Abstract
Extracellular vesicles (EV) are abundant, lipid-enclosed vectors that contain nucleic acids and proteins, they can be secreted from donor cells and freely circulate, and they can be engulfed by recipient cells thus enabling systemic communication between heterotypic cell types. However, genetic tools for labeling, isolating, and auditing cell type-specific EVs in vivo, without prior in vitro manipulation, are lacking. We have used CRISPR-Cas9-mediated genome editing to generate mice bearing a CD63-emGFPloxP/stop/loxP knock-in cassette that enables the specific labeling of circulating CD63+ vesicles from any cell type when crossed with lineage-specific Cre recombinase driver mice. As proof-of-principle, we have crossed these mice with Cdh5-CreERT2 mice to generate CD63emGFP+ vasculature. Using these mice we show that developing vasculature is marked with emerald GFP (emGFP) following tamoxifen administration to pregnant females. In adult mice, quiescent vasculature and angiogenic vasculature (in tumors) is also marked with emGFP. Moreover, whole plasma-purified EVs contain a subpopulation of emGFP+ vesicles that are derived from the endothelium, co-express additional EV (e.g. CD9 and CD81) and endothelial cell (e.g. CD105) markers, and they harbor specific miRNAs (e.g. miR-126, miR-30c, and miR-125b). This new mouse strain should be a useful genetic tool for generating cell type-specific, CD63+ EVs that freely circulate in serum and can subsequently be isolated and characterized using standard methodologies.
Keywords: exosomes, extracellular vesicles, endothelial lineage tracing, tumor microenvironment, cancer, angiogenesis
Introduction
EVs are abundant 30-1000 nm membrane-bound particles found in virtually all biological fluids (Kalra, Drummen, & Mathivanan, 2016; Zhang et al., 2015). They are further classified as exosomes (shed from multi-vesicular bodies, MVBs) or microvesicles (MVs, shed from plasma membranes) (Kalra et al., 2016; Thery, Zitvogel, & Amigorena, 2002). Due to their high concentration in the circulation and ability to systemically transfer protein and nucleic acid cargoes between different tissue and organ micro-environments, EVs are suggested to play important roles in diverse physiological and pathophysiological states; especially cancer (Mathieu, Martin-Jaular, Lavieu, & Thery, 2019).
In cancer, EVs have been shown to promote tumor progression, direct organotropic metastasis, and suppress tumor immune surveillance (Bobrie & Thery, 2013). Circulating EVs can also be probed for their RNA cargo which provides diagnostic information to predict, for example, tumor recurrence and progression-free survival (Peinado et al., 2012; Schwarzenbach, Nishida, Calin, & Pantel, 2014). Cancer cell-derived EVs, enriched in proteins, lipids, and nucleic acids “re-educate” different cellular recipients to promote tumor growth and invasion through diverse mechanisms (Fong et al., 2015; Hoshino et al., 2015; Suetsugu et al., 2013; Zhou et al., 2014; Zomer et al., 2015). However, since the cargo contained within EVs mirrors the cell of origin, and tumors are comprised of mixtures of heterogeneous cell types (e.g. cancer cells, immune cells, fibroblasts, and endothelial cells, ECs), it has been challenging to assign specific functions to EVs derived from different cell types or to audit the composition of individual circulating EVs in the cancer setting.
To meet this challenge, we have taken advantage of the selective tropism and eviction of fusion proteins between emGFP and the tetraspanin CD63 into EVs to generate a new knock-in mouse. These mice (CD63-emGFPloxP/stop/loxP mice), after crossing with a lineage-specific Cre driver mouse, enable the genetic labeling, isolation, and characterization of cell type-specific EVs in vivo. As proof-of-principle, we have crossed CD63-emGFPloxP/stop/loxP mice with Cdh5-CreERT2 mice to generate CD63emGFP+ vasculature in developing, adult, and pathophysiological tissues. The compound mice evict CD63emGFP+ fusion proteins into circulating EVs that can be captured and assayed using multiple standard methodologies.
Methods
Construction of Rosa26 donor vector
The repair cassette was constructed as a donor vector for homologous recombination into the safe harbor locus Rosa26. Briefly, a CD63-emerald GFP (emGFP) fusion protein was cloned into the EcoRI site of a modified Rosa26 targeting vector by isothermal assembly (Gibson, 2011). The EcoRI-linearized intermediate donor vector was purified by gel extraction over a silica column using Buffer QG (Cat Nr. 19063, Qiagen, Hilden, Germany). The CD63emGFP was amplified by PCR using a proofreading DNA polymerase and purified over a silica column using three volumes of Buffer PB (Cat Nr. 19066, Qiagen, Hilden, Germany). The final donor vector was maxi-prepped (HiSpeed Plasmid Maxi Kit, Cat Nr. 12663, Qiagen, Hilden, Germany), dialyzed in microinjection buffer (5mM Tris pH 7.4, 0.1mM EDTA) and sequenced to verify correct assembly. An in silico map and vector sequence is provided in the data supplement. Emerald GFP was derived from Aequorea victoria GFP with the following engineered mutations: S72A/N149K/M153T/I167T.
Genome engineering with CRISPR-Cas9
CRISPR Reagents:
A codon-optimized, His-tagged Cas9 was produced in expression host E. coli BL21(DE3) and purified by IMAC followed by size exclusion chromatography by the University of North Carolina at Chapel Hill Protein Expression and Purification core. Cas9 truncated guide RNAs were subcloned by an oligo annealing strategy, sequenced for verification, produced by in vitro transcription (HiScribe™ T7 Quick High Yield RNA Synthesis Kit, Cat Nr. E20250S, New England Biolabs, Ipswich, MA) and purified over a silica column (RNeasy Mini Kit, Cat Nr. 74106, Qiagen, Hilden, Germany). The use of truncated guides can reduce off-target mutagenesis (Fu, Sander, Reyon, Cascio, & Joung, 2014). Primers to subclone R26-sg75B protospacer 5’-GGAGTTGCAGATCACGA-3’ are described in the data supplement.
