In the title compound, the dihedral angle between the fused pyrazole and pyridine rings is 1.76 (7)°. The benzene and methoxy phenyl rings make dihedral angles of 44.8 (5) and 63.86 (5)°, respectively, with the pyrazolo[3,4-b] pyridine moiety. An intramolecular short S⋯O contact [3.215 (2) Å] is observed. The crystal packing features C—H⋯π interactions.
Keywords: crystal structure; pyrazolopyridine; pyrazolo[3,4-b]pyridine; C—H⋯π interactions
Abstract
In the title compound, C24H23N3O3S, the dihedral angle between the fused pyrazole and pyridine rings is 1.76 (7)°. The benzene and methoxy phenyl rings make dihedral angles of 44.8 (5) and 63.86 (5)°, respectively, with the pyrazolo[3,4-b] pyridine moiety. An intramolecular short S⋯O contact [3.215 (2) Å] is observed. The crystal packing features C—H⋯π interactions.
Chemical context
Pyrazolopyridines, in which a group of three nitrogen atoms is incorporated into a bicyclic heterocycle, are privileged medicinal scaffolds, often utilized in drug design and discovery regimes (Kumar et al., 2019 ▸). Owing to the possibilities of the easy synthesis of a literally unlimited number of a combinatorial library of small organic molecules with a pyrazolopyridine scaffold, there has been enormous interest in these molecules among medicinal chemists (Kumar et al. 2019 ▸; Pinheiro et al. 2019 ▸; Hardy, 1984 ▸). Indeed, molecules with pyrazolopyridine in the core structure exhibit multifaceted medicinal properties such as anti-microbial, anti-viral, anti-fungal, anti-hypertensive, analgesic, antiquorum-sensing, anti-cancer, anti-inflammatory, anti-Alzheimer’s, anti-diabetic, anti-nociceptive, anti-tuberculosis and anti-leishmanial activities (Hardy, 1984 ▸; Hawas et al., 2019 ▸; de Mello et al., 2004 ▸; El-Gohary et al., 2019 ▸; El-Gohary & Shaaban, 2018 ▸). Moreover, pyrazolopyridine-derived drug molecules exhibit anti-cancer properties (Huang et al., 2007 ▸; Ye et al., 2009 ▸). They are inhibitors of several important proteins, namely cycline-dependent kinase1, HIV reverse transcriptase, leucine zipper kinase, protein kinase, xanthine oxidase, interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), phosphodiesterase-4, NAD(P)H oxidases (Kumar et al., 2019 ▸; Gökhan-Kelekçi et al., 2007 ▸; Fathy et al., 2015 ▸; Park et al., 2017 ▸). A recent study reported that they could be promising inhibitors against the enzyme pantothenate synthetase from Mycobacterium tuberculosis (Amaroju et al., 2017 ▸). FDA-approved drugs incorporating the pyrazolopyridine scaffold include cartazolate, tracazolate and etazolate (Hawas et al., 2019 ▸). Among pyrazolopyridines, pyrazolo[3,4-b]pyridines are medicinally important because of the ease of combinatorial library synthesis, adherence to the Lipinski rule and favourable ADMET properties. (Chauhan & Kumar, 2013 ▸; Zhai et al., 2019 ▸). Based on the importance of pyrazolo[3,4-b]pyridine-containing molecules, we have undertaken a single-crystal X-ray diffraction study of the title compound. We have recently analyzed the solid-state structure of a pyrazolo[3,4-b]pyridine-containing molecule, ethyl 3-(4-chlorophenyl)-1,6-dimethyl-4-methylsulfanyl-1H-pyrazolo[3,4-b]pyridine-5-carboxylate (NUDWOB; Rao et al. 2020 ▸), but the title compound exhibits a very different conformational structure of the substituents and supramolecular structure, as discussed here.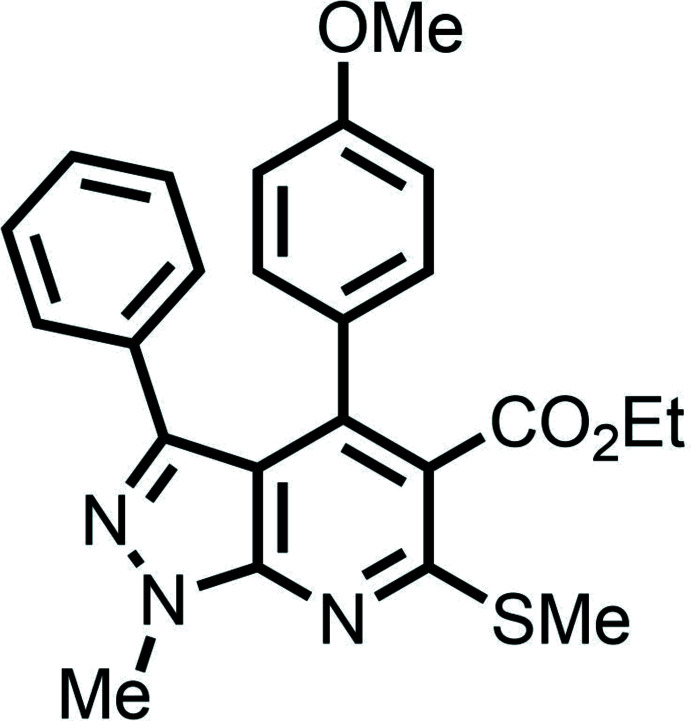
Structural commentary
In the title compound (Fig. 1 ▸), the phenyl (–C6H5) group attached to the pyrazolopyridine moiety exhibits a (+)anti-periplanar conformation [C12—C4—N1—N2 = 178.47 (14)°] whereas the methoxyphenyl (H3COC6H4–) group exhibits a (−)anti-clinal conformation [C3—C5—C18—C23 = −114.30 (19)°]. The thiomethyl (–SCH3) group fused to the pyrazolopyridine unit exhibits a (+)anti-periplanar conformation [C8—S1—C7—C6 = 175.47 (15)°]. The torsion angles involving the –SCH3 group differ from those for NUDWOB (Rao et al. 2020 ▸) because of the presence of the methoxyphenyl (H3COC6H4–) group. The –COOC2H5 group attached to the pyrazolopyridine moiety has a (+)anti-periplanar conformation [N3—C7—C6—C9 = 176.44 (16)°]. Further, the methyl group attached to the pyrazole subunit is (+)anti-periplanar [C1—N2—N1—C4 = 178.78 (17)°] and it is attached to the pyridine ring showing a (+)syn-periplanar conformation [C1—N2—C2—N3 = 1.3 (3)°]. The fused pyrazole and pyridine rings in the title compound are not exactly planar, as in NUDWOB (Rao et al. 2020 ▸), subtending a dihedral angle of 1.76 (7)°. The dihedral angle between the planes of the benzene and pyrazolo[3,4-b]pyridine rings is 44.8 (5)° and that between the methoxyphenyl and pyrazolo[3,4-b]pyridine rings is 63.86 (5)°.
