019;365(6455):780‐785. 10.1126/science.aaw5122. [DOI] [PubMed] [Google Scholar]
- 41. Younis S, Taj A, Zia R, et al. Nanosensors for the detection of viruses. In: Han B, Tomer VK, Nguyen TA, Farmani A, Singh PK, eds. Nanosensors for Smart Cities, Cambridge, MA: Elsevier; 2020:327‐338. [Google Scholar]
- 42. Qiu G, Gai Z, Tao Y, Schmitt J, Kullak‐Ublick GA, Wang J. Dual‐functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268‐5277. 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 43. Gao J, Shimada N, Maruyama A. MNAzyme‐catalyzed nucleic acid detection enhanced by a cationic copolymer. Biomater Sci. 2015;3(5):716‐720. 10.1039/C4BM00449C. [DOI] [PubMed] [Google Scholar]
- 44. Haveri A, Smura T, Kuivanen S, et al. Serological and molecular findings during SARS‐CoV‐2 infection: the first case study in Finland, January to February 2020. Euro Surveill. 2020;25(11), 1–6. 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. USFDA . 2020. Emergency use authorization (EUA) information, and list of all current EUAs. Emergency Use Authorization. Retrieved from https://www.fda.gov/emergency‐preparedness‐and‐response/mcm‐legal‐regulatory‐and‐policy‐framework/emergency‐use‐authorization#covidinvitrodev
- 46. Xiang J, Yan M, Li H, et al. Evaluation of enzyme‐linked immunoassay and colloidal gold‐ Immunochromatographic assay kit for detection of novel coronavirus (SARS‐Cov‐2) causing an outbreak of pneumonia (COVID‐19). medRxiv. 2020;2020.02.27.20028787. 10.1101/2020.02.27.20028787. [DOI] [Google Scholar]
- 47. Bookstaver ML, Tsai SJ, Bromberg JS, Jewell CM. Improving vaccine and immunotherapy design using biomaterials. Trends Immunol. 2018;39(2):135‐150. 10.1016/j.it.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giri B, Liu Y, Nchocho FN, Corcorana RC, Dutta D. Microfluidic ELISA employing an enzyme substrate and product species with similar detection properties. Analyst. 2018;143:989‐998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thaitrong N, Charlermroj R, Himananto O, Seepiban C, Karoonuthaisiri N. Implementation of microfluidic sandwich ELISA for superior detection of plant pathogens. PLoS ONE. 2013;8(12):e83231. 10.1371/journal.pone.0083231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun J, Jiang X. Chapter 15 ‐ microfluidic devices for viral detection. In: Li X, Zhou Y, eds. Microfluidic Devices for Biomedical Applications, Philadelphia, PA: Woodhead Publishing; 2013:527‐556. [Google Scholar]
- 51. Ackerman CM, Myhrvold C, Thakku SG, et al. Massively multiplexed nucleic acid detection using Cas13. Nature. 2020;582(7811):277‐282. 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saylan Y, Erdem Ö, Ünal S, Denizli A. An alternative medical diagnosis method: biosensors for virus detection. Biosensors (Basel). 2019;9(2):65‐86. 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Santiago I. Trends and innovations in biosensors for COVID‐19 mass testing. ChemBioChem. 2020. 10.1002/cbic.202000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chowdhury AD, Takemura K, Li TC, Suzuki T, Park EY. Electrical pulse‐induced electrochemical biosensor for hepatitis E virus detection. Nat Commun. 2019;10(1):3737. 10.1038/s41467-019-11644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chamorro‐Garcia A, Merkoci A. Nanobiosensors in diagnostics. Nanobiomedicine (Rij). 2016;3:1849543516663574. 10.1177/1849543516663574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cesewski E, Johnson BN. Electrochemical biosensors for pathogen detection. Biosens Bioelectron. 2020;159(1):112214. 10.1016/j.bios.2020.112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Seo G, Lee G, Kim MJ, et al. And others. Rapid detection of COVID‐19 causative virus (SARS‐CoV‐2) in human nasopharyngeal swab specimens using field‐effect transistor‐based biosensor. ACS Nano. 2020;14(4):5135‐5142. 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 58. Clercq EE. Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev Anti‐Infect Ther. 2006;4(2):291‐302. 10.1586/14787210.4.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kumar V, Jung YS, Liang PH. Anti‐SARS coronavirus agents: a patent review (2008 ‐ present). Expert Opin Ther Pat. 2013;23(10):1337‐1348. 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- 60. Pant S, Singh M, Ravichandiran V, Murty USN, Srivastava HK. Peptide‐like and small‐molecule inhibitors against Covid‐19. J Biomol Struct Dyn. 2020;1‐10. 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rider TH, Zook CE, Boettcher TL, Wick ST, Pancoast JS, Zusman BD. Broad‐spectrum antiviral therapeutics. PLoS ONE. 2011;6(7):e22572. 10.1371/journal.pone.0022572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dyall J, Gross R, Kindrachuk J, et al. Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs. 2017;77(18):1935‐1966. 10.1007/s40265-017-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hou T, Zeng W, Yang M, et al. Development and evaluation of a CRISPR‐based diagnostic for 2019‐novel coronavirus. medRxiv. 2020;2020.02.22.20025460. 10.1101/2020.02.22.20025460. [DOI] [Google Scholar]