Microinjection:
Guide RNAs were heat denatured at 95°C for three minutes and then immediately cooled on ice because the heating process may decrease hairpins which could lead to low or inactive gRNA structures (Thyme, Akhmetova, Montague, Valen, & Schier, 2016). Cas9 ribonucleoproteins (RNPs) were produced by incubating 400 nM Cas9 protein with 25 ng/μl R26-sg75B truncated guide RNA for 5 mins at 37°C. Twenty ng/μl supercoiled double-stranded donor vector was added and the mixture was injected into the pronucleus of C57BL/6J embryos produced by standard superovulation and mating protocols.
Southern blotting
Samples for the isolation of genomic DNA from 0.5 cm tail snips were prepared. Briefly, tail snips were enzymatically digested with 0.5 mg/ml proteinase K with activity greater than 600 mAU/ml in a tail lysis buffer containing SDS detergent. A saturated solution of sodium chloride was added to a final concentration of 25% v/v to salt out protein and then centrifuged for 15 min at 10,000x rcf. The genomic DNA remained in the supernatant and was transferred to a new tube for isopropanol precipitation, then washed in 70% ethanol to desalt. The DNA sample was resuspended in 150 μl TE with low EDTA (10 mM Tris, 0.1 mM EDTA, pH 8.0) and allowed to solubilize overnight. Typically, 8-10 micrograms of genomic DNA was digested at 37°C overnight with 50 U of restriction endonuclease, namely EcoRI-HF or MfeI-HF (Cat. Nrs. R3101L and R3589L, New England Biolabs, Ispswich, MA). To ensure complete digestion of the genomic DNA, a pilot gel containing 0.7% agarose and TAE was run at 50 V/cm to visualize EcoRI-HF/MfeI-HFcut genomic DNA that appeared as a smear with high-molecular banding. In cases where digestion was incomplete, the DNA was resuspended in 3 volumes of Buffer PB (Cat Nr. 19066, Qiagen, Hilden, Germany) purified over a silica-column, washed with WS Buffer twice (Cat. Nr. B404-400, Epoch Life Science, Missouri City, Texas) and eluted in TE with low 0.1 mM EDTA. The incompletely digested DNA was incubated 37°C overnight with 2-3 μl containing 10-15 units of EcoRI-HF or MfeI-HF in a 50 μl reaction to completion. A 15 x 10 cm Southern gel was cast with 0.7% TAE agarose and electrophoresed overnight at 20 V/cm to achieve high resolution and efficient separation. The Southern gel was processed as follows: depurination in 0.1 M HCl for 10 min, denaturation twice in 0.1 M NaOH for 15 min, neutralization twice with 100 mM Tris, 100 mM NaCl pH 7.5 for 15 min, and transferred to a wicking apparatus using 1x SSC to a 0.45 μm microporous nylon 66 membrane on a polyester support, carrying positively charged quaternary ammonium groups (Cat. Nr. 11417240001, Roche, Indianapolis, Indiana). A 20x SSC buffer contains 3M NaCl in 0.3M sodium citrate (pH 7.0). After approximately 16-24 h of transfer, the wicking apparatus was deconstructed and the transferred DNA was cross-linked to the nylon membrane with 1200 joules of UV light for 90 s. The nylon membrane was blocked by an initial pre-hybridization step with 30-50 ml of DIG Easy Hyb Granules (Cat. Nr. 1179689001, Roche, Indianapolis, Indiana) for 30 minutes according to the manufacturer’s recommendation. DIG-UTP labeled probes were denatured five min at 70°C and then immediately applied to pre-blocked nylon membrane in a rotisserie oven and hybridization continued overnight with constant agitation. A low and high-stringency wash was performed at 65°C (0.5x SSC low-stringency, then 0.5x SSC; 0.1% SDS high-stringency) for 30 min. Membrane blocking and immunological detection used the DIG Wash and Block Buffer Set (Cat Nr. 11585762001, Roche, Indianapolis, Indiana) per the manufacturer’s recommendations with 1:10,000 dilution of alkaline phosphatase-conjugated anti-DIG antibody and with 1:100 dilution of chemiluminescent substrate CDP-Star (Anti-digoxigenin-AP conjugate, Fab frag, Cat Nr. 11093274910; CDP-Star, Cat. Nr 11759051001, Roche, Indianapolis, Indiana). Southern blotting probes were labeled by PCR with a dNTP mix containing 200 μM dATP, dGTP, dCTP; 130 μM dTTP and 14-70 μM alkali-labile digoxigenin-dUTP according to the manufacturer’s recommendations (PCR DIG Probe Synthesis Kit, Cat Nr. 11636090910, Roche, Indianapolis, Indiana). Primers for probe synthesis are described in the data supplement.
Genotyping of founders
Toe samples were used for genotyping and prepared by the HotShot DNA method.
In vitro studies using HEK293T cells
HEK293T cells were transfected with CD63emGFP or empty emGFP plasmid (addgene #53976) using Lipofectamine 3000 and grown in HG-DMEM with 1% exosome-free FBS (Systems Biosciences, LLC). Cells were grown for 48 hrs and then EVs/exosomes were isolated from the media using high speed centrifugation. EVs were then lysed in RIPA buffer for WB analysis, resuspended in PBS for Zetaview analysis, or resuspended in HG-DMEM with 1% exosome-free FBS for EV transfer experiments. For transfer experiments, EVs were incubated with unlabeled cells for 24 hrs and then imaged using fluorescence microscopy or analyzed using FACS.
Tamoxifen induction of Cre recombinase expression
Vascular-specific induction of Cre recombinase was carried out as we described previously (McCann, Xiao, et al., 2019). Pregnant mice received two consecutive daily intraperitoneal injections of tamoxifen from embryonic day E13.5. Next, E15.5 embryos were harvested, fixed, and 40μm frozen sections were prepared. Images were taken using a ZEISS LSM 980 Confocal Laser Scanning Microscope with 20x objective.