Figure 1.
The molecular structure of the title compound with the atom-numbering scheme and displacement ellipsoids drawn at the 50% probability level. The short intramolecular S⋯O interaction is indicated by a dashed line.
Supramolecular features
The cohesion of the crystal packing is influenced by two weak C—H⋯π (C13—H13⋯Cg and C23—H23⋯Cg) interactions (Table 1 ▸, Fig. 2 ▸) with distances of 2.86 and 3.02 Å, respectively. These distances agree with those described by Nishio (2011 ▸). A short intermolecular C⋯O carbon=bonding stabilizing interaction [C1⋯O1(x + 1, y, z) = 3.291 (2) Å] may also exist (Fig. 2 ▸) between the electrophilic carbon atom of the methyl group connected to the electronegative nitrogen atom of the pyrazolo ring and the nucleophilic oxygen atom of the ester group of a neighbouring molecule. However, the distance between C1 and O1 is marginally higher than the carbon-bonding distances (less than 3.22 Å, the sum of the van der Waals radii of carbon and oxygen atoms) proposed by Guru Row and co-workers (Thomas et al., 2014 ▸). The distances between the methyl hydrogen atoms and the acceptor oxygen atom are C—H1A⋯O1 = 3.06, C—H1B⋯O1 = 3.04 and C—H1C⋯O1 = 3.22 Å, much longer than the hydrogen-bonding interactions (C—H⋯O = 2.90, 2.84 and 2.86 Å) noted by Thomas et al. (2014 ▸). Based on these observations, in addition to the C—H⋯π interactions, the short intermolecular C⋯O carbon-bonding interaction may also contribute to the cohesion of the title compound in the solid state.
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the N1/N2/C2–C4 and the N3/C2/C3/C5–C7 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C13—H13⋯Cg1i | 0.93 | 2.85 | 3.455 (2) | 124 |
| C23—H23⋯Cg2ii | 0.93 | 3.02 | 3.738 (2) | 136 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 2.
A view of the weak intermolecular C13—H13⋯Cg(N1/N2/C2–C4), C23—H13⋯Cg(N3/C2/C3/C5–C7) and C1⋯O1 interactions in the title compound.
Database survey
A similarity search of the Cambridge Structural Database (CSD, Version 5.40, update of March 2020; Groom et al., 2016 ▸) was performed. The title compound, along with related structures obtained from the database search could be used for further structure-based virtual screening, ligand-based virtual screening, pharmacophore-based virtual screening and drug repurposing against various drug target proteins. The molecules showing strong binding affinity towards drug target proteins might be considered potential lead candidates. In this study, the CSD search found five molecules that are similar to title compound, namely FIZLEI (ethyl 2,7-diamino-3,4-dicyano-5-phenylpyrazolo[1,5-a]pyridine-6-carboxylate; Naik et al., 2019 ▸), ALAFID [7-(2-methoxyphenyl)-2-phenylpyrazolo[1,5-a]pyridine; Wu et al., 2016 ▸], DAWKAQ [2-(4-chlorophenyl)pyrazolo[1,5-a]pyridin-3-yl(phenyl)methanone; Ravi et al., 2017 ▸], NADPIU [3-(4-chlorophenyl)pyrazolo[1,5-a]pyridine; Wu et al., 2016 ▸] and NUDWOB [ethyl 3-(4-chlorophenyl)-1,6-dimethyl-4-(methylsulfanyl)-1H-pyrazolo[3,4-b]pyridine-5-carboxylate; Rao et al., 2020 ▸]. The geometrical parameters of the –COOCH2CH3 substituent in the title compound are comparable with those reported for FIZLEI and NUDWOB. The bond distances for the thiomethyl and aryl moieties in the title compound are comparable with those of NUDWOB. Moreover, the bond lengths in the pyrazolo[3,4-b]pyridine unit of the title compound are comparable with those in NUDWOB, FIZLEI, ALAFID, DAWKAQ and NADPIU. The pyrazolo[3,4-b]pyridine moiety (N1–N3/C2–C4/C5–C7) of the title compound is approximately planar, as is also observed for NUDWOB, FIZLEI, ALAFID, DAWKAQ and NADPIU. In the title compound, a short intramolecular S⋯O contact of 3.215 (2) Å occurs, but this is not observed in FIZLEI, ALAFID, DAWKAQ, NADPIU and NUDWOB. Moreover, the interaction distance [3.291 (2) Å] of the short intermolecular C⋯O contact in the title compound is comparable with the C⋯O interaction [3.424 (2) Å] in the structure of NUDWOB. Furthermore, as in the title compound, C—H⋯π interactions are observed in the crystal structures of ALAFID, DAWKAQ and NADPIU. The interaction distance of these related structures ranges from 2.89 to 3.23 Å, comparable with the C—H⋯π interactions observed in the title compound.