- 64. Cherian SS, Agrawal M, Basu A, Abraham P, Gangakhedkar RR, Bhargava B. Perspectives for repurposing drugs for the coronavirus disease 2019. Indian J Med Res. 2020;151(2):160‐171. 10.4103/ijmr.IJMR_585_2010.4103/ijmr.IJMR_585_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shukla AM, Archibald LK, Wagle Shukla A, Mehta HJ, Cherabuddi K. Chloroquine and hydroxychloroquine in the context of COVID‐19. LID ‐ 10.7573/dic.2020-4-5 LID ‐ 2020‐4‐5. (1745‐1981 [Print]). [DOI] [PMC free article] [PubMed]
- 66. Yazdany J, Kim AHJ. Use of hydroxychloroquine and chloroquine during the COVID‐19 pandemic: what every clinician should know. (1539–3704 [Electronic]). [DOI] [PMC free article] [PubMed]
- 67. Ferner RE, Aronson JK. 2020. Chloroquine and Hydroxychloroquine in Covid‐19, London, UK: Electronic:1756‐1833. 10.1136/bmj.m1432. [DOI] [PubMed] [Google Scholar]
- 68. Grein J, Ohmagari N, Shin D, et al. 2020. Compassionate Use of Remdesivir for Patients with Severe Covid‐19. New England Journal of Medicine, 382, (24), 2327–2336. 10.1056/nejmoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Beigel JA‐O, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AA‐O, Hohmann E, Chu HY, Luetkemeyer A, Kline S and et al. Remdesivir for the Treatment of Covid‐19 ‐ Preliminary Report. LID ‐ 10.1056/NEJMoa2007764 LID ‐ NEJMoa2007764. (1533–4406 [Electronic]). [DOI]
- 70. Selvaraj V, Dapaah‐Afriyie K, Finn A, Flanigan TP. Short‐term dexamethasone in Sars‐CoV‐2 patients. (2327–2228 [Electronic]). [PubMed]
- 71. Mahase E. Covid‐19: demand for dexamethasone surges as RECOVERY trial publishes preprint. BMJ. 2020;369:m2512. 10.1136/bmj.m2512. [DOI] [PubMed] [Google Scholar]
- 72. Giudicessi JR, Noseworthy PA, Friedman PA, Ackerman MJ. Urgent guidance for navigating and circumventing the QTc‐prolonging and torsadogenic potential of possible pharmacotherapies for coronavirus disease 19 (COVID‐19). Mayo Clin Proc. 2020;95(6):1213‐1221. 10.1016/j.mayocp.2020.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lentini G, Cavalluzzi MM, Habtemariam S. COVID‐19, chloroquine repurposing, and cardiac safety concern: chirality might help. Molecules. 2020;25(8), 1–4. 10.3390/molecules25081834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Borba MGS, Val FFA, Sampaio VS, et al. 2020. Effect of High vs Low Doses of Chloroquine Diphosphate as Adjunctive Therapy for Patients Hospitalized With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) Infection. JAMA Network Open, 3, (4), e208857. 10.1001/jamanetworkopen.2020.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Borba MGS, Val FdA , Sampaio VS, et al. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection: a randomized clinical trial. JAMA Netw Open. 2020;3(4):e208857. 10.1001/jamanetworkopen.2020.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sheihet L, Garbuzenko OB, Bushman J, Gounder MK, Minko T, Kohn J. Paclitaxel in tyrosine‐derived nanospheres as a potential anti‐cancer agent: in vivo evaluation of toxicity and efficacy in comparison with paclitaxel in Cremophor. Eur J Pharm Sci. 2012;45(3):320‐329. 10.1016/j.ejps.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sheihet L, Dubin RA, Devore D, Kohn J. Hydrophobic drug delivery by self‐assembling triblock copolymer‐derived nanospheres. Biomacromolecules. 2005;6:2726‐2731. [DOI] [PubMed] [Google Scholar]
- 78. Sheihet L, Piotrowska K, Dubin RA, Kohn J, Devore D. Effect of tyrosine derived triblock copolymer compositions on nanosphere self‐assembly and drug delivery. Biomacromolecules. 2007;8:998‐1003. [DOI] [PubMed] [Google Scholar]
