The Diels–Alder cycloaddition of cycloheptatriene and maleic anhydride produces the title carboxylic anhydride; reaction of this anhydride with 4-bromophenylaniline forms the corresponding tetracyclic imide. The anhydride features C—H⋯O hydrogen bonds in the solid state, while the imide also features C—H⋯O hydrogen bonds as well as C—H⋯π and lone pair–π interactions.
Keywords: crystal structure, C-H⋯O hydrogen bond, C-H⋯π interaction, lone pair–π interaction, bicyclo[2.2.2]octene
Abstract
The syntheses and crystal structures of the two title compounds, C11H10O3 (I) and C17H14BrNO2 (II), both containing the bicyclo[2.2.2]octene ring system, are reported here [the structure of I has been reported previously: White & Goh (2014 ▸). Private Communication (refcode HOKRIK). CCDC, Cambridge, England]. The bond lengths and angles of the bicyclo[2.2.2]octene ring system are similar for both structures. The imide functional group of II features carbonyl C=O bond lengths of 1.209 (2) and 1.210 (2) Å, with C—N bond lengths of 1.393 (2) and 1.397 (2) Å. The five-membered imide ring is nearly planar, and it is positioned exo relative to the alkene bridgehead carbon atoms of the bicyclo[2.2.2]octene ring system. Non-covalent interactions present in the crystal structure of II include a number of C—H⋯O interactions. The extended structure of II also features C—H⋯O hydrogen bonds as well as C—H⋯π and lone pair–π interactions, which combine together to create supramolecular sheets.
Chemical context
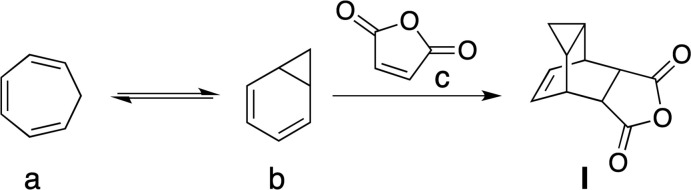
Cycloheptatriene, a, exhibits valence isomerism with norcaradiene, b, in solution (Fig. 1 ▸). The norcaradiene isomer readily reacts with maleic anhydride, c, to form the unique tricyclic anhydride, I (White & Goh, 2014 ▸). This reaction has been known since 1939 (Kohler et al., 1939 ▸), but the structure of the major product was not determined until 1953, when it was elucidated that the product contained a cyclopropane ring (Alder & Jacobs, 1953 ▸). The combination of a rigid tricyclic structure with alkene, anhydride and cyclopropane functional groups makes this structure interesting as a scaffold for drug design because of the ability to specifically place groups in molecular space and thus design molecules to interact selectively with protein active sites.
Figure 1.
Valence isomerism of cycloheptatriene a with norcaradiene b, then the Diels–Alder reaction with maleic anhydride c to give the title anhydride I.
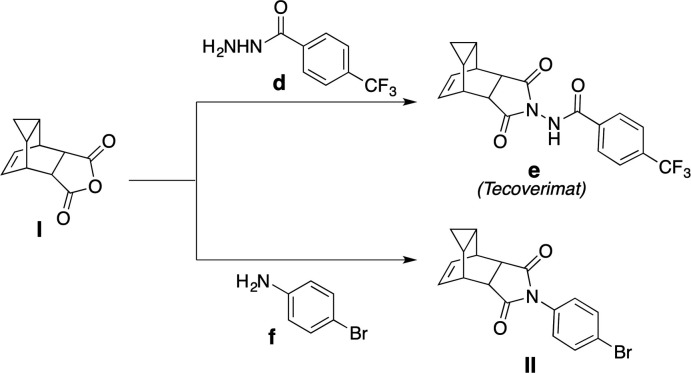
In a high-throughput screen of 356,000 compounds for activity against vaccinia and cowpox viruses, Bailey et al. (2007 ▸) discovered antiviral activity of imide derivatives related to I, including e (tecovirimat, C19H15F3N2O3; Fig. 2 ▸). SAR studies showed that this derivative was the most active of the entire library, and its mode of action was to inhibit extracellular virus formation. Interestingly, hydrogenation of the alkene had little effect on the activity of the compound. Tecoviramat has been approved as a treatment for smallpox, and the United States has created a stockpile of two million doses stored at the US Strategic National Stockpile (Hughes, 2019 ▸).
Figure 2.
Synthesis of the smallpox antiviral compound Tecovirimat, and the title imide II, which both use anhydride I as the starting material.
Substituted anilines, such as p-bromoaniline f, have also been reacted with the anhydride I to form imides that show insecticidal activity (Fig. 2 ▸, Brechbuhler & Petitpierre, 1975 ▸). A wide range of imides were synthesized, including compound II, and were shown to protect crops by inhibiting the growth of lepidoptera. Finally, we note that all of these imide derivatives will undergo a retro-Diels–Alder cycloaddition to form cycloheptatriene and a substituted maleimide. Structural investigations have shown that there is an increase in the length of the C—C bonds that are involved in the retro-Diels–Alder reaction relative to the other C—C bonds in the molecule (Birney et al., 2002 ▸; Pool et al., 2000 ▸). Herein we report the syntheses and crystal structures of the anhydride I and imide II. The structure of the anhydride was previously reported as a Private Communication to the CSD (refcode HOKRIK; White & Goh, 2014 ▸).
Structural commentary
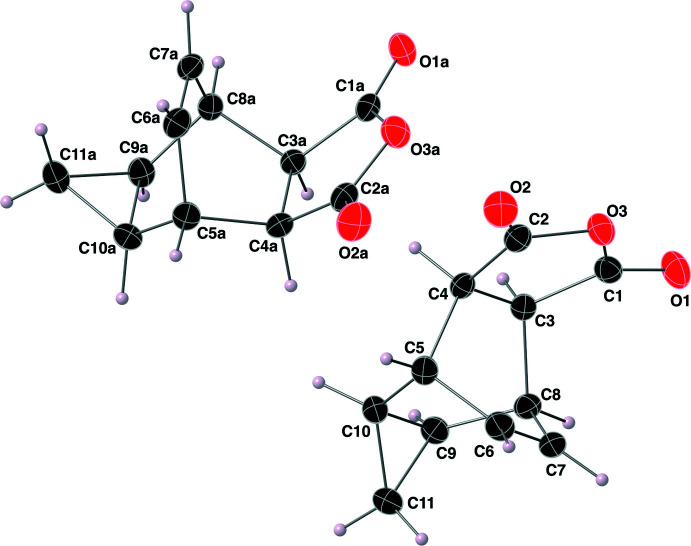
The structure of the title anhydride I was solved in the monoclinic space group P21/n with two molecules in the asymmetric unit. The atom labeling scheme (starting with C1 and C1a for the two molecules) is shown in Fig. 3 ▸. This structure is quite similar with respect to the bond lengths and angles described below for the imide II. The bond lengths of the carbonyl groups of the anhydride are shorter than the imide, as expected, with C1=O1 = 1.1943 (18), C2=O2 = 1.1904 (17), C1—O3 = 1.3868 (17) and C2—O3 = 1.3978 (16) Å. The corresponding data for the C1a molecule are 1.1913 (17), 1.1871 (18), 1.3855 (17) and 1.3905 (18) Å, respectively. The configurations of the stereogenic centres in the arbitrarily chosen asymmetric molecules are: C3 S, C4 R, C5 R, C8 S, C9 S, C10 R and C3a R, C4a S, C5a S, C8a R, C9a R, C10a S: crystal symmetry generates a racemic mixture in the bulk.
Figure 3.
The molecular structure of the anhydride I, with the atom-labeling scheme for both crystallographically unique molecules. Displacement ellipsoids are shown at the 40% probability level using standard CPK colors.