Synthesis and crystallization
To a solution of 1-methyl-3-phenyl-1H-pyrazol-5-amine (100 mg, 0.57 mmol) and ethyl 2-(4-methoxybenzoyl)-3,3-bis(methylthio)acrylate (188 mg, 0.57 mmol) in toluene (3 mL), a catalytic amount of TFA (trifluoroacetic acid) 30 mol % in toluene (3 mL) was added under an N2 atmosphere. The reaction mixture was refluxed for 24 h in an oil bath, the progress of the reaction being monitored by TLC using a mixture of hexane and ethyl acetate (9.9:0.1). After completion of the reaction, the mixture was loaded on a silica gel column (100–200 mesh, 15 cm × 1 cm) and eluted with increasing amounts of ethyl acetate in hexanes (1% to 5%) to obtain 186 mg (yield = 75%) of ethyl 6-(4-methoxyphenyl)-1-methyl-4-(methylthio)-3-phenyl-1H-pyrazolo[3,4-b]pyridine-5-carboxylate as a colourless crystalline solid; m.p. 406 K; R f = 0.3 cm (hexane: ethyl acetate 9.9:0.1). A sample suitable for single-crystal X-ray analysis was obtained by recrystallization from 2 mL of dry methanol.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The hydrogen atoms were placed in calculated positions, with C—H = 0.93–0.97 Å and included in the final cycles of refinement using a riding model with U iso(H) = 1.2U eq(C) or 1.5U eq(C-methyl).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C24H23N3O3S |
| M r | 433.51 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 298 |
| a, b, c (Å) | 10.1911 (4), 10.7274 (6), 11.7692 (6) |
| α, β, γ (°) | 98.550 (5), 105.632 (4), 111.015 (4) |
| V (Å3) | 1112.75 (10) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.18 |
| Crystal size (mm) | 0.75 × 0.44 × 0.42 |
| Data collection | |
| Diffractometer | Agilent Xcalibur, Eos |
| Absorption correction | Multi-scan (CrysAlis PRO; Agilent, 2014 ▸) |
| T min, T max | 0.959, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 13297, 5161, 3672 |
| R int | 0.026 |
| (sin θ/λ)max (Å−1) | 0.686 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.048, 0.168, 1.06 |
| No. of reflections | 5161 |
| No. of parameters | 284 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.20, −0.26 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989020008841/dx2029sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020008841/dx2029Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989020008841/dx2029Isup3.cml
CCDC reference: 2005976
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
HSPRao thanks DST–FIST Single Crystal XRD facility at Department of Chemistry, Pondicherry University for the diffraction data. RG thanks the Department of Chemistry for facilities, and the UGC and CSIR for a fellowship. JM thanks Dr Clara Gomes (FCT–UNL, Portugal) for the CSD database survey and Dr Amit Kumar Singh (Sharda University, India) for support.