Cryosections and immunohistochemistry
Heart, liver, brain, mammary tumors, and other tissues were resected and placed in 4% PFA in PBS for 24 hours at 4°C. Tumors were then transferred to 30% sucrose in PBS overnight. Tumors or tissues were embedded in optimal cutting temperature compound (OCT) for sectioning. Slides were fixed in ice cold methanol for 20 minutes at −20°C and then rinsed three times with PBS. Blocking buffer (PBS + 5% BSA + 5% goat serum) was added for 30 minutes at room temperature. The primary antibody was then diluted in blocking buffer and added to the slides. Rat anti-mouse CD31 (BD Bioscience Cat#550274) was used at 1:100 dilution. Slides were then incubated overnight at 4°C and then washed three times with PBS. Goat anti-rat cy3 secondary antibody was diluted 1:200 in blocking buffer and incubated at room temperature for one hr. The slides were washed three times with PBS and then stained with DAPI. Coverslips were then mounted to the slide using VectaShield Mounting Media Hard Set.
EV isolation (in vitro)
HEK293T cells were grown as described above. Conditioned Media was spun at 250 g for 5 minutes, 2,000 g for 10 minutes, and then 100,000 g for 1.5 hrs. All centrifugation steps were performed at 4C in a Beckman Coulter Optima LE-80K Ultracentrifuge with SW-41TI rotor in Beckman Coulter thin wall, polypropylene centrifuge tubes. The EV pellet was resuspended in Dulbecco’s phosphate buffered saline (DPBS) and stored at −80C until characterization or immediately lysed with RIPA buffer. Media only was also centrifuged and run as a control in all experiments.
EV isolation (in vivo)
Blood was harvested from mice via cardiac puncture and spun at 4,000 g for 10 minutes at room temperature to isolate platelet-free plasma. Plasma was carefully removed and spun again at 4,000 g to ensure no cell or platelet contamination. The remaining platelet-free plasma was diluted in 10mL of DPBS and spun at 100,000 g for 1.5 hrs at 4C.
ZetaView analysis
Brightfield and fluorescent particles were quantified using Zetaview NTA. Conditioned medium and plasma-derived EVs were diluted in water (1:250 to 1:1000 for media samples and 1:250 to 1:5 for plasma samples). EVs were measured both in bright field (total) and using the 488 nm filter. Samples were read alongside buffer only controls to ensure no EV contamination from buffers or other reagents.
FACS
FACS analysis of isolated EVs from cell culture or plasma was performed on a BD Influx Cell Sorter. Buffer only was run prior to each sample to measure background and to ensure there was no contamination between samples. Samples were run with 70 μM nozzle at 33 psi with a drop frequency of 72. EmGFP was detected with a band-path filter of 540/40 with a threshold of 0.65.
ExoView™ EV analysis
ExoView mouse custom kit (Nanoview Biosciences, Cat. No. EV-CUST-5) was generated with minor modifications using anti-CD9 (Clone MZ3), anti-CD81 (Clone Eat2), anti-CD45 (Clone 30F11), anti-CD105 (Clone MJ7/18), streptavidin (Sigma, Cat. No. S0677) and isotype control antibodies. Anti-CD63 antibody was biotinylated according to manufacturer’s instructions using Pierce Antibody Biotinylation Kit for IP (Thermo Scientific, Cat. No. 90407). Degree of biotinylation was assessed to be less than an average of four molecules per antibody using Pierce Biotin Quantification Kit (Thermo Scientific, Cat. No. 28005). To immobilize biotinylated anti-CD63 onto the streptavidin spots on the chip, 35 μL of antibody (0.2 mg/mL) was incubated on the chip for 16 h at 4°C. Chips were washed three times with Solution A for three mins in an orbital shaker to remove unbound antibody. The chips were subsequently rinse using filtered DI water, dried and stored at 4°C for subsequent use. Purified and unpurified mouse plasma samples were individually incubated on the custom chips placed in a sealed 24-well plate for 16 hrs at room temperature. The chips were then washed three times in solution A for three min each on an orbital shaker, and subsequently incubated with a cocktail of fluorescently labeled antibodies: anti-CD105 CF647 and anti-CD45 CF555, or anti-CD63 CF647 and anti-CD105 CF555. Antibody incubation was for one hr at room temperature with mild shaking. The chips were then washed three times in solution A for three mins each on an orbital shaker, twice in Solution B, and rinsed once in DI water. After drying, chip imaging was carried out using the ExoView R100 reader (software nScan v2.8.7.3) with enabled fluorescence acquisition. The data were then analyzed using nanoViewer v2.8.8. All samples analyzed by nanoview were normalized to a total area of 150 μM to avoid including antibody binding bias in our analysis.