- 79. Bushman J, Vaughan A, Sheihet L, Zhang Z, Costache M, Kohn J. Functionalized nanospheres for targeted delivery of paclitaxel. J Control Release. 2013;171(3):315‐321. 10.1016/j.jconrel.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 80. Goyal R, Macri L, Kohn J. Formulation strategy for the delivery of cyclosporine a: comparison of two polymeric nanospheres. Sci Rep. 2015;5:13065. 10.1038/srep13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Qi F, Wu J, Li H, Ma G. Recent research and development of PLGA/PLA microspheres/nanoparticles: a review in scientific and industrial aspects. Front Chem Sci Eng. 2018;13(1):14‐27. 10.1007/s11705-018-1729-4. [DOI] [Google Scholar]
- 82. Sheihet L, Chandra P, Batheja P, Devore D, Kohn J, Michniak B. Tyrosine‐derived nanospheres for enhanced topical skin penetration. Int J Pharm. 2008;350(1–2):312‐319. 10.1016/j.ijpharm.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 83. Kohn J, Zeltinger J. Degradable, drug‐eluting stents: a new frontier for the treatment of coronary artery disease. Expert Rev Med Devices. 2014;2(6):667‐671. 10.1586/17434440.2.6.667. [DOI] [PubMed] [Google Scholar]
- 84. Macri LK, Sheihet L, Singer AJ, Kohn J, Clark RA. Ultrafast and fast bioerodible electrospun fiber mats for topical delivery of a hydrophilic peptide. J Control Release. 2012;161(3):813‐820. 10.1016/j.jconrel.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 85. Thakur RA, Florek CA, Kohn J, Michniak BB. Electrospun nanofibrous polymeric scaffold with targeted drug release profiles for potential application as wound dressing. Int J Pharm. 2008;364(1):87‐93. 10.1016/j.ijpharm.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 86. Muralidharan P, Malapit M, Mallory E, Hayes D, Mansour HM. Inhalable nanoparticulate powders for respiratory delivery. Nanomedicine. 2015;11(5):1189‐1199. 10.1016/j.nano.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 87. Lee W‐H, Loo C‐Y, Traini D, Young PM. Inhalation of nanoparticle‐based drug for lung cancer treatment: advantages and challenges. Asian J Pharma Sci. 2015;10(6):481‐489. 10.1016/j.ajps.2015.08.009. [DOI] [Google Scholar]
- 88. El‐Sherbiny IM, El‐Baz NM, Yacoub MH. Inhaled nano‐ and microparticles for drug delivery. Global Cardiol Sci Pract. 2015;2015:2‐2. 10.5339/gcsp.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ammar AA, Gruber M, Martin P, et al. Local delivery of mometasone furoate from an eluting endotracheal tube. J Control Release. 2018;272(28):54‐61. 10.1016/j.jconrel.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 90. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Batlle D, Wysocki J, Satchell K. Soluble angiotensin‐converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci. 2020;134(5):543‐545. 10.1042/cs20200163. [DOI] [PubMed] [Google Scholar]
- 92. Mishra V, Svennevig JL, Bugge JF, et al. Cost of extracorporeal membrane oxygenation: evidence from the Rikshospitalet university hospital, Oslo, Norway. Eur J Cardiothorac Surg. 2010;37(2):339‐342. 10.1016/j.ejcts.2009.06.059. [DOI] [PubMed] [Google Scholar]
- 93. Patel AR, Patel AR, Singh S, Singh S, Khawaja I. Applied uses of extracorporeal membrane oxygenation therapy. Cureus. 2019;11(7), 1–8. 10.7759/cureus.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tao Z, Ghoroghchian PP. Microparticle, nanoparticle, and stem cell‐based oxygen carriers as advanced blood substitutes. Trends Biotechnol. 2014;32(9):466‐473. 10.1016/j.tibtech.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 95. Taniguchi T. Cytokine adsorbing columns. Contrib Nephrol. 2010;166:134‐141. 10.1159/000314863. [DOI] [PubMed] [Google Scholar]
- 96. Han J, Zhao D, Li D, Wang X, Jin Z, Zhao K. Polymer‐based nanomaterials and applications for vaccines and drugs. Polymers (Basel). 2018;10(1):31‐44. 10.3390/polym1001003110.3390/polym10010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Miller BLK, Garg P, Bronstein B, et al. Extracorporeal stromal cell therapy for subjects with dialysis‐dependent acute kidney injury. Kidney Int Rep. 2018;3(5):1119‐1127. 10.1016/j.ekir.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Parekkadan B, van Poll D, Suganuma K, et al. Mesenchymal stem cell‐derived molecules reverse fulminant hepatic failure. PLoS ONE. 2007;2(9):e941. 10.1371/journal.pone.0000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yagi H, Parekkadan B, Suganuma K, et al. Long‐term superior performance of a stem cell/hepatocyte device for the treatment of acute liver failure. Tissue Eng Part A. 2009;15(11):3377‐3388. 10.1089/ten.TEA.2008.0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Seffer M‐T, Martens‐Lobenhoffer J, Schmidt JJ, Eden G, Bode‐Böger SM, Kielstein JT. Clearance of chloroquine and hydroxychloroquine by the seraph® 100 microbind® affinity blood filter ‐ approved for the treatment of COVID‐19 patients. Ther Apher Dial. 2020. 