supplementary crystallographic information
Crystal data
| C24H23N3O3S | Z = 2 |
| Mr = 433.51 | F(000) = 456 |
| Triclinic, P1 | Dx = 1.294 Mg m−3 |
| a = 10.1911 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.7274 (6) Å | Cell parameters from 3586 reflections |
| c = 11.7692 (6) Å | θ = 3.8–29.1° |
| α = 98.550 (5)° | µ = 0.18 mm−1 |
| β = 105.632 (4)° | T = 298 K |
| γ = 111.015 (4)° | Block, colorless |
| V = 1112.75 (10) Å3 | 0.75 × 0.44 × 0.42 mm |
Data collection
| Agilent Xcalibur, Eos diffractometer | 5161 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 3672 reflections with I > 2σ(I) |
| Detector resolution: 15.9821 pixels mm-1 | Rint = 0.026 |
| ω scans | θmax = 29.2°, θmin = 3.8° |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2014) | h = −13→12 |
| Tmin = 0.959, Tmax = 1.000 | k = −13→14 |
| 13297 measured reflections | l = −15→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.048 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.168 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.1P)2] where P = (Fo2 + 2Fc2)/3 |
| 5161 reflections | (Δ/σ)max < 0.001 |
| 284 parameters | Δρmax = 0.20 e Å−3 |
| 0 restraints | Δρmin = −0.26 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.67085 (6) | 0.56012 (6) | 0.41095 (5) | 0.0647 (2) | |
| O2 | 0.38290 (15) | 0.56954 (14) | 0.22189 (15) | 0.0618 (4) | |
| N3 | 0.72006 (14) | 0.40967 (14) | 0.23884 (14) | 0.0456 (4) | |
| O3 | −0.16778 (14) | 0.10375 (17) | −0.27340 (14) | 0.0716 (4) | |
| O1 | 0.26375 (16) | 0.36781 (15) | 0.25921 (15) | 0.0716 (4) | |
| N2 | 0.74888 (15) | 0.26571 (16) | 0.08007 (15) | 0.0507 (4) | |
| N1 | 0.66463 (15) | 0.17775 (16) | −0.03455 (15) | 0.0524 (4) | |
| C6 | 0.46748 (17) | 0.39796 (17) | 0.18528 (16) | 0.0426 (4) | |
| C18 | 0.25937 (17) | 0.25136 (17) | −0.01408 (15) | 0.0403 (4) | |
| C5 | 0.41691 (17) | 0.30623 (16) | 0.07038 (15) | 0.0397 (4) | |
| C23 | 0.2031 (2) | 0.33918 (19) | −0.06467 (18) | 0.0509 (4) | |
| H23 | 0.262169 | 0.434277 | −0.040467 | 0.061* | |
| C19 | 0.16737 (17) | 0.11066 (18) | −0.05037 (16) | 0.0450 (4) | |
| H19 | 0.203616 | 0.050794 | −0.017572 | 0.054* | |
| C4 | 0.52914 (18) | 0.17844 (18) | −0.06206 (17) | 0.0440 (4) | |
| C3 | 0.52331 (17) | 0.26645 (16) | 0.03830 (16) | 0.0403 (4) | |
| C12 | 0.41458 (18) | 0.09398 (18) | −0.18366 (16) | 0.0443 (4) | |
| C21 | −0.02998 (18) | 0.1451 (2) | −0.18636 (17) | 0.0507 (4) | |
| C20 | 0.02258 (18) | 0.05683 (19) | −0.13439 (16) | 0.0491 (4) | |
| H20 | −0.038626 | −0.037581 | −0.155630 | 0.059* | |
| C7 | 0.61907 (18) | 0.44751 (18) | 0.26597 (17) | 0.0451 (4) | |
| C17 | 0.3250 (2) | 0.1467 (2) | −0.25396 (17) | 0.0518 (4) | |
| H17 | 0.335347 | 0.236158 | −0.223765 | 0.062* | |
| C22 | 0.0606 (2) | 0.2860 (2) | −0.15032 (19) | 0.0575 (5) | |
| H22 | 0.024819 | 0.345484 | −0.184333 | 0.069* | |
| C1 | 0.9027 (2) | 0.2863 (3) | 0.1384 (2) | 0.0737 (6) | |
| H1A | 0.954270 | 0.370449 | 0.204079 | 0.110* | |
| H1B | 0.952102 | 0.292919 | 0.079218 | 0.110* | |
| H1C | 0.903503 | 0.209297 | 0.170462 | 0.110* | |
| C2 | 0.66831 (17) | 0.32140 (18) | 0.12734 (17) | 0.0435 (4) | |
| C16 | 0.2213 (2) | 0.0669 (2) | −0.36804 (19) | 0.0632 (5) | |
| H16 | 0.161631 | 0.102596 | −0.414423 | 0.076* | |
| C13 | 0.3980 (2) | −0.0386 (2) | −0.23167 (19) | 0.0583 (5) | |
| H13 | 0.458611 | −0.074552 | −0.186802 | 0.070* | |