Results and discussion
Generation and characterization of a CD63emGFP construct
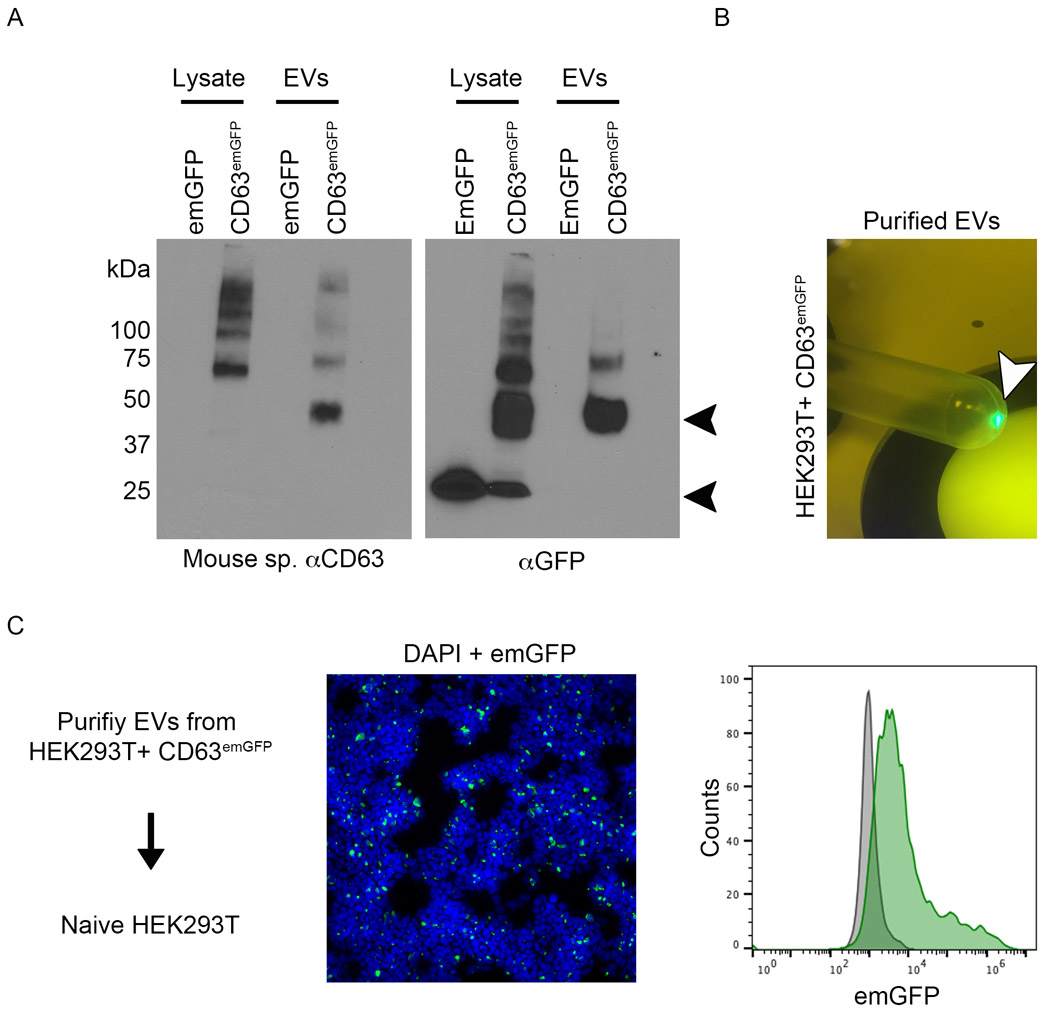
Previously, it was shown that fusions between CD63 and fluorescent or pH sensitive optical reporters are secreted from transfected cells via EVs and this has enabled pulse-chase type experiments to examine the loading of fluorophore-tagged EVs into unlabeled cell types (Verweij et al., 2018). Using a similar approach, we cloned full-length mouse CD63 as an N-terminal, in frame-fusion with emGFP (emGFP was used because several point mutations in the coding regions prevent aggregation/dimerization resulting in a GFP monomer that is less likely to impact the function of the fusion protein product). We transfected HEK293T cells with emGFP empty vector or CD63emGFP and then performed western blotting using an antibody raised against murine CD63 or GFP. Using anti-CD63 antibody, the results show a migrating band at ~ 50 kDa corresponding to the expected molecular weight of the fusion protein in the EV fraction (fig 1, A). The slightly higher MW bands in the lysate are likely glycosylation products since CD63 is well-known be post-translationally modified. With anti-GFP antibody, we detect a clear 50 kDa band in the EV fraction. The ~ 25 kDa band found in the lysate is likely a GFP cleavage product based on its ~ 25 KDa molecular mass which corresponds with cells transfected with emGFP vector alone. Purified EVs were brightly fluorescent under a 488 nm lamp, suggesting that CD63emGFP fusion proteins are in-frame and correctly folded thereby maintaining their fluorescent properties (fig 1, B). Indeed, purified EVs from the conditioned media of CD63emGFP transfectants were engulfed by HEK293T cells resulting in a right-shifted histogram using flow cytometry (fig 1, C). Thus, the construct we have generated has been used as a guide to create a targeting vector to generate the CD63-emGFPl/s/l knock-in mice described herein.
Fig 1. Generation and characterization of a CD63emGFP construct.
(A) Western blotting using a mouse specific CD63 antibody or an anti-GFP antibody in HEK293T lysates or EV fractions. Cells were transfected with emGFP vector only or with a vector expressing full-length murine CD63 cloned in-frame with emGFP. Arrow heads indicate the expected positions of CD63 (upper) and emGFP (lower). (B) Purified EVs from HEK293T transfectants are brightly illuminated under a 488 nm wavelength lamp. (C) Purified EVs from CD63emGFP transfectants are engulfed by naive HEK293T cells as determined using immunofluorescence and flow cytometry. In the immunofluorescence image, nuclei are labeled with 4′,6-diamidino-2-phenylindole (DAPI).