10.1111/1744-9987.13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Schmidt JJ, Eden G, Seffer M‐T, Winkler M, Kielstein JT. In vitro elimination of anti‐infective drugs by the seraph® 100 microbind® affinity blood filter. Clin Kidney J. 2020). 13, 421–424. 10.1093/ckj/sfaa063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McCrea K, Ward R, LaRosa SP. Removal of Carbapenem‐resistant Enterobacteriaceae (CRE) from blood by heparin‐functional hemoperfusion media. PLoS ONE. 2014;9(12):e114242‐e114242. 10.1371/journal.pone.0114242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. LaRosa S, McCrea K, Ward R. 984: removal of cytomegalovirus from blood by heparin‐functional hemoperfusion media. Crit Care Med. 2014;42(12):A1597. 10.1097/01.ccm.0000458481.24778.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Liu C, Zhou Q, Li Y, et al. Research and development on therapeutic agents and vaccines for COVID‐19 and related human coronavirus diseases. ACS Cent Sci. 2020;6(3):315‐331. 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chen WH, Strych U, Hotez PJ, Bottazzi ME. The SARS‐CoV‐2 vaccine pipeline: an overview. Curr Trop Med Rep. 2020;7:61‐64. 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yenkoidiok‐Douti L, Jewell CM. Integrating biomaterials and immunology to improve vaccines against infectious diseases. ACS Biomater Sci Eng. 2020;6(2):759‐778. 10.1021/acsbiomaterials.9b01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hotaling NA, Tang L, Irvine DJ, Babensee JE. Biomaterial strategies for immunomodulation. Annu Rev Biomed Eng. 2015;17:317‐349. 10.1146/annurev-bioeng-071813-104814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Skountzou I, Compans RW. Skin immunization with influenza vaccines. Curr Top Microbiol Immunol. 2015;386:343‐369. 10.1007/82_2014_407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kaushik S, Hord AH, Denson DD, et al. Lack of pain associated with microfabricated microneedles. Anesth Analg. 2001;92(2):502‐504. 10.1213/00000539-200102000-00041. [DOI] [PubMed] [Google Scholar]
- 110. Kolluru C, Gomaa Y, Prausnitz MR. Development of a thermostable microneedle patch for polio vaccination. (2190–3948 [Electronic]). [DOI] [PMC free article] [PubMed]
- 111. Kim MC, Kim KH, Lee JW, Lee YN, Choi HA‐O, Jung YJ, Kim YJ, Compans RW, Prausnitz MA‐O, Kang SM. Co‐Delivery of M2e Virus‐Like Particles with Influenza Split Vaccine to the Skin Using Microneedles Enhances the Efficacy of Cross Protection. LID ‐ 10.3390/pharmaceutics11040188 LID – 188. (1999–4923 [Print]). [DOI] [PMC free article] [PubMed]
- 112. Choi YH, Perez‐Cuevas MB, Kodani M, Zhang X, Prausnitz MR, Kamili S, O'Connor SM. Feasibility of Hepatitis B Vaccination by Microneedle Patch: Cellular and Humoral Immunity Studies in Rhesus Macaques. (1537–6613 [Electronic]). [DOI] [PMC free article] [PubMed]
- 113. Kim E, Erdos G, Huang S, et al. Microneedle array delivered recombinant coronavirus vaccines: immunogenicity and rapid translational development. EBioMedicine. 2020;55:102743. 10.1016/j.ebiom.2020.102743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Park J, Babensee JE. Differential functional effects of biomaterials on dendritic cell maturation. Acta Biomater. 2012;8(10):3606‐3617. 10.1016/j.actbio.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kohn J, Niemi SM, Albert EC, Murphy JC, Langer R, Fox JG. Single‐step immunization using a controlled release, biodegradable polymer with sustained adjuvant activity. J Immunol Methods. 1986;95(1):31‐38. 10.1016/0022-1759(86)90314-5. [DOI] [PubMed] [Google Scholar]
- 116. Morawska L, Cao J. Airborne transmission of SARS‐CoV‐2: the world should face the reality. Environ Int. 2020;139:105730. 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. van Doremalen N, Morris DH, Holbrook MG, Gamble A, Williamson BN. Aerosol and surface stability of SARS‐CoV‐2 as compared with SARS‐CoV‐1. N Engl J Med. 2020;382(16):1564‐1567. 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hasan J, Xu Y, Yarlagadda T, Schuetz M, Spann K, Yarlagadda PKDV. Antiviral and antibacterial nanostructured surfaces with excellent mechanical properties for hospital applications. ACS Biomater Sci Eng. 2020;6(6):3608‐3618. 10.1021/acsbiomaterials.0c00348. [DOI] [PubMed] [Google Scholar]