| C9 | 0.35959 (19) | 0.44074 (19) | 0.22766 (17) | 0.0477 (4) | |
| C10 | 0.2937 (3) | 0.6327 (2) | 0.2651 (3) | 0.0740 (7) | |
| H10A | 0.202428 | 0.560492 | 0.264182 | 0.089* | |
| H10B | 0.265806 | 0.685559 | 0.210502 | 0.089* | |
| C15 | 0.2054 (3) | −0.0656 (2) | −0.4137 (2) | 0.0716 (6) | |
| H15 | 0.135537 | −0.119091 | −0.490858 | 0.086* | |
| C8 | 0.8584 (2) | 0.5829 (3) | 0.4825 (2) | 0.0821 (7) | |
| H8A | 0.859059 | 0.495191 | 0.489817 | 0.123* | |
| H8B | 0.900377 | 0.646868 | 0.562541 | 0.123* | |
| H8C | 0.917043 | 0.619009 | 0.433710 | 0.123* | |
| C11 | 0.3801 (3) | 0.7253 (3) | 0.3911 (3) | 0.0868 (8) | |
| H11A | 0.402829 | 0.671847 | 0.445827 | 0.130* | |
| H11B | 0.321725 | 0.769022 | 0.416900 | 0.130* | |
| H11C | 0.471812 | 0.795051 | 0.392304 | 0.130* | |
| C24 | −0.2684 (2) | −0.0390 (3) | −0.3089 (3) | 0.0881 (8) | |
| H24A | −0.228284 | −0.093559 | −0.348676 | 0.132* | |
| H24B | −0.363754 | −0.052474 | −0.364364 | 0.132* | |
| H24C | −0.280979 | −0.067455 | −0.237465 | 0.132* | |
| C14 | 0.2926 (3) | −0.1182 (2) | −0.3453 (2) | 0.0756 (7) | |
| H14 | 0.280866 | −0.208232 | −0.375564 | 0.091* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0602 (3) | 0.0687 (4) | 0.0507 (3) | 0.0333 (3) | −0.0005 (2) | −0.0037 (3) |
| O2 | 0.0566 (7) | 0.0531 (8) | 0.0840 (11) | 0.0294 (6) | 0.0309 (7) | 0.0137 (7) |
| N3 | 0.0379 (7) | 0.0429 (8) | 0.0482 (9) | 0.0157 (6) | 0.0063 (6) | 0.0105 (7) |
| O3 | 0.0436 (7) | 0.0890 (11) | 0.0611 (10) | 0.0256 (7) | −0.0054 (7) | 0.0109 (8) |
| O1 | 0.0659 (9) | 0.0703 (10) | 0.0955 (12) | 0.0307 (7) | 0.0459 (9) | 0.0313 (9) |
| N2 | 0.0348 (7) | 0.0594 (9) | 0.0538 (10) | 0.0211 (6) | 0.0110 (7) | 0.0093 (8) |
| N1 | 0.0436 (8) | 0.0610 (10) | 0.0527 (10) | 0.0239 (7) | 0.0166 (7) | 0.0112 (8) |
| C6 | 0.0380 (8) | 0.0409 (9) | 0.0459 (10) | 0.0173 (7) | 0.0096 (7) | 0.0109 (8) |
| C18 | 0.0347 (8) | 0.0456 (9) | 0.0397 (9) | 0.0177 (7) | 0.0101 (7) | 0.0123 (7) |
| C5 | 0.0366 (8) | 0.0395 (8) | 0.0419 (9) | 0.0163 (6) | 0.0099 (7) | 0.0144 (7) |
| C23 | 0.0478 (9) | 0.0459 (10) | 0.0541 (11) | 0.0218 (8) | 0.0078 (8) | 0.0129 (8) |
| C19 | 0.0405 (9) | 0.0471 (9) | 0.0474 (10) | 0.0181 (7) | 0.0137 (8) | 0.0167 (8) |
| C4 | 0.0400 (8) | 0.0466 (9) | 0.0459 (10) | 0.0178 (7) | 0.0155 (7) | 0.0141 (8) |
| C3 | 0.0359 (8) | 0.0391 (8) | 0.0440 (10) | 0.0155 (6) | 0.0101 (7) | 0.0137 (7) |
| C12 | 0.0414 (8) | 0.0489 (10) | 0.0404 (10) | 0.0150 (7) | 0.0174 (7) | 0.0098 (8) |
| C21 | 0.0370 (9) | 0.0682 (12) | 0.0419 (10) | 0.0220 (8) | 0.0081 (8) | 0.0120 (9) |
| C20 | 0.0388 (9) | 0.0520 (10) | 0.0486 (11) | 0.0124 (7) | 0.0131 (8) | 0.0130 (8) |
| C7 | 0.0411 (8) | 0.0406 (9) | 0.0452 (10) | 0.0154 (7) | 0.0059 (7) | 0.0097 (7) |
| C17 | 0.0512 (10) | 0.0547 (11) | 0.0459 (11) | 0.0210 (8) | 0.0145 (8) | 0.0111 (9) |
| C22 | 0.0532 (10) | 0.0623 (12) | 0.0574 (12) | 0.0329 (9) | 0.0073 (9) | 0.0182 (10) |
| C1 | 0.0404 (10) | 0.0916 (16) | 0.0792 (16) | 0.0328 (10) | 0.0088 (10) | 0.0052 (13) |
| C2 | 0.0370 (8) | 0.0442 (9) | 0.0484 (10) | 0.0180 (7) | 0.0112 (7) | 0.0149 (8) |
| C16 | 0.0566 (11) | 0.0755 (14) | 0.0472 (12) | 0.0219 (10) | 0.0108 (9) | 0.0167 (11) |
| C13 | 0.0737 (12) | 0.0529 (11) | 0.0523 (12) | 0.0292 (9) | 0.0241 (10) | 0.0152 (9) |
| C9 | 0.0413 (9) | 0.0508 (10) | 0.0460 (10) | 0.0199 (7) | 0.0098 (8) | 0.0077 (8) |