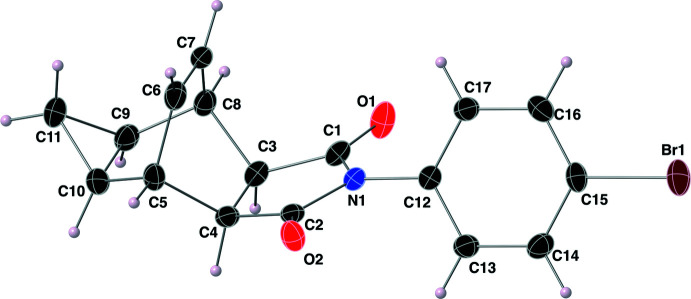
The structure of the imide II was solved in the monoclinic space group P21/n, and its atom labeling scheme is shown in Fig. 4 ▸. The imide functional group of this structure has C=O bond lengths of 1.209 (2) and 1.210 (2) Å, with C—N bond lengths of 1.393 (2) and 1.397 (2) Å. The O—C—N bond angles of the imide functional group are 123.98 (17) and 123.97 (17)°. The aromatic ring, C12–C17, is oriented nearly perpendicular to the plane containing the atoms of the imide functional group with a C1—N1—C12—C17 torsion angle of 65.0 (2)°. The five-membered ring that contains the imide functional group (–C1—N1—C2—C4—C3–) is close to planar with a Cremer–Pople τ value of 2.8 (Cremer & Pople, 1975 ▸). When considering the bicyclo[2.2.2]octene ring system (C3–C10), both C11 and the atoms of the imide functional group are oriented exo relative to the bridgehead alkene carbon atoms C6–C7. The length of the C6=C7 double bond is 1.324 (3) Å, and the cyclopropyl ring C9–C11 has C—C—C bond angles ranging from 59.89 (13)–60.14 (14)°. The stereogenic centres in the asymmetric molecule of II are C3 R, C4 S, C5 S, C8 R, C9 R and C10 S; again, crystal symmetry generates a racemic mixture.
Figure 4.
The molecular structure of the imide II, with the atom-labeling scheme. Displacement ellipsoids are shown at the 40% probability level using standard CPK colors.
Supramolecular features
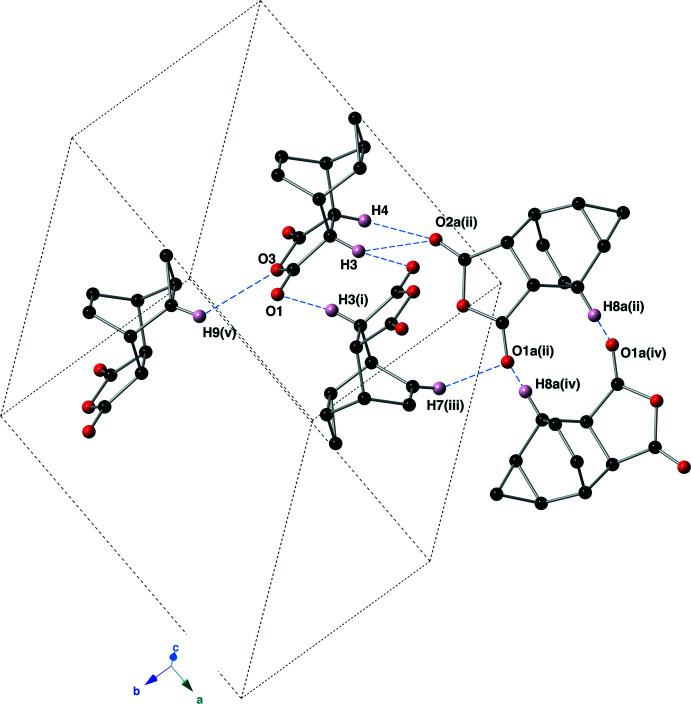
The extended structure of the anhydride I is dominated by C—H⋯O hydrogen bonds (Sutor, 1962 ▸, 1963 ▸; Steiner, 1996 ▸) involving both carbonyl groups as acceptors (Table 1 ▸, Fig. 5 ▸). The D⋯A distances range from 3.1897 (16) to 3.4882 (17) Å with D—H⋯A angles ranging from 119 to 159°; the C9 bond is likely very weak based on its H⋯A distance of 2.73 Å. Combined together, these interactions create supramolecular sheets that lie in the ab plane.
Table 1. Hydrogen-bond geometry (Å, °) for I .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3⋯O1i | 1.00 | 2.64 | 3.4882 (17) | 143 |
| C3—H3⋯O2A ii | 1.00 | 2.54 | 3.2487 (17) | 128 |
| C4—H4⋯O2A ii | 1.00 | 2.43 | 3.1897 (16) | 133 |
| C7—H7⋯O1A iii | 0.95 | 2.56 | 3.4652 (18) | 159 |
| C8A—H8A⋯O1A iv | 1.00 | 2.59 | 3.2521 (17) | 123 |
| C9—H9⋯O3v | 1.00 | 2.73 | 3.3409 (17) | 119 |
Symmetry codes: (i) 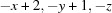 ; (ii)
; (ii) 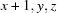 ; (iii)
; (iii) 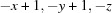 ; (iv)
; (iv) 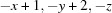 ; (v)
; (v) 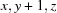 .
.
Figure 5.
Depiction of the C—H⋯O hydrogen bonds (blue, dashed lines) present in the crystal of anhydride I, using a ball-and-stick model. For clarity, only those hydrogen atoms involved in an interaction are shown. Symmetry codes: (i) −x + 2, −y + 1, −z; (ii) x + 1, y, z; (iii) −x + 1, −y + 1, −z; (iv) −x + 1, −y + 2, −z; (v) x, y + 1, z.