- 119. Haldar J, Weight AK, Klibanov AM. Preparation, application and testing of permanent antibacterial and antiviral coatings. Nat Protoc. 2007;2(10):2412‐2417. 10.1038/nprot.2007.353. [DOI] [PubMed] [Google Scholar]
- 120. Campoccia D, Montanaro L, Arciola CR. A review of the biomaterials technologies for infection‐resistant surfaces. Biomaterials. 2013;34(34):8533‐8554. 10.1016/j.biomaterials.2013.07.089. [DOI] [PubMed] [Google Scholar]
- 121. Li Y, Pi Q‐m, You H‐h, et al. A smart multi‐functional coating based on anti‐pathogen micelles tethered with copper nanoparticles via a biosynthesis method using l‐vitamin C. RSC Adv. 2018;8(33):18272‐18283. 10.1039/C8RA01985A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Anti‐Viral Surface Coating to Prevent the Spread of COVID‐19. Focus Powder Coat. 2020;2020(7):5‐5. 10.1016/j.fopow.2020.05.062. [DOI] [Google Scholar]
- 123. Malmsten M. Antimicrobial and antiviral hydrogels. Soft Matter. 2011;7(19):8725‐8736. 10.1039/C1SM05809F. [DOI] [Google Scholar]
- 124. Park D, Larson AM, Klibanov AM, Wang Y. Antiviral and antibacterial polyurethanes of various modalities. Appl Biochem Biotechnol. 2013;169(4):1134‐1146. 10.1007/s12010-012-9999-7. [DOI] [PubMed] [Google Scholar]
- 125. Tuladhar E, de Koning MC, Fundeanu I, Beumer R, Duizer E. Different virucidal activities of hyperbranched quaternary ammonium coatings on poliovirus and influenza virus. Appl Environ Microbiol. 2012;78(7):2456‐2458. 10.1128/AEM.07738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wong SY, Li Q, Veselinovic J, Kim B‐S, Klibanov AM, Hammond PT. Bactericidal and virucidal ultrathin films assembled layer by layer from polycationic N‐alkylated polyethylenimines and polyanions. Biomaterials. 2010;31(14):4079‐4087. 10.1016/j.biomaterials.2010.01.119. [DOI] [PubMed] [Google Scholar]
- 127. Dolez PI, Mlynarek J. 22 ‐ smart materials for personal protective equipment: tendencies and recent developments. In: Koncar V, ed. Smart Textiles and their Applications. Oxford: Woodhead Publishing; 2016:497‐517. [Google Scholar]
- 128. Grass G, Rensing C, Solioz M. Metallic copper as an antimicrobial surface. Appl Environ Microbiol. 2011;77(5):1541‐1547. 10.1128/aem.02766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Borkow G, Gabbay J. Putting copper into action: copper‐impregnated products with potent biocidal activities. FASEB J. 2004;18(14):1728‐1747. 10.1096/fj.04-2029fje. [DOI] [PubMed] [Google Scholar]
- 130. Minoshima M, Lu Y, Kimura T, et al. Comparison of the antiviral effect of solid‐state copper and silver compounds. J Hazard Mater. 2016;312(15):1‐7. 10.1016/j.jhazmat.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. NIH . 2017. Colloidal silver. Retrieved from https://www.nccih.nih.gov/health/colloidal-silver
- 132. USFDA . 2020. Coronavirus update: FDA and FTC warn seven companies selling fraudulent products that claim to treat or prevent COVID‐19. Retrieved from https://www.fda.gov/news-events/press-announcements/coronavirus-update-fda-and-ftc-warn-seven-companies-selling-fraudulent-products-claim-treat-or
- 133. Lara HH, Ayala‐Nuñez NV, Ixtepan‐Turrent L, Rodriguez‐Padilla C. Mode of antiviral action of silver nanoparticles against HIV‐1. J Nanobiotechnol. 2010;8(1):1. 10.1186/1477-3155-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Galdiero S, Falanga A, Vitiello M, Cantisani M, Marra V, Galdiero M. Silver nanoparticles as potential antiviral agents. Molecules. 2011;16(10):8894‐8918. 10.3390/molecules16108894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Vazquez‐Munoz R, Borrego B, Juarez‐Moreno K, et al. Toxicity of silver nanoparticles in biological systems: does the complexity of biological systems matter? Toxicol Lett. 2017;276:11‐20. 10.1016/j.toxlet.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 136. Broglie JJ, Alston B, Yang C, et al. Antiviral activity of gold/copper sulfide core/shell nanoparticles against human norovirus virus‐like particles. PLoS ONE. 2015;10(10):e0141050. 10.1371/journal.pone.0141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Mori Y, Ono T, Miyahira Y, Nguyen VQ, Matsui T, I M. Antiviral activity of silver nanoparticle:chitosan composites against H1N1 influenza a virus. Nanoscale Res Lett. 2013;8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Thurman RB, Gerba CP, Bitton G. The molecular mechanisms of copper and silver ion disinfection of bacteria and viruses. Crit Rev Environ Control. 1989;18(4):295‐315. 10.1080/10643388909388351. [DOI] [Google Scholar]