The vasculature in Cdh5-CreERT2:CD63-emGFPl/s/l mice is labeled with emGFP
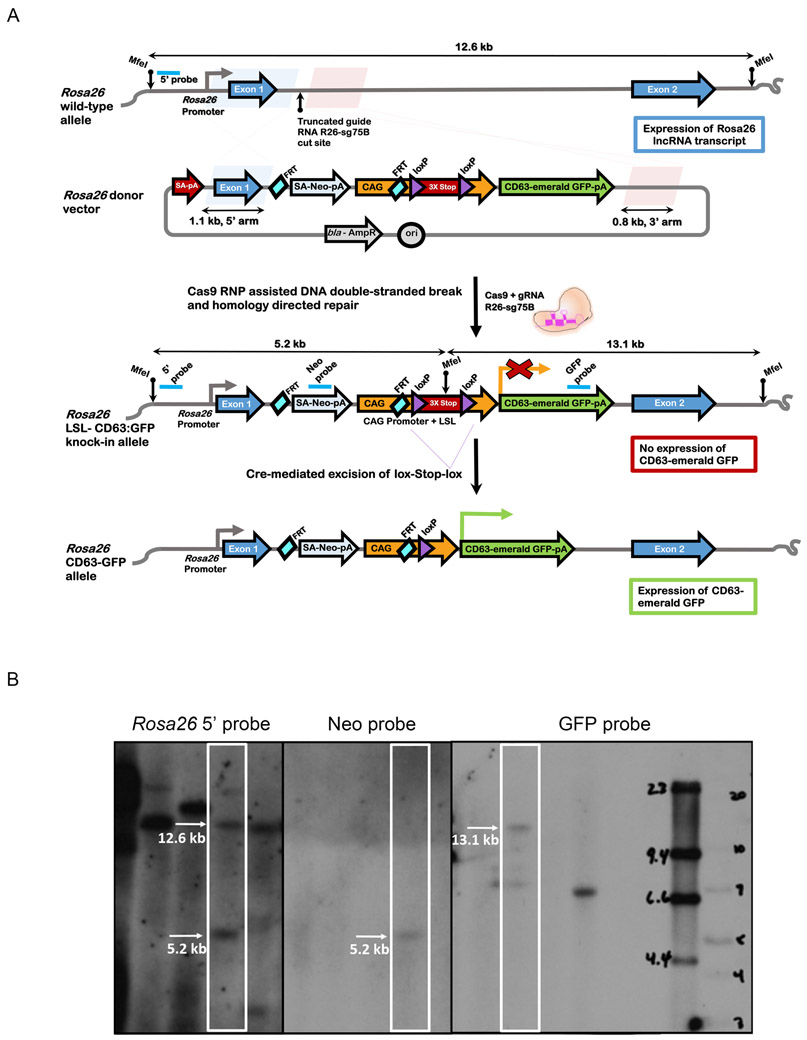
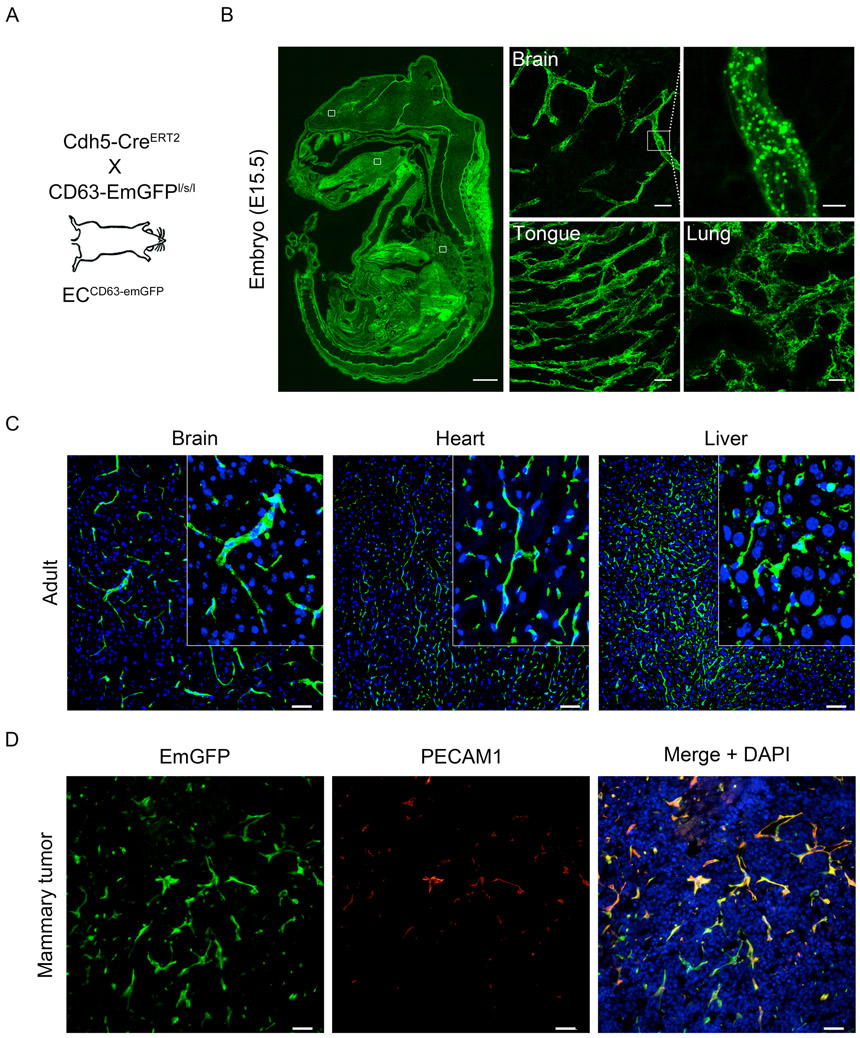
First, we used the CD63emGFP vector described above to clone CD63emGFP into the EcoRI site of a modified Rosa26 targeting vector bearing a LoxP-flanked triple polyA STOP signal (generating CD63-emGFPl/s/l mice). Following genome engineering with CRISPR-Cas9, microinjection of guide RNAs, and Southern blotting to verify creation of the knock-in, we genotyped the founders and expanded the mouse colony for further testing (fig 2, A-B). As proof-of-principle, we crossed CD63-emGFPl/s/l mice with Cdh5-CreERT2 mice (herein called ECCD63-emGFP mice) to enable endothelial cell-specific expression of CD63emGFP (fig 3, A). After tamoxifen administration to pregnant females, we assed the vasculature in various tissues harvested from E15.5 embryos. As expected, the results show efficient labeling of the vasculature in all tissues examined including, for example, brain, tongue, and lung (fig 3, B). These results suggest that it is possible to genetically mark the developing vasculature with the CD63emGFP reporter, thereby enabling one to assess vascular-derived EVs in developing tissues and organs. In adult mice administered tamoxifen at ~ 8 weeks of age, fresh cryosections from brain, heart, and liver also demonstrate the vascular localization and expression of the emGFP reporter (fig 3, C). Next, we orthotopically engrafted EO771 mammary tumors in ECCD63-emGFP mice to test whether angiogenic/pathogenically-generated vasculature also expressed emGFP. Again, the results showed that tumor blood vessels strongly expressed the emGFP fluorophore and that emGFP expression co-localized with the pan endothelial cell marker PECAM1 (fig 3, D). Together, these results suggest that we have successfully generated reporter mice that enable cell type-specific labeling and localization of CD63emGFP in vivo in developing and adult tissues and organs.
Fig 2. Generation of Rosa26LSL-CD63emGFP mice.