| C10 | 0.0638 (13) | 0.0697 (14) | 0.1010 (19) | 0.0400 (11) | 0.0353 (13) | 0.0147 (13) |
| C15 | 0.0773 (14) | 0.0637 (14) | 0.0447 (12) | 0.0073 (11) | 0.0135 (11) | 0.0049 (10) |
| C8 | 0.0566 (12) | 0.0955 (18) | 0.0603 (15) | 0.0251 (12) | −0.0075 (11) | −0.0043 (13) |
| C11 | 0.1061 (19) | 0.0984 (19) | 0.0812 (19) | 0.0613 (16) | 0.0479 (16) | 0.0204 (16) |
| C24 | 0.0442 (11) | 0.1039 (19) | 0.0718 (17) | 0.0056 (11) | −0.0057 (11) | 0.0133 (15) |
| C14 | 0.1098 (18) | 0.0521 (12) | 0.0520 (13) | 0.0261 (12) | 0.0237 (13) | 0.0066 (10) |
Geometric parameters (Å, º)
| S1—C7 | 1.7590 (19) | C12—C17 | 1.392 (3) |
| S1—C8 | 1.777 (2) | C21—C22 | 1.383 (3) |
| O2—C9 | 1.331 (2) | C21—C20 | 1.385 (3) |
| O2—C10 | 1.460 (2) | C20—H20 | 0.9300 |
| N3—C7 | 1.328 (2) | C17—C16 | 1.378 (3) |
| N3—C2 | 1.340 (2) | C17—H17 | 0.9300 |
| O3—C21 | 1.364 (2) | C22—H22 | 0.9300 |
| O3—C24 | 1.423 (3) | C1—H1A | 0.9600 |
| O1—O1 | 0.000 (4) | C1—H1B | 0.9600 |
| O1—C9 | 1.199 (2) | C1—H1C | 0.9600 |
| N2—C2 | 1.356 (2) | C16—C15 | 1.377 (3) |
| N2—N1 | 1.365 (2) | C16—H16 | 0.9300 |
| N2—C1 | 1.450 (2) | C13—C14 | 1.378 (3) |
| N1—C4 | 1.334 (2) | C13—H13 | 0.9300 |
| C6—C5 | 1.388 (2) | C10—C11 | 1.481 (3) |
| C6—C7 | 1.429 (2) | C10—H10A | 0.9700 |
| C6—C9 | 1.503 (2) | C10—H10B | 0.9700 |
| C18—C19 | 1.385 (2) | C15—C14 | 1.366 (4) |
| C18—C23 | 1.393 (2) | C15—H15 | 0.9300 |
| C18—C5 | 1.483 (2) | C8—H8A | 0.9600 |
| C5—C3 | 1.416 (2) | C8—H8B | 0.9600 |
| C23—C22 | 1.377 (3) | C8—H8C | 0.9600 |
| C23—H23 | 0.9300 | C11—H11A | 0.9600 |
| C19—C20 | 1.386 (2) | C11—H11B | 0.9600 |
| C19—H19 | 0.9300 | C11—H11C | 0.9600 |
| C4—C3 | 1.427 (2) | C24—H24A | 0.9600 |
| C4—C12 | 1.479 (2) | C24—H24B | 0.9600 |
| C3—C2 | 1.409 (2) | C24—H24C | 0.9600 |
| C12—C13 | 1.381 (3) | C14—H14 | 0.9300 |
| C7—S1—C8 | 102.08 (11) | H1A—C1—H1B | 109.5 |
| C9—O2—C10 | 118.64 (16) | N2—C1—H1C | 109.5 |
| C7—N3—C2 | 114.13 (14) | H1A—C1—H1C | 109.5 |
| C21—O3—C24 | 117.99 (17) | H1B—C1—H1C | 109.5 |
| C2—N2—N1 | 111.38 (13) | N3—C2—N2 | 125.09 (15) |
| C2—N2—C1 | 127.81 (17) | N3—C2—C3 | 128.10 (15) |
| N1—N2—C1 | 120.77 (16) | N2—C2—C3 | 106.80 (15) |
| C4—N1—N2 | 106.91 (14) | C15—C16—C17 | 120.3 (2) |
| C5—C6—C7 | 120.93 (15) | C15—C16—H16 | 119.9 |
| C5—C6—C9 | 119.54 (14) | C17—C16—H16 | 119.9 |
| C7—C6—C9 | 119.47 (16) | C14—C13—C12 | 120.77 (19) |
| C19—C18—C23 | 118.28 (15) | C14—C13—H13 | 119.6 |
| C19—C18—C5 | 120.91 (14) | C12—C13—H13 | 119.6 |
| C23—C18—C5 | 120.70 (15) | O1—C9—O2 | 124.91 (16) |
| C6—C5—C3 | 116.49 (14) | O1—C9—O2 | 124.91 (16) |
| C6—C5—C18 | 121.92 (14) | O1—C9—C6 | 124.73 (17) |
| C3—C5—C18 | 121.59 (15) | O1—C9—C6 | 124.73 (17) |
| C22—C23—C18 | 120.38 (17) | O2—C9—C6 | 110.33 (15) |
| C22—C23—H23 | 119.8 | O2—C10—C11 | 110.43 (19) |
| C18—C23—H23 | 119.8 | O2—C10—H10A | 109.6 |
| C18—C19—C20 | 121.62 (15) | C11—C10—H10A | 109.6 |
| C18—C19—H19 | 119.2 | O2—C10—H10B | 109.6 |
| C20—C19—H19 | 119.2 | C11—C10—H10B | 109.6 |
| N1—C4—C3 | 110.06 (15) | H10A—C10—H10B | 108.1 |
| N1—C4—C12 | 118.69 (16) | C14—C15—C16 | 119.8 (2) |
| C3—C4—C12 | 131.25 (14) | C14—C15—H15 | 120.1 |
| C2—C3—C5 | 116.67 (15) | C16—C15—H15 | 120.1 |
| C2—C3—C4 | 104.82 (14) | S1—C8—H8A | 109.5 |
| C5—C3—C4 | 138.47 (15) | S1—C8—H8B | 109.5 |
| C13—C12—C17 | 118.49 (17) | H8A—C8—H8B | 109.5 |