- 139. Kim J, Yeom M, Lee T, et al. Song D and others. Porous gold nanoparticles for attenuating infectivity of influenza a virus. J Nanobiotechnol. 2020;18(54):54. 10.1186/s12951-020-00611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Melendez‐Villanueva MA, Moran‐Santibanez K, Martinez‐Sanmiguel JJ, et al. Virucidal activity of gold nanoparticles synthesized by green chemistry using garlic extract. Viruses. 2019;11(12), 1–13. 10.3390/v11121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ishida T. Antiviral activities of Cu2+ ions in viral prevention, replication, RNA degradation, and for antiviral efficacies of lytic virus, ROS‐mediated virus, copper chelation. World Scien News. 2018;99:148‐168. [Google Scholar]
- 142. Bahadar H, Maqbool F, Niaz K, Abdollahi M. Toxicity of nanoparticles and an overview of current experimental models. Iran Biomed J. 2016;20(1):1‐11. 10.7508/ibj.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Mohammadyari A, Razavipour ST, Mohammadbeigi M, Negahdary M, Ajdary M. Exploring vivo toxicity assessment of copper oxide nanoparticle in Wistar rats. J Biol Today World. 2014;3(6):124‐128. 10.15412/j.Jbtw. [DOI] [Google Scholar]
- 144. Singh G, Beddow J, Mee C, Maryniak L, Joyce EM, Mason TJ. Cytotoxicity study of textile fabrics impregnated with CuO nanoparticles in mammalian cells. Int J Toxicol. 2017;36(6):478‐484. 10.1177/1091581817736712. [DOI] [PubMed] [Google Scholar]
- 145. Sundberg K, Champagne V, McNally B, Helfritch D, Sisson R. Effectiveness of nanomaterial copper cold spray surfaces on inactivation of influenza a virus. J Biotechnol Biomater. 2015;5(4):1000205. 10.4172/2155-952x.1000205. [DOI] [Google Scholar]
- 146. Imai K, Ogawa H, Bui VN, et al. Inactivation of high and low pathogenic avian influenza virus H5 subtypes by copper ions incorporated in zeolite‐textile materials. Antivir Res. 2012;93(2):225‐233. 10.1016/j.antiviral.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 147. Iyigundogdu ZU, Demir O, Asutay AB, Sahin F. Developing novel antimicrobial and antiviral textile products. Appl Biochem Biotechnol. 2017;181(3):1155‐1166. 10.1007/s12010-016-2275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Modak SM, Sampath L; The Trustees of Columbia University in the City of New York, assignee. Antiviral glove. US patent US005133090A. 1990.
- 149. Nakama Y. Chapter 15 ‐ surfactants. In: Sakamoto K, Lochhead RY, Maibach HI, Yamashita TYBT‐CS, eds. Cosmetic Science and technology. Amsterdam: Elsevier; 2017:231‐244. [Google Scholar]
- 150. Deleu M, Paquot M. From renewable vegetables resources to microorganisms: new trends in surfactants. C R Chim. 2004;7(6–7):641‐646. 10.1016/j.crci.2004.04.002. [DOI] [Google Scholar]
- 151. Lin Q, Lim JYC, Xue K, et al. Sanitizing agents for virus inactivation and disinfection. Viewpoints. 2020;1(2):e16. 10.1002/viw2.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lucassen J, Giles D. Dynamic surface properties of nonionic surfactant solutions. J Chem Soc Faraday Trans. 1974: Physical Chemistry in Condensed Phases, 71:217‐232. [Google Scholar]
- 153. Mertens BS, Velev OD. Characterization and control of surfactant‐mediated norovirus interactions. Soft Matter. 2015;11(44):8621‐8631. 10.1039/c5sm01778e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Merchel Piovesan Pereira B, Tagkopoulos I. Benzalkonium chlorides: uses, regulatory status, and microbial resistance. Appl Environ Microbiol. 2019;85(13):e00377‐e00319. 10.1128/AEM.00377-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Piret J, Roy S, Gagnon M, et al. Comparative study of mechanisms of herpes simplex virus inactivation by sodium lauryl sulfate and n‐lauroylsarcosine. Antimicrob Agents Chemother. 2002;46(9):2933‐2942. 10.1128/aac.46.9.2933-2942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Popkin DL, Zilka S, Dimaano M, et al. Cetylpyridinium chloride (CPC) exhibits potent, rapid activity against influenza viruses in vitro and in vivo. Pathog Immun. 2017;2(2):252‐269. 10.20411/pai.v2i2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Sirisattha S, Momose Y, Kitagawa E, Iwahashi H. Toxicity of anionic detergents determined by Saccharomyces cerevisiae microarray analysis. Water Res. 2004;38(1):61‐70. 10.1016/j.watres.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 158. Tsujimura K, Murase H, Bannai H, Nemoto M, Yamanaka T, Kondo T. Efficacy of five commercial disinfectants and one anionic surfactant against equine herpesvirus type 1. J Vet Med Sci. 2015;77(11):1545‐1548. 