(A) Strategy for knock-in of a conditional CD63-emGFP transgene into a genomic safe harbor site of mouse zygotes. Schematic of wild-type Rosa26 locus and donor vector are shown. The donor vector is designed for CD63:emGFP transgene expression driven by the CAG promoter (CAG) after Cre-mediated excision of the floxed triple SV40 polyA sites. To facilitate insertion of CAG-loxPSTOPloxP-CD63:emGFP cassette into the Rosa26 locus by homology directed repair, ribonucleoprotein complexes containing Cas9 and truncated guide R26-sg75B were directly microinjected into the pronucleus of mouse C57BL/6J zygotes to stimulate a double-stranded DNA break between the 1.1 kb and 0.8 kb homology arms in the donor vector. The locations of MfeI restriction sites and the Southern hybridization probes for detection of the wildtype and knock-in alleles are indicated. (B) Southern blotting analysis from MfeI-digested tail genomic DNA of a representative Rosa26LSL-CD63emGFP founder. An external 5’ hybridization probe detects the Rosa26 wildtype and knock-in alleles of 12.6 kb and 5.2 kb respectively, and indicates that the founder contains both the targeted and wildtype alleles. An internal probe that hybridizes to Neomycin resistance gene (Neo) detects the 5’5.2 kb band for the targeted allele. An internal GFP probe detects the 3’ 13.1 kb band from the knock-in allele.
Fig 3. The vasculature in Cdh5-CreERT2:CD63-emGFPl/s/l mice is labeled with GFP.
(A) Schematic summarizing the mouse cross. (B) Blood vessels are labeled in pregnant mice that receive tamoxifen to induce emGFP expression in the developing vasculature of embryonic tissues are organs. The inset at right of the brain section shows emGFP+ puncta that are likely CD63+ endosomes. Scale bar: whole embryo = 1mm, organs = 25 μm, and enlarged brain vessels = 5 μm. (C) Native emGFP fluorescence using fresh cryosections from brain, heart, and liver. The inset shows clear vascular expression. Nuclei are labeled with DAPI. (D) Angiogenic vasculature in EO771 mammary tumors is also emGFP+ and co-localizes with the pan EC marker PECAM1. (D)
EmGFP+ particles are detectable in plasma from ECCD63-emGFP mice and they co-localize with EV tetraspanins
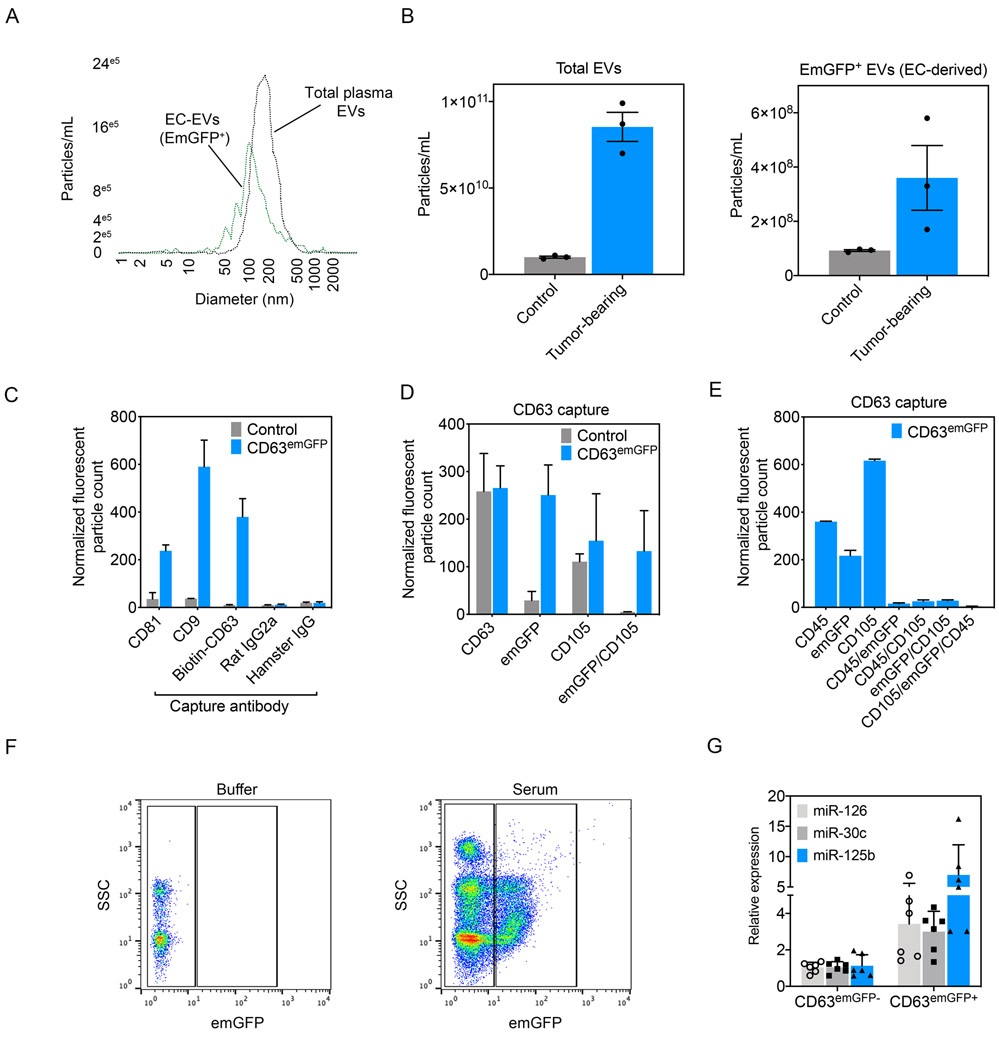
Next, we collected plasma via intracardiac punctures from ECCD63-emGFP mice. Using a Zetaview Nanoparticle Tracking (NTA) system equipped with a 488 nm wavelength laser, we could detect both total plasma EVs (non fluorescent) and vascular-derived EVs (fluorescent) in the circulation (fig 4, A). Total plasma EVs had an ~ median vesicle size of 200.2 +/−148.4 nm whereas the emGFP+ vesicles were slightly left-shifted on the histogram indicating the smaller overall median diameter of 158.8 +/−120.7 nm. Consistent with our previous study, total circulating EVs and vascular-derived EVs were elevated in the circulation of tumor-bearing mice (McCann, Liu, et al., 2019) (fig 4, B). To further characterize EVs using our mouse model, we turned to the Exoview platform which uses arrayed capture antibodies to immunophenotype individual EVs (Daaboul et al., 2016). Mouse plasma-derived EVs from tamoxifen-treated mice produced EVs that were positive for emGFP and they also expressed the tetraspanins and EV markers CD81, CD9, and CD63, suggesting the presence of the fusion protein within bona fide EVs (fig 4, C). As expected, EVs derived from the plasma of control mice were positive for the same tetraspanins, but negative for emGFP. Next, we further characterized CD63+ vesicles from control versus ECCD63-emGFP mice using Exoview and found they had similar number of CD63+ vesicles and CD105+ vesicles (a selective endothelial marker) but only EVs from ECCD63-emGFP mice had emGFP colocalized with CD63 and CD105, suggesting emGFP+ vesicles are derived from ECs (fig 4, D). Given the close developmental relationships between ECs and hematopoietic precursors and the recently reported deletion of the Shb gene in small number of hematopoietic cells using Cdh5-CreERT2 mice, plasma-purified EVs were further characterized for the presence of CD45 (a pan hematopoietic cell marker) (He et al., 2019). CD63 EVs were immobilized and subsequently immunolabeled with anti-CD45 and anti-CD105. The results show that control samples lacked the presence of emGFP+ vesicles, as well as absence of co-localization with emGFP, CD105, and CD45 in individual EVs (data not shown). Conversely, plasma-derived EVs from ECCD63-emGFP mice contained GFP+ vesicles co-localized with CD105; however, no CD105+/CD45+/emGFP+ EVs were detected (fig 4, E). These data suggest that the CD63emGFP+ vesicles observed using Exoview are produced by cells of the EC lineage and not by hematopoietic/immune cells. However, in the present study, we cannot entirely rule out the possibility that a minor fraction of CD63emGFP+ vesicles might emanate from hematopoietic precursors which are then detectable in the circulation. Finally, we used FACS to isolate emGFP+ vesicles from the serum of ECCD63-emGFP mice (fig 4, F). Compared to buffer controls, serum from these mice produced a GFP+ fraction that could be isolated for RNA purification. Focusing on three miRNAs that we recently showed are enriched in endothelial cell-derived EVs, we found a 4-10 fold enrichment in expression of miR-126, miR-30c, and miR-125b in CD63emGFP+ EVs compared to emGFP− counterparts (fig 4, G) (McCann, Liu, et al., 2019; McCann, Xiao, et al., 2019). Taken together, these result suggest that, using CD63emGFP reporter mice, EC-derived EVs can be captured from the serum and then interrogated for their nucleic acid cargoes; especially miRNAs.