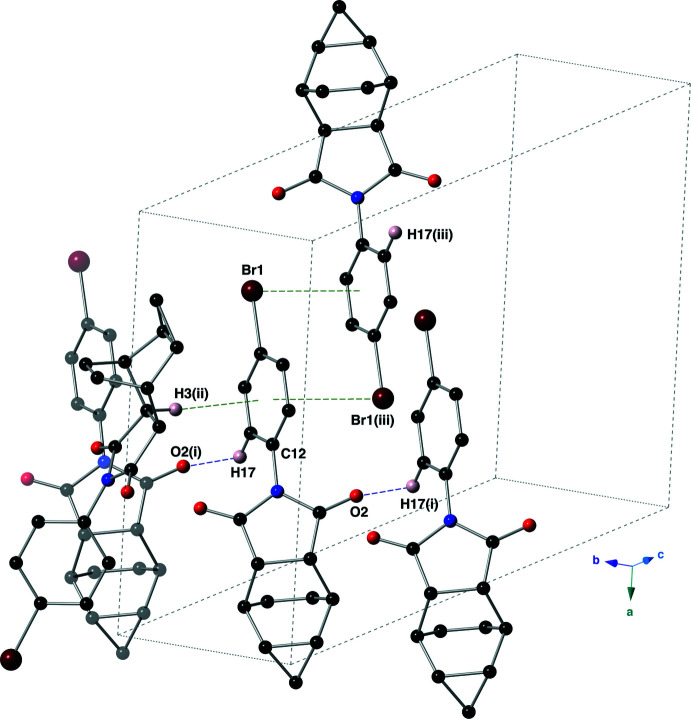
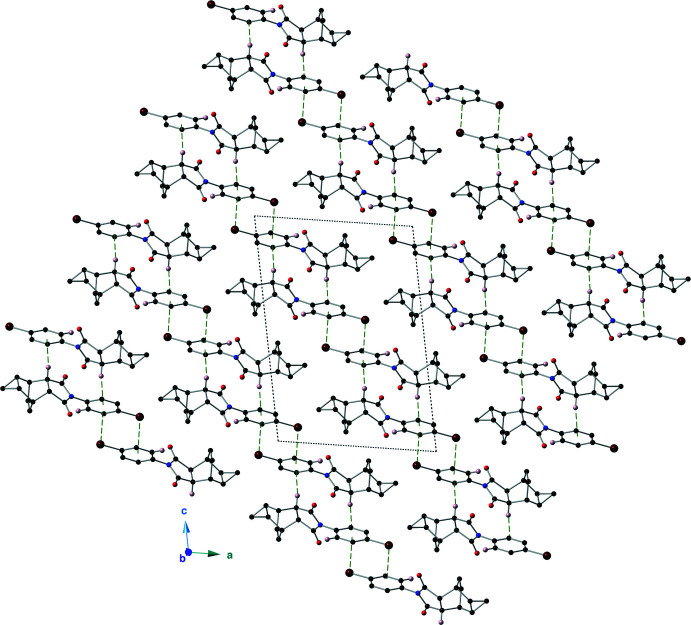
In the crystal of the imide II, the molecules are linked by C—H⋯O hydrogen bonds as well as C—H⋯π and C—Br⋯π interactions (Table 2 ▸, Fig. 6 ▸). The C—H⋯O hydrogen bond is between C17—H17 of the aromatic ring and O2 of an imide carbonyl group. This hydrogen bond has a D⋯A distance of 3.175 (2) Å with a D—H⋯A angle of 139°. The C—H⋯π interaction is between C3—H3, which is α to the carbonyl group C1(O1), and the aromatic ring C12–C17. This interaction has a H⋯Cg distance of 3.801 (2) Å (where Cg is the centroid of the C12–C17 ring), with a C—H⋯Cg angle of 165°. The aromatic ring C12–C17 bears an electron-withdrawing bromine atom, and accepts a lone pair(LP)–π interaction from the bromine atom of a nearby molecule (Mooibroek, et al., 2008 ▸). This LP–π interaction has a Br⋯Cg distance of 3.5854 (8) Å with a C15—Br1⋯Cg angle of 87.43 (6)°. Dimers of imide II are formed via the Br⋯π interactions, and these dimers are linked into supramolecular sheets that lie along (010) by the C—H⋯O and C—H⋯π interactions (Fig. 7 ▸).
Table 2. Hydrogen-bond geometry (Å, °) for II .
Cg1 is the centroid of the C12–C17 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C17—H17⋯O2i | 0.95 | 2.40 | 3.175 (2) | 139 |
| C3—H3⋯Cg1ii | 1.00 | 2.83 | 3.801 (2) | 165 |
Symmetry codes: (i)  ; (ii)
; (ii) 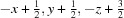 .
.
Figure 6.
Non-covalent interactions present in the crystal of imide II, using a ball-and-stick model. Only those hydrogen atoms involved in an interaction are shown for clarity. C—H⋯O hydrogen bonds are shown with purple, dashed lines, while C—H⋯π and C—Br⋯π interactions are shown with green, dashed lines. Symmetry codes: (i) x, y − 1, z; (ii) −x + 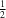 , y +
, y + 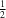 , −z +
, −z +  .
.
Figure 7.
A view of the packing in the crystal of imide II, as viewed down the b axis. C—H⋯O hydrogen bonds are shown with purple, dashed lines, while C—H⋯π and C—Br⋯π interactions are shown with green, dashed lines. For clarity, only those hydrogen atoms involved in a non-covalent interaction are shown.
Database survey
The structure of the anhydride I has been deposited in the Cambridge Structural Database (CSD, Version 5.41, November, 2019; Groom et al., 2016 ▸) as a Private Communication from White & Goh (2014 ▸, refcode HOKRIK). The acquisition temperature for this data set was 130 K, versus 173 K for the structure reported here. Other than this, the structures are nearly identical. A search of the CSD for structures containing the same bicyclo[2.2.2]octene ring system bearing a cyclic anhydride shows 52 hits (including HOKRIK). Of these, an interesting structure is FAXPAV (Coxon et al., 1986 ▸), which bears a very complex fused-ring system in the place of the cyclopropane ring on anhydride I.
A search of the CSD for structures containing a bicyclo[2.2.2]octene ring system fused to a cyclic imide resulted in 125 structures related to imide II. Structure COZMAH (Wu et al., 2014 ▸) also bears a p-bromobenzene ring bonded to the imide nitrogen atom, but is derivatized with two esters and an indole ring on the octene portion of the ring system. The structure of tecovirimat (e, Fig. 2 ▸) has been deposited as SOKVIY (Bailey et al., 2007 ▸). Finally, structure HARNEV bears two cyclic imide groups on either side of the octene ring system (Song et al., 2012 ▸).
Synthesis and crystallization
Synthesis of the anhydride (I):
Cycloheptatriene (1.38 g, 15 mmol) and maleic anhydride (1.37 g, 14 mmol) were added to an oven-dried round-bottom flask containing 10 ml of xylene and the mixture was refluxed for 1.5 h. Approximately half of the xylenes were distilled off via short-path distillation and the reaction mixture was left to cool at room temperature. The round-bottom flask was fitted with a stopper and left to recrystallize for 48 h to afford large, cream-colored needles. The product was recrystallized once more by dissolving in 8 ml of xylene: after a week at room temperature, the pure product I was obtained in the form of large colorless crystals (1.13 g, 40%, m.p. = 372–374 K). 1H NMR (400 MHz, chloroform-d) δ 5.88 (dd, J = 4.8, 3.2 Hz, 2H), 3.46 (dh, J = 6.6, 2.1 Hz, 2H), 3.23 (dd, J = 2.1, 1.6 Hz, 2H), 1.17–1.04 (m, 4H). 13C NMR (101 MHz, chloroform-d) δ 172.45, 128.55, 45.88, 33.65, 9.56, 5.24.
Synthesis of the imide (II):
Compound I (0.28 g, 1.47 mmol) and p-bromoaniline (0.25 g, 1.45 mmol) were added to a vial containing 5 ml of xylene and the mixture was refluxed for 5 min. The mixture was then cooled to room temperature and left for 5 days in a sealed vial. The precipitate was recrystallized from ethanol solution to yield colorless needle-like crystals of II (0.27 g, 52% yield, m.p. = 465–467 K). 1H NMR (400 MHz, chloroform-d) δ 7.54 (d, J = 8.7 Hz, 1H), 7.06 (d, J = 8.7 Hz, 1H), 5.84 (dd, J = 4.7, 3.4 Hz, 1H), 3.48 (s, 1H), 3.12 (s, 1H), 1.14 (s, 1H), 0.38–0.21 (m, 1H). 13C NMR (101 MHz, chloroform-d) δ 177.42, 132.34, 130.88, 128.11, 127.92, 122.47, 45.40, 33.90, 9.97, 4.80.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. For both structures, hydrogen atoms bonded to carbon atoms were placed in calculated positions and refined to ride on their parent atoms: C—H = 0.95–1.00 Å with U iso(H) = 1.2U eq(C).