| C13—C12—C4 | 120.04 (16) | S1—C8—H8C | 109.5 |
| C17—C12—C4 | 121.44 (16) | H8A—C8—H8C | 109.5 |
| O3—C21—C22 | 115.77 (17) | H8B—C8—H8C | 109.5 |
| O3—C21—C20 | 124.66 (17) | C10—C11—H11A | 109.5 |
| C22—C21—C20 | 119.56 (16) | C10—C11—H11B | 109.5 |
| C21—C20—C19 | 119.30 (17) | H11A—C11—H11B | 109.5 |
| C21—C20—H20 | 120.4 | C10—C11—H11C | 109.5 |
| C19—C20—H20 | 120.4 | H11A—C11—H11C | 109.5 |
| N3—C7—C6 | 123.66 (17) | H11B—C11—H11C | 109.5 |
| N3—C7—S1 | 118.89 (13) | O3—C24—H24A | 109.5 |
| C6—C7—S1 | 117.42 (13) | O3—C24—H24B | 109.5 |
| C16—C17—C12 | 120.30 (19) | H24A—C24—H24B | 109.5 |
| C16—C17—H17 | 119.9 | O3—C24—H24C | 109.5 |
| C12—C17—H17 | 119.9 | H24A—C24—H24C | 109.5 |
| C23—C22—C21 | 120.80 (17) | H24B—C24—H24C | 109.5 |
| C23—C22—H22 | 119.6 | C15—C14—C13 | 120.3 (2) |
| C21—C22—H22 | 119.6 | C15—C14—H14 | 119.8 |
| N2—C1—H1A | 109.5 | C13—C14—H14 | 119.8 |
| N2—C1—H1B | 109.5 | ||
| C2—N2—N1—C4 | 0.92 (19) | C5—C6—C7—S1 | −178.50 (13) |
| C1—N2—N1—C4 | 178.78 (17) | C9—C6—C7—S1 | −1.3 (2) |
| C7—C6—C5—C3 | −0.5 (2) | C8—S1—C7—N3 | −2.36 (18) |
| C9—C6—C5—C3 | −177.72 (15) | C8—S1—C7—C6 | 175.47 (15) |
| C7—C6—C5—C18 | 178.87 (14) | C13—C12—C17—C16 | 0.8 (3) |
| C9—C6—C5—C18 | 1.6 (2) | C4—C12—C17—C16 | 178.66 (16) |
| C19—C18—C5—C6 | −117.54 (18) | C18—C23—C22—C21 | 0.9 (3) |
| C23—C18—C5—C6 | 66.4 (2) | O3—C21—C22—C23 | −178.79 (18) |
| C19—C18—C5—C3 | 61.8 (2) | C20—C21—C22—C23 | 0.9 (3) |
| C23—C18—C5—C3 | −114.30 (19) | C7—N3—C2—N2 | −178.44 (16) |
| C19—C18—C23—C22 | −1.3 (3) | C7—N3—C2—C3 | 0.3 (2) |
| C5—C18—C23—C22 | 174.93 (17) | N1—N2—C2—N3 | 178.93 (15) |
| C23—C18—C19—C20 | −0.2 (3) | C1—N2—C2—N3 | 1.3 (3) |
| C5—C18—C19—C20 | −176.43 (16) | N1—N2—C2—C3 | 0.00 (19) |
| N2—N1—C4—C3 | −1.47 (18) | C1—N2—C2—C3 | −177.67 (18) |
| N2—N1—C4—C12 | 178.47 (14) | C5—C3—C2—N3 | −1.5 (3) |
| C6—C5—C3—C2 | 1.5 (2) | C4—C3—C2—N3 | −179.75 (16) |
| C18—C5—C3—C2 | −177.89 (14) | C5—C3—C2—N2 | 177.41 (14) |
| C6—C5—C3—C4 | 178.94 (18) | C4—C3—C2—N2 | −0.86 (17) |
| C18—C5—C3—C4 | −0.4 (3) | C12—C17—C16—C15 | −0.2 (3) |
| N1—C4—C3—C2 | 1.46 (18) | C17—C12—C13—C14 | −1.6 (3) |
| C12—C4—C3—C2 | −178.47 (17) | C4—C12—C13—C14 | −179.45 (18) |
| N1—C4—C3—C5 | −176.19 (19) | O1—O1—C9—O2 | 0.00 (13) |
| C12—C4—C3—C5 | 3.9 (3) | O1—O1—C9—C6 | 0.00 (9) |
| N1—C4—C12—C13 | 42.4 (2) | C10—O2—C9—O1 | 4.7 (3) |
| C3—C4—C12—C13 | −137.69 (19) | C10—O2—C9—O1 | 4.7 (3) |
| N1—C4—C12—C17 | −135.43 (18) | C10—O2—C9—C6 | −177.05 (17) |
| C3—C4—C12—C17 | 44.5 (3) | C5—C6—C9—O1 | 73.3 (2) |
| C24—O3—C21—C22 | −176.7 (2) | C7—C6—C9—O1 | −104.0 (2) |
| C24—O3—C21—C20 | 3.6 (3) | C5—C6—C9—O1 | 73.3 (2) |
| O3—C21—C20—C19 | 177.29 (17) | C7—C6—C9—O1 | −104.0 (2) |
| C22—C21—C20—C19 | −2.4 (3) | C5—C6—C9—O2 | −104.97 (18) |
| C18—C19—C20—C21 | 2.1 (3) | C7—C6—C9—O2 | 77.8 (2) |
| C2—N3—C7—C6 | 0.9 (2) | C9—O2—C10—C11 | 99.1 (2) |
| C2—N3—C7—S1 | 178.58 (12) | C17—C16—C15—C14 | 0.3 (3) |
| C5—C6—C7—N3 | −0.8 (3) | C16—C15—C14—C13 | −1.0 (4) |
| C9—C6—C7—N3 | 176.44 (16) | C12—C13—C14—C15 | 1.7 (3) |
Hydrogen-bond geometry (Å, º)
Cg1 and Cg2 are the centroids of the N1/N2/C2–C4 and the N3/C2/C3/C5–C7 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C13—H13···Cg1i | 0.93 | 2.85 | 3.455 (2) | 124 |
| C23—H23···Cg2ii | 0.93 | 3.02 | 3.738 (2) | 136 |
Symmetry codes: (i) −x+1, −y, −z; (ii) −x+1, −y+1, −z.