Fig 4. EmGFP+ particles are detectable in plasma from ECCD63-emGFP mice and they co-localize with EV tetraspanins.
(A) Zetaview analysis of mouse plasma detects two peaks: one representing total circulating EVs and a smaller, left-shifted peak representing EC-derived, CD63emGFP+ EVs. (B) Quantification of total circulating EVs (emGFP−) versus EC-derived EVs (emGFP+) in control versus tumor-bearing mice, n=3 mice per group. Data are presented as mean +/− SEM. (C) Exoview analysis of mouse plasma isolated from ECCD63-emGFP mice. Capture antibodies are indicated on the x-axis. Data are presented as mean +/− SEM. (D) CD63 antibody capture of EVs using Exoview analysis. (E) CD63 antibody capture using Exoview shows absence of CD45 detection in CD63emGFP+ EVs. (F) Representative plots showing detection and sorting of emGFP particles in serum using FACS in ECCD63-emGFP reporter mice. (G) qPCR analysis of EC-selective miRNAs in the emGFP− (non EC-derived) and emGFP+ (EC-derived) particles.
Summary and perspective
Given their high concentration in the circulation and their ability to protect and deliver nucleic acid cargoes systemically into different organ and tissue microenvironments, EVs have the potential to regulate multiple physiological and pathophysiological processes at the whole-organism level. However, a challenge in the field has been assigning and characterizing the cargoes and functions of EVs that are derived from specific cell types. Underlying this challenge has been the lack of genetic tools to genetically mark EVs in vivo without prior in vitro manipulation (i.e., labeling with a lipophilic dye) followed by re-injection into a host animal. While it is known that ectopically-expressed fluorescent proteins can be misfolded, directed into aggresomes, and evicted from cells via a non-classical secretory pathway, this process is inefficient and misfolded fluorophores such as GFP may lose their fluorescent properties (Tanudji, 2002). In contrast, fusions between a fluorophore (e.g., pH-sensitive GFP, pHluorin) and a tetraspanin such as CD63 are typically directed into MVBs and eventually evicted via EVs/exosomes which has enabled elegant in vitro studies to analyze MVB/membrane fusion events and EV release (Verweij et al., 2018). Other than one recent study showing that cancer cells secrete Cre recombinase via EVs/exosomes into recipient cells leading to activation of a fluorescent reporter, few to no other studies have demonstrated the transfer of EVs between two cell types in vivo (Zomer et al., 2015; Zomer, Steenbeek, Maynard, & van Rheenen, 2016). The recently described Transgenic Inducible GFP EV Reporter mice (TIGER mice) that use a CD9-TurboGFP fusion as the reporter in addition to work from our own lab, and others, showing that the ZsGreen fluorophore is efficiently but non-selectively evicted into EVs and other large particles may help to resolve long-standing questions related to the origin, fates, and functions of cell type-specific EVs in vivo (Headley et al., 2016; McCann, Liu, et al., 2019; Neckles et al., 2019). In addition, the CD63emGFP reporter mice described herein should serve as a compliment to these other in vivo models to comprehensively address questions related to EV heterogeneity and the functions and organ/tissue distribution of individual EV subpopulations. However, a few limitations of the model should be considered. For example, CD63emGFP reporter mice will only allow for labeling and capture of those EVs that have evicted the fusion protein; thus, EVs that are negative for emGFP will be excluded from the analysis. Also, in the tumor models presented herein, the miRNA signatures we report in circulating EC-derived EVs are generated from mixtures of both normal and tumor endothelium because we do not yet have the methodology to specifically enrich for those EVs that are exclusively derived from the tumor vasculature. Finally, while our mice are adaptable to numerous disease models and could theoretically be used to genetically mark EVs derived from any cell type, we have chosen here to focus especially on EC-derived EVs given their proximity to the circulation. It is possible, therefore, that the vascular endothelium is an enriched source for circulating EVs that serve is sentinels to predict disease status or act as vectors to promote or inhibit disease progression; particularly in diseases characterized by dysfunctional vasculature such as diabetes, Alzheimer’s disease, cardiovascular disease, and cancer.