Table 3. Experimental details.
| I | II | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C11H10O3 | C17H14BrNO2 |
| M r | 190.19 | 344.20 |
| Crystal system, space group | Monoclinic, P21/n | Monoclinic, P21/n |
| Temperature (K) | 173 | 173 |
| a, b, c (Å) | 11.3538 (3), 7.4062 (2), 20.5398 (5) | 12.49907 (16), 6.41302 (8), 17.8772 (2) |
| β (°) | 92.6226 (15) | 99.8083 (6) |
| V (Å3) | 1725.35 (8) | 1412.04 (3) |
| Z | 8 | 4 |
| Radiation type | Cu Kα | Cu Kα |
| μ (mm−1) | 0.88 | 4.00 |
| Crystal size (mm) | 0.53 × 0.32 × 0.22 | 0.42 × 0.12 × 0.04 |
| Data collection | ||
| Diffractometer | Bruker APEXII CCD | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Bruker, 2013 ▸) | Multi-scan (SADABS; Bruker, 2013 ▸) |
| T min, T max | 0.675, 0.754 | 0.578, 0.753 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 13466, 3353, 3070 | 23789, 2683, 2441 |
| R int | 0.028 | 0.039 |
| (sin θ/λ)max (Å−1) | 0.618 | 0.610 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.045, 0.116, 1.06 | 0.027, 0.072, 1.04 |
| No. of reflections | 3353 | 2683 |
| No. of parameters | 253 | 190 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.22, −0.40 | 0.56, −0.48 |
Supplementary Material
Crystal structure: contains datablock(s) global, II, I. DOI: 10.1107/S2056989020009512/hb7931sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020009512/hb7931Isup3.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989020009512/hb7931IIsup4.hkl
Supporting information file. DOI: 10.1107/S2056989020009512/hb7931Isup4.cml
Supporting information file. DOI: 10.1107/S2056989020009512/hb7931IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are grateful to Pfizer, Inc. for the donation of the Varian INOVA 400 F T NMR spectrometer. We thank the MSU Chemistry Department for purchasing/upgrading the CCD-based X-ray diffractometers, and the National Science Foundation for support from the MRI program to purchase the Rigaku Synergy S. Diffractometer (MSU).
supplementary crystallographic information
4-Oxatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (I) . Crystal data
| C11H10O3 | F(000) = 800 |
| Mr = 190.19 | Dx = 1.464 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54178 Å |
| a = 11.3538 (3) Å | Cell parameters from 8934 reflections |
| b = 7.4062 (2) Å | θ = 3.9–72.4° |
| c = 20.5398 (5) Å | µ = 0.88 mm−1 |
| β = 92.6226 (15)° | T = 173 K |
| V = 1725.35 (8) Å3 | Chunk, colourless |
| Z = 8 | 0.53 × 0.32 × 0.22 mm |
4-Oxatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (I) . Data collection
| Bruker APEXII CCD diffractometer | 3070 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.028 |
| Absorption correction: multi-scan (SADABS; Bruker, 2013) | θmax = 72.4°, θmin = 4.3° |
| Tmin = 0.675, Tmax = 0.754 | h = −14→13 |
| 13466 measured reflections | k = −9→9 |
| 3353 independent reflections | l = −25→25 |
4-Oxatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (I) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.045 | H-atom parameters constrained |
| wR(F2) = 0.116 | w = 1/[σ2(Fo2) + (0.0673P)2 + 0.5724P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max < 0.001 |
| 3353 reflections | Δρmax = 0.22 e Å−3 |
| 253 parameters | Δρmin = −0.40 e Å−3 |
| 0 restraints |
4-Oxatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (I) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-Oxatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (I) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.92952 (11) | 0.22515 (15) | 0.01539 (5) | 0.0378 (3) | |
| O2 | 0.91944 (10) | 0.21601 (15) | 0.23113 (5) | 0.0354 (3) | |
| O3 | 0.92868 (9) | 0.18344 (13) | 0.12312 (5) | 0.0287 (2) | |
| C1 | 0.92625 (12) | 0.29232 (19) | 0.06814 (7) | 0.0249 (3) | |
| C2 | 0.92145 (12) | 0.28818 (19) | 0.17941 (7) | 0.0243 (3) | |
| C3 | 0.92092 (11) | 0.48779 (18) | 0.08716 (6) | 0.0212 (3) | |
| H3 | 0.9931 | 0.5521 | 0.0734 | 0.025* | |
| C4 | 0.91806 (11) | 0.48510 (18) | 0.16185 (6) | 0.0206 (3) | |
| H4 | 0.9892 | 0.5477 | 0.1814 | 0.025* | |
| C5 | 0.80361 (12) | 0.57992 (19) | 0.18403 (6) | 0.0240 (3) | |
| H5 | 0.7976 | 0.5761 | 0.2324 | 0.029* | |
| C6 | 0.70138 (12) | 0.4864 (2) | 0.14922 (7) | 0.0284 (3) | |
| H6 | 0.6400 | 0.4300 | 0.1717 | 0.034* | |
| C7 | 0.70364 (12) | 0.4889 (2) | 0.08459 (8) | 0.0288 (3) | |
| H7 | 0.6438 | 0.4348 | 0.0573 | 0.035* | |
| C8 | 0.80824 (12) | 0.58391 (19) | 0.05799 (6) | 0.0245 (3) | |
| H8 | 0.8054 | 0.5833 | 0.0093 | 0.029* | |