10.1292/jvms.15-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wydro P. The influence of the size of the hydrophilic group on the miscibility of zwitterionic and nonionic surfactants in mixed monolayers and micelles. J Colloid Interface Sci. 2007;316(1):107‐113. 10.1016/j.jcis.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 160. Hsu BB, Wong SY, Hammond PT, Chen J, Klibanov AM. Mechanism of inactivation of influenza viruses by immobilized hydrophobic polycations. Proc Natl Acad Sci U S A. 2011;108(1):61‐66. 10.1073/pnas.1017012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Rodrigues L, Banat IM, Teixeira J, Oliveira R. Biosurfactants: potential applications in medicine. J Antimicrob Chemother. 2006;57(4):609‐618. 10.1093/jac/dkl024. [DOI] [PubMed] [Google Scholar]
- 162. Makkar RS, Rockne KJ. Comparison of synthetic surfactants and biosurfactants in enhancing biodegradation of polycyclic aromatic hydrocarbons. Environ Taxicol Chem. 2002;22(10):2280‐2292. [DOI] [PubMed] [Google Scholar]
- 163. Fakruddin . Biosurfactant: production and application. J Pet Environ Biotechnol. 2012;3(4), 1–5. 10.4172/2157-7463.1000124. [DOI] [Google Scholar]
- 164. Banat IM, Franzetti A, Gandolfi I, et al. Microbial biosurfactants production, applications and future potential. Appl Microbiol Biotechnol. 2010;87(2):427‐444. 10.1007/s00253-010-2589-0. [DOI] [PubMed] [Google Scholar]
- 165. Vollenbroich D, Özel M, Vater J, Kamp RM, Pauli G. Mechanism of inactivation of enveloped viruses by the biosurfactant surfactin from Bacillus subtilis. Biologicals. 1997;25:289‐297. [DOI] [PubMed] [Google Scholar]
- 166. Fracchia L, Banat J, Cavallo M, Ceresa C, Banat MI. Potential therapeutic applications of microbial surface‐active compounds. AIMS Bioengine. 2015;2(3):144‐162. 10.3934/bioeng.2015.3.144. [DOI] [Google Scholar]
- 167. Raghav PK, Agarwal N, Saini M. Edible coating of fruits vegetables: a review. Int J Scient Res Mod Educ. 2016;1(1):188‐204. [Google Scholar]
- 168. Randazzo W, Fabra MJ, Falcó I, López‐Rubio A, Sánchez G. Polymers and biopolymers with antiviral activity: potential applications for improving food safety. Compr Rev Food Sci Food Saf. 2018;17(3):754‐768. 10.1111/1541-4337.12349. [DOI] [PubMed] [Google Scholar]
- 169. Hassan B, Chatha SAS, Hussain AI, Zia KM, Akhtar N. Recent advances on polysaccharides, lipids and protein based edible films and coatings: a review. Int J Biol Macromol. 2018;109:1095‐1107. 10.1016/j.ijbiomac.2017.11.097. [DOI] [PubMed] [Google Scholar]
- 170. Amankwaah C. Incorporation Of Selected Plant Extracts into Edible Chitosan Films and the Effect on the Antiviral, Antibacterial and Mechanical Properties of the Material. Vol 167, Columbus, OH: Ohio State University; 2013. [Google Scholar]
- 171. Fabra MJ, Castro‐Mayorga JL, Randazzo W, et al. Efficacy of cinnamaldehyde against enteric viruses and its activity after incorporation into biodegradable multilayer systems of interest in food packaging. Food Environ Virol. 2016;8(2):125‐132. 10.1007/s12560-016-9235-7. [DOI] [PubMed] [Google Scholar]
- 172. Ng YC, Kim YW, Ryu S, Lee A, Lee J‐S, Song MJ. Suppression of norovirus by natural phytochemicals from Aloe vera and Eriobotryae folium. Food Control. 2017;73:1362‐1370. 10.1016/j.foodcont.2016.10.051. [DOI] [Google Scholar]
- 173. Falcó I, Flores‐Meraz PL, Randazzo W, Sánchez G, López‐Rubio A, Fabra MJ. Antiviral activity of alginate‐oleic acid based coatings incorporating green tea extract on strawberries and raspberries. Food Hydrocoll. 2019;87:611‐618. 10.1016/j.foodhyd.2018.08.055. [DOI] [Google Scholar]
- 174. Xu J, Xu Z, Zheng W. A review of the antiviral role of green tea catechins. Molecules. 2017;22(8):1337. 10.3390/molecules22081337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Jones ST, Cagno V, Janeček M, et al. Modified cyclodextrins as broad‐spectrum antivirals. Sci Adv. 2020;6(5):eaax9318. 10.1126/sciadv.aax9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Leung NHL, Chu DKW, Shiu EYC, et al. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat Med. 2020;26:676‐680. 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Balazy A, Toivola M, Adhikari A, Sivasubramani SK, Reponen T, Grinshpun SA. Do N95 respirators provide 95% protection level against airborne viruses, and how adequate are surgical masks? Am J Infect Control. 2006;34(2):51‐57. 10.1016/j.ajic.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 178. USFDA 2020. N95 respirators and surgical masks (face masks). Retrieved from https://www.fda.gov/medical‐devices/personal‐protective‐equipment‐infection‐control/n95‐respirators‐and‐surgical‐masks‐face‐masks