Supplementary Material
Acknowledgements
ACD is supported by grants from the NIH (RO1-CA177874) and the American Cancer Society (129755-RSG-16-176-01-DDC). Other funding support includes a pre-doctoral fellowship to JVM (F31-CA213793-03). Portions of this work were supported by a core grant to the Emily Couric Cancer Center at The University of Virginia (P30CA044579). We thank the staff of the UNC Animal Models Core Facility for their work to generate the reporter mice. Requests for the reporter mice described in this study should be made to the corresponding author.
Bibliography
- Bobrie A, & Thery C (2013). Exosomes and communication between tumours and the immune system: are all exosomes equal? Biochemical Society Transactions, 41(1), 263–267. doi: 10.1042/BST20120245 [DOI] [PubMed] [Google Scholar]
- Daaboul GG, Gagni P, Benussi L, Bettotti P, Ciani M, Cretich M, … Chiari M (2016). Digital Detection of Exosomes by Interferometric Imaging. Sci Rep, 6, 37246. doi: 10.1038/srep37246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, … Wang SE (2015). Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nature cell biology, 17(2), 183–194. doi: 10.1038/ncb3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, & Joung JK (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol, 32(3), 279–284. doi: 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG (2011). Enzymatic assembly of overlapping DNA fragments. Methods Enzymol, 498, 349–361. doi: 10.1016/B978-0-12-385120-8.00015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Li X, Singh K, Luo Z, Meija-Cordova M, Jamalpour M, … Welsh M (2019). The Cdh5-CreERT2 transgene causes conditional Shb gene deletion in hematopoietic cells with consequences for immune cell responses to tumors. Sci Rep, 9(1), 7548. doi: 10.1038/s41598-019-44039-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headley MB, Bins A, Nip A, Roberts EW, Looney MR, Gerard A, & Krummel MF (2016). Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature, 531(7595), 513–517. doi: 10.1038/nature16985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Mark MT, … Lyden D (2015). Tumour exosome integrins determine organotropic metastasis. Nature, 527(7578), 329–335. doi: 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H, Drummen G, & Mathivanan S (2016). Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. International Journal of Molecular Sciences, 17(2), 170–129. doi: 10.3390/ijms17020170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Martin-Jaular L, Lavieu G, & Thery C (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol, 21(1), 9–17. doi: 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- McCann JV, Liu A, Musante L, Erdbrugger U, Lannigan J, & Dudley AC (2019). A miRNA signature in endothelial cell-derived extracellular vesicles in tumor-bearing mice. Sci Rep, 9(1), 16743. doi: 10.1038/s41598-019-52466-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann JV, Xiao L, Kim DJ, Khan OF, Kowalski PS, Anderson DG, … Dudley AC (2019). Endothelial miR-30c suppresses tumor growth via inhibition of TGF-beta-induced Serpine1. J Clin Invest, 130(3), 1654–1670. doi: 10.1172/JCI123106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckles VN, Morton MC, Holmberg JC, Sokolov AM, Nottoli T, Liu D, & Feliciano DM (2019). A transgenic inducible GFP extracellular-vesicle reporter (TIGER) mouse illuminates neonatal cortical astrocytes as a source of immunomodulatory extracellular vesicles. Sci Rep, 9(1), 3094. doi: 10.1038/s41598-019-39679-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, … Lyden D (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Medicine, 18(6), 883–891. doi: 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Nishida N, Calin GA, & Pantel K (2014). Clinical relevance of circulating cell-free microRNAs in cancer. Nature Publishing Group, 11(3), 145–156. doi: 10.1038/nrclinonc.2014.5 [DOI] [PubMed] [Google Scholar]
- Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, & Hoffman RM (2013). Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Advanced Drug Delivery Reviews, 65(3), 383–390. doi: 10.1016/j.addr.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Tanudji M (2002). Improperly folded green fluorescent protein is secreted via a non-classical pathway. Journal of cell science, 115(19), 3849–3857. doi: 10.1242/jcs.00047 [DOI] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, & Amigorena S (2002). Exosomes: composition, biogenesis and function. Nat Rev Immunol, 2(8), 569–579. doi: 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- Thyme SB, Akhmetova L, Montague TG, Valen E, & Schier AF (2016). Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat Commun, 7, 11750. doi: 10.1038/ncomms11750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij FJ, Bebelman MP, Jimenez CR, Garcia-Vallejo JJ, Janssen H, Neefjes J, … Pegtel DM (2018). Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol, 217(3), 1129–1142. doi: 10.1083/jcb.201703206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Li L, Li M, Guo C, Yao J, & Mi S (2015). Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genomics, Proteomics & Bioinformatics, 13(1), 17–24. doi: 10.1016/j.gpb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, … Wang SE (2014). Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. 25(4), 501–515. doi: 10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, Maynard C, Verweij FJ, Kamermans A, Schafer R, Beerling E, … van Rheenen J (2015). In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell, 161(5), 1046–1057. doi: 10.1016/j.cell.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, Steenbeek SC, Maynard C, & van Rheenen J (2016). Studying extracellular vesicle transfer by a Cre-loxP method. Nat Protoc, 11(1), 87–101. doi: 10.1038/nprot.2015.138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.