| C9 | 0.81906 (12) | 0.77608 (19) | 0.08545 (7) | 0.0259 (3) | |
| H9 | 0.8782 | 0.8575 | 0.0659 | 0.031* | |
| C10 | 0.81589 (12) | 0.77348 (19) | 0.15890 (7) | 0.0255 (3) | |
| H10 | 0.8732 | 0.8536 | 0.1835 | 0.031* | |
| C11 | 0.71901 (13) | 0.8646 (2) | 0.11912 (8) | 0.0317 (3) | |
| H11C | 0.7163 | 0.9982 | 0.1197 | 0.038* | |
| H11D | 0.6410 | 0.8046 | 0.1159 | 0.038* | |
| O1A | 0.45014 (10) | 0.77462 (14) | 0.02985 (5) | 0.0328 (3) | |
| O2A | 0.12278 (10) | 0.77025 (15) | 0.13936 (6) | 0.0386 (3) | |
| O3A | 0.28206 (9) | 0.73501 (13) | 0.08039 (5) | 0.0313 (3) | |
| C1A | 0.36811 (12) | 0.84253 (19) | 0.05447 (6) | 0.0241 (3) | |
| C2A | 0.19782 (12) | 0.8398 (2) | 0.10974 (7) | 0.0270 (3) | |
| C3A | 0.34016 (11) | 1.03897 (17) | 0.06434 (6) | 0.0206 (3) | |
| H3A | 0.3307 | 1.1025 | 0.0215 | 0.025* | |
| C4A | 0.22267 (11) | 1.03737 (18) | 0.09904 (6) | 0.0219 (3) | |
| H4A | 0.1585 | 1.0939 | 0.0710 | 0.026* | |
| C5A | 0.23788 (12) | 1.13775 (19) | 0.16575 (6) | 0.0239 (3) | |
| H5A | 0.1634 | 1.1373 | 0.1899 | 0.029* | |
| C6A | 0.33646 (12) | 1.04165 (19) | 0.20315 (6) | 0.0254 (3) | |
| H6A | 0.3267 | 0.9881 | 0.2446 | 0.031* | |
| C7A | 0.43832 (12) | 1.03759 (19) | 0.17356 (6) | 0.0240 (3) | |
| H7A | 0.5066 | 0.9791 | 0.1919 | 0.029* | |
| C8A | 0.43686 (11) | 1.13289 (18) | 0.10893 (6) | 0.0216 (3) | |
| H8A | 0.5158 | 1.1289 | 0.0893 | 0.026* | |
| C9A | 0.39235 (12) | 1.32691 (19) | 0.11555 (7) | 0.0250 (3) | |
| H9A | 0.4001 | 1.4080 | 0.0771 | 0.030* | |
| C10A | 0.27558 (12) | 1.32973 (19) | 0.14820 (7) | 0.0260 (3) | |
| H10A | 0.2131 | 1.4120 | 0.1293 | 0.031* | |
| C11A | 0.38257 (13) | 1.4167 (2) | 0.18102 (7) | 0.0307 (3) | |
| H11A | 0.4195 | 1.3554 | 0.2196 | 0.037* | |
| H11B | 0.3854 | 1.5503 | 0.1826 | 0.037* |
4-Oxatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (I) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0524 (7) | 0.0336 (6) | 0.0279 (6) | 0.0065 (5) | 0.0060 (5) | −0.0079 (4) |
| O2 | 0.0469 (7) | 0.0310 (6) | 0.0285 (5) | −0.0024 (5) | 0.0018 (5) | 0.0078 (4) |
| O3 | 0.0367 (6) | 0.0208 (5) | 0.0286 (5) | 0.0005 (4) | 0.0026 (4) | −0.0004 (4) |
| C1 | 0.0235 (6) | 0.0260 (7) | 0.0255 (7) | 0.0018 (5) | 0.0032 (5) | −0.0010 (5) |
| C2 | 0.0223 (6) | 0.0261 (7) | 0.0246 (7) | −0.0028 (5) | 0.0009 (5) | 0.0001 (5) |
| C3 | 0.0198 (6) | 0.0226 (6) | 0.0212 (6) | −0.0012 (5) | 0.0027 (5) | −0.0005 (5) |
| C4 | 0.0186 (6) | 0.0226 (6) | 0.0205 (6) | −0.0031 (5) | 0.0007 (5) | −0.0005 (5) |
| C5 | 0.0220 (6) | 0.0265 (7) | 0.0238 (6) | −0.0006 (5) | 0.0050 (5) | −0.0031 (5) |
| C6 | 0.0192 (6) | 0.0274 (7) | 0.0391 (8) | −0.0028 (5) | 0.0066 (6) | −0.0035 (6) |
| C7 | 0.0210 (7) | 0.0274 (7) | 0.0375 (8) | −0.0006 (5) | −0.0039 (6) | −0.0077 (6) |
| C8 | 0.0253 (7) | 0.0258 (7) | 0.0220 (6) | 0.0028 (5) | −0.0025 (5) | −0.0014 (5) |
| C9 | 0.0263 (7) | 0.0232 (7) | 0.0281 (7) | 0.0024 (5) | −0.0008 (5) | 0.0013 (5) |
| C10 | 0.0245 (7) | 0.0241 (7) | 0.0279 (7) | 0.0000 (5) | 0.0004 (5) | −0.0051 (5) |
| C11 | 0.0281 (7) | 0.0269 (7) | 0.0398 (8) | 0.0063 (6) | −0.0011 (6) | −0.0046 (6) |
| O1A | 0.0345 (6) | 0.0312 (5) | 0.0329 (5) | 0.0046 (4) | 0.0041 (4) | −0.0083 (4) |
| O2A | 0.0344 (6) | 0.0365 (6) | 0.0454 (7) | −0.0156 (5) | 0.0066 (5) | 0.0019 (5) |
| O3A | 0.0321 (6) | 0.0218 (5) | 0.0399 (6) | −0.0027 (4) | 0.0022 (4) | −0.0014 (4) |
| C1A | 0.0268 (7) | 0.0251 (7) | 0.0201 (6) | −0.0015 (5) | −0.0031 (5) | −0.0024 (5) |
| C2A | 0.0242 (7) | 0.0278 (7) | 0.0287 (7) | −0.0053 (5) | −0.0024 (5) | −0.0007 (6) |
| C3A | 0.0226 (6) | 0.0217 (6) | 0.0175 (6) | 0.0003 (5) | 0.0003 (5) | 0.0014 (5) |
| C4A | 0.0188 (6) | 0.0245 (7) | 0.0221 (6) | −0.0008 (5) | −0.0009 (5) | 0.0019 (5) |
| C5A | 0.0211 (6) | 0.0275 (7) | 0.0236 (6) | −0.0012 (5) | 0.0050 (5) | −0.0008 (5) |
| C6A | 0.0284 (7) | 0.0299 (7) | 0.0180 (6) | −0.0031 (6) | 0.0009 (5) | 0.0014 (5) |
| C7A | 0.0237 (6) | 0.0266 (7) | 0.0215 (6) | 0.0004 (5) | −0.0031 (5) | −0.0011 (5) |
| C8A | 0.0194 (6) | 0.0239 (7) | 0.0218 (6) | −0.0021 (5) | 0.0031 (5) | −0.0018 (5) |
| C9A | 0.0260 (7) | 0.0225 (7) | 0.0267 (7) | −0.0031 (5) | 0.0046 (5) | −0.0005 (5) |
| C10A | 0.0261 (7) | 0.0239 (7) | 0.0282 (7) | 0.0029 (5) | 0.0043 (5) | −0.0021 (5) |
| C11A | 0.0328 (8) | 0.0265 (7) | 0.0330 (8) | −0.0023 (6) | 0.0037 (6) | −0.0070 (6) |
4-Oxatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (I) . Geometric parameters (Å, º)
| O1—C1 | 1.1943 (18) | O1A—C1A | 1.1913 (17) |