References
- Agilent (2014). CrysAlis PRO. Agilent Technologies Ltd, Yarnton, England.
- Amaroju, S., Kalaga, M. N., Srinivasarao, S., Napiórkowska, A., Augustynowicz-Kopeć, E., Murugesan, S., Chander, S., Krishnan, R. & Chandra Sekhar, K. V. G. (2017). New J. Chem. 41, 347–357.
- Chauhan, M. & Kumar, R. (2013). Bioorg. Med. Chem. 21, 5657–5668. [DOI] [PubMed]
- El-Gohary, N. S., Gabr, M. T. & Shaaban, M. I. (2019). Bioorg. Chem. 89, 1–13. [DOI] [PubMed]
- El-Gohary, N. S. & Shaaban, M. I. (2018). Eur. J. Med. Chem. 152, 126–136. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Fathy, U., Younis, A. & Awad, H. M. (2015). J. Chem. Pharm. Res. 7, 4–12.
- Gökhan-Kelekçi, N., Yabanoğlu, S., Küpeli, E., Salgin, U., Ozgen, O., Uçar, G., Yeşilada, E., Kendi, E., Yeşilada, A. & Bilgin, A. A. A. (2007). Bioorg. Med. Chem. 15, 5775–5786. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hardy, C. R. (1984). In Advances in Heterocyclic Chemistry, Vol. 36, pp. 343–409. New York: Academic Press.
- Hawas, S. S., El-Gohary, N. S., Gabr, M. T., Shaaban, M. I. & El-Ashmawy, M. B. (2019). Synth. Commun. 49, 2466–2487.
- Huang, S., Lin, R., Yu, Y., Lu, Y., Connolly, P. J., Chiu, G., Li, S., Emanuel, S. L. & Middleton, S. A. (2007). Bioorg. Med. Chem. Lett. 17, 1243–1245. [DOI] [PubMed]
- Kumar, S. V., Muthusubramanian, S. & Perumal, S. (2019). Org. Prep. Proced. Int. 51, 1–89.
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Mello, H. de, Echevarria, A., Bernardino, A. M., Canto-Cavalheiro, M. & Leon, L. L. (2004). J. Med. Chem. 47, 5427–5432. [DOI] [PubMed]
- Naik, N. S., Shastri, L. A., Shastri, S. L., Chougala, B. M., Shaikh, F., Madar, J. M., Kulkarni, R. C., Dodamani, S., Jalalpure, S., Joshi, S. D. & Sunagar, V. (2019). Chemistry Select, 4, 285–297.
- Nishio, M. (2011). Phys. Chem. Chem. Phys. 13, 13878–13900.
- Park, C. M., Jadhav, V. B., Song, J. H., Lee, S., Won, H. Y., Choi, S. U. & Son, Y. H. (2017). Bull. Korean Chem. Soc. 38, 595–602.
- Pinheiro, L. C. S., Feitosa, L. M., Gandi, M. O., Silveira, F. F. & Boechat, N. (2019). Molecules, 24, 4095.
- Rao, H. S. P., Gunasundari, R. & Muthukumaran, J. (2020). Acta Cryst. E76, 443–445. [DOI] [PMC free article] [PubMed]
- Ravi, C., Samanta, S., Mohan, D. C., Reddy, N. N. K. & Adimurthy, S. (2017). Synthesis, 49, 2513–2522.
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Thomas, S. P., Pavan, M. S. & Guru Row, T. N. (2014). Chem. Commun. 50, 49–51. [DOI] [PubMed]
- Wu, H.-C., Chu, J.-H., Li, C.-W., Hwang, L.-C. & Wu, M.-J. (2016). Organometallics, 35, 288–300.
- Ye, Q., Cao, J., Zhou, X., Lv, D., He, Q., Yang, B. & Hu, Y. (2009). Bioorg. Med. Chem. 17, 4763–4772. [DOI] [PubMed]
- Zhai, M., Liu, S., Gao, M., Wang, L., Sun, J., Du, J., Guan, Q., Bao, K., Zuo, D., Wu, Y. & Zhang, W. (2019). Eur. J. Med. Chem. 168, 426–435. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989020008841/dx2029sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020008841/dx2029Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989020008841/dx2029Isup3.cml
CCDC reference: 2005976
Additional supporting information: crystallographic information; 3D view; checkCIF report