- 179. F2100‐19e1 A . Standard specification for performance of materials used in medical face masks. Retrieved from http://www.astm.org/cgi-bin/resolver.cgi?F2100
- 180. Tiliket G, Sage DL, Moules V, et al. A new material for airborne virus filtration. Chem Eng J. 2011;173(2):341‐351. 10.1016/j.cej.2011.07.059. [DOI] [Google Scholar]
- 181. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Das O, Neisiany RE, Capezza AJ, et al. The need for fully bio‐based facemasks to counter coronavirus outbreaks: a perspective. Sci Total Environ. 2020;736:139611. 10.1016/j.scitotenv.2020.139611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Allison AL, Ambrose‐Dempster E, Domenech Aparsi T, et al. The environmental dangers of employing single‐use face masks as part of a COVID‐19 exit strategy. Environment. 2020. [Google Scholar]
- 184. Borkow G, Zhou SS, Page T, Gabbay J. A novel anti‐influenza copper oxide containing respiratory face mask. PLoS ONE. 2010;5(6):e11295. 10.1371/journal.pone.0011295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Mitsui K, Yuji K, Kazunari Y, Kazukiyo K, Yasuo S. Virus‐removing filter. Europe. 1997. 1–8. [Google Scholar]
- 186. Nashimoto K, Tashiro T, Kosaka Y, Hara Y; Matushita Seiko Co., Ltd. Mitsui Norin Co., Ltd., assignee. Antiviral filter air cleaner impregnated with tea extract. US patent US5747053. 1998.
- 187. Nashimoto K, Tashiro Y; Matsushita Seiko Co., Ltd. Mitsui Norin Co., Ltd., assignee. Gargling cup, antiviral mask, antiviral filter, antifungal, antibacterial, and antiviral filter air cleaner and air‐cleaner humidifier. US patent US5888527. 1997.
- 188. Oxford JS, Lambkin R, Guralnik M, et al. Preclinical in vitro activity of QR‐435 against influenza a virus as a virucide and in paper masks for prevention of viral transmission. Am J Ther. 2007;14(5):455‐461. [DOI] [PubMed] [Google Scholar]
- 189. Konda A, Prakash A, Moss GA, Schmoldt M, Grant GD, Guha S. Aerosol filtration efficiency of common fabrics ised in respiratory cloth masks. ACS Nano. 2020;14(5):6339‐6347. 10.1021/acsnano.0c03252. [DOI] [PubMed] [Google Scholar]
- 190. Heuer JF, Crozier TA, Howard G, Quintel M. Can breathing circuit filters help prevent the spread of influenza A (H1N1) virus from intubated patients? GMS Hyg Infect Control. 2013;8(1):Doc09. 10.3205/dgkh000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Ari A, Fink JB, Pilbeam S. Secondhand aerosol exposure during mechanical ventilation with and without expiratory filters: an in‐vitro study. Indian J Respirat Care. 2016;5(1):677‐682. [Google Scholar]
- 192. Bolashikov ZD, Melikov AK. Methods for air cleaning and protection of building occupants from airborne pathogens. Build Environ. 2009;44(7):1378‐1385. 10.1016/j.buildenv.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Hyun J, Lee SG, Hwang J. Application of corona discharge‐generated air ions for filtration of aerosolized virus and inactivation of filtered virus. J Aerosol Sci. 2017;107:31‐40. 10.1016/j.jaerosci.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Kim J, Jang J. Inactivation of airborne viruses using vacuum ultraviolet photocatalysis for a flow‐through indoor air purifier with short irradiation time. Aerosol Sci Technol. 2018;52(5):557‐566. 10.1080/02786826.2018.1431386. [DOI] [Google Scholar]
- 195. Zerda KS, Gerba CP, Hou KC, Goyal SM. Adsorption of viruses to charge‐modified silica. Appl Environ Microbiol. 1985;49(1):91‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Han B, Kim H‐J, Kim Y‐J, Sioutas C. Unipolar charging of fine and ultra‐fine particles using carbon fiber ionizers. Aerosol Sci Technol. 2008;42(10):793‐800. 10.1080/02786820802339553. [DOI] [Google Scholar]
- 197. Hagbom M, Nordgren J, Nybom R, Hedlund K‐O, Wigzell H, Svensson L. Ionizing air affects influenza virus infectivity and prevents airborne‐transmission. Sci Rep. 2015;5(1):11431. 10.1038/srep11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Bodzek M, Konieczny K, Rajca M. Membranes in water and wastewater disinfection – review. Archives Environ Protect. 2019;45(1):3‐18. 10.24425/aep.2019.126419. [DOI] [Google Scholar]
- 199. Sinclair TR, Patil A, Raza BG, et al. Cationically modified membranes using covalent layer‐by‐layer assembly for antiviral applications in drinking water. J Membr Sci. 2019;570‐571(15):494‐503. 10.1016/j.memsci.2018.10.081. [DOI] [Google Scholar]