| O2—C2 | 1.1904 (17) | O2A—C2A | 1.1871 (18) |
| O3—C1 | 1.3868 (17) | O3A—C1A | 1.3855 (17) |
| O3—C2 | 1.3978 (16) | O3A—C2A | 1.3905 (18) |
| C1—C3 | 1.5014 (18) | C1A—C3A | 1.5048 (18) |
| C2—C4 | 1.5024 (18) | C2A—C4A | 1.5080 (19) |
| C3—H3 | 1.0000 | C3A—H3A | 1.0000 |
| C3—C4 | 1.5360 (17) | C3A—C4A | 1.5409 (17) |
| C3—C8 | 1.5596 (18) | C3A—C8A | 1.5607 (17) |
| C4—H4 | 1.0000 | C4A—H4A | 1.0000 |
| C4—C5 | 1.5631 (17) | C4A—C5A | 1.5616 (18) |
| C5—H5 | 1.0000 | C5A—H5A | 1.0000 |
| C5—C6 | 1.5040 (19) | C5A—C6A | 1.5068 (19) |
| C5—C10 | 1.5321 (19) | C5A—C10A | 1.5326 (19) |
| C6—H6 | 0.9500 | C6A—H6A | 0.9500 |
| C6—C7 | 1.329 (2) | C6A—C7A | 1.3312 (19) |
| C7—H7 | 0.9500 | C7A—H7A | 0.9500 |
| C7—C8 | 1.504 (2) | C7A—C8A | 1.5028 (18) |
| C8—H8 | 1.0000 | C8A—H8A | 1.0000 |
| C8—C9 | 1.5337 (19) | C8A—C9A | 1.5313 (19) |
| C9—H9 | 1.0000 | C9A—H9A | 1.0000 |
| C9—C10 | 1.5110 (19) | C9A—C10A | 1.5131 (18) |
| C9—C11 | 1.5066 (19) | C9A—C11A | 1.5091 (19) |
| C10—H10 | 1.0000 | C10A—H10A | 1.0000 |
| C10—C11 | 1.500 (2) | C10A—C11A | 1.507 (2) |
| C11—H11C | 0.9900 | C11A—H11A | 0.9900 |
| C11—H11D | 0.9900 | C11A—H11B | 0.9900 |
| C1—O3—C2 | 110.55 (11) | C1A—O3A—C2A | 110.90 (11) |
| O1—C1—O3 | 119.75 (13) | O1A—C1A—O3A | 119.95 (13) |
| O1—C1—C3 | 129.86 (13) | O1A—C1A—C3A | 129.73 (13) |
| O3—C1—C3 | 110.39 (11) | O3A—C1A—C3A | 110.31 (11) |
| O2—C2—O3 | 119.55 (13) | O2A—C2A—O3A | 120.26 (14) |
| O2—C2—C4 | 130.45 (13) | O2A—C2A—C4A | 129.74 (14) |
| O3—C2—C4 | 110.00 (11) | O3A—C2A—C4A | 109.97 (11) |
| C1—C3—H3 | 110.1 | C1A—C3A—H3A | 110.6 |
| C1—C3—C4 | 104.49 (10) | C1A—C3A—C4A | 104.31 (11) |
| C1—C3—C8 | 112.48 (11) | C1A—C3A—C8A | 111.28 (11) |
| C4—C3—H3 | 110.1 | C4A—C3A—H3A | 110.6 |
| C4—C3—C8 | 109.58 (10) | C4A—C3A—C8A | 109.43 (10) |
| C8—C3—H3 | 110.1 | C8A—C3A—H3A | 110.6 |
| C2—C4—C3 | 104.53 (10) | C2A—C4A—C3A | 104.27 (11) |
| C2—C4—H4 | 110.0 | C2A—C4A—H4A | 110.7 |
| C2—C4—C5 | 112.25 (11) | C2A—C4A—C5A | 110.36 (11) |
| C3—C4—H4 | 110.0 | C3A—C4A—H4A | 110.7 |
| C3—C4—C5 | 109.95 (10) | C3A—C4A—C5A | 109.80 (10) |
| C5—C4—H4 | 110.0 | C5A—C4A—H4A | 110.7 |
| C4—C5—H5 | 111.9 | C4A—C5A—H5A | 111.8 |
| C6—C5—C4 | 106.76 (11) | C6A—C5A—C4A | 105.79 (10) |
| C6—C5—H5 | 111.9 | C6A—C5A—H5A | 111.8 |
| C6—C5—C10 | 110.54 (12) | C6A—C5A—C10A | 110.44 (11) |
| C10—C5—C4 | 103.44 (10) | C10A—C5A—C4A | 104.84 (10) |
| C10—C5—H5 | 111.9 | C10A—C5A—H5A | 111.8 |
| C5—C6—H6 | 122.6 | C5A—C6A—H6A | 122.6 |
| C7—C6—C5 | 114.75 (12) | C7A—C6A—C5A | 114.74 (12) |
| C7—C6—H6 | 122.6 | C7A—C6A—H6A | 122.6 |
| C6—C7—H7 | 122.6 | C6A—C7A—H7A | 122.6 |
| C6—C7—C8 | 114.89 (12) | C6A—C7A—C8A | 114.71 (12) |
| C8—C7—H7 | 122.6 | C8A—C7A—H7A | 122.6 |
| C3—C8—H8 | 111.8 | C3A—C8A—H8A | 111.7 |
| C7—C8—C3 | 107.11 (11) | C7A—C8A—C3A | 106.74 (10) |
| C7—C8—H8 | 111.8 | C7A—C8A—H8A | 111.7 |
| C7—C8—C9 | 110.60 (11) | C7A—C8A—C9A | 110.65 (11) |
| C9—C8—C3 | 103.41 (10) | C9A—C8A—C3A | 104.13 (10) |
| C9—C8—H8 | 111.8 | C9A—C8A—H8A | 111.7 |
| C8—C9—H9 | 117.1 | C8A—C9A—H9A | 116.9 |
| C10—C9—C8 | 110.50 (11) | C10A—C9A—C8A | 110.59 (11) |
| C10—C9—H9 | 117.1 | C10A—C9A—H9A | 116.9 |
| C11—C9—C8 | 121.54 (12) | C11A—C9A—C8A | 122.06 (12) |
| C11—C9—H9 | 117.1 | C11A—C9A—H9A | 116.9 |
| C11—C9—C10 | 59.62 (9) | C11A—C9A—C10A | 59.82 (9) |
| C5—C10—H10 | 116.9 | C5A—C10A—H10A | 117.1 |
| C9—C10—C5 | 110.79 (11) | C9A—C10A—C5A | 110.56 (11) |
| C9—C10—H10 | 116.9 | C9A—C10A—H10A | 117.1 |
| C11—C10—C5 | 121.90 (12) | C11A—C10A—C5A | 121.31 (12) |
| C11—C10—C9 | 60.05 (9) | C11A—C10A—C9A | 59.96 (9) |
| C11—C10—H10 | 116.9 | C11A—C10A—H10A | 117.1 |
| C9—C11—H11C | 117.7 | C9A—C11A—H11A | 117.7 |
| C9—C11—H11D | 117.7 | C9A—C11A—H11B | 117.7 |
| C10—C11—C9 | 60.33 (9) | C10A—C11A—C9A | 60.22 (9) |
| C10—C11—H11C | 117.7 | C10A—C11A—H11A | 117.7 |
| C10—C11—H11D | 117.7 | C10A—C11A—H11B | 117.7 |
| H11C—C11—H11D | 114.9 | H11A—C11A—H11B | 114.9 |
4-Oxatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (I) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3···O1i | 1.00 | 2.64 | 3.4882 (17) | 143 |
| C3—H3···O2Aii | 1.00 | 2.54 | 3.2487 (17) | 128 |
| C4—H4···O2Aii | 1.00 | 2.43 | 3.1897 (16) | 133 |
| C7—H7···O1Aiii | 0.95 | 2.56 | 3.4652 (18) | 159 |
| C8A—H8A···O1Aiv | 1.00 | 2.59 | 3.2521 (17) | 123 |
| C9—H9···O3v | 1.00 | 2.73 | 3.3409 (17) | 119 |
Symmetry codes: (i) −x+2, −y+1, −z; (ii) x+1, y, z; (iii) −x+1, −y+1, −z; (iv) −x+1, −y+2, −z; (v) x, y+1, z.
4-(4-Bromophenyl)-4-azatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (II) . Crystal data
| C17H14BrNO2 | F(000) = 696 |
| Mr = 344.20 | Dx = 1.619 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54178 Å |
| a = 12.49907 (16) Å | Cell parameters from 9902 reflections |
| b = 6.41302 (8) Å | θ = 4.7–70.1° |
| c = 17.8772 (2) Å | µ = 4.00 mm−1 |
| β = 99.8083 (6)° | T = 173 K |
| V = 1412.04 (3) Å3 | Needle, colourless |
| Z = 4 | 0.42 × 0.12 × 0.04 mm |
4-(4-Bromophenyl)-4-azatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (II) . Data collection
| Bruker APEXII CCD diffractometer | 2441 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.039 |
| Absorption correction: multi-scan (SADABS; Bruker, 2013) | θmax = 70.2°, θmin = 4.0° |
| Tmin = 0.578, Tmax = 0.753 | h = −15→15 |
| 23789 measured reflections | k = −7→7 |
| 2683 independent reflections | l = −21→21 |
4-(4-Bromophenyl)-4-azatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (II) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.027 | H-atom parameters constrained |
| wR(F2) = 0.072 | w = 1/[σ2(Fo2) + (0.033P)2 + 1.1866P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.003 |
| 2683 reflections | Δρmax = 0.56 e Å−3 |
| 190 parameters | Δρmin = −0.48 e Å−3 |
| 0 restraints |
4-(4-Bromophenyl)-4-azatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (II) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Bromophenyl)-4-azatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (II) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.63472 (2) | −0.24792 (4) | 0.56752 (2) | 0.03728 (10) | |
| O1 | 0.18672 (12) | −0.1467 (3) | 0.72460 (9) | 0.0360 (4) | |
| O2 | 0.19184 (11) | 0.3903 (2) | 0.55825 (8) | 0.0307 (3) | |
| N1 | 0.21210 (12) | 0.1061 (2) | 0.63812 (8) | 0.0204 (3) | |
| C1 | 0.16094 (14) | 0.0182 (3) | 0.69397 (10) | 0.0237 (4) | |
| C2 | 0.16488 (15) | 0.2941 (3) | 0.61018 (11) | 0.0218 (4) | |
| C3 | 0.07071 (14) | 0.1617 (3) | 0.70770 (10) | 0.0233 (4) | |
| H3 | 0.0846 | 0.2102 | 0.7616 | 0.028* | |
| C4 | 0.07607 (14) | 0.3487 (3) | 0.65452 (10) | 0.0229 (4) | |
| H4 | 0.0962 | 0.4777 | 0.6851 | 0.028* | |
| C5 | −0.03628 (15) | 0.3789 (3) | 0.60269 (11) | 0.0257 (4) | |
| H5 | −0.0355 | 0.4979 | 0.5667 | 0.031* | |
| C6 | −0.06246 (15) | 0.1766 (3) | 0.56169 (11) | 0.0288 (4) | |
| H6 | −0.0753 | 0.1670 | 0.5079 | 0.035* | |
| C7 | −0.06639 (15) | 0.0122 (3) | 0.60590 (12) | 0.0280 (4) | |
| H7 | −0.0819 | −0.1243 | 0.5864 | 0.034* | |
| C8 | −0.04426 (14) | 0.0588 (3) | 0.68951 (11) | 0.0258 (4) | |
| H8 | −0.0490 | −0.0694 | 0.7206 | 0.031* | |
| C9 | −0.12081 (16) | 0.2311 (3) | 0.70832 (12) | 0.0285 (4) | |
| H9 | −0.1212 | 0.2589 | 0.7633 | 0.034* | |
| C10 | −0.11491 (16) | 0.4179 (3) | 0.65832 (12) | 0.0300 (4) | |
| H10 | −0.1116 | 0.5579 | 0.6832 | 0.036* | |
| C11 | −0.21970 (16) | 0.2988 (4) | 0.65312 (14) | 0.0348 (5) | |
| H11A | −0.2427 | 0.2114 | 0.6076 | 0.042* | |
| H11B | −0.2797 | 0.3652 | 0.6741 | 0.042* | |
| C12 | 0.31028 (14) | 0.0235 (3) | 0.61886 (10) | 0.0209 (4) | |
| C13 | 0.40447 (15) | 0.1405 (3) | 0.63508 (11) | 0.0262 (4) | |
| H13 | 0.4030 | 0.2752 | 0.6569 | 0.031* | |
| C14 | 0.50106 (16) | 0.0594 (3) | 0.61918 (11) | 0.0296 (4) | |
| H14 | 0.5662 | 0.1383 | 0.6299 | 0.035* | |
| C15 | 0.50157 (15) | −0.1369 (3) | 0.58771 (10) | 0.0254 (4) | |
| C16 | 0.40808 (18) | −0.2541 (3) | 0.57108 (11) | 0.0284 (4) | |
| H16 | 0.4098 | −0.3887 | 0.5491 | 0.034* | |
| C17 | 0.31120 (15) | −0.1728 (3) | 0.58690 (11) | 0.0253 (4) | |
| H17 | 0.2461 | −0.2516 | 0.5758 | 0.030* |
4-(4-Bromophenyl)-4-azatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (II) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.03088 (14) | 0.04690 (17) | 0.03609 (15) | 0.01410 (9) | 0.01146 (10) | −0.00083 (10) |
| O1 | 0.0294 (7) | 0.0422 (9) | 0.0382 (8) | 0.0087 (6) | 0.0107 (6) | 0.0211 (7) |
| O2 | 0.0312 (7) | 0.0263 (7) | 0.0371 (8) | 0.0002 (6) | 0.0131 (6) | 0.0095 (6) |
| N1 | 0.0175 (7) | 0.0233 (8) | 0.0205 (7) | −0.0015 (6) | 0.0031 (6) | 0.0013 (6) |
| C1 | 0.0184 (8) | 0.0322 (10) | 0.0197 (8) | −0.0015 (7) | 0.0008 (7) | 0.0046 (8) |
| C2 | 0.0201 (8) | 0.0194 (8) | 0.0252 (9) | −0.0045 (7) | 0.0019 (7) | −0.0007 (7) |
| C3 | 0.0197 (8) | 0.0319 (10) | 0.0184 (8) | −0.0016 (7) | 0.0034 (7) | 0.0010 (8) |
| C4 | 0.0221 (9) | 0.0213 (9) | 0.0257 (9) | −0.0033 (7) | 0.0049 (7) | −0.0033 (7) |
| C5 | 0.0235 (9) | 0.0243 (9) | 0.0295 (9) | 0.0043 (7) | 0.0049 (7) | 0.0054 (8) |
| C6 | 0.0211 (9) | 0.0397 (11) | 0.0239 (9) | 0.0038 (8) | −0.0010 (7) | −0.0059 (9) |
| C7 | 0.0180 (8) | 0.0260 (10) | 0.0394 (11) | −0.0015 (7) | 0.0028 (8) | −0.0097 (9) |
| C8 | 0.0191 (9) | 0.0276 (10) | 0.0315 (10) | −0.0018 (7) | 0.0063 (7) | 0.0049 (8) |
| C9 | 0.0210 (9) | 0.0346 (11) | 0.0312 (10) | −0.0008 (8) | 0.0083 (8) | −0.0033 (8) |
| C10 | 0.0241 (9) | 0.0272 (10) | 0.0400 (11) | 0.0023 (8) | 0.0093 (8) | −0.0043 (9) |
| C11 | 0.0203 (10) | 0.0382 (11) | 0.0465 (12) | 0.0036 (9) | 0.0074 (9) | −0.0021 (10) |
| C12 | 0.0201 (8) | 0.0247 (9) | 0.0180 (8) | 0.0000 (7) | 0.0037 (7) | 0.0022 (7) |
| C13 | 0.0248 (9) | 0.0266 (10) | 0.0284 (9) | −0.0035 (8) | 0.0080 (7) | −0.0061 (8) |
| C14 | 0.0215 (9) | 0.0362 (11) | 0.0322 (10) | −0.0042 (8) | 0.0082 (8) | −0.0057 (9) |
| C15 | 0.0245 (9) | 0.0315 (10) | 0.0213 (8) | 0.0084 (8) | 0.0071 (7) | 0.0025 (8) |
| C16 | 0.0358 (11) | 0.0239 (10) | 0.0253 (9) | 0.0045 (8) | 0.0042 (8) | −0.0017 (8) |
| C17 | 0.0244 (9) | 0.0246 (9) | 0.0259 (9) | −0.0023 (7) | 0.0011 (7) | 0.0013 (8) |
4-(4-Bromophenyl)-4-azatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (II) . Geometric parameters (Å, º)
| Br1—C15 | 1.9004 (18) | C8—H8 | 1.0000 |
| O1—C1 | 1.210 (2) | C8—C9 | 1.536 (3) |
| O2—C2 | 1.209 (2) | C9—H9 | 1.0000 |
| N1—C1 | 1.393 (2) | C9—C10 | 1.504 (3) |
| N1—C2 | 1.397 (2) | C9—C11 | 1.508 (3) |
| N1—C12 | 1.432 (2) | C10—H10 | 1.0000 |
| C1—C3 | 1.508 (3) | C10—C11 | 1.506 (3) |
| C2—C4 | 1.511 (2) | C11—H11A | 0.9900 |
| C3—H3 | 1.0000 | C11—H11B | 0.9900 |
| C3—C4 | 1.539 (3) | C12—C13 | 1.384 (3) |
| C3—C8 | 1.564 (2) | C12—C17 | 1.383 (3) |
| C4—H4 | 1.0000 | C13—H13 | 0.9500 |
| C4—C5 | 1.557 (3) | C13—C14 | 1.388 (3) |
| C5—H5 | 1.0000 | C14—H14 | 0.9500 |
| C5—C6 | 1.499 (3) | C14—C15 | 1.379 (3) |
| C5—C10 | 1.533 (3) | C15—C16 | 1.379 (3) |
| C6—H6 | 0.9500 | C16—H16 | 0.9500 |
| C6—C7 | 1.324 (3) | C16—C17 | 1.391 (3) |
| C7—H7 | 0.9500 | C17—H17 | 0.9500 |
| C7—C8 | 1.503 (3) | ||
| C1—N1—C2 | 112.82 (15) | C9—C8—H8 | 111.8 |
| C1—N1—C12 | 122.65 (15) | C8—C9—H9 | 116.8 |
| C2—N1—C12 | 124.12 (15) | C10—C9—C8 | 110.33 (16) |
| O1—C1—N1 | 123.98 (17) | C10—C9—H9 | 116.8 |
| O1—C1—C3 | 127.52 (17) | C10—C9—C11 | 59.97 (14) |
| N1—C1—C3 | 108.50 (15) | C11—C9—C8 | 122.38 (18) |
| O2—C2—N1 | 123.97 (17) | C11—C9—H9 | 116.8 |
| O2—C2—C4 | 127.59 (17) | C5—C10—H10 | 116.9 |
| N1—C2—C4 | 108.43 (15) | C9—C10—C5 | 110.92 (16) |
| C1—C3—H3 | 109.6 | C9—C10—H10 | 116.9 |
| C1—C3—C4 | 105.22 (14) | C9—C10—C11 | 60.14 (14) |
| C1—C3—C8 | 113.26 (16) | C11—C10—C5 | 121.61 (18) |
| C4—C3—H3 | 109.6 | C11—C10—H10 | 116.9 |
| C4—C3—C8 | 109.59 (15) | C9—C11—H11A | 117.8 |
| C8—C3—H3 | 109.6 | C9—C11—H11B | 117.8 |
| C2—C4—C3 | 104.86 (15) | C10—C11—C9 | 59.89 (13) |
| C2—C4—H4 | 109.9 | C10—C11—H11A | 117.8 |
| C2—C4—C5 | 112.66 (15) | C10—C11—H11B | 117.8 |
| C3—C4—H4 | 109.9 | H11A—C11—H11B | 114.9 |
| C3—C4—C5 | 109.55 (14) | C13—C12—N1 | 118.82 (16) |
| C5—C4—H4 | 109.9 | C17—C12—N1 | 120.27 (16) |
| C4—C5—H5 | 111.8 | C17—C12—C13 | 120.88 (17) |
| C6—C5—C4 | 106.45 (15) | C12—C13—H13 | 120.2 |
| C6—C5—H5 | 111.8 | C12—C13—C14 | 119.50 (18) |
| C6—C5—C10 | 110.37 (16) | C14—C13—H13 | 120.2 |
| C10—C5—C4 | 104.30 (15) | C13—C14—H14 | 120.3 |
| C10—C5—H5 | 111.8 | C15—C14—C13 | 119.34 (18) |
| C5—C6—H6 | 122.4 | C15—C14—H14 | 120.3 |
| C7—C6—C5 | 115.13 (17) | C14—C15—Br1 | 118.97 (15) |
| C7—C6—H6 | 122.4 | C16—C15—Br1 | 119.49 (15) |
| C6—C7—H7 | 122.7 | C16—C15—C14 | 121.54 (18) |
| C6—C7—C8 | 114.59 (18) | C15—C16—H16 | 120.4 |
| C8—C7—H7 | 122.7 | C15—C16—C17 | 119.14 (18) |
| C3—C8—H8 | 111.8 | C17—C16—H16 | 120.4 |
| C7—C8—C3 | 107.32 (15) | C12—C17—C16 | 119.60 (18) |
| C7—C8—H8 | 111.8 | C12—C17—H17 | 120.2 |
| C7—C8—C9 | 110.18 (16) | C16—C17—H17 | 120.2 |
| C9—C8—C3 | 103.63 (15) |
4-(4-Bromophenyl)-4-azatetracyclo[5.3.2.02,6.08,10]dodec-11-ene-3,5-dione (II) . Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the C12–C17 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C17—H17···O2i | 0.95 | 2.40 | 3.175 (2) | 139 |
| C3—H3···Cg1ii | 1.00 | 2.83 | 3.801 (2) | 165 |
Symmetry codes: (i) x, y−1, z; (ii) −x+1/2, y+1/2, −z+3/2.
Funding Statement
This work was funded by National Science Foundation grants MRI CHE-1725699, MRI CHE-1919817, and MRI CHE-1919565.
References
- Alder, K. & Jacobs, G. (1953). Chem. Ber. 86, 1528–1539.
- Bailey, T. R., Rippin, S. R., Opsitnick, E., Burns, C. J., Pevear, D. C., Collett, M. S., Rhodes, G., Tohan, S., Huggins, J. W., Baker, R. O., Kern, E. R., Keith, K. A., Dai, D., Yang, G., Hruby, D. & Jordan, R. (2007). J. Med. Chem. 50, 1442–1444. [DOI] [PMC free article] [PubMed]
- Birney, D., Lim, T. K., Koh, J. H. P., Pool, B. R. & White, J. M. (2002). J. Am. Chem. Soc. 124, 5091–5099. [DOI] [PubMed]
- Bourhis, L. J., Dolomanov, O. V., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2015). Acta Cryst. A71, 59–75. [DOI] [PMC free article] [PubMed]
- Brechbuhler, H. U. & Petitpierre, C. (1975). US Patent 3,993,662.
- Bruker (2013). APEX2, SAINT and SADABS. Bruker AXS Inc. Madison, Wisconsin, USA.
- Coxon, J. M., O’Connell, M. J. & Steel, P. J. (1986). Acta Cryst. C42, 1773–1777.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hughes, D. L. (2019). Org. Process Res. Dev. 23, 1298–1307.
- Kohler, E. P., Tishler, M., Potter, H. & Thompson, H. (1939). J. Am. Chem. Soc. 61, 1057–1061.
- Mooibroek, T. J., Gamez, P. & Reedijk, J. (2008). CrystEngComm, 10, 1501–1515.
- Palmer, D. (2007). CrystalMaker. CrystalMaker Software, Bicester, Oxfordshire, England.
- Pool, B. R. & White, J. M. (2000). Org. Lett. 2, 3505–3507. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Song, X.-Z., Qin, C., Guan, W., Song, S.-Y. & Zhang, H.-J. (2012). New J. Chem. 36, 877–882.
- Steiner, T. (1996). Crystallogr. Rev. 6, 1–51.
- Sutor, D. J. (1962). Nature, 195, 68–69.
- Sutor, D. J. (1963). J. Chem. Soc. pp. 1105–1110.
- White, J. M. & Goh, R. Y. W. (2014). Private Communication (refcode HOKRIK). CCDC, Cambridge, England.
- Wu, X., Huang, J., Guo, B., Zhao, L., Liu, Y., Chen, J. & Cao, W. (2014). Adv. Synth. Catal. 356, 3377–3382.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, II, I. DOI: 10.1107/S2056989020009512/hb7931sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020009512/hb7931Isup3.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989020009512/hb7931IIsup4.hkl
Supporting information file. DOI: 10.1107/S2056989020009512/hb7931Isup4.cml
Supporting information file. DOI: 10.1107/S2056989020009512/hb7931IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report